Abstract
We report the isolation and complete genome sequencing of a new Mimiviridae family member, infecting Acanthamoeba castellanii, from sewage in Mumbai, India. The isolated virus has a particle size of about 435 nm and a 1,182,200-bp genome. A phylogeny based on the DNA polymerase sequence placed the isolate as a new member of the Mimiviridae family lineage A and was named as Mimivirus bombay. Extensive presence of Mimiviridae family members in different environmental niches, with remarkably similar genome size and genetic makeup, point towards an evolutionary advantage that needs to be further investigated. The complete genome sequence of Mimivirus bombay was deposited at GenBank/EMBL/DDBJ under the accession number KU761889.
Keywords: NCLDV, Giant virus, Mimivirus bombay, Amoeba, CRISPR
Specifications
| Organism/cell line/tissue | Mimivirus bombay |
| Sex | NA |
| Sequencer or array type | Illumina MiSeq v2 150 x 2 PE |
| Data format | analyzed, complete genome FASTA sequence |
| Experimental factors | virus grown in Acanthamoeba castellanii |
| Experimental features | de novo genome assembly and annotation |
| Consent | not applicable |
| Sample source location | Mumbai, India, City, 19.180158 N, 72.848614 E |
Direct link to deposited data
NCBI Sequence graphics
https://www.ncbi.nlm.nih.gov/nuccore/1020265955?report=graph
Experimental design, materials and methods
Environmental sample processing and virus isolation
Water (50 ml) from sewage was filtered through a 20 μm Whatman filter paper, incubated overnight incubation at 4 °C in 8% w/v PEG and 0.4% w/v NaCl (pH 7.2) and centrifuged at 500 × g for 15 min at 4 °C. The pellet was re-suspended in 1 ml PBS and centrifuged again at 500 × g for 15 min at 4 °C and the supernatant obtained was centrifuged at 5000 × g for 45 min at 4 °C. Both supernatant and pellet (re-suspended in 50 μl PBS) from the final round of centrifugation were tested for infection of Acantamoeba castellanii as per the previously described protocol (17). Infection of A. castellanii cells with pellet resulted in lysis of amoeba cells in 48 h. Cell debris and un-infected host cells were removed by centrifugation at 500 × g for 10 min and the virus particles were pelleted by centrifugation at 1500 × g for 30 min (12). The pellet was re-suspended in PBS and was used to infect a fresh culture of A. castellani. Virus particles, obtained after three rounds of infection, were used for infecting A. castellanii in T-75 flasks and the virus particles were purified using sucrose gradient as reported earlier (12).
DNA extraction
DNA was extracted from the density-gradient purified virus particles using phenol-chloroform protocol followed by ethanol precipitation (17). DNA quality and quantity was ascertained by spectrophotometric and electrophoretic methods.
Whole genome shotgun sequencing
Library preparation was performed at the Genotypic Technology's (Bengaluru, India) Genomics facility according to the SureSelectQXT Library Prep protocol outlined in the Sure SelectQXT whole genome library prep for Illumina multiplexed sequencing protocol (Cat #5500–0121). Twenty five nanogram of genomic DNA was fragmented and the adapter-tag was added using Sure SelectQXT. Amplified adapter-tagged libraries were purified using high prep beads clean up kit (MAGBIO, USA). The libraries were quantified using Qubit flourometer and quality validated by running an aliquot on D1000 Tape (Cat# 5067–5582) using D1000 Tape Station Kit (Agilent, Cat# 5067–5583). After quality check, the library was sequenced using IlluminaMiSeq v2 2 × 150 bp paired-end sequencing.
Genome assembly and annotation
Adapter trimming and read filtering for QV > 30 was performed using Agilent SureCall suite. De novo assembly was performed using multiple assemblers including SOAPdenovo2 (15), A5-miseq (5), Velvet (18) and SPAdes (3), and were evaluated using QUAST (10). MAUVE (6) was used to reorder the contigs and generate consensus FASTA. Open reading frames (ORFs) were predicted with GeneMarkS (4), individually annotated using Blastp (2) and the results were retrieved using custom Python scripts. Phylogenetic analysis was performed using MEGA-CC Linux distribution (11). A5 miseq provided the best assembly parameters with a median coverage of 714 × and N50 of 906,835. All contigs were aligned to BLAST NR database using MEGABLAST (16) and the consensus FASTA was generated by reordering the 7 contigs using MAUVE (6). MVB has a genome size of 1,182,200 bp with 898 predicted ORFs. The annotated genome was uploaded to NCBI using BankIt web based submission tool.
Data description
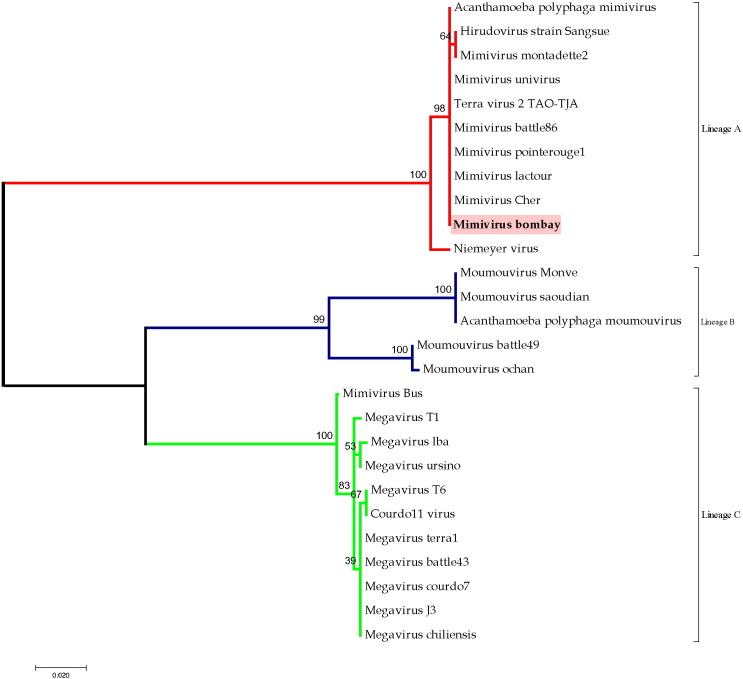
Transmission electron microscopy revealed virus particles of about 435 nm in size (Fig. 1), similar to some recently reported giant viruses known as Nucleo-Cytoplasmic Large DNA Viruses (NCLDV) (17). Illumina Basespace web tool (Kraken metagenomics) taxonomically classified 98% of the total 3,017,739 reads (the trimmed and QC filtered) as Mimiviridae. Hence the isolate was named as Mimivirus bombay (MVB). Further, a Maximum Likelihood (ML) based phylogeny of DNA polymerase showed close identity of MVB with lineage A of Mimiviridae (Fig. 2). The GC content of MVB (28%) is also comparable to other mimiviruses (1).
Fig. 1.

Transmission electron micrograph of Mimivirus bombay (MVB).
Fig. 2.
Amino acid sequence of MVB ORF#318, annotated as DNA polymerase, was used as input sequence for blastp query against non-redundant protein sequence database. Aligned sequences with a cut-off criteria for sequence selection included an E-value threshold = 0.0 with a minimum sequence coverage of greater than 40% and sequence identity of greater than 60% were retrieved for phylogentic analysis. Alignment was performed using Clustal algorithm within the MEGACC Linux distribution framework (11) with the following parameters: Substitution matrix: BLOSUM; Gap open penalty = 3.0; Gap extend penalty = 1.8. Rest of the parameters were used as default. Un-rooted Maximum Likelihood based phylogeny was plotted with 1000 bootstraps iterations. Bootstrap values are labelled at the nodes of the tree. The sequences used are: YP_003986825.1 [Acanthamoeba polyphaga mimivirus], AHA45542.1 [Hirudovirus strain Sangsue], CRK54683.1 [Mimivirus montadette2], CRK54684.1 [Mimivirus univirus], ADC39049.1 [Terra virus 2 TAO-TJA], CRI62815.1 [Mimivirus battle86], AFM52353.1 [Mimivirus pointerouge1], AFM52359.1 [Mimivirus lactour], AFM52352.1 [Mimivirus Cher], ALR83823.1 [Niemeyer virus], AEX62677.1 [Moumouvirus Monve], CRI62819.1 [Moumouvirus saoudian], YP_007354477.1 [Acanthamoeba polyphaga moumouvirus], CRI62820.1 [Moumouvirus battle49], AEY99267.1 [Moumouvirus ochan], AFM52363.1 [Mimivirus Bus], CRI62804.1 [Megavirus T1], AGD92513.1 [Megavirus lba], CRI62807.1 [Megavirus ursino], CRI62806.1 [Megavirus T6], AFM52349.1 [Courdo11 virus], AFM52356.1 [Megavirus terra1], CRI62802.1 [Megavirus battle43], AEX61758.1 [Megavirus courdo7], CRI62803.1 [Megavirus J3], YP_004894633.1 [Megavirus chiliensis].
tRNAscan-SE search server (14) showed the presence of 6 tRNAs in the MVB genome. Further, 9 transposons (http://transposonpsi.sourceforge.net/) and 6 Mimiviral CRISPR-like elements (Clustered Regularly Interspaced Short Palindromic Repeat, [7], [8], [9]) that have been recently attributed to impart immunity to virophage infection (13) were found in the MVB genome. The discovery of the first Mimivirus from India indicates the pan-geographic presence of large DNA viruses, and warrants a thorough study of their ecological and evolutionary significance.
Nucleotide accession number
The assembled complete genome was deposited to NCBI under accession number KU761889.1.
Acknowledgements
This work is supported by IIT Bombay Seed grant (11IRCCG004) to KK. AC is supported by IIT Bombay Post-Doctoral Fellowship. FA and DB were supported by Department of Biotechnology (DBT) Masters Program.
References
- 1.Aherfi S., Colson P., La Scola B., Raoult D. Giant viruses of amoebas: an update. Front. Microbiol. 2016;7:349. doi: 10.3389/fmicb.2016.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., Pyshkin A.V., Sirotkin A.V., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Besemer J., Lomsadze A., Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coil D., Jospin G., Darling A.E. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 2015;31:587–589. doi: 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- 6.Darling A.C., Mau B., Blattner F.R., Perna N.T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grissa I., Vergnaud G., Pourcel C. CRISPRcompar: a website to compare clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2008;36:W145–W148. doi: 10.1093/nar/gkn228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grissa I., Vergnaud G., Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinf. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grissa I., Vergnaud G., Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar S., Stecher G., Peterson D., Tamura K. MEGA-CC: computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis. Bioinformatics. 2012;28:2685–2686. doi: 10.1093/bioinformatics/bts507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Legendre M., Lartigue A., Bertaux L., Jeudy S., Bartoli J., Lescot M., Alempic J.M., Ramus C., Bruley C., Labadie K., Shmakova L., Rivkina E., Coute Y., Abergel C., Claverie J.M. In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E5327–E5335. doi: 10.1073/pnas.1510795112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levasseur A., Bekliz M., Chabriere E., Pontarotti P., La Scola B., Raoult D. MIMIVIRE is a defence system in mimivirus that confers resistance to virophage. Nature. 2016;531:249–252. doi: 10.1038/nature17146. [DOI] [PubMed] [Google Scholar]
- 14.Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., Chen Y., Pan Q., Liu Y., Tang J., Wu G., Zhang H., Shi Y., Yu C., Wang B., Lu Y., Han C., Cheung D.W., Yiu S.M., Peng S., Xiaoqian Z., Liu G., Liao X., Li Y., Yang H., Wang J., Lam T.W. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morgulis A., Coulouris G., Raytselis Y., Madden T.L., Agarwala R., Schaffer A.A. Database indexing for production MegaBLAST searches. Bioinformatics. 2008;24:1757–1764. doi: 10.1093/bioinformatics/btn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raoult D., Audic S., Robert C., Abergel C., Renesto P., Ogata H., La Scola B., Suzan M., Claverie J.M. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 18.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]