Abstract.
Two-photon excited fluorescence (TPEF) imaging of the cellular cofactors nicotinamide adenine dinucleotide and oxidized flavin adenine dinucleotide is widely used to measure cellular metabolism, both in normal and pathological cells and tissues. When dual-wavelength excitation is used, ratiometric TPEF imaging of the intrinsic cofactor fluorescence provides a metabolic index of cells—the “optical redox ratio” (ORR). With increased interest in understanding and controlling cellular metabolism in cancer, there is a need to evaluate the performance of ORR in malignant cells. We compare TPEF metabolic imaging with seahorse flux analysis of cellular oxygen consumption in two different breast cancer cell lines (MCF-7 and MDA-MB-231). We monitor metabolic index in living cells under both normal culture conditions and, for MCF-7, in response to cell respiration inhibitors and uncouplers. We observe a significant correlation between the TPEF-derived ORR and the flux analyzer measurements (, ). Our results confirm that the ORR is a valid dynamic index of cell metabolism under a range of oxygen consumption conditions relevant for cancer imaging.
Keywords: optical redox ratio, two-photon excited fluorescence, breast cancer, dynamic imaging
Two-photon excited fluorescence (TPEF) microscopy is widely used to quantify cellular metabolism by assessing the intrinsic fluorescence originating from two fluorescent cofactors: reduced nicotinamide adenine dinucleotide (NADH) and oxidized flavin adenine dinucleotide ().1 Because of their role in mitochondrial electron transport, their relative fluorescence intensity, defined as the “optical redox ratio (ORR),” can be used to estimate the contribution of oxidative phosphorylation versus glycolysis in cellular adenosine triphosphate (ATP) production.2,3 Since the first publication by Chance et al.,4 the ORR has been used in a broad range of applications spanning from cancer detection and diagnosis to predicting drug response to monitoring cellular function and stem cell differentiation.5–9 Unlike competing quantitative techniques with which the cells have to be lyzed,10–12 TPEF microscopy provides a nondestructive, real-time, and label-free method for quantifying cellular metabolism. It enables the spatial mapping of metabolic rate at the submicrometer scale with minimum interruption of normal cell function.13,14
However, quantifying and interpreting the ORR is challenging. The measured fluorescence intensity overlaps with the emission of additional fluorophores. Nicotinamide adenine dinucleotide phosphate and NADH, in particular, have similar excitation and emission spectra and cannot be differentiated easily.15 Moreover, the fluorescence quantum yields of NADH and are affected by their binding state to other metabolic enzymes.16,17 In order to test the robustness of the ORR for measuring cell metabolism, a number of studies have attempted to validate the correlation between ORR and the oxidation–reduction ratio of .18,19 Among them, Quinn et al.20 used liquid chromatography/mass spectrometry and found that while the fluorescence intensities of and NADH are not correlated with their actual intracellular concentrations, the ORR is significantly correlated with . However, no work has been reported to evaluate the stability and accuracy of ORR in living, dynamic biological samples.
In this work, we use TPEF to measure ORR in living MDA-MB-231 breast cancer cells under normal culture conditions and in MCF-7 breast cancer cells to dynamically measure metabolic changes in response to a series of mitochondrial inhibitors and uncouplers [oligomycin, carbonyl cyanide-4-phenylhydrazone (FCCP) and rotenone/antimycin A]. The ORR, defined as , was calculated from dual-wavelength excitation TPEF images, as discussed below. The ORR reflects the joint effects of oxidative phosphorylation and glycolysis on cellular metabolism. Higher ORR values indicate that cells are more oxidative, whereas lower ORR values reflect greater glycolytic metabolism. TPEF results were compared with seahorse flux analysis (Seahorse , Seahorse Bioscience) of the cellular oxygen consumption and proton production rates (OCR and PPR, respectively). Seahorse flux analysis is a standard metric that probes average cell oxidative phosphorylation and glycolysis rates separately.21 To compare the results from the seahorse flux analyzer to the ORR, we used a normalized oxygen consumption rate (defined as ), which evaluates the ratio between oxidative phosphorylation and glycolysis.
MDA-MB-231 and MCF-7 breast cancer cells were cultured in stable isotope labeling by amino acids in cell culture Dulbecco’s modified eagle medium flex medium (A2493901, Life Technologies) supplemented with 6-mM glucose, 0.85-mM L-arginine, 0.6-mM L-lysine, 5% fetal bovine serum, and 1% glutaMAX (35050-061, Life Technologies). Cells were detached from plates using 2% ethylenediaminetetraacetic acid in phosphate-buffered saline, counted with a hemocytometer, and then plated 16 h prior to experiment. We plated in XF24 cell culture microplates (#V7-PS, Seahorse Bioscience) for seahorse analysis and on imaging dishes (fd35-100, World Precision Instrument) for ORR measurements. An hour ahead of the seahorse experiment, cells were washed twice with 1 mL of XF base medium (#102353-100, Seahorse Bioscience) before adding XF complete medium (base sodium pyruvate) and incubated at 37°C with 0% . Standard protocols for seahorse flux assay calibration and analysis were employed. All seahorse flux data were normalized to total protein content measured by bicinchoninic acid assay (#23225, Thermo Scientific) at the end of the experiment, assuming tight correlation between the protein content and cell numbers per well. The ORR of the cancer cells was measured with a commercial Zeiss LSM510 microscope equipped with a tunable ultrafast laser source (Chameleon, Coherent Inc.). In each dish, three locations were randomly selected and imaged. The fluorescence of NADH (excited at 740 nm and collected at ) and (excited at 900 nm and collected at ) was excited and collected in the epi-direction sequentially. Two imaging dishes were prepared for each cell line. We used oligomycin, FCCP, and antimycin A/rotenone in sequence on live MCF-7 cells and studied the dynamic changes of cell metabolism by the ORR and seahorse flux analysis on individual cell lines employing the same protocol. Both ORR and n-OCR were acquired every 5 min for a total of 10 measurements in 45 min. Following baseline measurements (), oligomycin was added and three measurements were performed (, and 15). FCCP and antimycin A/rotenone were added in a similar manner, right after and at , respectively, and following completion of three ORR/n-OCR measurements. Identical image planes and constant laser power were maintained for all measurements. We used a freshly prepared fluorescein solution ( at pH 7) as a reference sample for calibrating all measurements. The image intensities were adjusted based on the fluorescein reference images and analyzed with ImageJ. The region of the cytoplasm of each cell was manually selected to calculate the ORR and compared with the n-OCR measured by flux analyzer. For statistical analysis, we included the data from 12 wells for the flux analyzer ( and three experiments for each cell line) and 25 cells for ORR.
Figure 1 shows typical images of NADH (1a) and (1b) of MDA-MB-231 cells in the same field. Average intensities within the cytoplasm region of interest (ROI) were used to calculate the ORR. Both seahorse flux analyzer (n-OCR) and ORR showed higher oxidative phosphorylation rates for MDA-MB-231 cells versus MCF-7 cells [tested with two-tailed analysis of variance (ANOVA), ] [Fig. 1(c)]. Similar observations for these two cell lines were reported by Walsh et al.18 using fluorescence lifetime-based ORR estimates. Differences in ORR are likely due to the cancer phenotype; highly metastatic MDA-MB-231 cells have relatively high mitochondrial biogenesis and oxidative phosphorylation rates mediated by .22
Fig. 1.
Metabolic measurements of tumor cells under normal culture conditions. (a) TPEF of MDA-MB-231 cells, NADH channel; (b) TPEF of MDA-MB-231 cells, channel. The outlined ROI selects cellular cytoplasm for calculation of the ORR. (c) n-OCR and the ORR for MCF-7 and MDA-MB-231 cells. The measurements from each cell line are compared with two-tailed ANOVA. Both measurements demonstrate statistically different metabolic rates for MCF-7 cells and MDA-MB-231 cells (). The error bars for n-OCR come from the standard deviation of 12 different wells (), and the error bars for ORR come from measurements on 25 different cells. The scale bar indicates .
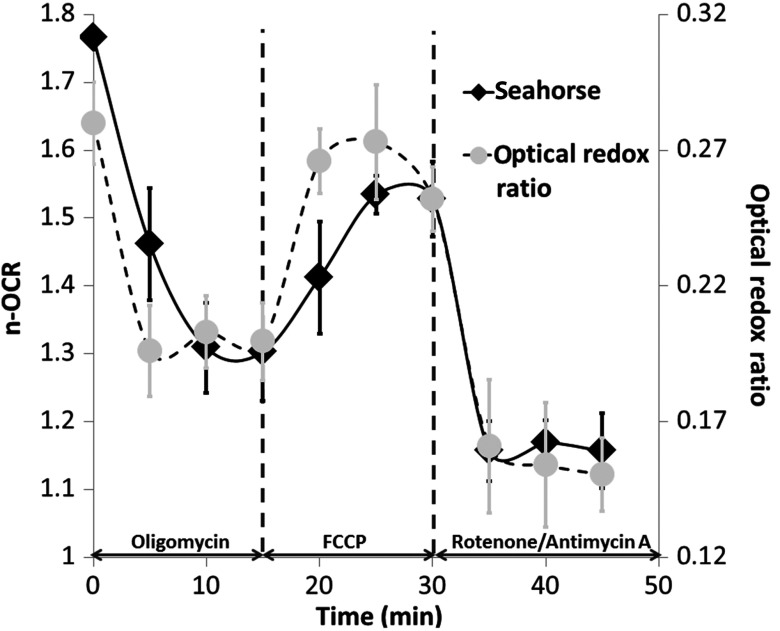
Figure 2 shows the metabolic response of MCF-7 cells to the mitochondrial inhibitors and uncouplers measured by both TPEF and seahorse flux analysis on the same day. The ORR changes are similar to the n-OCR ratio measured independently with seahorse experiments. After adding oligomycin, the cell mitochondrial proton channels are blocked and energy is provided primarily by glycolysis. The inhibition of oxidative phosphorylation is reflected by the substantial reduction in n-OCR and ORR. The subsequent addition of FCCP uncouples ATP synthesis from electron transport and recovers the cells from the effects of oligomycin. Both n-OCR and ORR return to values that are close to baseline. Finally, rotenone/antimycin A was added to stop the passing of protons to coenzyme Q, completely shutting down oxidative phosphorylation and dropping n-OCR and ORR to their lowest levels. Similar results were observed in three independent experiments that were carried out on different days.
Fig. 2.
Dynamic monitoring of MCF-7 cell metabolism by TPEF microscopy (circles, dashed line) and seahorse flux analyzer (diamonds, solid line). Cells in regular culture conditions were measured at time and then treated with oligomycin, FCCP, and rotenone/antimycin A at , and 30 min in sequence. The n-OCR and ORR were measured every 5 min. The error bars for n-OCR come from the standard deviation of 12 measurements, and the error bars for ORR come from measurements on 25 different cells.
To evaluate the correlation between the ORR and n-OCR, we pooled the data acquired from measurements of the two cell lines under both static (Fig. 1) and dynamic (Fig. 2) conditions and evaluated it using a linear regression model (Fig. 3). Elevated ORR values are correlated with higher cellular n-OCR. Under these conditions, we expect that the relative contribution of oxidative phosphorylation to cellular energy production is higher than that of glycolysis. A decrease of ORR implies a metabolic shift from oxidative phosphorylation toward aerobic glycolysis in cancer cells. In our experiments, pure anaerobic glycolysis under rotenone/antimycin A treated condition is accompanied by the lowest n-OCR and ORR. The overall correlation is highly significant (, ).
Fig. 3.
The ORR is significantly correlated with the n-OCR (, ). The circles are from the dynamic study of MCF-7 cells; the squares and triangles come from the measurements of MDA-MB-231 and MCF-7 cells, respectively, under normal culture conditions. The black line is the general linear fit with all the data points. Each data point represents an average value from 12 flux analyzer measurements and 25 cells from TPEF. The error bars are calculated as the standard deviation of corresponding measurements.
In summary, we demonstrate that the ORR derived from dual-wavelength TPEF of intrinsic NADH and cofactors strongly correlates with seahorse flux analysis of the cellular metabolic rate of oxygen consumption. Our results, obtained from two different breast cancer cell lines under both static conditions and dynamic perturbations of mitochondrial function, suggest that the ORR is a simple yet robust imaging index of cancer cell metabolism that can be used across a broad range of oxygen consumption conditions.
Acknowledgments
This research was supported in part by the National Institutes of Health, National Institute of Biomedical Imaging and Bioengineering, Laser Microbeam and Medical Program (LAMMP, P41-EB015890), National Cancer Institute R01CA142989, the Chao Family Comprehensive Cancer Center (P30CA62203), and programmatic support from the Arnold and Mabel Beckman Foundation.
References
- 1.Zipfel W. R., Williams R. M., Webb W. W., “Nonlinear magic: multiphoton microscopy in the biosciences,” Nat. Biotechnol. 21(11), 1369–1377 (2003). 10.1038/nbt899 [DOI] [PubMed] [Google Scholar]
- 2.Rice W. L., Kaplan D. L., Georgakoudi I., “Two-photon microscopy for non-invasive, quantitative monitoring of stem cell differentiation,” PLoS One 5(4), e10075 (2010). 10.1371/journal.pone.0010075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scholz R., et al. , “Flavin and pyridine nucleotide oxidation-reduction changes in perfused rat liver. I. Anoxia and subcellular localization of fluorescent flavoproteins,” J. Biol. Chem. 244(9), 2317–2324 (1969). [PubMed] [Google Scholar]
- 4.Chance B., et al. , “Oxidation–reduction ratio studies of mitochondria in freeze-trapped samples. NADH and flavoprotein fluorescence signals,” J. Biol. Chem. 254(11), 4764–4771 (1979). [PubMed] [Google Scholar]
- 5.Walsh A. J., et al. , “Quantitative optical imaging of primary tumor organoid metabolism predicts drug response in breast cancer,” Cancer Res. 74(18), 5184–5194 (2014). 10.1158/0008-5472.CAN-14-0663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barhoumi R., et al. , “Multiphoton spectral analysis of benzo[a]pyrene uptake and metabolism in breast epithelial cell lines,” J. Toxicol. Sci. 34(1), 13–25 (2009). 10.2131/jts.34.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ostrander J. H., et al. , “Optical redox ratio differentiates breast cancer cell lines based on estrogen receptor status,” Cancer Res. 70(11), 4759–4766 (2010). 10.1158/0008-5472.CAN-09-2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varone A., et al. , “Endogenous two-photon fluorescence imaging elucidates metabolic changes related to enhanced glycolysis and glutamine consumption in precancerous epithelial tissues,” Cancer Res. 74(11), 3067–3075 (2014). 10.1158/0008-5472.CAN-13-2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu H. N., et al. , “Quantitative mitochondrial redox imaging of breast cancer metastatic potential,” J. Biomed. Opt. 15(3), 036010 (2010). 10.1117/1.3431714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chance B., Estabrook R. W., Ghosh A., “Damped sinusoidal oscillations of cytoplasmic reduced pyridine nucleotide in yeast cells,” Proc. Natl. Acad. Sci. U. S. A. 51, 1244–1251 (1964). 10.1073/pnas.51.6.1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veech R. L., “The determination of the redox states and phosphorylation potential in living tissues and their relationship to metabolic control of disease phenotypes,” Biochem. Mol. Biol. Educ. 34(3), 168–179 (2006). 10.1002/(ISSN)1539-3429 [DOI] [PubMed] [Google Scholar]
- 12.Ji J., et al. , “Mitochondria-related miR-141-3p contributes to mitochondrial dysfunction in HFD-induced obesity by inhibiting PTEN,” Sci. Rep. 5, 16262 (2015). 10.1038/srep16262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiara Stringari R. A. E., et al. , “Metabolic trajectory of cellular differentiation in small intestine by phasor fluorescence lifetime microscopy of NADH,” Sci. Rep. 2, 9 (2012). 10.1038/srep00568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skala M., Ramanujam N., “Multiphoton redox ratio imaging for metabolic monitoring in vivo,” Methods Mol. Biol. 594, 155–162 (2010). 10.1007/978-1-60761-411-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rocheleau J. V., Head W. S., Piston D. W., “Quantitative NAD(P)H/flavoprotein autofluorescence imaging reveals metabolic mechanisms of pancreatic islet pyruvate response,” J. Biol. Chem. 279(30), 31780–31787 (2004). 10.1074/jbc.M314005200 [DOI] [PubMed] [Google Scholar]
- 16.Huang S., Heikal A. A., Webb W. W., “Two-photon fluorescence spectroscopy and microscopy of NAD(P)H and flavoprotein,” Biophys. J. 82(5), 2811–2825 (2002). 10.1016/S0006-3495(02)75621-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunz W. S., Kunz W., “Contribution of different enzymes to flavoprotein fluorescence of isolated rat liver mitochondria,” Biochim. Biophys. Acta 841(3), 237–246 (1985). 10.1016/0304-4165(85)90064-9 [DOI] [PubMed] [Google Scholar]
- 18.Walsh A. J., et al. , “Optical metabolic imaging identifies glycolytic levels, subtypes, and early-treatment response in breast cancer,” Cancer Res. 73(20), 6164–6174 (2013). 10.1158/0008-5472.CAN-13-0527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chance B., Thorell B., “Localization and kinetics of reduced pyridine nucleotide in living cells by microfluorometry,” J. Biol. Chem. 234, 3044–3050 (1959). [PubMed] [Google Scholar]
- 20.Quinn K. P., et al. , “Quantitative metabolic imaging using endogenous fluorescence to detect stem cell differentiation,” Sci. Rep. 3, 3432 (2013). 10.1038/srep03432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernyhough P., “Mitochondrial dysfunction in diabetic neuropathy: a series of unfortunate metabolic events,” Curr. Diab. Rep. 15(11), 89 (2015). 10.1007/s11892-015-0671-9 [DOI] [PubMed] [Google Scholar]
- 22.LeBleu V. S., et al. , “PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis,” Nat. Cell Biol. 16(10), 992–1003 (2014). 10.1038/ncb3039 [DOI] [PMC free article] [PubMed] [Google Scholar]