Abstract
Hepatitis C is a major worldwide disease and health hazard, affecting ∼3% of the world population. The p7 protein of hepatitis C virus (HCV) is an intracellular ion channel and pH regulator that is involved in the viral replication cycle. It is targeted by various classical ion channel blockers. Here, we generated p7 constructs corresponding to HCV genotypes 1a, 2a, 3a, and 4a for recombinant expression in HEK293 cells, and studied p7 channels using patch-clamp recording techniques. The pH50 values for recombinant p7 channels were between 6.0 and 6.5, as expected for proton-activated channels, and current-voltage dependence did not show any differences between genotypes. Inhibition of p7-mediated currents by amantadine, however, exhibited significant, genotype-specific variation. The IC50 values of p7-1a and p7-4a were 0.7 ± 0.1 nM and 3.2 ± 1.2 nM, whereas p7-2a and p7-3a had 50- to 1000-fold lower sensitivity, with IC50 values of 2402 ± 334 nM and 344 ± 64 nM, respectively. The IC50 values for rimantadine were low across all genotypes, ranging from 0.7 ± 0.1 nM, 1.6 ± 0.6 nM, and 3.0 ± 0.8 nM for p7-1a, p7-3a, and p7-4a, respectively, to 24 ± 4 nM for p7-2a. Results from patch-clamp recordings agreed well with cellular assays of p7 activity, namely, measurements of intracellular pH and hemadsorption assays, which confirmed the much reduced amantadine sensitivity of genotypes 2a and 3a. Thus, our results establish patch-clamp studies of recombinant viroporins as a valid analytical tool that can provide quantitative information about viroporin channel properties, complementing established techniques.
Introduction
Hepatitis C is a major cause of acute and chronic liver disease, affecting ∼3% of the world population (1). The prevalence rate in Egypt is the highest worldwide, with ∼18% of the population infected and 100,000–200,000 new cases reported every year (1, 2, 3).
To date, six major genotypes of hepatitis C virus (HCV) have been considered, although different classification systems were used in early studies (1, 4). Genotypes 1–3 are distributed worldwide, genotype 4 prevails in North Africa and the Middle East, genotype 5 is almost exclusive to South Africa, and genotype 6, with several subgenotypes, is found in Asia (1). Generally, genotypes 2 and 3 respond best to therapy. Genotype 4, which has a prevalence of >95% in Egypt (5), has an intermediate prognosis, and genotype 1 has the poorest response to classic interferon-based therapy (4). The most widely used treatment is a double therapy of pegylated interferon and ribavirin, which causes notable side effects and provides a sustained response in only 16% of patients. Triple therapy (pegylated interferon, ribivarin, and the channel blocker amantadine) has been shown to provide an up to 24% sustained response, but still results in a rate of ∼50% nonresponders (6, 7), highlighting the need for novel effective antiviral therapies. Numerous new approaches to treat hepatitis C have been developed, with the most notable developments being the introduction of specific anti-HCV antibodies (8, 9) and the discovery of novel direct antiviral agents (DAAs), such as sofosbuvir, as a better tolerated replacement for or complement to interferon-ribavirin therapy (10, 11, 12, 13, 14, 15, 16). Indeed, these novel therapies show great promise, albeit at a high cost of treatment, and their efficacy against all major genotypes of hepatitis C has yet to be established.
HCV is an enveloped, single-stranded RNA+ virus of the Flaviviridae family, genus Hepacivirus (17, 18). The genome of 9.6 kb is flanked by 5′ and 3′ untranslated regions. The open reading frame (ORF) translates to a single polyprotein of 3010–3033 amino acid residues (strain specific), which is cut by several proteases into 10 individual proteins (18).
The p7 protein of HCV is a 63 residue transmembrane protein located in the endoplasmic reticulum (ER). The N- and C-termini of p7 protrude into the ER lumen, and the short loop connecting the transmembrane helices is on the cytosolic side (19). p7 was shown to form channels in artificial bilayer lipid membranes, likely conducting H+, K+, Na+, and Ca2+ (20, 21). Heptameric (22) as well as hexameric (23) p7 complexes have been reported. NMR solution structures (24) and molecular modeling studies (25) suggest an arrangement of largely parallel helix bundles for p7 subunits (26, 27), although such structures are predicted to vary between genotypes, as shown in a modeling study that compared genotypes 1a, 1b, and 5a (27). A recent NMR structure suggests a different arrangement of short helical segments of p7, predicting the formation of a binding pocket for rimantadine outside of the channel pore, where it acts as an allosteric inhibitor (23). An NMR-based determination of solvent accessibility confirmed this architecture of the p7 protein and the allosteric mechanism of rimantadine inhibition (28). The activity of p7 as an intracellular pH shunt is believed to be involved in critical steps of the viral replication cycle, controlling acidification of endosomes and virion-loaded particles, and promoting vesicle trafficking and assembly and release of virions (29). Thus, p7 is a promising target for antiviral therapies. Indeed, p7 inhibitors have been identified as antiviral agents, including the channel blockers amantadine and rimantadine, as well as iminosugars and hexamethylene amiloride (HMA) (20, 21, 30, 31), although mechanistic data regarding the function of p7 are still sparse.
Here, we present a novel, to our knowledge, combination of methods for studying the ion channel function of the p7 protein directly. Genotype-specific p7 constructs were generated (Table 1; Fig. S1 in the Supporting Material), cloned into a eukaryotic expression vector, and recombinantly expressed in HEK293 cells. Whole-cell, patch-clamp recording techniques were used to study ion channel activation and inhibition. The results were validated against cellular assays of p7 activity. We show that p7 is a pH-gated proton channel that regulates intracellular pH. Inhibition by amantadine and rimantadine is strongly genotype dependent. The technique allows for specific and detailed dissection of p7 function, which should aid in the development of novel HCV therapies.
Table 1.
p7 Consensus Sequences for HCV Genotypes 1–4
| Genotype | p7 Protein Sequence |
|---|---|
| All | ALENLVVLNAASAAGTHGILWFLVFFCAAWYVKGRLVPGATYSLLGLWPLLLLLLALPQRAYA |
| 1 | ALENLVLLNAASLAGTHGLVSFLVFFCFAWYLKGRWVPGAAYAVYGMWPLLLLLLALPQRAYA |
| 1aa | ALENLVILNAASLAGTHGLVSFLVFFCFAWYLKGRWVPGAVYAFYGMWPLLLLLLALPQRAYA |
| 1b | ALENLVVLNAASLAGAHGILSFLVFFCAAWYIKGRLVPGAAYALYGVWPLLLLLLALPPRAYA |
| 1c | ALENLIVLNAASLVGTHGIVPFFIFFCAAWYLKGKWAPGLAYSVYGMWPLLLLLLALPQRAYA |
| 2 | ALEKLVILHAASAASANGPLWFFIFFVAAWYIKGRVVPLATYSLLGLWSFLLLVLALPQQAYA |
| 2aa | ALEKLVVLHAASAASCNGFLYFVIFFVAAWYIKGRVVPLATYSLTGLWSFGLLLLALPQQAYA |
| 2b | ALEKLIILHSASAASANGPLWFFIFFTAAWYLKGRVVPVATYSVLGLWSFLLLVLALPQQAYA |
| 2c | ALEKLVILHAASAASSNGLLYFILFFVAAWCIKGRAVPMVTYTLLGCWSFVLLLMALPHQAYA |
| 3 | ALENLVTLNAVAAAGTHGIGWYLVAFCAAWYVRGKLVPLVTYSLTGLWSLALLVLLLPQRAYA |
| 3aa | ALENLVTLNAVAAAGTHGIGWYLVAFCAAWYVRGKLVPLVTYSLTGLWSLALLVLLLPQRAYA |
| 3b | AMENLVMLNALSAAGQQGYVWYLVAFCAAWHIRGKLVPLITYGLTGLWPLALLDLLLPQRAYA |
| 4 | ALSNLININAASAAGTHGFWYAIFFICIAWHVKGRLPAAATYAACGMWPLLLLLLMLPERAYA |
| 4aa | ALSNLININAASAAGAQGFWYAILFICIVWHVKGRFPAAAAYAACGLWPCFLLLLMLPYRAYA |
p7 protein sequences after Carrère-Kremer et al. (19). Consensus sequences are shown in bold.
Sequences used in this study.
Materials and Methods
Generation of p7 expression constructs
We prepared genotype-specific p7 constructs (Table 1) using an overlap extension PCR protocol and finally cloned the p7 sequence into the eukaryotic expression vector pRK5. All constructs contained a membrane-directing signal peptide taken from the inhibitory glycine receptor, generated with and without the c-myc tag for Western blot analysis, and sequenced to verify successful generation (LGCgenomics, Berlin, Germany). See the Supporting Materials and Methods for details.
Cell culture and transfection
HEK293 cells (kindly provided by Prof. Cord-Michael Becker, Department of Biochemistry, University of Erlangen, Erlangen, Germany) were cultured in 10 cm tissue culture petri dishes in minimum essential medium (Sigma-Aldrich, Deisenhofen, Germany) supplemented with 10% fetal bovine serum (Invitrogen, Karlsruhe, Germany) and penicillin/streptomycin (Sigma-Aldrich) at 5% CO2 and 37°C in a water-saturated atmosphere (32). For experiments, cells were plated on poly-L-lysine-treated glass coverslips in 6 cm dishes and transfected 1 day after passage using 1.5 μg of p7 cDNA, 1.5 μg of green fluorescent protein cDNA, and 3 μL of GenCarrier (Epoch Life Science, Sugar Land, TX). Measurements were performed 2–5 days after transfection.
Membrane preparation and Western blot analysis
HEK293 cells were harvested 3 days after transfection and a crude membrane fraction was prepared and subjected to SDS-PAGE and Western blotting (see the Supporting Materials and Methods for details). An alkaline phosphatase-conjugated anti-c-myc antibody (Santa Cruz, Heidelberg, Germany) was used and the blot was visualized using 0.03% nitro blue tetrazolium and 0.02% 5-bromo-4-chloro-3-indolyl-phosphate in substrate buffer (100 mM Tris-HCl, pH 9.5; 100 mM NaCl; 5 mM MgCl2).
Live-cell imaging of vesicular pH
For live-cell imaging of vesicular pH (33), HEK293 cells were grown overnight on poly-L-lysine-coated coverslips and transfected as described above. At 24 h posttransfection, the cells were washed in HEPES buffer (10 mM HEPES, 133.5 mM NaCl, 2.0 mM CaCl2, 4.0 mM KCl, 1.2 mM MgSO4, 1.2 mM NaH2PO4, 11 mM glucose, pH 7.4) and loaded with 2 μM LysoSensor Green DND-189 (Life Technologies, Frankfurt, Germany) in HEPES buffer for 30 min at 37°C. The cells were washed twice with HEPES and immediately imaged at wavelengths of λ = 443 nm (excitation) and λ = 500 nm (emission) on an AxioCam Erc5s imaging system (Carl Zeiss, Jena, Germany). Fluorescence intensity was determined using the Carl Zeiss Zen 2012 lite-blue edition software package. For each cell, the area of interest, which included the cytosolic space of the cell and the corresponding background, was delineated. Noise values were determined from a region on the same image that was nearby but did not contain any cells. Fluorescence intensities from 20 individual determinations were averaged for analysis.
Hemadsorption assay
The method used to prepare cells for hemadsorption assays was adapted from a published protocol (34). Briefly, HEK293-FT cells were grown in poly-L-lysine-treated six-well plates as described above. Cells were transfected using 3 μL GenCarrier-2, 1.5 μL p7 DNA, and 1.5 μL M2-dependent hemagglutinin (HA-cDNA, kindly donated by Prof. Wendy S. Barclay, Imperial College, London). At 48 h posttransfection, the cells were washed twice with PBS, treated with 1 mL/well of 5.5 mU/mL Vibrio cholerae neuraminidase (Roche, Mannheim, Germany) in serum-free minimum essential medium for 1 h at 37°C, and then washed three times with PBS. Then, 1 mL of a 0.5% (w/v) suspension of human red blood cells was added to each well and the cells were incubated for 2 h. After incubation, the cells were washed three times with PBS and lysed using 1 mL Triton X-100 lysis buffer (1% Triton X-100, 150 mM NaCl, 50 mM Tris, pH 8.0) for 5 min. The lysate was centrifuged at 13,000 rpm for 5 min and absorbance was recorded at λ = 540 nm using a V-530 UV/Vis spectrophotometer (Jasco, Easton, MD). Triplicate determinations from two independent experiments per construct were averaged. The means and standard deviation (SD) were calculated using GraphPad InStat software (version 3.1 for Microsoft Windows). One-way analysis of variance (ANOVA) was used to determine the differences between various groups. Tukey’s pair comparison test was used to determine significant differences (p < 0.05) between means.
Electrophysiological recordings and data analysis
HEK293 cells were plated in 6 cm tissue culture dishes and transfected 2–4 days before electrophysiological recordings were obtained. For experiments, cells were kept in a bath solution containing 135 mM NaCl, 5.5 mM KCl, 2 mM CaCl2, 1.0 mM MgCl2, and 10 mM HEPES (pH 7.4 with NaOH). Current responses were measured at room temperature (21–23°C) at a holding potential of −60 mV. Whole-cell recordings were performed using a HEKA EPC10 amplifier (HEKA Electronics, Lambrecht, Germany) controlled by Pulse software (HEKA Electronics). Recording pipettes were pulled from borosilicate glass (World Precision Instruments, Berlin, Germany) using a Sutter P-97 horizontal puller (Sutter, Novato, CA). Solutions were applied using an Octaflow system (NPI Electronics, Tamm, Germany) in which the cells were bathed in a laminar flow of buffer, giving a time resolution for solution exchange and reequilibration of ∼100 ms. The external buffer consisted of 90 mM N-methyl-D-glucamine (NMDG), 3 mM CaCl2, 90 mM HEPES, and 90 mM 2-(N-morpholino)ethanesulfonic acid (MES). The pH of the external buffer was adjusted to 8.5, 7.5, 6.5, 6.0, 5.5, 5.0, and 4.5 using NaOH or HCl. The internal buffer was 90 mM NMDG, 10 mM EGTA, 180 mM HEPES, pH 7.5 (NaOH). Buffers were adapted from a similar buffer system developed for the study of recombinant influenza virus M2 proton channels (35). To avoid changes in buffer composition, the mixture of 90 mM HEPES and 90 mM MES was used for the entire pH range. After the pH was set, the buffers were salt adjusted to ensure the same osmolarity at all pH values. Between experiments, HEK293 cells were held in the standard bath solution. For measurement of whole-cell p7 currents, cells were perfused continuously with recording buffer during 1-min recording intervals. Inhibitors were diluted from stock solutions (10 mM) into pH 5.5 buffer. Inhibition measurements were started with baseline recordings of extracellular buffer pH 7.5 (4–5 s), followed by pH 5.5 (to induce channel opening). Then inhibitors (at pH 5.5) were applied, followed by final control applications. Sometimes, control solution (pH 5.5 without inhibitor) was perfused between inhibitor applications. Typical current amplitudes were between 100 and 400 pA. Dose-response curves were constructed from four to five cells per each genotype (Table 2), and inhibitor and IC50 values were determined using a nonlinear fit to the equation Iobs = Imax / [1 + ([I]/IC50)], where Iobs is the observed current at any given concentration of inhibitor, Imax is the maximum current amplitude observed in the absence of inhibitor, and [I] is the concentration of inhibitor.
Table 2.
Summary of Activation and Inhibition of p7-Mediated Whole-Cell Currents
| pH50 | nH | I-V-Dependent Zero Crossing/Slope (pA)/(pA/mV) | IC50 (nM) Rimantadine | No. of Cells | IC50 (nM) Amantadine | No. of Cells | |
|---|---|---|---|---|---|---|---|
| p7-1a | 6.2 ± 0.4 | 0.77 ± 0.04 | −5.0 ± 3/1.5 ± 0.1 | 0.7 ± 0.1 | 5 | 0.7 ± 0.1 | 5 |
| p7-2a | 6.0 ± 0.9 | 1.3 ± 0.2 | −3.6 ± 4/1.5 ± 0.1 | 24 ± 4 | 5 | 2402 ± 334 | 5 |
| p7-3a | 6.3 ± 0.9 | 1.0 ± 0.2 | −1.2 ± 7/1.4 ± 0.1 | 1.6 ± 0.6 | 5 | 344 ± 64 | 4 |
| p7-4a | 6.5 ± 1.2 | 1.0 ± 0.2 | −8.3 ± 8/1.6 ± 0.2 | 3.0 ± 0.8 | 4 | 3.2 ± 1.2 | 4 |
Results
p7 protein generation and expression
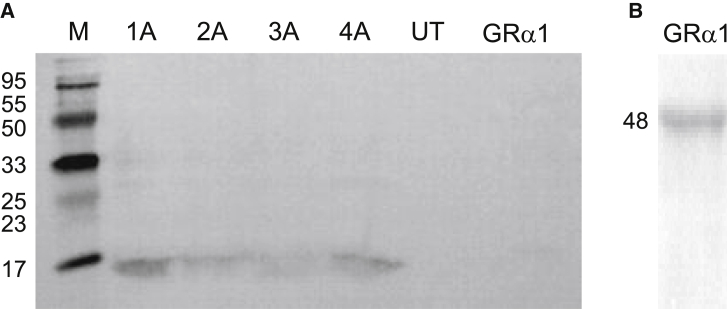
Genotype-specific constructs corresponding to the p7 sequence of genotypes 1a–4a were generated by overlap extension PCR (Table 1). Each p7 construct contained a membrane-directing signal peptide (taken from glycine receptor constructs in the same expression vector). A c-myc tag (EQKLISEEDL) was introduced for Western blot detection (Fig. S1), and p7 constructs without myc-tag were used for functional assays. All constructs were sequenced to verify successful generation. Plasma membrane expression was verified by Western blotting of membrane preparations from HEK293 cells transfected with cDNA constructs of each of the four genotypes. Detection with an anti-myc antibody revealed a specific band at ∼12 kD, indicating that p7 protein was present in the membrane fraction of transfected HEK293 cells (Fig. 1). Whole-cell patch-clamp experiments confirmed successful plasma membrane expression of recombinant p7.
Figure 1.
Western blot analysis of p7 proteins expressed in HEK293 cells. (A) Membrane preparations of p7 constructs corresponding to genotypes 1a–4a were subjected to SDS-PAGE and Western blotting using an anti-c-myc primary antibody. Left panel, p7 constructs; right panel, membrane preparation from untransfected HEK293 cells (UT) and cells transfected with GRα1 subunits. No myc signal was detected using the same anti-c-myc antibody. p7 proteins including the myc tag and signal peptide have a theoretical size of ∼11.5 kD. Markers are 6 kD and 20 kD. (B) Test for transfection of GRα1 subunits. A membrane preparation of HEK293 cells transfected with GRα1 cDNA was probed with a commercial GRα antibody, showing the expected band at 48 kD.
Live-cell imaging of the vesicular pH
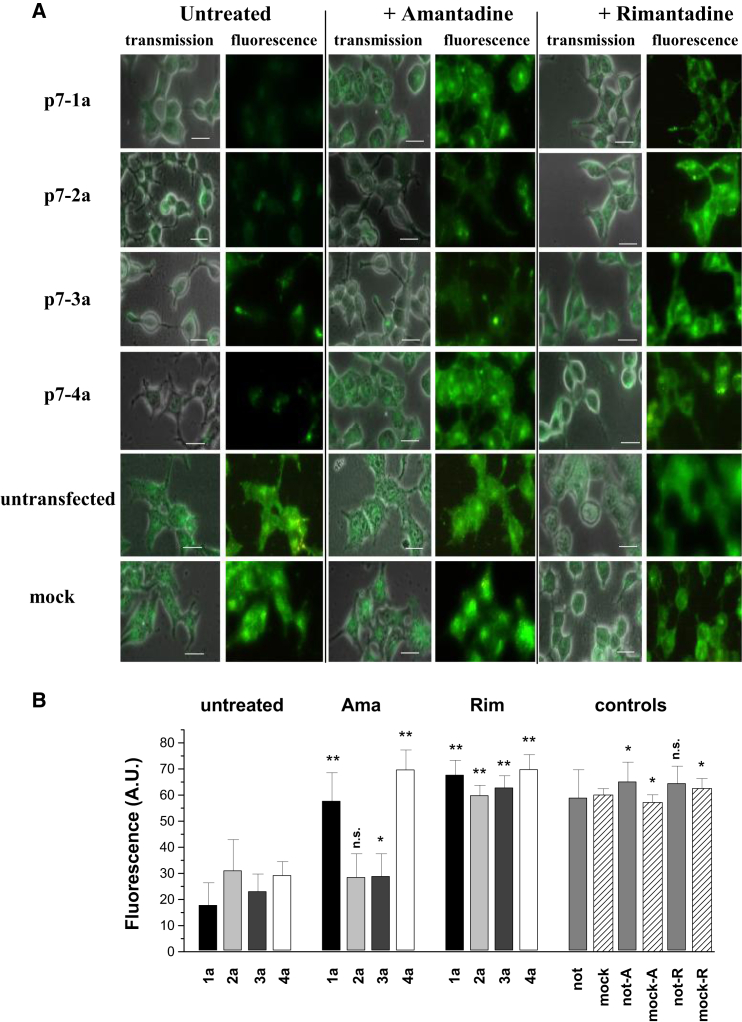
It has been suggested that p7 functions as a proton channel to modulate pH in artificial lipid membranes and liposomes. To verify the effect of p7 activity on the acidity of intracellular vesicles such as lysosomes, we expressed p7 constructs of the four different HCV genotypes (1a, 2a, 3a, and 4a) in HEK293 cells and determined the intracellular pH using the pH-sensitive fluorescent probe LysoSensor Green DND-189. Expression of p7 in HEK293 cells significantly reduced the mean cellular fluorescence intensity for all genotypes compared with untransfected controls (p < 0.001), with mean fluorescence intensity values of 18 ± 2, 31 ± 12, 23 ± 2, 29 ± 3, and 59 ± 4 for genotypes 1a, 2a, 3a, 4a, and untransfected control, respectively (Fig. 2).
Figure 2.
Recombinant p7 constructs modulate intracellular pH. HEK293 cells, transfected with p7 of the HCV genotypes 1a, 2a, 3a, and 4a, were loaded with the pH-sensitive fluorescent probe LysoSensor Green DND-189 (2 μM in HEPES buffer) for 30 min and imaged with excitation at λ = 443 nm and emission at λ = 500 nm. Amantadine and rimantadine (100 μM) in HEPES buffer were added 10 min before imaging. (A) Images show transmission overlaid with the fluorescence channel (transmission) and the fluorescence channel alone. Genotypes and treatments are indicated. Mock transfection used glycine receptor constructs in the same vector. Scale bar, 10 μm. (B) Mean of LysoSensor fluorescence intensity for each treatment. Data are the average of 20 determinations from a total of three independent cell preparations. Black columns, p7-1a; light gray, p7-2a; dark gray, p7-3a; white, p7-4a. Controls: medium gray, not transfected; dashes, mock transfected. Significance (one-way ANOVA) is indicated; ∗p < 0.05, ∗∗p < 0.01). To see this figure in color, go online.
Mock transfection with glycine receptor α1 (GRα1) cDNA had no effect on intracellular pH (mean fluorescence intensity 61 ± 4; Fig. 2). The effect of the channel blockers amantadine and rimantadine on p7 activity was examined by adding the inhibitors to transfected cells at a concentration of 100 μM 10 min before cell imaging. The fluorescence intensities of rimantadine-treated cells, untransfected, or mock-transfected controls were in the same range (p > 0.05). The mean fluorescence values for 1a, 2a, 3a, and 4a constructs after rimantadine treatment were 68 ± 6, 60 ± 4, 63 ± 5, and 70 ± 1, respectively (p < 0.001), indicating restoration of vesicular acidity to control values. In contrast, the effect of amantadine on p7 activity in transfected HEK293 cells varied considerably between genotypes. Genotypes 2a and 3a were insensitive to 100 μM amantadine, with mean fluorescence intensities remaining low, at values of 29 ± 3 and 29 ± 4, respectively. For genotypes 1a and 4a, p7 activity was blocked by 100 μM amantadine, with mean fluorescence intensities of 58 ± 3 and 70 ± 7 respectively, i.e., restoration to control levels (Fig. 2 B).
Hemadsorption assay
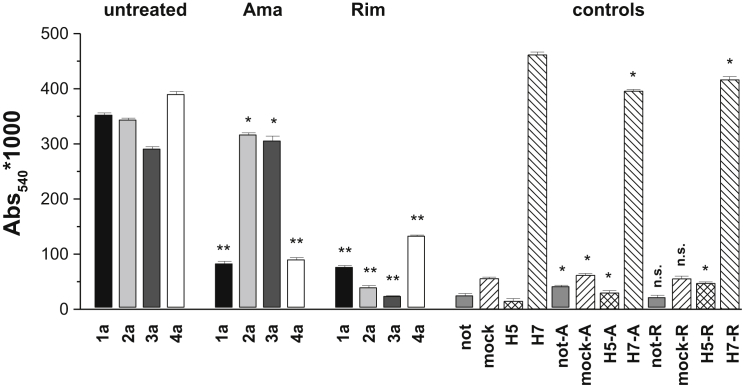
As a functional assay of recombinant p7 proteins, we investigated the ability of p7 channels to mediate transport of multibasic HA protein (H5HA) to the cell surface. This transport is sensitive to vesicular pH, requiring alkalization of exocytic vesicles. H5HA at the cell surface binds to sialic acid present on erythrocytes, and this can be assayed spectrophotometrically through the hemoglobin absorption at λ = 540 nm. Originally developed for the M2 channel of influenza virus, the assay was later adapted to characterize p7 activity (34). HEK293-FT cells were cotransfected with p7 and HA cDNA as described above. Coexpression of p7 and H5HA resulted in hemadsorption levels comparable to those observed for M2-independent HA (H7HA) as a positive control (34), with mean hemadsorption values (absorbance units) 0.352 ± 0.009, 0.343 ± 0.025, 0.291 ± 0.043, 0.390 ± 0.005, and 0.462 ± 0.029 for genotypes 1a, 2a, 3a, 4a, and H7HA, respectively. In control cells expressing H5HA alone, or in mock-transfected cells (H5HA plus GRα1) or untransfected cells, the hemadsorption values were >10-fold lower (p < 0.005), with mean hemadsorption values of 0.015 ± 0.002, 0.056 ± 0.006, and 0.025 ± 0.010, respectively (Fig. 3). For inhibition studies, amantadine (100 μM) and rimantadine (100 μM) were added 10 min before cell lysis and assay. Rimantadine inhibited p7 activity, reducing hemadsorption values for all four genotypes to control levels, with mean hemadsorption values of 0.08 ± 0.02, 0.040 ± 0.003, 0.030 ± 0.004, and 0.13 ± 0.05 for genotypes 1a, 2a, 3a, and 4a, respectively (Fig. 3). Consistent with results from cellular pH imaging assays, inhibition by amantadine was genotype dependent. Hemadsorption values were unaffected by amantadine in the case of genotypes 2a and 3a (mean hemadsorption values of 0.32 ± 0.01 and 0.31 ± 0.03), whereas for genotypes 1a and 4a, p7 activity was abolished by amantadine, as evident from significantly reduced hemadsorption values of 0.08 ± 0.03 and 0.090 ± 0.008. It is noted that errors in the assay were small, so sometimes values that clearly were in the same range and of similar physiological relevance were calculated to be significantly different.
Figure 3.
Hemadsorption assay of p7 activity. Hemoglobin binding to HEK293-FT cells was determined 48 h after transfection with HCV p7-1a, 2a, 3a, 4a, and M2-dependent H5HA in the presence and absence of amantadine and rimantadine (100 μM). Controls include untransfected cells, cells transfected with glycine receptor and H5HA (mock), H5HA only, and M2-independent H7HA. Data represent mean absorbance at λ = 540 nm of six data points (triplicate determinations from two independent experiments).
Characterization of p7 channels by electrophysiological methods
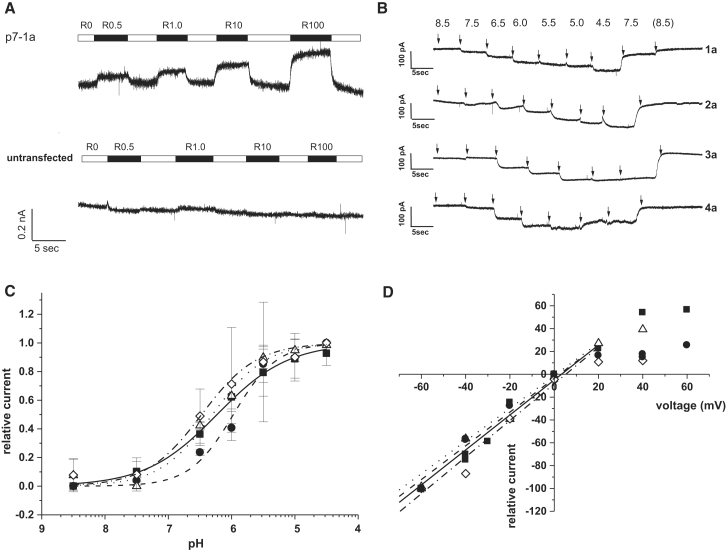
HEK293 cells were transfected with p7 constructs corresponding to the four different genotypes and measurements were performed 2–4 days after transfection. Since the p7 channel is considered a proton channel, salts were excluded from the recording buffer, with the exception of 3 mM CaCl2, which was needed to stabilize the seal. Cells were voltage clamped at −60 mV, where p7 activation caused an inward current. Care was taken to isolate p7-specific responses. Changes in extracellular pH also induced currents, even in untransfected cells. Currents mediated by p7 activity, however, were sensitive to rimantadine inhibition (Fig. 4 A). Only currents that could be blocked by rimantadine were used for analysis.
Figure 4.
Characterization of recombinant p7 channels by patch-clamp electrophysiology. HEK293 cells were transfected with genotype-specific p7 constructs 48–72 h before experiments. (A) Control traces at pH 5.5 from untransfected HEK293 cells (bottom) and HEK203 cells transfected with p7-1a cDNA constructs (top). Only currents that were blocked by rimantadine were used for analysis. (B) Traces were measured at a constant voltage of −60 mV. Cells were superfused with solutions of varying pH for ∼5 s; switches are indicated by arrows. Note the increased inward currents (downward deflection) at more acidic pH. (C) pH50 curves for p7 proteins of genotypes 1a, 2a, 3a, and 4a. Data points from at least two cells per genotype were used. p7-1a: solid line, solid squares; p7-2a: dashed line, solid circles; p7-3a: dotted line, open triangles; p7-4a: dash-dotted line, open diamonds. pH50 values were between 6.0 and 6.5 for all genotypes; see Table 2 for constants. (D) I-V dependence of recombinant p7 channels. p7-1a: solid line, solid squares; p7-2a: solid circles; p7-3a: dotted line, open triangles; p7-4a: dash-dotted line, open diamonds. All p7 channels showed ohmic behavior at negative voltage and inward rectification at voltages of >20 mV.
pH dependence
The pH dependence of proton-mediated currents from cells transfected with p7 constructs was determined for each individual genotype. We chose a minimum of 15 data points from at least two cells to assess the pH dependence of each genotype. Maximal current responses were observed at pH 5.5 or pH 4.5 (Fig. 4). The pH50 values were 6.2 ± 0.4, 6.0 ± 0.9, 6.3 ± 0.9, and 6.5 ± 1.2 for 1a, 2a, 3a, and 4a, respectively; nH was ∼1 for all genotypes (Fig. 4, A and B; Table 2).
Current-voltage dependence
The maximum currents of different p7 proteins were recorded at voltages between −60 mV and +60 mV. Currents from open p7 channels (at pH 5.5) and rimantadine-blocked channels at the same pH were compared to determine the maximum amplitude (Imax) of p7-mediated currents. All recorded responses were normalized to the response at −60 mV, which was set to −100%. Current-voltage (I-V) data were linear in the range of −60 mV to +20 mV. At higher voltages, p7 currents showed inward rectification (Fig. 4 C). Regression of the linear portion of the I-V curves gave zero crossings (0 mV) of −5% ± 3%, −4% ± 4%, −1% ± 7%, and −8% ± 8% for genotypes 1a, 2a, 3a, and 4a, respectively. The slope conductances of genotypes 1a, 2a, 3a, and 4a were 1.5 ± 0.1 pS, 1.5 ± 0.1 pS, 1.4 ± 0.1 pS, and 1.6 ± 0.2 pS, respectively (Table 2; Fig. 4 C). As with the pH dependence, the I-V relationships and zero potential currents were quite similar between genotypes.
Inhibition by rimantadine and amantadine
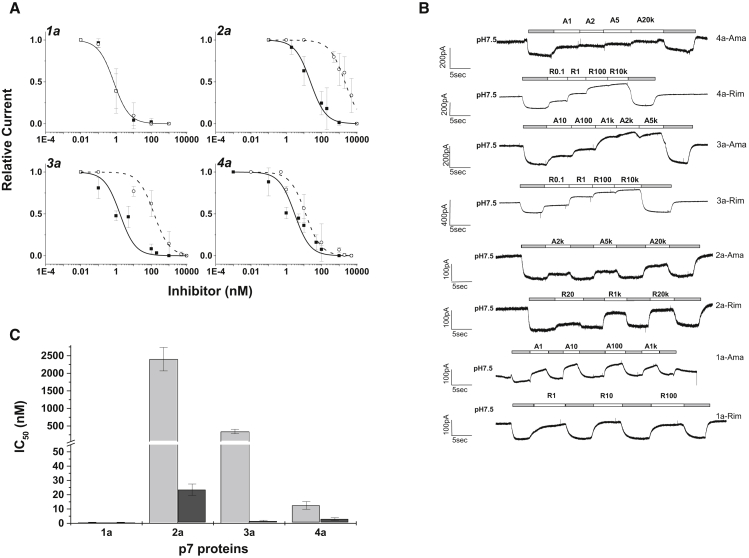
We found robust inhibition of p7-mediated currents for the channel blockers amantadine and rimantadine (Fig. 5). The IC50 values derived from these curves were approximately 0.7 ± 0.1 nM (1a), 1.6 ± 0.6 nM (3a), and 3.0 ± 0.8 nM (4a) for rimantadine (Fig. 5 A; Table 2). Channels of the p7 genotype 2a showed lower sensitivity, with an IC50 value for rimantadine of 24 ± 4 nM. Inhibition by amantadine was less potent, with IC50 values of 0.7 ± 0.1 nM (1a) and 3.2 ± 1.2 nM (4a). For genotypes 2 and 3, the inhibitory activity of amantadine was dramatically lower, with IC50 values of 2402 ± 334 nM and 344 ± 64 nM for 2a and p7-3a channels, respectively (Fig. 5 C).
Figure 5.
Inhibition of recombinant p7 channels by amantadine and rimantadine. Genotype-specific constructs were expressed in HEK293 cells 2–4 days before experiments. (A) Dose-response curves for inhibition of p7-1a, p7-2a, p7-3a, and p7-4a channels. Rimantadine: solid line, solid squares; amantadine: dashed line, open circles. Data points from at least four cells were used to construct the inhibition curves. (B) Whole-cell current traces show inhibition by rimantadine and amantadine for all four genotypes. Inhibitors were applied at pH 5.5; upward deflection indicates reduced p7-mediated current. (C) IC50 values of rimantadine (light gray) and amantadine (dark gray). Note the reduced amantadine sensitivity of genotypes 2a and 3a. See Table 2 for constants.
Discussion
Hepatitis C is one of the most widespread virus diseases. The situation in Egypt is particularly serious since the HCV infection rate (∼18% of the population) is the highest worldwide, making hepatitis C one of the leading causes of morbidity (2). In Egypt, genotype 4 is predominant, being found in more than 90% of patients (5). To date, a number of full-length clones of HCV have been made available for studies (36, 37, 38, 39). Infectious HCV particles have been obtained from the prototype strains HCV-1 (genotype 1a) (40), JFH-1, and H-77 (genotype 2a) (41). Recently, a replicon for genotype 3a was reported (42), and cell culture methods for the production of high-titer HCV particles of genotypes 1–6 were developed (43). The p7 protein, forming an intracellular ion channel, has been identified as a crucial member of the HCV replication machinery (21, 29, 44, 45) and thus is a viable therapeutic target. Here, we prepared genotype-specific constructs for the expression of isolated p7 in a recombinant system to analyze the function and inhibition of p7 channels.
p7 protein generation and expression
We generated cDNA constructs of the p7 protein of the HCV genotypes 1a, 2a, 3a, and 4a to study the channel activity of p7 in isolation but in a well-defined in vivo system. Sequences were selected according to alignments published by Carrère-Kremer et al. (19) (Table 1). In contrast to published HCV strains, in our p7 protein sequences there is one amino acid exchange for genotype 1a (p7-1a-H77: V41A) and two exchanges for genotype 3a (p7-3a-452: A13V and Y31H). In the case of genotype 2a, the p7 sequences of two HCV strains (J6 and JFH-1) were previously published (22). The chimeric strain JC-1 has the same sequence for the p7 protein as J6 and p7-2a used here. The p7 sequence of JFH-1 (also classified as genotype 2a) differs from our 2a, J6, and JC-1 in 12 amino acids. The well-established HEK293 system was chosen for cellular assays and electrophysiological measurements of p7 ion channels (46, 47). We generated genotype-specific p7 constructs carrying an N-terminal signal peptide that was taken from the inhibitory glycine receptor, a plasma membrane ion channel. This signal peptide is expected to transport the protein to the ER, from which it will be distributed to both the outer membrane and intracellular membranes. Indeed, expression in HEK293 cells yielded an active protein that was found in membrane fractions of the cell. We could identify p7 channel activity by whole-cell, patch-clamp measurements, which are directed toward proteins in the outer plasma membrane, and hemadsorption assays and measurements of intracellular vesicular activity indicated that p7 activity was also present at intracellular membranes.
Cellular pH imaging and hemadsorption assay
To validate our patch-clamp results, we performed two different cell-based assays of p7 function in living HEK293 cells: one to measure acidification of the cellular organelles and endosomes, and one to test the transportation of H5HA to the cell surface after p7 protein expression in exocytic vesicles. Both assays were performed on each of the four genotypes in the absence and presence of the inhibitors amantadine and rimantadine. Activity of p7 protein in HEK293 cells was completely inhibited by rimantadine in all four genotypes. Amantadine, on the other hand, showed significant differences between genotypes. Genotypes 1a and 4a were sensitive to amantadine, whereas genotypes 2a and 3a were not. Data from both cellular assays agreed well with results from electrophysiological measurements. Overall, the cellular assays confirmed that the artificial p7 channels were expressed in the recombinant system, forming functional intracellular regulators of vesicular pH. Our findings reveal a genotype-specific pharmacology of the p7 ion channel.
Characterization of p7 proteins by electrophysiological methods
We used a membrane-directing signal peptide and expressed SP-p7 chimeric constructs in HEK293 cells. This system is widely and successfully used for the characterization of neuronal ion channels. The signal peptide was taken from the inhibitory glycine receptor, a plasma membrane ion channel, and is primarily expected to transport the protein to the ER, from which it will be distributed to the outer and intracellular membranes. Patch-clamp recording conditions for recombinant p7 channels had to be optimized, and considerable effort was spent searching for the ideal buffer system for patch-clamp recordings. A buffer system that was previously used for electrophysiological measurements of M2 channels of the influenza virus (35) was adapted, and it provided stable and reproducible currents for recombinant p7 channels.
To date, several studies on viral ion channels in black lipid membranes have been published in which synthesized or recombinant protein was allowed to self-assemble in artificial phospholipid membranes (20, 30, 31). Only a few electrophysiological studies have been performed in recombinant systems such as oocytes and mammalian cell lines. Some pioneering work was done on the M2 channel of the influenza A virus, which showed amantadine and rimantadine inhibition of the channel expressed in oocytes (48, 49, 50) or host cells (51). Studies of the p7 ion channel in liposomes (52) and artificial bilayer membranes (26) showed that p7 mediates pH-sensitive channel activity. Several studies described p7 activity as an intracellular pH shunt that is required for viral assembly and release (20, 21, 26, 29, 31, 34, 44, 53, 54). Recently, data from an NMR structure determination were correlated with the function of p7 channels as determined from I-V relationships in p7-transfected oocytes (23). Likely, regulation of intracellular pH (33) is one relevant function of p7 in the replication cycle of HCV. Taken together, these results indicate that p7 is a pH-sensitive proton and cation channel whose inhibition interferes with virus assembly and release (20, 26, 29, 44, 54, 55). Genotype-specific variation in p7 activity and virus infectivity has been shown (27, 44, 55), and our data confirm the genotype-specific activity and drug sensitivity of recombinant p7 channel function.
pH and I-V dependence
We found pH50 values between 6.0 and 6.5, and similar I-V relationships for all tested genotypes (Fig. 4; Table 2). To our knowledge, no pH50 values have been previously determined for the p7 protein. Similar properties for p7 channels of genotypes 1–4 would be expected, as they represent p7 from active virus subtypes. The pH dependence we found for p7 is similar to that found for the M2 channel of the influenza A virus by patch-clamp electrophysiology (35, 51) and is consistent with a proton-gated channel.
Inhibition by rimantadine and amantadine
Various studies have examined the effect of amantadine and rimantadine on HCV p7 channels, usually focusing on genotypes 1a, 1b, and 2a. Robust inhibition by rimantadine was observed throughout (23, 33, 55). In the case of amantadine, the results published in the literature are controversial. Some reports described amantadine as a p7 inhibitor with potency similar to that of rimantadine (21, 34, 52), whereas other studies found no inhibition at all (54) or even augmentation (26) of p7 function by amantadine. Similar ambiguity exists in clinical situations, where amantadine has often been reported to have no effect in triple therapy (7, 56, 57). Furthermore, it was shown that HCV constructs corresponding to genotypes 1a, 1b, and 3 were amantadine sensitive, whereas results from clones of genotype 2a were ambiguous, showing inhibition of JC1, but not JFH-1, by amantadine (55).
In our experiments, all genotypes were robustly inhibited by rimantadine, with IC50 values in the nanomolar range and the lowest potency observed for genotype 2a. The sensitivities of genotypes 1a and 4a for amantadine were in the same range as those for rimantadine (2.4 nM and 12 nM, respectively). The IC50 values for genotypes 2a and 3a, on the other hand, were 50- to 1000-fold higher. Thus, 2a and 3a p7 channels were essentially insensitive to amantadine in patch-clamp experiments. In agreement with published data (55), we observed the least inhibitory effect of amantadine with genotype 2a. Surprisingly, the p7-2a sequence used here (amantadine insensitive) is identical to that of strains J6 and JC-1, which were reported to be amantadine sensitive (55). Nevertheless, the results indicate that p7 channels of genotype 2a have variable responses and show the least sensitivity to amantadine. In our hands, p7-3a channels also were less sensitive to amantadine (Fig. 5 C), with IC50 values being ∼10-fold higher than that of 2a but still 50- to 100-fold lower than those observed for genotypes 1 and 4 (Table 2). The results from patch-clamp recordings were consistent with those obtained from cellular assays of p7 activity, with both approaches showing independently that genotypes 2 and 3 are less sensitive to amantadine than genotypes 1 and 4.
Notably, the IC50 values from patch-clamp recordings were in the nanomolar range, whereas in cell-based assays the inhibitors were applied in micromolar concentrations. This numerical difference is likely due to the fact that in patch-clamp recordings, the p7 channel is directly exposed to the drug, which does not need to enter the cell and diffuse to its target. Thus, the numerical values of inhibition constants from different assays cannot be directly equated, but the trends between genotypes and inhibitors that we found were robust and consistent between different assay methods and with published data.
Conclusions
Here, we have presented a novel, to our knowledge, combination of techniques for studying the function of HCV p7 channels by directly recording their intrinsic transmembrane currents. Constructs of hepatitis C p7 protein corresponding to genotypes 1–4 were generated, expressed in HEK293 cells, and studied using patch-clamp techniques. All genotypes had p7 channels of similar permeation properties and pH dependence. The p7 channels of genotypes 1–4 were sensitive to rimantadine, and the amantadine sensitivity of genotypes 2 and 3 was 30- to 200-fold lower than that of genotypes 1 and 4.
The patch-clamp data were confirmed by established cellular assays of p7 function. All tests consistently showed a decreased sensitivity of genotypes 2 and 3 to amantadine, whereas rimantadine robustly inhibited p7 channels of all genotypes studied. Our results are in agreement with published findings that show a greater variety of amantadine sensitivity among p7 channels from different genotypes. Patch-clamp studies of recombinant viroporins complement the existing array of functional assays and may be a valuable tool for studying p7 channel function and its sequence and genotype dependency, as well as for developing novel therapeutic approaches.
Author Contributions
U.B. and H.-G.B. designed the research and wrote the manuscript. U.B., N.S.F., and N.K.M.A. performed the research. U.B., N.S.F., N.K.M.A., and H.-G.B. analyzed the data.
Acknowledgments
We thank Prof. Stephen Griffin (University of Leeds) and Prof. Wendy S. Barclay (Imperial College London) for the gift of HA-cDNA and Prof. Cord-Michael Becker (University of Erlangen) for the donation of HEK293 cells.
This work was supported by the Science and Technology Development Fund in Egypt (grant 1785 to H.-G.B.).
Editor: Joseph Mindell.
Footnotes
Supporting Materials and Methods and one figure are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30213-2.
Supporting Material
References
- 1.Messina J.P., Humphreys I., Barnes E. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology. 2015;61:77–87. doi: 10.1002/hep.27259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mohamoud Y.A., Mumtaz G.R., Abu-Raddad L.J. The epidemiology of hepatitis C virus in Egypt: a systematic review and data synthesis. BMC Infect. Dis. 2013;13:288. doi: 10.1186/1471-2334-13-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lauer G.M., Walker B.D. Hepatitis C virus infection. N. Engl. J. Med. 2001;345:41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 4.Zein N.N. Clinical significance of hepatitis C virus genotypes. Clin. Microbiol. Rev. 2000;13:223–235. doi: 10.1128/cmr.13.2.223-235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamal S.M., Nasser I.A. Hepatitis C genotype 4: what we know and what we don’t yet know. Hepatology. 2008;47:1371–1383. doi: 10.1002/hep.22127. [DOI] [PubMed] [Google Scholar]
- 6.El-Zayadi A.R., Attia M., Saied A. Response of hepatitis C genotype-4 naïve patients to 24 weeks of Peg-interferon-alpha2b/ribavirin or induction-dose interferon-alpha2b/ribavirin/amantadine: a non-randomized controlled study. Am. J. Gastroenterol. 2005;100:2447–2452. doi: 10.1111/j.1572-0241.2005.00253.x. [DOI] [PubMed] [Google Scholar]
- 7.Maynard M., Pradat P., Trépo C., French Multicenter Group Amantadine triple therapy for non-responder hepatitis C patients. Clues for controversies (ANRS HC 03 BITRI) J. Hepatol. 2006;44:484–490. doi: 10.1016/j.jhep.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 8.Carlsen T.H., Pedersen J., Bukh J. Breadth of neutralization and synergy of clinically relevant human monoclonal antibodies against HCV genotypes 1a, 1b, 2a, 2b, 2c, and 3a. Hepatology. 2014;60:1551–1562. doi: 10.1002/hep.27298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meuleman P., Bukh J., Leroux-Roels G. In vivo evaluation of the cross-genotype neutralizing activity of polyclonal antibodies against hepatitis C virus. Hepatology. 2011;53:755–762. doi: 10.1002/hep.24171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhopale G.M., Nanda R.K. Emerging drugs for chronic hepatitis C. Hepatol. Res. 2005;32:146–153. doi: 10.1016/j.hepres.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Colombo M. Interferon-free therapy for hepatitis C: the hurdles amid a golden era. Dig. Liver Dis. 2015;47:727–733. doi: 10.1016/j.dld.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 12.El-Zayadi A.R., Attia M., Saied A. Non-interferon-based therapy: an option for amelioration of necro-inflammation in hepatitis C patients who cannot afford interferon therapy. Liver Int. 2005;25:746–751. doi: 10.1111/j.1478-3231.2005.01110.x. [DOI] [PubMed] [Google Scholar]
- 13.Ferenci P. Treatment of hepatitis C in difficult-to-treat patients. Nat. Rev. Gastroenterol. Hepatol. 2015;12:284–292. doi: 10.1038/nrgastro.2015.53. [DOI] [PubMed] [Google Scholar]
- 14.Kwo P.Y., Badshah M.B. New hepatitis C virus therapies: drug classes and metabolism, drug interactions relevant in the transplant settings, drug options in decompensated cirrhosis, and drug options in end-stage renal disease. Curr. Opin. Organ Transplant. 2015;20:235–241. doi: 10.1097/MOT.0000000000000198. [DOI] [PubMed] [Google Scholar]
- 15.Noell B.C., Besur S.V., deLemos A.S. Changing the face of hepatitis C management—the design and development of sofosbuvir. Drug Des. Devel. Ther. 2015;9:2367–2374. doi: 10.2147/DDDT.S65255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stahmeyer J.T., Rossol S., Krauth C. Outcomes, costs and cost-effectiveness of treating hepatitis C with direct acting antivirals. J. Comp. Eff. Res. 2015;11:1–11. doi: 10.2217/cer.15.13. [DOI] [PubMed] [Google Scholar]
- 17.De Francesco R. Molecular virology of the hepatitis C virus. J. Hepatol. 1999;31(Suppl 1):47–53. doi: 10.1016/s0168-8278(99)80374-2. [DOI] [PubMed] [Google Scholar]
- 18.Dubuisson J. Hepatitis C virus proteins. World J. Gastroenterol. 2007;13:2406–2415. doi: 10.3748/wjg.v13.i17.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrère-Kremer S., Montpellier-Pala C., Dubuisson J. Subcellular localization and topology of the p7 polypeptide of hepatitis C virus. J. Virol. 2002;76:3720–3730. doi: 10.1128/JVI.76.8.3720-3730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Premkumar A., Wilson L., Gage P.W. Cation-selective ion channels formed by p7 of hepatitis C virus are blocked by hexamethylene amiloride. FEBS Lett. 2004;557:99–103. doi: 10.1016/s0014-5793(03)01453-4. [DOI] [PubMed] [Google Scholar]
- 21.Griffin S.D., Beales L.P., Rowlands D.J. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, Amantadine. FEBS Lett. 2003;535:34–38. doi: 10.1016/s0014-5793(02)03851-6. [DOI] [PubMed] [Google Scholar]
- 22.Clarke D., Griffin S., Rowlands D. Evidence for the formation of a heptameric ion channel complex by the hepatitis C virus p7 protein in vitro. J. Biol. Chem. 2006;281:37057–37068. doi: 10.1074/jbc.M602434200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.OuYang B., Xie S., Chou J.J. Unusual architecture of the p7 channel from hepatitis C virus. Nature. 2013;498:521–525. doi: 10.1038/nature12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cook G.A., Opella S.J. NMR studies of p7 protein from hepatitis C virus. Eur. Biophys. J. 2010;39:1097–1104. doi: 10.1007/s00249-009-0533-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patargias G., Zitzmann N., Fischer W.B. Protein-protein interactions: modeling the hepatitis C virus ion channel p7. J. Med. Chem. 2006;49:648–655. doi: 10.1021/jm050721e. [DOI] [PubMed] [Google Scholar]
- 26.Montserret R., Saint N., Penin F. NMR structure and ion channel activity of the p7 protein from hepatitis C virus. J. Biol. Chem. 2010;285:31446–31461. doi: 10.1074/jbc.M110.122895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalita M.M., Griffin S., Fischer W.B. Genotype-specific differences in structural features of hepatitis C virus (HCV) p7 membrane protein. Biochim. Biophys. Acta. 2015;1848:1383–1392. doi: 10.1016/j.bbamem.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dev J., Brüschweiler S., Chou J.J. Transverse relaxation dispersion of the p7 membrane channel from hepatitis C virus reveals conformational breathing. J. Biomol. NMR. 2015;61:369–378. doi: 10.1007/s10858-015-9912-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinmann E., Penin F., Pietschmann T. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 2007;3:e103. doi: 10.1371/journal.ppat.0030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pavlović D., Neville D.C., Zitzmann N. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc. Natl. Acad. Sci. USA. 2003;100:6104–6108. doi: 10.1073/pnas.1031527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pavlovic D., Fischer W., Zitzmann N. Long alkylchain iminosugars block the HCV p7 ion channel. Adv. Exp. Med. Biol. 2005;564:3–4. doi: 10.1007/0-387-25515-X_2. [DOI] [PubMed] [Google Scholar]
- 32.Breitinger U., Breitinger H.G., Becker C.M. Conserved high affinity ligand binding and membrane association in the native and refolded extracellular domain of the human glycine receptor alpha1-subunit. J. Biol. Chem. 2004;279:1627–1636. doi: 10.1074/jbc.M303811200. [DOI] [PubMed] [Google Scholar]
- 33.Wozniak A.L., Griffin S., Weinman S.A. Intracellular proton conductance of the hepatitis C virus p7 protein and its contribution to infectious virus production. PLoS Pathog. 2010;6:e1001087. doi: 10.1371/journal.ppat.1001087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griffin S.D., Harvey R., Rowlands D.J. A conserved basic loop in hepatitis C virus p7 protein is required for amantadine-sensitive ion channel activity in mammalian cells but is dispensable for localization to mitochondria. J. Gen. Virol. 2004;85:451–461. doi: 10.1099/vir.0.19634-0. [DOI] [PubMed] [Google Scholar]
- 35.Chizhmakov I.V., Ogden D.C., Hay A.J. Differences in conductance of M2 proton channels of two influenza viruses at low and high pH. J. Physiol. 2003;546:427–438. doi: 10.1113/jphysiol.2002.028910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gottwein J.M., Scheel T.K., Bukh J. Development and characterization of hepatitis C virus genotype 1-7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology. 2009;49:364–377. doi: 10.1002/hep.22673. [DOI] [PubMed] [Google Scholar]
- 37.Lindenbach B.D., Meuleman P., Rice C.M. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc. Natl. Acad. Sci. USA. 2006;103:3805–3809. doi: 10.1073/pnas.0511218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pietschmann T., Kaul A., Bartenschlager R. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. USA. 2006;103:7408–7413. doi: 10.1073/pnas.0504877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinmann E., Pietschmann T. Cell culture systems for hepatitis C virus. Curr. Top. Microbiol. Immunol. 2013;369:17–48. doi: 10.1007/978-3-642-27340-7_2. [DOI] [PubMed] [Google Scholar]
- 40.Li Y.P., Ramirez S., Bukh J. Efficient infectious cell culture systems of the hepatitis C virus (HCV) prototype strains HCV-1 and H77. J. Virol. 2015;89:811–823. doi: 10.1128/JVI.02877-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Date T., Kato T., Wakita T. Novel cell culture-adapted genotype 2a hepatitis C virus infectious clone. J. Virol. 2012;86:10805–10820. doi: 10.1128/JVI.07235-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim S., Date T., Wakita T. Development of hepatitis C virus genotype 3a cell culture system. Hepatology. 2014;60:1838–1850. doi: 10.1002/hep.27197. [DOI] [PubMed] [Google Scholar]
- 43.Mathiesen C.K., Jensen T.B., Gottwein J.M. Production and characterization of high-titer serum-free cell culture grown hepatitis C virus particles of genotype 1-6. Virology. 2014;458-459:190–208. doi: 10.1016/j.virol.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Griffin S. Inhibition of HCV p7 as a therapeutic target. Curr. Opin. Investig. Drugs. 2010;11:175–181. [PubMed] [Google Scholar]
- 45.Pawlotsky J.M. Pathophysiology of hepatitis C virus infection and related liver disease. Trends Microbiol. 2004;12:96–102. doi: 10.1016/j.tim.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 46.Breitinger H.G., Becker C.M. The inhibitory glycine receptor-simple views of a complicated channel. ChemBioChem. 2002;3:1042–1052. doi: 10.1002/1439-7633(20021104)3:11<1042::AID-CBIC1042>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 47.Breitinger H.G. Fast kinetic analysis of ligand-gated ion channels. Neuroscientist. 2001;7:95–103. doi: 10.1177/107385840100700204. [DOI] [PubMed] [Google Scholar]
- 48.Wang C., Lamb R.A., Pinto L.H. Direct measurement of the influenza A virus M2 protein ion channel activity in mammalian cells. Virology. 1994;205:133–140. doi: 10.1006/viro.1994.1628. [DOI] [PubMed] [Google Scholar]
- 49.Wang C., Takeuchi K., Lamb R.A. Ion channel activity of influenza A virus M2 protein: characterization of the amantadine block. J. Virol. 1993;67:5585–5594. doi: 10.1128/jvi.67.9.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang C., Lamb R.A., Pinto L.H. Activation of the M2 ion channel of influenza virus: a role for the transmembrane domain histidine residue. Biophys. J. 1995;69:1363–1371. doi: 10.1016/S0006-3495(95)80003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chizhmakov I.V., Geraghty F.M., Hay A.J. Selective proton permeability and pH regulation of the influenza virus M2 channel expressed in mouse erythroleukaemia cells. J. Physiol. 1996;494:329–336. doi: 10.1113/jphysiol.1996.sp021495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.StGelais C., Tuthill T.J., Griffin S. Inhibition of hepatitis C virus p7 membrane channels in a liposome-based assay system. Antiviral Res. 2007;76:48–58. doi: 10.1016/j.antiviral.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Griffin S., Trowbridge R., Bright H. Chimeric GB virus B genomes containing hepatitis C virus p7 are infectious in vivo. J. Hepatol. 2008;49:908–915. doi: 10.1016/j.jhep.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.StGelais C., Foster T.L., Griffin S. Determinants of hepatitis C virus p7 ion channel function and drug sensitivity identified in vitro. J. Virol. 2009;83:7970–7981. doi: 10.1128/JVI.00521-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Griffin S., Stgelais C., Harris M. Genotype-dependent sensitivity of hepatitis C virus to inhibitors of the p7 ion channel. Hepatology. 2008;48:1779–1790. doi: 10.1002/hep.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.von Wagner M., Hofmann W.P., Zeuzem S. Placebo-controlled trial of 400 mg amantadine combined with peginterferon alfa-2a and ribavirin for 48 weeks in chronic hepatitis C virus-1 infection. Hepatology. 2008;48:1404–1411. doi: 10.1002/hep.22483. [DOI] [PubMed] [Google Scholar]
- 57.van Soest H., van der Schaar P.J., Boland G.J. No beneficial effects of amantadine in treatment of chronic hepatitis C patients. Dig. Liver Dis. 2010;42:496–502. doi: 10.1016/j.dld.2009.10.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.