Abstract
Life arose in a world without oxygen and the first organisms were anaerobes. Here we investigate the gene repertoire of the prokaryote common ancestor, estimating which genes it contained and to which lineages of modern prokaryotes it was most similar in terms of gene content. Using a phylogenetic approach we found that among trees for all 8779 protein families shared between 134 archaea and 1847 bacterial genomes, only 1045 have sequences from at least two bacterial and two archaeal groups and retain the ancestral archaeal–bacterial split. Among those, the genes shared by anaerobes were identified as candidate genes for the prokaryote common ancestor, which lived in anaerobic environments. We find that these anaerobic prokaryote common ancestor genes are today most frequently distributed among methanogens and clostridia, strict anaerobes that live from low free energy changes near the thermodynamic limit of life. The anaerobic families encompass genes for bifunctional acetyl-CoA-synthase/CO-dehydrogenase, heterodisulfide reductase subunits C and A, ferredoxins, and several subunits of the Mrp-antiporter/hydrogenase family, in addition to numerous S-adenosyl methionine (SAM) dependent methyltransferases. The data indicate a major role for methyl groups in the metabolism of the prokaryote common ancestor. The data furthermore indicate that the prokaryote ancestor possessed a rotor stator ATP synthase, but lacked cytochromes and quinones as well as identifiable redox-dependent ion pumping complexes. The prokaryote ancestor did possess, however, an Mrp-type H+/Na+ antiporter complex, capable of transducing geochemical pH gradients into biologically more stable Na+-gradients. The findings implicate a hydrothermal, autotrophic, and methyl-dependent origin of life. This article is part of a Special Issue entitled ‘EBEC 2016: 19th European Bioenergetics Conference, Riva del Garda, Italy, July 2–6, 2016’, edited by Prof. Paolo Bernardi.
Abbreviations: HCO, heme–copper oxygen reductase, cytochrome c oxidase, complex IV; LGT, lateral gene transfer; NOR, nitric oxide reductase; WL, Wood–Ljungdahl; SAM, S-adenosyl methionine; SLP, substrate level phosphorylation
Keywords: Early evolution, Geochemistry, Methanogens, Acetogens, Anaerobes, Autotrophy
Highlights
-
•
Life arose without oxygen, the universal ancestor (Luca) was an anaerobe.
-
•
We used phylogenetic and physiological criteria to identify genes present in Luca.
-
•
An ancient core of 65 metabolic genes shed light on Luca's anaerobic lifestyle.
-
•
Ancient core genes are most widespread among modern methanogens and clostridia.
-
•
The data implicate a major role for methyl groups in Luca's anaerobic metabolism.
“It must be clear that all the changes and the original life forms are dependent upon energy as well as material capture and flow.”
Williams and Fraústo da Silva [113]
1. Introduction
One of the more intriguing enterprises in comparative genomics is to infer the nature of the last universal common ancestor, also called Luca, on the basis of gene content [46], [57], [73], [6], [74], [75], [81], [112]. The standard approach to the problem is to generate a reference tree – sometimes called a backbone tree or species tree – and then to plot the distribution of gene families, usually the COGs, or clusters of orthologous groups [110] onto the leaves of the tree and then to infer presence and absence patterns along the inner branches and nodes of the tree, right down to its root, the presence and absence patterns at the root giving an estimate of Luca's gene content [6], [27], [46], [73], [74], [75], [81], [112]. The models that one assumes for gene gain and loss have a considerable impact on the inferred genome of Luca [23], [24], [46], [73], [100] as does the selected reference tree [31] and the genome collection of the study.
In early investigations of Luca gene content, Luca was considered as the last common ancestor of bacteria, archaea and eukaryotes [115]. More recent findings have eukaryotic ribosomes branching within the archaea, rather than as their sister [22], [87], [108], [114], such that in the more modern “two domain” trees, Luca is the last common ancestor of prokaryotes. To stress that, one could introduce the term last prokaryotic common ancestor, or Lpca. But new terms for established concepts are seldom helpful, and Luca means different things to different people anyway. Here we stick to the term Luca, but we use it here to mean the last common ancestor of prokaryotes, which in our view of early evolution was not a free living cell, but rather was an entity that had the genetic code, that had proteins, that had ribosomes and an ATPase [106], that could make DNA as a stable repository for retrievable information, but probably could not replicate DNA as chromosomes [51], [53], and that – we posit – probably was contained within naturally forming inorganic compartments as chemical confines of a geological structure like a hydrothermal vent, which supplied the reduced carbon and continuous chemical disequilibrium (energy supply) that Luca needed to get organized in the first place [65], [66], [59], [107]. But irrespective of where and how it arose, newer phylogenetic data indicate that eukaryotes need to be excluded when it comes to estimating the gene content of Luca.
Excluding eukaryotes has an immense effect on Luca gene content estimation. This is because current views and current data have it that eukaryotes arose from a symbiosis of two prokaryotes, the bacterial ancestor of mitochondria and its archaeal host [1], [22], [54], [58], [64], [90], and there are only about 2585 gene families that eukaryotes share widely with prokaryotes [55]. By including eukaryotes in Luca gene content estimation, one would be excluding all enzymes specific to anaerobic chemolithoautotrophy, all enzymes specific to anoxygenic photosynthesis, all enzymes specific to sulfate reduction, and all enzymes specific to all biochemical pathways that eukaryotes do not possess, which comprises the vast majority of genes distributed across prokaryotic genomes. Eukaryotes possess only a very, very small sample of prokaryotic energy metabolic diversity [120] and an even smaller sample of prokaryotic gene diversity in general [55]. It is thus important to estimate Luca gene content based upon all prokaryotic genes, not just the narrow sample of genes that eukaryotes inherited from prokaryotes at eukaryotic (and plastid) origin [55].
Removing the restriction that the inclusion of eukaryotes introduces into Luca gene content estimation is easy, one just excludes eukaryotic gene from the set to be considered for Luca inference. Far more problematic, however, is the issue of lateral gene transfer. This is because – even in studies that exclude eukaryotes from Luca inference – many studies score genes as present in Luca if the genes are present in several archaea and one (or more) bacterium, or present in several bacteria and one (or more) archaeon [9]. But such genes could easily be transdomain lateral gene transfers and not holdovers from Luca at all. In haloarchaea alone, there are more than 1000 well-documented cases of genes that were acquired from bacteria via transdomain lateral gene transfers [76]. In a broader sample of archaeal lineages, Nelson-Sathi et al. [77] identified more than 6000 cases of transdomain lateral gene transfers. In prokaryotes, LGT is not only frequent [10], [45], [50], [61] but it also played an important role in prokaryote lineage diversification [77]. Transdomain LGT generates gene distribution patterns that complicate the inference of Luca's gene content.
In an insightful paper, Kannan et al. [46] clearly outlined the problems that LGT introduces regarding Luca: If a gene family was invented relatively late in evolution, in a particular bacterial lineage, and then transferred across broad taxonomic boundaries (for example from bacteria to archaea or vice versa), then its phylogenetic distribution would erroneously mimic presence in Luca. If not recognized as LGTs, such genes lead to a vastly (and artefactually) inflated Luca genome content. If such interdomain LGT is widespread, reconstructing Luca's gene content becomes tedious. How to identify transdomain LGTs so as to remove their inflating effects upon Luca gene content estimation? We have a suggestion.
We recently reported a clustering and phylogenetic analysis of over 6 million genes from 1891 prokaryotic genomes, focusing on genes shared by archaea and bacteria [77]. We found that interdomain LGTs from bacteria to archaea vastly outnumbered gene transfers in the other direction and that gene acquisitions from bacteria correspond to the origin of several major archaeal groups [77]. In that study, 4705 protein families showed extensive interdomain LGT, and another 4397 protein families were identified where archaea and bacteria are monophyletic in the corresponding phylogenetic tree. Gene presence in archaea and bacteria, in addition to monophyly of archaea and bacteria, is the minimal condition that should be fulfilled for genes that were present in Luca but not subject to interdomain LGT. Among the 4397 protein families reported in which archaea and bacteria are monophyletic, 3347 cases represent fairly obvious interdomain LGTs in that the genes are widespread in bacteria but present in only one archaeal lineage [77]. The remaining 1045 genes show archaea and bacteria to be monophyletic but show no obvious signs of interdomain LGT. This set of genes is, in principle, a candidate list for genes present in Luca but not transferred between domains since the divergence of bacteria and archaea. These 1045 genes are therefore of interest and compose our starting point for the functional analysis of how the primordial ancestor of bacteria and archaea made a living.
Yet there still might be some gene families among those 1045 that, despite bi-domain presence and domain monophyly, were subject to interdomain LGTs that went undetected in our earlier report. For example, oxygen dependent enzymes can hardly have existed in Luca because life arose in a world without oxygen [42], [56], [67], but they might have been passed around promiscuously after the advent of oxygenated environments. Because Luca had to be an anaerobe (oxygen being a biological product), we can introduce one more criterion for Luca presence: oxygen dependent enzymes and pathways cannot have been present in Luca, such that enzymes and pathways specific to, or typically found among, aerobes (but not in anaerobes) can be excluded from Luca's gene set. To gain insights on the primordial metabolism of the common ancestor of bacteria and archaea before its diversification into the bacterial and archaea lineages, we set out here to identify protein families that span the archaeal–bacterial division, that were not subject to interdomain LGT, and that are preferentially found within the genomes of anaerobes. Of course, we cannot exclude the existence of other proteins in Luca, such as the ones widely shared by aerobic and anaerobic organisms, or some of the ones whose evolutionary history involved interdomain LGT events.
2. Methods
2.1. Aerotolerance profiling
The method described in Sousa et al. [105] was employed to identify and classify heme–copper oxygen reductases (HCOs) and nitric oxide reductases (NORs) across the genomes used in this study. Briefly, a manually curated database of 1225 sequences classified as being A1, A2, B and C type heme–copper oxygen reductases [82] or NORs was download from HCO database and used to query the genomes of 1981 prokaryotes from our dataset (blast cut-of 25% identity, E-value 10− 10, alignment coverage of at least 300 amino acids). In a second step, the obtained hits were classified as belonging to one of the five recognized enzyme types. An organism is considered aerobic if it contains genes coding for A1, A2, B or C type enzymes and anaerobic otherwise. Protein families were classified as aerobic or anaerobic if 90% of their representatives and, at least 85% of archaeal organisms and, at least 85% of bacterial organisms belong to the same classification. The remaining cases were classified as mixed families. Results are summarized in Supplemental Table A1.
2.2. Functional characterization of protein families
Functional annotations were retrieved from COG [37], KEGG [44], and when justified, Biocyc [17] and Brenda [95] databases. The combined information is available as Supplemental Table A2. In the wordle representation, common words such as hypothetical-protein, protein or subunit were removed as well as the superphyla Proteobacteria, Firmicutes, Crenarchaeota and Euryarchaeota taxonomic descriptions.
2.3. Distribution of O2 dependent reactions
Reactions, reaction directionality (according to metabolic pathways), KEGG orthology and Brite hierarchy were parsed from KEGG's database (June 2015) [44]. This allowed the mapping onto KEGG genomes of the reactions depending on O2 as in [88].
3. Results and discussion
3.1. Universal (or nearly so) genes
Genes that are widely distributed across prokaryotic domains were either present in Luca before the divergence of bacteria and archaea [27], [52] or were subject to interdomain LGT [46]. We previously clustered 134 archaeal genomes into 25,762 protein families and identified their corresponding homologs among 1847 bacterial genomes [77]. In that dataset, the genomes span, according to prokaryote systematics, 13 archaeal and 23 bacterial higher taxonomic groups respectively, which roughly correspond to phyla (or class) and are designated for convenience henceforth as phyla here.
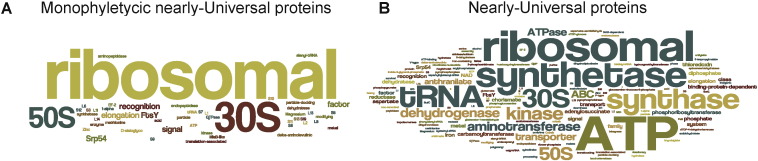
If we search for nearly universal protein families using the criteria of i) presence in at least 22 bacterial phyla (missing in only one phylum) and at least 12 archaeal phyla (missing in only one phylum) and ii) monophyly of the domains within the corresponding maximum likelihood tree, we end up with a set of 27 nearly-universal protein families (Fig. 1a), corresponding to the familiar set of 30–35 “core” genes for (mostly ribosomal) proteins that are now commonly used to infer lineage relationships in place of rRNA alone [20], [21], [22], [39], [108], [114].
Fig. 1.
Wordle representation of the most frequent functional descriptions within a) 27 interdomain nearly universal monophyletic protein families and b) 109 nearly universal protein families. The size of the words relates to the number of times the word appears within the annotations. The larger the word, the higher its frequency.
If we allow for some gene loss during evolution (or rapid sequence divergence in some lineages) and thus opt for a less stringent distribution criterion, and furthermore relax the criterion for domain monophyly, our extended protein set consists of 109 protein families that include 9 aminoacyl-tRNA synthetase families, whose complex evolutionary history is well known [116], [117], several enzymes involved in amino acid biosynthesis and ATP synthase subunits (Fig. 1b, Table 1). This extended, or nearly universal, set corresponds very closely in content to the 102 nearly universal trees, or “nuts”, reported by Puigbò et al. [84]), in that sense we could independently reproduce (109 genes) their nearly universal tree set (102 genes). The core and the extended core thus indicate the (obvious) presence of ribosomes in Luca [33], [74], an ion-gradient-dependent energy harvesting machinery [59], and the presence of some amino acid biosynthesis.
Table 1.
Functional category of the nearly universal protein families and the 1045 families that retain the archaeal bacterial division.
| Nearly universal mono. | Nearly universal | 1045 mono. families | Anaerobic | Aerobic | Mixed | |
|---|---|---|---|---|---|---|
| Cellular processes and signaling | ||||||
| Cell cycle control, division, chromos part. | 0 | 1 | 9 | 0 | 0 | 9 |
| Cell motility | 0 | 0 | 9 | 0 | 0 | 9 |
| Cell wall/membrane/envelope biogenesis | 0 | 5 | 50 | 0 | 1 | 49 |
| Defense mechanisms | 0 | 1 | 49 | 4 | 1 | 44 |
| Extracellular structures | 0 | 0 | 1 | 0 | 0 | 1 |
| Intra traff., secretion, and vesicular transport | 2 | 2 | 14 | 0 | 2 | 12 |
| Mobilome: prophages, transposons | 0 | 0 | 21 | 1 | 0 | 20 |
| Nuclear structure | 0 | 0 | 0 | 0 | 0 | 0 |
| Post-transl. mod., protein turnover, chaperones | 1 | 3 | 33 | 1 | 4 | 28 |
| Signal transduction mechanisms | 0 | 0 | 25 | 0 | 2 | 23 |
| Information, storage and processing | ||||||
| Chromatin structure and dynamics | 0 | 0 | 0 | 0 | 0 | 0 |
| Replication, recombination and repair | 0 | 3 | 30 | 1 | 2 | 27 |
| RNA processing and modification | 0 | 0 | 0 | 0 | 0 | 0 |
| Transcription | 0 | 0 | 46 | 2 | 5 | 39 |
| Translation, ribosomal struct. and biogenesis | 23 | 34 | 91 | 6 | 0 | 85 |
| Metabolism | ||||||
| Amino acid transport and metabolism | 0 | 22 | 68 | 3 | 5 | 60 |
| Carbohydrate transport and metabolism | 0 | 5 | 75 | 4 | 3 | 68 |
| Coenzyme transport and metabolism | 1 | 7 | 53 | 3 | 1 | 49 |
| Energy production and conversion | 0 | 5 | 91 | 10 | 11 | 70 |
| Inorganic ion transport and metabolism | 0 | 4 | 56 | 2 | 6 | 48 |
| Lipid transport and metabolism | 0 | 1 | 41 | 0 | 7 | 34 |
| Nucleotide transport and metabolism | 0 | 13 | 15 | 1 | 2 | 12 |
| Synth., transp. cat. metabolites | 0 | 1 | 23 | 0 | 7 | 16 |
| Poorly characterized | ||||||
| Function unknown | 0 | 0 | 68 | 5 | 4 | 59 |
| General function prediction only | 0 | 2 | 117 | 7 | 12 | 98 |
| Not found | 0 | 0 | 60 | 12 | 4 | 44 |
| Total | 27 | 109 | 1045 | 62 | 79 | 904 |
However, neither the core nor the expanded core (or nearly universal set) deliver information regarding the type of carbon and energy metabolism of primordial cells, because microbial metabolism has diversified within and across lineages during 3.5 billion years of microbial evolution. But the size of both the core (~ 30 genes) and the extended core (~ 100 genes) indicates that the clusters that we are using [77] deliver universal and nearly universal gene family distributions that correspond very well with what others have found independently using smaller genome samples and different methods [39], [84]. From this point on, we will focus on the remaining non-universal protein families that might have been present in Luca.
3.2. Distinction between recent and ancient protein families
In a previous study [77] we identified 4397 protein families that retained the monophyly of archaea and bacteria in maximum likelihood trees (Fig. 2a). However 3347 of those correspond to protein families in which either i) several bacteria and only one archaeal lineage or ii) several archaea and only one bacterial lineage group is represented. We further include in this group, for thoroughness, five protein families in which the archaeal representatives belong to Desulfurococcales and Fervidicoccus fontis Kam984 (here it is grouped with Desulfurococcales). The narrow phylogenetic distributions of these 3352 protein families (present in only one group in one of the domains) indicate that they correspond to interdomain LGT events that occurred after the prokaryotic domains had already diversified into major lineages. As such, they contain information about LGT frequencies, which is not our focus here, but do not contain direct clues about early metabolism and were excluded from our present analysis.
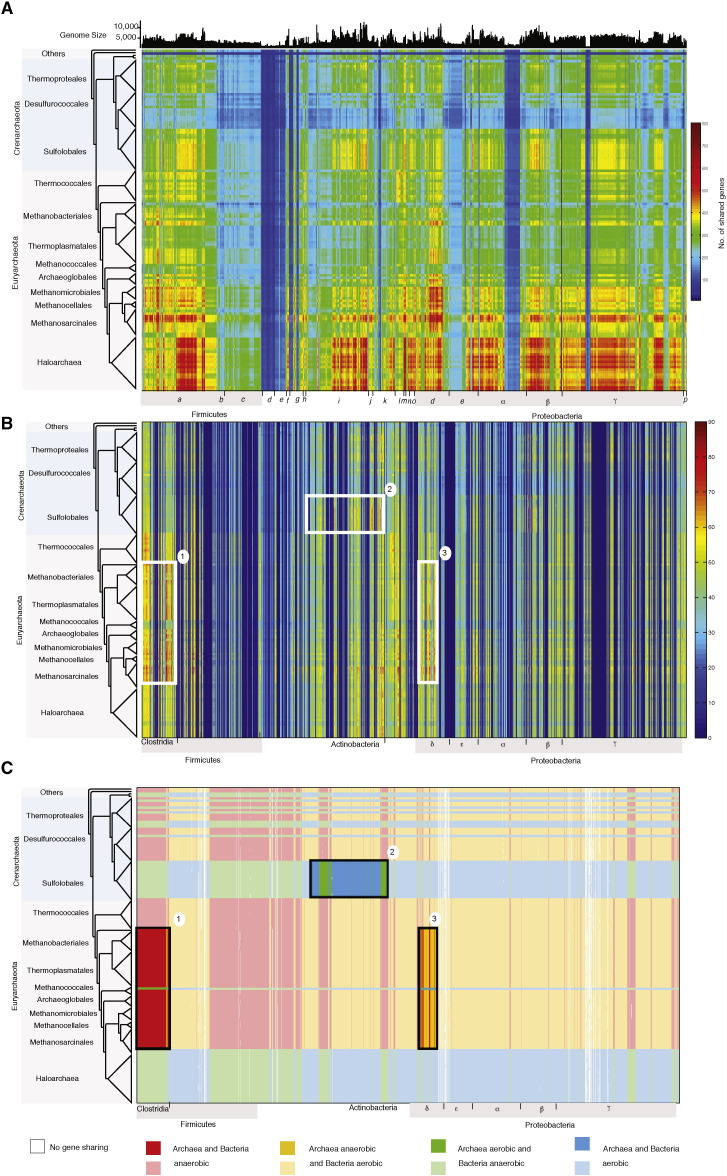
Fig. 2.
Inter-domain gene sharing and aerobic profile network of families that retain the archaeal bacteria division. Number of genes shared between archaeal and bacterial organisms A) within the monophyletic 4397 protein families and B) within the remaining 1045 monophyletic protein families after removal of obvious interdomain LGTs. Each cell in the matrix indicates the number of genes (E-value ≤ 10− 10 and ≥ 25% global identity) shared between the protein families of 134 archaeal and 1847 bacterial genomes whose tree retain the archaea–bacteria division (scale bar at right). C) Inter-domain gene sharing network in terms of the aerobic classification of the organism pairs. Panel A is adapted from [77].
When we exclude those interdomain LGTs, following the suggestion of Kannan et al. [46], what remains is a set of 1045 protein families, containing sequences from at least two archaeal groups and at least two bacterial groups are present in the families and where the domains are monophyletic in phylogenetic trees. Fig. 2b shows which archaeal lineages and which bacterial lineages harbor these Luca genome candidates. Their patterns of gene sharing are not randomly distributed among either archaea or bacteria, rather they are preferentially distributed in pairs of lineages (taxa) from each domain. These taxon pairs are boxed and labeled with numbers in Fig. 2b: 1) clostridial and methanogenic lineages, 2) actinobacterial and Sulfolobales lineages, and 3) deltaproteobacterial and methanogen lineages. The boxed taxon pairs identify archaeal and bacterial lineages that share Luca candidate genes, that is, genes that are i) present in archaea and bacteria, but ii) not present in all bacteria and archaea (which we expect for Luca's genes, because Luca's habitat was different from today's), iii) where the domains do not interleave in the 1045 maximum likelihood trees, and iv) where the gene family is present in more than one archaeal lineage and more than one bacterial lineage (that is, lineage specific interdomain LGTs have been filtered out). Within the Luca candidate genes that identify the lineage pairs 1–3, the COG categories “energy production and conversion” and “carbohydrate metabolism” are among the most prominently represented (Table 1).
3.3. Ancient means anaerobic
However, even for these Luca candidate gene protein families, it is still possible that domain monophyly stems from lineage specific interdomain LGT and subsequent within domain transfer. This mechanism of distribution could apply both to ancient genes present in Luca and to later lineage specific inventions and/or later environment specific genes, for example oxygenic environments. Here we seek to identify ancient proteins. Because Luca arose in anaerobic environments [26], [67], proteins that arose in, or are typical for, oxygenated environments cannot be ancient, hence we would like to exclude them from the Luca candidate gene set. If we understand the literature of geochemists correctly, nobody can say for sure at the moment when the first oxygen arose [62], but we can be reasonably sure that it was present in the atmosphere roughly 2.5 billion years ago [42], [99] and accumulated in the oceans roughly 600 million years ago [62], [109]. A few might disagree [79] and argue for the presence of O2 since the early Archaean. Yet despite some uncertainty about when oxygen arose, we can be relatively sure that Luca arose in a world without appreciable amounts of oxygen [56], [67], because oxygen is a product of cyanobacterial photosynthesis involving two photosystems, which is a highly derived form of microbial physiology, having arisen after anoxygenic photosynthesis, cytochrome dependent respirations, fermentations and autotrophy [26], [67], [96].
Thus, in the search for a list of bona fide Luca candidate genes, the next pruning step is to look for the protein families shared only by anaerobic organisms, meaning that we filter aerobes and proteins typical of aerobes from the data. For this, we have to ascertain the oxygen tolerance or oxygen requirements of the 1981 organisms within our dataset. How to do this in the absence of specific growth information for each genome, and taking facultative aerobes into account?
Microbial physiology can help. To reduce O2 to water, prokaryotes use two evolutionary unrelated membrane complexes, the bd oxygen reductase and the heme–copper oxygen reductases (HCOs, also known as the complex IV or cytochrome c oxidase). While the bd oxygen reductase is generally associated with oxygen detoxification or very low oxygen environments [11], the HCOs are incorporated in prokaryotic electron transfer chains that are much more diverse than the canonical mitochondrial one, but, as in the case of eukaryotes, that also promote the establishment of an electrochemical cation gradient across the membrane to feed the universal ATP synthase [82], [29]. Since organisms that express bd oxygen reductases usually also possess heme–copper oxygen reductases [11], one way to assess the oxygen requirements of the organisms present in our dataset is simply to look for the presence of HCOs in their genome. This is not trivial, though, because HCOs are related (structurally and sequence-wise) with nitric oxide reductases (NORs), with which they share the same general structural core of subunit I, the presence of a low-spin heme and a similar catalytic center composed of a high-spin heme and a metal ion — copper in the case of HCOs and iron in the case of NORs [18], [25], [105], [29]. However, instead of reducing oxygen to water, NORs perform the two-electron reduction of NO to N2O and are not related with aerobic respiration.
Thus, if we can effectively distinguish between HCOs (aerobic) and NORs (anaerobic) we can distinguish at the genome level between organisms that regularly deal with O2 (aerobes, facultative aerobes, having HCOs) and those that shun it (anaerobes, lacking HCO while possessing NOR, or lacking both). For this we used tools developed elsewhere [105] and adapted to this specific problem (see Methods). In the present genome sample, ~ 67% (1332 out of 1981) of the genomes contain one or more HCOs, revealing the adaptation to oxic habitats, while ~ 33% (649 out of 1981) of the genomes are devoid of HCOs (Table 2). Our method sorted 18 organisms (genomes) that only contain NORs into the anaerobic category, which is important, because of the possible existence of NO dependent chemistry in early earth [12] and the presence of NORs at the onset of bioenergetic processes as argued by some [78], [29]. NORs are divided into two main groups, according to the nature of their electron donor. Thus, cNORs represent the enzymes that use soluble electron donors such as cytochrome c, HiPIPs or cupredoxins and qNORs represent the enzymes which oxidize quinols [29]. qNORs have representatives in the two prokaryotic domains although their presence in Archaea can be both attributed exclusively to two interdomain LGT events, one to Crenarchaeota and one to halobacteria [14], [38] or vertical inherence, multiple losses (except in some Crenarchaeota organisms), and an additional LGT of bacterial qNORs to halobacteria [29]. Regarding cNORs, only one sequence has been identified so far within Archaea [29] and the phylogenies of this subfamily, as the authors recognize, are prone to change over time.
Table 2.
Taxonomic distribution of HCOs within 1981 genomes.
| Groups | Aerobic | Anaerobic | N. genomes |
|---|---|---|---|
| Archaea | |||
| Others | |||
| Korarchaeota | 0 | 1 | 1 |
| Thaumarchaeota | 2 | 0 | 2 |
| Nanoarchaeota | 0 | 1 | 1 |
| 2 | 2 | 4 | |
| Crenarchaeota | |||
| Thermoproteales | 6 | 7 | 13 |
| Desulfurococcalesa | 1 | 13 | 14 |
| Sulfolobales | 16 | 0 | 16 |
| 23 | 20 | 43 | |
| Euryarchaeota | |||
| Thermococcales | 0 | 14 | 14 |
| Thermoplasmatales | 1 | 3 | 4 |
| Archaeoglobales | 0 | 4 | 4 |
| Methanobacteriales | 0 | 8 | 8 |
| Methanococcales | 0 | 15 | 15 |
| Methanomicrobiales | 0 | 6 | 6 |
| Methanocellales | 0 | 3 | 3 |
| Methanosarcinales | 0 | 10 | 10 |
| Halobacteriales | 23 | 0 | 23 |
| 24 | 63 | 87 | |
| Bacteria | |||
| Clostridia | 4 | 105 | 109 |
| Bacilli | 137 | 151 | 288 |
| Negativicutes | 0 | 6 | 6 |
| Tenericutes | 0 | 47 | 47 |
| Planctomycetes | 6 | 0 | 6 |
| Chlamydiae | 4 | 34 | 38 |
| Spirochaetes | 7 | 39 | 46 |
| Bacteroidetes | 55 | 22 | 77 |
| Actinobacteria | 166 | 41 | 207 |
| Chlorobi | 5 | 6 | 11 |
| Fusobacteria | 0 | 5 | 5 |
| Thermotogae | 0 | 15 | 15 |
| Aquificae | 8 | 2 | 10 |
| Chloroflexi | 10 | 6 | 16 |
| Deinococcus–Thermus | 17 | 0 | 17 |
| Cyanobacteria | 44 | 0 | 44 |
| Acidobacteria | 8 | 0 | 8 |
| Deltaproteobacteria | 31 | 17 | 48 |
| Epsilonproteobacteria | 72 | 2 | 74 |
| Alphaproteobacteria | 204 | 4 | 208 |
| Betaproteobacteria | 120 | 3 | 123 |
| Gammaproteobacteria | 374 | 41 | 415 |
| Other bacteria | 11 | 18 | 29 |
| 1283 | 564 | 1847 | |
| Total | 1332 | 649 | 1981 |
Fervidicoccus fontis Kam984 (Fervidicoccales order) is grouped within Desulfurococcales order.
By the HCO criterion, aerobes are present in three archaeal phyla in our sample (Thaumarchaeota, Crenarchaeota, and Euryarchaeota). HCOs are widely distributed among crenarchaeotes, being present in all 16 members of the Sulfolobales sampled, among Thermoproteales (6 out of 13 Pyrobaculum species) and one member of the Desulfurococcales. By contrast, among the euryarchaeotic genomes sampled, only Halobacteriales and one member of the Thermoplasmatales (Picrophilus torridus DSM9790) contain genes coding for HCO (Table 2). The existence of functional HCOs in halobacteria is well documented [121], [122] as is the identification of large influx of gene transfers from bacteria to the halophiles [89] and to the halophile common ancestor [76], [118], that transformed an ancient methanogen into an oxygen-respiring heterotroph. Interestingly, a strictly anaerobic, acetate-oxidizing S0-reducing haloarchaeon has been sequenced [104], showing that aerobic respiration is not anymore a universal feature of the haloarchaea and underscoring ongoing metabolic diversification and gene loss within haloarchaea.
Within bacteria, HCOs are present in genomes from 19 out of the 23 bacterial groups, although with different densities of distribution. While aerobic (and facultative aerobic) organisms dominate the proteobacterial, cyanobacterial and actinobacterial lineages, the majority of the bacterial genomes sampled belonging to the fusobacteria, thermotogae, aquificae, negativicutes, tenericutes, chlamydiae and spirochaetes groups do not contain detectable HCOs.
Having a genomic proxy for oxygen tolerance among organisms (genome lineages) within the present sample allows us to classify the domain pairs as anaerobes, oxygen tolerant, or mixed (Fig. 2c, Table 2). Of the three most frequent domain pairs, pair 1 contains methanogen (anaerobes) or anaerobic lineages derived from methanogens together with clostridia (anaerobes), pair 2 contains Sulfolobales (aerotolerant) and Actinobacteria (aerotolerant), while pair 3 contains deltaproteobacterial (mixed aerotolerant and anaerobes) and methanogen (anaerobes) or anaerobic lineages derived from methanogens. The pairs also allow us to separate the protein families into aerobic (recent), anaerobic (ancient) or mixed protein families.
3.4. Who is new (aerotolerant), who is old (anaerobic)?
HCOs allow us to sort genomes into categories of aerotolerant (having HCOs) or not (lacking HCOs). But the exercise here is to classify protein families as being typical for aerobes or anaerobes. In a simple scheme, we could just say that proteins encoded in genomes labeled as anaerobic should correspond to anaerobic protein families and those families having members that occur in aerotolerant genomes (having HCOs) should be excluded from the LucaGC set. But that criterion is too strict and will falsely exclude many anaerobic protein families, because i) they might be present in facultative anaerobes, and ii) they might have been subject to recent LGTs into aerotolerant species but have not yet been lost.
This presents a difficult problem that readily lends itself to years of parameter space exploration, recurrent adjustments, and getting bogged down in endless minutiae of calculations. We took a pragmatic, albeit in some aspects somewhat arbitrary, approach. For a protein family to qualify for designation as an anaerobic family, we chose as our first (arbitrary) criterion that least 90% of all genes present in the family belong to organisms devoid of HCOs. To ensure a similar representation of archaea and bacteria in terms of anaerobic organisms, we included an additional filtering to consider only the protein families where at least 85% of the archaeal organisms represented are anaerobic and at least 85% of the bacterial organisms are anaerobic. This has the effect of excluding protein families whose high anaerobic score is due to an overrepresentation of anaerobic organism from one domain versus the other. Conversely, the criteria of at least 85% of archaeal organisms and 85% of bacterial organisms represented containing HCO were used to identify protein families as aerotolerant. What about the other protein families beyond these thresholds? Could they also be Luca candidate genes? Some might. We recognize that we might be excluding both “aerotolerant” and “anaerobic” protein families from these groups, this initial approach to filtering LGTs can probably be improved upon in future applications.
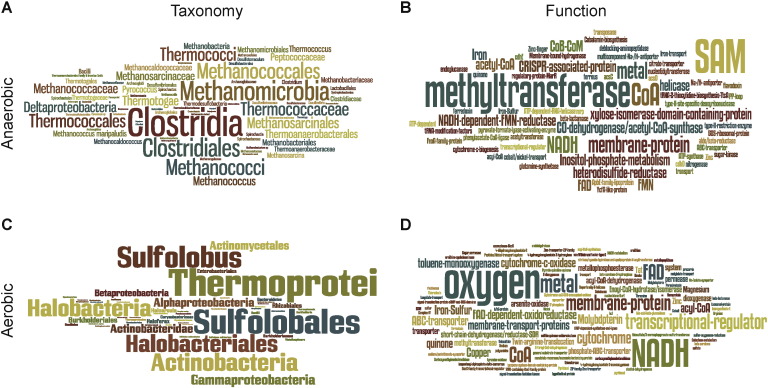
Using these criteria, 79 protein families were classified as “aerotolerant”. The majority of the taxonomic distribution of the Luca candidate genes that identify aerotolerant taxon pairs identify Sulfolobales, Halobacteriales, Actinobacteria and Proteobacteria (Fig. 3b). In terms of functional annotation, these Luca candidate genes consist of mainly cytochrome and copper-containing related proteins, dioxygenases, and some NADH- and FAD-dependent oxidoreductases (Fig 3d). That is, the Luca candidate genes that link these aerobe-containing interdomain taxon pairs are inventions that arose after the cyanobacterial innovation of oxygenic photosynthesis [2]. Their distributions are not likely the result of ancient vertical inheritance, as Luca arose long before oxygen did.
Fig. 3.
Wordle representation of the most frequent taxonomic and functional descriptions within the 79 aerobic and the 62 anaerobic protein families. The size of the words relates to the number of times the word appears within the annotations. The larger the word, the higher its frequency.
Perhaps as a slight disappointment, but also not entirely as a surprise, 904 of the 1045 families were classified as “mixed” with respect to occurrence within aerotolerant lineages. This has to do with LGTs and facultative anaerobes and these families deserve further inspection in future studies. What, on the bottom line, did this investigation uncover? We found two things, which we will summarize in the final two sections. 1) Perhaps more important than its role as a terminal acceptor, oxygen is an outstanding oxidant that microbes learned to use widely in many pathways. 2) At the anaerobic core of genes that reflect vertical inheritance from the prokaryotic common ancestor, we find evidence for antiporter-dependent ion gradient conversion, ATP synthase ion-gradient harnessing, FeS-cluster dependent soluble electron transport, and methyl-group dependent metabolism.
3.5. What did O2 do for metabolism?
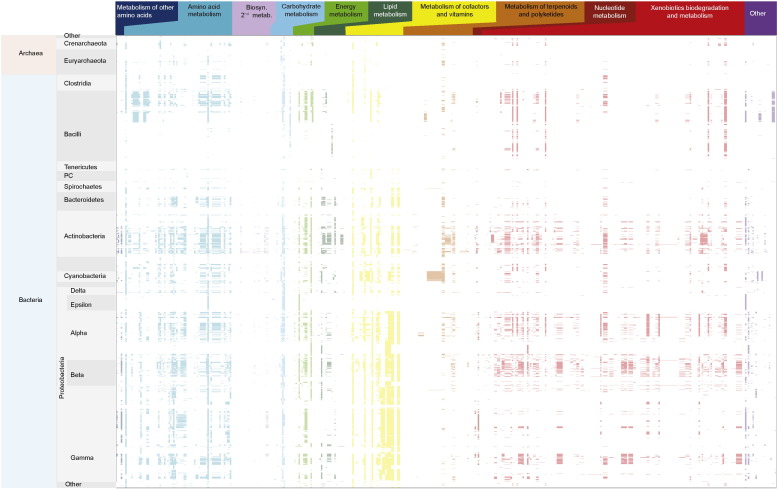
The transition from an anoxic world to the emergence of oxic environments promoted overall modifications in the environment redox potential [26], [48], changing the metal availability and leading to the invention of new proteins and folds [49], [43] that allowed the expansion of the existing metabolic pathways to include oxygen-dependent reactions [19], [43], [88], [99]. Moreover, the irreversibility of oxygen consuming reactions promoted a positive selection pressure for the transfer and maintenance of oxygen related metabolism throughout prokaryotic organisms. Modern genomes harbor many O2-dependent reactions distributed across 11 metabolic pathway categories (Fig. 4).
Fig. 4.
Distribution of oxygen dependent reaction within prokaryotic genomes grouped by the 11 Kegg metabolic pathway maps.
In addition to its availability as a terminal acceptor for pre-existing (anaerobic) respiratory chains, O2 allowed several energetically demanding reactions to occur more readily, for example the oxidation and cleavage of aromatic compounds by various dioxygenases [35]. Many existing biosynthetic pathways were also affected, as for example, the O2 independent (more ancient) and alternative O2-dependent (derived) biosynthetic pathways for heme [13], [34], [40], cobalamine [91], [92], and chlorophyll [80] to name a few. About half of the O2-dependent enzymes that we identified in the current sample of 1821 genomes are involved in the degradation of isoprenoids and xenobiotics. Without question, O2 expanded the realm of anabolic and catabolic pathways across genomes [88], but it did not alter the nature of the basic building blocks of life, nor did it fundamentally alter their biosynthesis. In the main, oxygen facilitated the oxidation of the building blocks of life, opening up new pathways of heterotrophic growth. A similar metabolic transition occurred at the origin of heterotrophy, as the first heterotrophs arose from autotrophs by learning to glean energy from the anaerobic oxidation of amino acids and bases at low H2 partial pressures [96].
Earth is now different from when life arose, the main difference for microbes being that today there are oxic habitats [42], [99]. Some have argued that O2 or high potential acceptors like O2 (for example NO), must have existed at life's origin, the argument being that the free energy changes associated with H2-dependent CO2 reduction were not sufficient to get life started [94]. O2 is indeed a strong terminal acceptor for electron transfer phosphorylation (hence valuable once life had already evolved respiratory chains). But is it too often overlooked that the use of oxygen comes at a huge cost. How so?
As it relates to organic compounds like amino acids and nucleic acid bases – the substance of life – it is a curious but significant observation that life with oxygen is energetically far more expensive than life without oxygen [5]. The energetic costs to synthesize the cellular building blocks under oxic conditions are on average 12.9 times higher than under anoxic conditions [60]. This is because the reactions that generate the basic building blocks of cells (amino acids in the main) are thermodynamically favorable in anoxic environments but thermodynamically unfavorable in oxidizing environments [60], [70]. The reason is that modest H2 partial pressures (≤ 10− 4–10− 3 atm, [111]) favor the reduction of CO2 to organic compounds (which is why acetogens and methanogens can grow) while low H2 partial pressures favor the slow oxidation of amino acids and bases back to ammonium and CO2 [96], whereas O2 allows fast and highly exergonic oxidations of even unfermentable organics, which is why wood, coal and oil burn well, releasing heat in the process.
3.6. The ancient metabolic core: small, but strictly anaerobic
Only 62 protein families that trace to the prokaryote common ancestor fulfilled the criteria set here to be designated as “anaerobic” families. Even though 62 is not a genome-sized number, it is still greater than ~ 30, the usual list of suspect genes used for the reconstruction of deep phylogeny, and importantly, our list of 62 does not include either the core (~ 30) or the extended core (~ 100). The 27 universal and the 102 nearly-universal protein families were filtered out from the ancient anaerobic core due to their presence in both aerobic and anaerobic organisms. At this point we have to make a distinction between prokaryotic interdomain monophyletic families (that is, vertical inherence from Luca) and universal proteins, whose wide distribution with or without interdomain LGT events (e.g. ATP synthases), were present in Luca.
The 62 anaerobic protein families identified above are the only remaining markers for ancient metabolism we can still retrieve from extant genomic data using this procedure. Of course, we cannot exclude the existence of other proteins in Luca, such as the ones widely shared by aerobic and anaerobic organisms, or some whose evolutionary history involved interdomain LGT events. But these are the ones we identify with this procedure. Yet these are new insights into Luca's gene complement. Furthermore, the functional classification of these 62 ancient anaerobic proteins goes beyond the traditionally identified informational genes, covering a number of functional categories not previously known about Luca (Table 1).
The Luca candidate genes shared by anaerobes are the genes that we sought to identify at the outset of this study. The protein families that i) still retain the ancient archaeal–bacterial division in phylogenies, and ii) that are preferentially encoded in the genomes of anaerobes, identify mainly methanogens and clostridia (Fig. 3a).
Here one might ask “identify them as what?” Methanogens (Methanomicrobia and Methanococci) and clostridia (Clostridiales) are the most frequent pairs within the 62 protein families that preserve the archaeal–bacterial split, hence were not obviously distributed via LGT, and that only rarely (if ever) occur in genomes that harbor HCOs. These protein families are typical of anaerobes, whereby Luca had to be an anaerobe. Put another way, if we acknowledge that Luca was an anaerobe, there are only 62 protein families that we can trace as probable vertical inheritances from Luca (as opposed to LGTs) that are typical of strict anaerobes in that they rarely, if ever, occur in genomes of aerotolerant (facultative anaerobic) prokaryotes, and these 62 proteins are mainly shared by clostridia and methanogens.
One might object that this observation results from the high number of clostridial and methanogenic organisms devoid of HCOs within our dataset. However, both archaea and bacteria contain other taxonomic groups without HCOs, but that have far fewer Luca candidate genes than clostridia and methanogens. The functional annotations of the anaerobic Luca candidate genes reveal numerous methyl transferases, subunits of the acetyl-CoA synthase complex, the soluble heterodisulfide reductase subunits C and A, several SAM-dependent methyltransferases and ferredoxin (Supplemental Table A2), in addition to several subunits of the H+/Na+-antiporter MRP/hydrogenases and related complexes within this group (Fig. 3c).
The Mrp antiporter is surprising and particularly interesting. Based purely on comparative physiology and theoretical considerations, it was recently proposed that a crucial step in bioenergetic evolution at an early stage prior to the emergence of free-living cells, was the advent of an Mrp-type H+/Na+ antiporter that could convert geochemical pH gradients into biologically more useful Na+ gradients [59]. This antiporter, the kind common to Ech and FeNi hydrogenases [63], was suggested to have been the first step en route to replacing geochemical ion gradients with biologically derived ones, to generate Na+ gradients that could be more readily harnessed by Luca's rotor–stator ATPase, yet prior to the invention of redox-chemistry-based (electron-transport-based) ion pumping systems [59]. Later work on simulated gradients provided support for that view [103]. Here, in a completely independent genome based approach, the Mrp-type antiporter suddenly turns up among 62 genes in the ancient anaerobic core. This is surprising, but was also predicted on the basis of bioenergetics by the theory that life arose at an alkaline hydrothermal vent. The present data also indicate the absence of redox-based ion-pumping systems in Luca (Table 3), as previously suggested [66], [59], [106], [107]. The 27 universal and the 102 nearly-universal protein families were filtered out from the ancient anaerobic core because they are present in both aerobic and anaerobic organisms.
Table 3.
Gene annotations of the 62 anaerobic core families.
| Cluster | Annotation | Comments |
|---|---|---|
| 6079 | radical_SAM_protein | SAM methyltransferase |
| 9248 | radical_SAM_protein | Pyruvate formate lyase activating enzyme EC:1.97.1.4; SAM methyltransferase |
| 21,920 | Radical_SAM_domain_protein | rlmN; 23S rRNA methyltransferase [EC:2.1.1.192]; SAM |
| 10,466 | type_11_methyltransferase | SAM methyltransferase |
| 21,520 | type_11_methyltransferase | SAM methyltransferase |
| 14,283 | Methylase_involved_in… | SAM methyltransferase |
| 1662 | 50S_ribosomal_protein_L29 | RP-L29, rpmC; large subunit ribosomal protein L29 |
| 1296 | acetyl-CoA_decarbonylase/synthase… | Acetyl-CoA decarbonylase/synthase complex subunit gamma [EC:2.1.1.245] |
| 1321 | acetyl-CoA_decarbonylase/synthase… | Acetyl-CoA decarbonylase/synthase complex subunit delta [EC:2.1.1.245] |
| 7851 | ATP_synthase_subunit_c | ATPVK, ntpK, atpK; V/A-type H +-transporting ATPase subunit K |
| 4404 | ferredoxin | fer; ferredoxin |
| 18,705 | flavodoxin | Flavodoxin |
| 13,779 | heterodisulfide_reductase_subunit_A… | Heterodisulfide-reductase CoB CoM |
| 14,009 | heterodisulfide_reductase_subunit_C… | Heterodisulfide reductase; CoM CoS |
| 4789 | membrane_bound_hydrogenase_subunit… | Multicomponent Na +:H + antiporter |
| 11,625 | Membrane-bound_hydrogenase_MBH… | Membrane-bound-hydrogenase antiporter subunit |
| 11,548 | L-glutamine_synthetase | glnA, GLUL; glutamine synthetase [EC:6.3.1.2]; glutamine-synthetase |
| 10,709 | nitrogenase_iron-iron_accessory_protein… | Iron–iron nitrogenase accessory protein AnfO |
| 5364 | acetyltransferase | CoA acyl-CoA acetyltransferase |
| 10,044 | phenylacetate–CoA_ligase | paaK; phenylacetate-CoA ligase [EC:6.2.1.30]; CoA phenylacetate-CoA-ligase |
| 23,640 | NADPH-dependent_FMN_reductase | FMN FAD NADH NADH-dependent-FMN-reductase |
| 10,494 | NADPH-dependent_FMN_reductase | NADH-dependent-FMN-reductase |
| 4774 | sugar_kinase | Sugar-kinase |
| 11,273 | FeoA_family_protein | feoA ferrous iron transport protein A |
| 11,762 | aldo/keto_reductase | K07079; NADH aldo/keto-reductase |
| 12,136 | putative_ABC_transporter | Putative ABC transport system permease protein |
| 6304 | citrate_transporter | Citrate_transporter |
| 16,010 | cobalamin_biosynthesis_protein | cbiN; cobalt/nickel transport protein; cobalamin |
| 14,243 | cytochrome_c_biogenesis… | Cytochrome c-biogenesis transmembrane protein |
| 1071 | ApbE_family_lipoprotein | K09740; hypothetical protein; ApbE_family_lipoprotein |
| 6375 | beta-lactamase_domain-containing_protein | Beta-lactamase metal dependent |
| 4311 | deblocking_aminopeptidase | E3.2.1.4; endoglucanase metal metallopeptidase |
| 22,518 | YcfA-like_protein | hicA; mRNA interferase HicA [EC:3.1.-.-]; YcfA-like-protein |
| 15,732 | nucleotidyltransferase | K07076; nucleotidyltransferase |
| 4155 | PP-loop_domain-containing… | ttcA; tRNA 2-thiocytidine biosynthesis protein TtcA; PP-loop |
| 9988 | regulatory_protein_MarR | Regulatory-protein-MarR transcriptional-regulator |
| 23,018 | type_II_site-specific_deoxyribonuclease | Type-II-restriction-enzyme type-II-site-specific-deoxyribonuclease |
| 16,528 | transposase,_IS4 | Transposase |
| 10,489 | ATP-dependent_RNA_helicase | CRISPR-associated-endonuclease/helicase Cas3; ATP-dependent |
| 15,539 | CRISPR-associated_protein… | CRISPR-associated protein Cmr3 |
| 14,799 | CRISPR-associated_protein… | CRISPR-associated protein Csm1 |
| 18,976 | helicase_domain_protein | Helicase |
| 20,220 | helicase-like_protein | Helicase |
| 2237 | xylose_isomerase_domain… | Metal-dependent; xylose-isomerase-domain-containing-protein |
| 20,694 | xylose_isomerase_domain… | Metal xylose-isomerase-domain-containing-protein |
| 7805 | putative_YgiT-type_zinc_finger | Zinc zinc-finger domain |
16 other protein families annotated as hypothetical proteins.
If Luca could synthesize ATP with the help of proteins that were able to harness and transduce a geochemical pH gradient, where did the biochemical energy come from that allowed Luca to perform protein synthesis in the absence of chemiosmotic harnessing? It remains true that there are only two ways in which cells conserve energy in the form of ATP: chemiosmotic coupling and substrate level phosphorylation (SLP) [98]. Before the origin of chemiosmotic coupling, SLP would thus have been the only option. In essence, there have been only two viable suggestions for how the first cells might have harnessed energy via SLP: a geochemical supply of carbon monoxide [32] or a geochemical supply of methyl groups [66], [68] — on the early Earth, oxidations of organics from space will not support energy metabolism [96]. Both proposals focus on the reactions catalyzed by bifunctional acetyl-CoA synthase/carbon monoxide dehydrogenase (CODH/ACS), which condenses a Ni-bound methyl group with Ni-bound carbon monoxide (carbonyl) to generate a Ni-bound acetyl group that is removed from the enzyme via thiolysis to generate a thioester, which is cleaved by phosphate to yield acetyl phosphate, which phosphorylates ADP via SLP [85], [106]. Consistent with both proposals [32], [66] CODH/ACS is contained within the anaerobic core (Table 3). Additional evidence for the existence of abiotic synthesis within alkaline vents systems comes from laboratory experiments showing that the direct reduction of CO2 by H2 in the presence of pH gradients or electric potential can lead to the formation of formaldehyde, acetate and pyruvate [41], [93], [102].
Though an excellent source of energy and electrons, carbon monoxide has a very restricted role in biochemistry outside of energy metabolism [86]. Studies of abiotic synthesis under the far-from-equilibrium electrochemical vent conditions will help to clarify the presence or absence of CO under these conditions [41]. Methyl groups, on the other hand, have a very central and general role in microbial metabolism, in particular in S-adenosyl methionine (SAM) and in radical SAM enzymes, which have a myriad of functions in biosyntheses, in particular in cofactor biosyntheses [101], [119]. Of the 62 proteins in the ancient core, six (~ 10%) are SAM-dependent enzymes, making these the most common class of proteins in the ancient core behind “unknown function”. Clearly, the ancient core points to a central involvement of methyl groups in ancient metabolism. This is very much in line with the predications from the theory that life arose at hydrothermal vents: “…the biochemical system proposed [] would remain strictly dependent upon geochemically provided methyl groups up until the advent of (protein dependent) chemiosmotic harnessing” ([66], p. 1912). Recent geochemical studies have uncovered evidence for disequilibrium of one carbon species in the Von Damm hydrothermal field, indicating that there are distinct kinetic barriers to methane synthesis in some hydrothermal systems [72], such that a model for the origin of life that requires a geochemical source of chemically accessible methyl moieties is not asking for too much. That methyl groups have such a prominent place in the ancient anaerobic core is interesting and possibly significant. If vent conditions allowed methyl synthesis, then the synthesis of thioesters and acetyl phosphate would also, in principle, be possible [41], [93], [102].
Other proteins in the ancient anaerobic core, besides Mrp subunits, ATP synthase subunits, CODH/ACS subunits and SAM dependent enzymes include heterodisulfide reductase, an electron bifurcating enzyme essential to energy metabolism in methanogens [47], ferredoxin, flavodoxin, an iron (II) transport protein, and a thiocytidine biosynthesis protein involved tRNA modification. Four proteins in the list are membrane transport proteins, which would indicate that Luca existed in an environment where hydrophobic layers corresponding to the thickness of a lipid bilayer existed. That makes sense because neither the Mrp antiporter nor the rotor stator ATP synthase can function without a membrane, though the membrane need not be genetically encoded [16], as the synthesis of hydrophobic compounds at vents is expected from thermodynamics [3] and is observed in some modern hydrothermal systems [83], [97]. Glutamine synthase, the enzyme that introduces nitrogen into metabolism, is in the list, as are CoA-ligases and NADPH dependent flavin reductases, transducers of one electron to two electron transport. The latter, together with ferredoxin, flavodoxin, electron bifurcating enzymes and radical SAM enzymes point to a major role of one electron transport in Luca's metabolism. But where would the reduced nitrogen come from? For life to get started, reduced nitrogen had to be (at least locally) available, and both transition metal catalysts [28] and hydrothermal vent conditions [12] could have been a local source of ammonium. Moreover, the presence of a nitrogenase accessory protein in the 62 anaerobic families list can be an indication for the existence of an ancient nitrogenase in Luca, as previously proposed [69].
Thus, if we i) remove interdomain LGTs as Kannan et al. [46] suggested, ii) identify lineages that have the most Luca candidate genes, and iii) distinguish between anaerobic Luca candidate genes in anaerobic lineages (might be ancient) and aerotolerant Luca candidate genes in aerotolerant lineages (cannot be ancient) we are left with the result in Fig. 3a and c, namely that Luca looks most similar to modern bacterial and archaeal lineages that harbor anaerobic chemolithoautotrophs: clostridial-type acetogens and methanogens. This is noteworthy in the respect that acetogens and methanogens are the groups of organisms that stand in the foreground of theories for the origin of life that are based in microbial physiology and that connect well to geochemistry [66], [8], [59], [106]. The idea that life started at hydrothermal vents has been around for 30 years [7], it continued to be further developed. In our inference, we have ended up with something that is half-alive, a curious but necessary intermediate in the transition from non-living to living things.
4. Conclusions
The overall picture of core physiology in Luca that we infer from genome sequences is almost indistinguishable from that in Fig. 1c of Lane and Martin [59] that was obtained from comparative physiology, bioenergetics, and theory. That two completely independent approaches converge on the same set of proteins and functions in early bioenergetics is noteworthy. It indicates that a version of the hydrothermal vent theory focusing on the acetyl-CoA pathway, methyl synthesis, acetogens and methanogens [66] possesses an element of robustness in that it interfaces well with the physiology of anaerobic prokaryotes [15], [98], with thermodynamics [3], [4], with findings from geochemistry about serpentinization and hydrothermal vents [71], [97], and as we have shown here, with comparative genomics. Even ribosomal phylogenies tend to agree with the predictions of the model in that newer metagenomic indicate a greater antiquity for methanogen-related metabolism within the archaea than previously assumed [30], and newer phylogenetic data put methanogenesis at the root of the archaeal domain [67], [87].
In contrast to today's oxidized and strongly oxidizing environment, the highly reducing hydrothermal setting on a young, metal-rich, anaerobic Earth that we have in mind as Luca's residence [106] offers very favorable thermodynamic conditions for the synthesis of Luca's building blocks [3], [5], [71], [97]. When we look at Luca as an anaerobe and as the common ancestor of prokaryotes, we obtain a picture of its genome that resembles clostridial acetogens and methanogens. With regard to the most primitive forms of microbial physiology, microbiologists reached the same conclusion 45 years ago [26], namely that methanogens and acetogens probably represent the most ancient lineages [36]. We required 2000 genomes and powerful computers for our conclusions, while Decker et al. just thought about it. Evidently, just thinking about things can be a source of scientific progress.
The following are the supplementary data related to this article.
Oxygen requirements of the organisms present in the 1045 protein families.
Functional annotation.
Transparency document
Transparency document.
Acknowledgments
We thank the ERC (grant 666053) for the funding.
Footnotes
The Transparency document associated with this article can be found, in online version.
References
- 1.Akanni W.A. Horizontal gene flow from Eubacteria to Archaebacteria and what it means for our understanding of eukaryogenesis. Philos. Trans. R. Soc. B. 2015;370:20140337. doi: 10.1098/rstb.2014.0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen J.F. A redox switch hypothesis for the origin of two light reactions in photosynthesis. FEBS Lett. 2005;579:963–968. doi: 10.1016/j.febslet.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 3.Amend J.P., LaRowe D.E., McCollom T.M., Shock E.L. The energetics of organic synthesis inside and outside the cell. Philos. Trans. R. Soc. B. 2013;368:20120255. doi: 10.1098/rstb.2012.0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amend JP, McCollom TM. 2009. Energetics of biomolecule synthesis on early Earth. In: Chemical Evolution II: From Origins of Life to Modern Society. American Chemical Society (L Zaikowski, et al., Ed.), pp. 63–94. (1025, Washington, DC).
- 5.Amend J.P., Shock E.L. Energetics of amino acid synthesis in hydrothermal ecosystems. Science. 1998;281:1659–1662. doi: 10.1126/science.281.5383.1659. [DOI] [PubMed] [Google Scholar]
- 6.Anantharaman V., Koonin E.V., Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002;30:1427–1464. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baross J.A., Hoffman S.E. Submarine hydrothermal vents and associated gradient environments as sites for the origin and evolution of life. Orig. Life Evol. Biosph. 1985;15:327–345. [Google Scholar]
- 8.Baross J.A., Martin W.F. The ribofilm as a concept for life's origins. Cell. 2015;162:13–15. doi: 10.1016/j.cell.2015.06.038. [DOI] [PubMed] [Google Scholar]
- 9.Baymann F. The redox protein construction kit: pre-last universal common ancestor evolution of energy-conserving enzymes. Philos. Trans. R. Soc. B. 2003;358:267–274. doi: 10.1098/rstb.2002.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beiko R.G., Harlow T.J., Ragan M.A. Highways of gene sharing in prokaryotes. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14332–14337. doi: 10.1073/pnas.0504068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borisov V.B., Gennis R.B., Hemp J., Verkhovsky M.I. The cytochrome bd respiratory oxygen reductases. BBA Bioenerg. 2011;1807:1398–1413. doi: 10.1016/j.bbabio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brandes J.A. Abiotic nitrogen reduction on the early Earth. Nature. 1998;395:365–367. doi: 10.1038/26450. [DOI] [PubMed] [Google Scholar]
- 13.Breckau D., Mahlitz E., Sauerwald A., Layer G., Jahn D. Oxygen-dependent coproporphyrinogen III oxidase (HemF) from Escherichia coli is stimulated by manganese. J. Biol. Chem. 2003;278:46625–46631. doi: 10.1074/jbc.M308553200. [DOI] [PubMed] [Google Scholar]
- 14.Brochier-Armanet C., Talla E., Gribaldo S. The multiple evolutionary histories of dioxygen reductases: implications for the origin and evolution of aerobic respiration. Mol. Biol. Evol. 2009;26:285–297. doi: 10.1093/molbev/msn246. [DOI] [PubMed] [Google Scholar]
- 15.Buckel W., Thauer R.K. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. BBA Bioenergetics. 2013;1827:94–113. doi: 10.1016/j.bbabio.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 16.Carbone V. Structure and evolution of the archaeal lipid synthesis enzyme sn-glycerol-1-phosphate dehydrogenase. J. Biol. Chem. 2015;290:21690–21704. doi: 10.1074/jbc.M115.647461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caspi R. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2015;42:D459–D471. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castresana J., Lübben M., Saraste M., Higgins D.G. Evolution of cytochrome oxidase, an enzyme older than atmospheric oxygen. EMBO J. 1994;13:2516–2525. doi: 10.1002/j.1460-2075.1994.tb06541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Catling D.C., Glein C.R., Zahnle K.J., McKay C.P. Why O2 is required by complex life on habitable planets and the concept of planetary ‘oxygenation time’. Astrobiology. 2005;5:415–438. doi: 10.1089/ast.2005.5.415. [DOI] [PubMed] [Google Scholar]
- 20.Charlebois R.L., Doolittle W.F. Computing prokaryotic gene ubiquity: rescuing the core from extinction. Genome Res. 2004;14:2469–2477. doi: 10.1101/gr.3024704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ciccarelli F.D., Doerks T., von Mering C., Creevey C.J., Snel B., Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 22.Cox C.J., Foster P.G., Hirt R.P., Harris S.R., Embley T.M. The archaebacterial origin of eukaryotes. Proc. Natl. Acad. Sci. U. S. A. 2008;105:20356–20361. doi: 10.1073/pnas.0810647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dagan T., Martin W. The tree of one percent. Genome Biol. 2006;7:118. doi: 10.1186/gb-2006-7-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dagan T., Martin W. Ancestral genome sizes specify the minimum rate of lateral gene transfer during prokaryote evolution. Proc. Natl. Acad. Sci. U. S. A. 2007;104:870–875. doi: 10.1073/pnas.0606318104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Vries S., Schonder I. Comparison between the nitric oxide reductase family and its aerobic relatives, the cytochrome oxidases. Biochem. Soc. Trans. 2002;30:662–667. doi: 10.1042/bst0300662. [DOI] [PubMed] [Google Scholar]
- 26.Decker K., Jungermann K., Thauer R.K. Energy production in anaerobic organisms. Angew. Chem. Int. Ed. 1970;9:138–158. doi: 10.1002/anie.197001381. [DOI] [PubMed] [Google Scholar]
- 27.Delaye L., Becerra A., Lazcano A. The last common ancestor: what's in a name? Orig. Life Evol. Biosph. 2005;35:537–554. doi: 10.1007/s11084-005-5760-3. [DOI] [PubMed] [Google Scholar]
- 28.Dorr M., Kässbohrer J., Grunert R., Kreisel G., Brand W.A., Werner R.A., Geilmann H., Apfel C., Robl C., Weigand W. A possible prebiotic formation of ammonia from dinitrogen on iron sulfide surfaces. Angew. Chem. Int. Ed. 2003;42:1540–1543. doi: 10.1002/anie.200250371. [DOI] [PubMed] [Google Scholar]
- 29.Ducluzeu A.-L., Schoepp-Cothenet v.L.R., Baymann F., MJ R., W. N. The evolution of respiratory O2/NO reductases: an out-of-phylogenetic-box perspective. J. R. Soc. Interface. 2014;11:20140196. doi: 10.1098/rsif.2014.0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans P.N. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science. 2015;350:434–438. doi: 10.1126/science.aac7745. [DOI] [PubMed] [Google Scholar]
- 31.Fang H. A daily-updated tree of (sequenced) life as a reference for genome research. Sci. Rep. 2013;3:2015. doi: 10.1038/srep02015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferry J.G., House C.H. The stepwise evolution of early life driven by energy conservation. Mol. Biol. Evol. 2006;23:1286–1292. doi: 10.1093/molbev/msk014. [DOI] [PubMed] [Google Scholar]
- 33.Fox G.E. Origin and evolution of the ribosome. Cold Spring Harb. Perspect. Biol. 2010;2:a003483. doi: 10.1101/cshperspect.a003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frankenberg N., Moser J., Jahn D. Bacterial heme biosynthesis and its biotechnological application. Appl. Microbiol. Biotechnol. 2003;63:115–127. doi: 10.1007/s00253-003-1432-2. [DOI] [PubMed] [Google Scholar]
- 35.Fuchs G., Boll M., Heider J. Microbial degradation of aromatic compounds — from one strategy to four. Nat. Rev. Microbiol. 2011;9:803–816. doi: 10.1038/nrmicro2652. [DOI] [PubMed] [Google Scholar]
- 36.Fuchs G. Alternative pathways of carbon dioxide fixation: insights into the early evolution of life? Annu. Rev. Microbiol. 2011;65:631–658. doi: 10.1146/annurev-micro-090110-102801. [DOI] [PubMed] [Google Scholar]
- 37.Galperin M.Y., Makarova K.S., Wolf Y.I., Koonin E.V. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015;43:D261–D269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gribaldo S., Talla E., Brochier-Armanet C. Evolution of the haem copper oxidases superfamily: a rooting tale. Trends Biochem. Sci. 2009;34:375–381. doi: 10.1016/j.tibs.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Hansmann S., Martin W. Phylogeny of 33 ribosomal and six other proteins encoded in an ancient gene cluster that is conserved across prokaryotic genomes: influence of excluding poorly alignable sites from analysis. Int. J. Syst. Evol. Microbiol. 2000;4:1655–1663. doi: 10.1099/00207713-50-4-1655. [DOI] [PubMed] [Google Scholar]
- 40.Heinemann I.U., Jahn M., Jahn D. The biochemistry of heme biosynthesis. Arch. Biochem. Biophys. 2008;474:238–251. doi: 10.1016/j.abb.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 41.Herschy B. An origin-of-life reactor to simulate alkaline hydrothermal vents. J. Mol. Evol. 2014;79:213–227. doi: 10.1007/s00239-014-9658-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holland H.D. The oxygenation of the atmosphere and oceans. Philos. Trans. R. Soc. B. 2006;361:903–915. doi: 10.1098/rstb.2006.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang Y.-Y. The impact of oxygen on metabolic evolution: a chemoinformatic investigation. PLoS Comput. Biol. 2012;8 doi: 10.1371/journal.pcbi.1002426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanehisa M., Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2002;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanhere A., Vingron A. Horizontal gene transfers in prokaryotes show differential preferences for metabolic and translation genes. BMC Evol. Biol. 2009;9:9. doi: 10.1186/1471-2148-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kannan L., Li H., Rubinstein B., Mushegian A. Models of gene gain and gene loss for probabilistic reconstruction of gene content in the last universal common ancestor of life. Biol. Direct. 2013;8:32. doi: 10.1186/1745-6150-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaster A.-K., Moll J., Parey K., Thauer R.K. Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea. Proc. Natl. Acad. Sci. U. S. A. 2011;108:2981–2986. doi: 10.1073/pnas.1016761108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kasting J.F., Siefert J.L. Life and the evolution of Earth's atmosphere. Science. 2002;296:1066–1068. doi: 10.1126/science.1071184. [DOI] [PubMed] [Google Scholar]
- 49.Kim Protein domain structure uncovers the origin of aerobic metabolism and the rise of planetary oxygen. Structure. 2012;20:67–86. doi: 10.1016/j.str.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 50.Koonin E.V., Makarova K.S., Aravind L. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koonin E.V., Martin F. On the origin of genomes and cells within inorganic compartments. Trends Genet. 2005;21:647–654. doi: 10.1016/j.tig.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koonin E.V. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat. Rev. Microbiol. 2003;1:127–136. doi: 10.1038/nrmicro751. [DOI] [PubMed] [Google Scholar]
- 53.Koonin E.V. FT Press; Upper Saddle River, New Jersey: 2011. The Logic of Chance: The Nature and Origin of Biological Evolution. [Google Scholar]
- 54.Koonin E.V. Darwinian evolution in the light of genomics. Nucleic Acids Res. 2009;37:1011–1034. doi: 10.1093/nar/gkp089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ku C. Endosymbiotic origin and differential loss of eukaryotic genes. Nature. 2015;524:427–432. doi: 10.1038/nature14963. [DOI] [PubMed] [Google Scholar]
- 56.Kump L.R. The rise of atmospheric oxygen. Nature. 2008;451:277–278. doi: 10.1038/nature06587. [DOI] [PubMed] [Google Scholar]
- 57.Kyrpides N., Overbeek R., Ouzounis C. Universal protein families and the functional content of the last universal common ancestor. J. Mol. Evol. 1999;49:413–423. doi: 10.1007/pl00006564. [DOI] [PubMed] [Google Scholar]
- 58.Lane N., Martin W. The energetics of genome complexity. Nature. 2010;467:929–934. doi: 10.1038/nature09486. [DOI] [PubMed] [Google Scholar]
- 59.Lane N., Martin W.F. The origin of membrane bioenergetics. Cell. 2012;151:1406–1416. doi: 10.1016/j.cell.2012.11.050. [DOI] [PubMed] [Google Scholar]
- 60.Lever M.A. Life under extreme energy limitation: a synthesis of laboratory- and field-based investigations. FEMS Microbiol. Rev. 2015;39 doi: 10.1093/femsre/fuv020. [DOI] [PubMed] [Google Scholar]
- 61.Lynch E.A. Sequencing of seven haloarchaeal genomes reveals patterns of genomic flux. PLoS One. 2012;7 doi: 10.1371/journal.pone.0041389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lyons T.W., Reinhard C.T., Planavsky N.J. The rise of oxygen in Earth's early ocean and atmosphere. Nature. 2014;506:307–315. doi: 10.1038/nature13068. [DOI] [PubMed] [Google Scholar]
- 63.Marreiros B.C., Batista A.P., Duarte A.M.S., Pereira M.M. A missing link between complex I and group 4 membrane-bound [NiFe] hydrogenases. BBA Bioenergetics. 2013;1827:198–209. doi: 10.1016/j.bbabio.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 64.Martin W., Müller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- 65.Martin W., Russell M.J. On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Philos. Trans. R. Soc. Lond. B. 2003;358:59–83. doi: 10.1098/rstb.2002.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martin W., Russell M.J. On the origin of biochemistry at an alkaline hydrothermal vent. Philos. Trans. R. Soc. Lond. B. 2007;362:1887–1925. doi: 10.1098/rstb.2006.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martin W.F., Sousa F.L. Cold Spring Harb Perspect; 2015. Early microbial evolution: the age of anaerobes. (⋯:⋯–⋯ in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin W.F. Hydrogen, metals, bifurcating electrons, and proton gradients: the early evolution of biological energy conservation. FEBS Lett. 2012;9:485–493. doi: 10.1016/j.febslet.2011.09.031. [DOI] [PubMed] [Google Scholar]
- 69.Mehta M.P., Baross J.A. Nitrogen fixation at 92 °C by a hydrothermal vent archaeon. Science. 2006;314:1783–1786. doi: 10.1126/science.1134772. [DOI] [PubMed] [Google Scholar]
- 70.McCollom T.M., Amend J.P. A thermodynamic assessment of energy requirements for biomass synthesis by chemolithoautotrophic micro-organisms in oxic and anoxic environments. Geobiology. 2005;3:135–144. [Google Scholar]
- 71.McCollom T.M., Seewald J.S. Serpentinites, hydrogen, and life. Elements. 2013;9:129–134. [Google Scholar]
- 72.McDermott J.M., Seewald J.S., German C.R., Sylva S.P. Pathways for abiotic organic synthesis at submarine hydrothermal fields. Proc. Natl. Acad. Sci. U. S. A. 2015;112:7668–7672. doi: 10.1073/pnas.1506295112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mirkin B.G., Fenner T.I., Galperin M.Y., Koonin E.V. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol. Biol. 2003;3:2. doi: 10.1186/1471-2148-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mushegian A. Gene content of LUCA, the last universal common ancestor. Front. Biosci. 2008;13:4657–4666. doi: 10.2741/3031. [DOI] [PubMed] [Google Scholar]
- 75.Mushegian A.R., Koonin E.V. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl. Acad. Sci. U. S. A. 1996;93:10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nelson-Sathi S. Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea. Proc. Natl. Acad. Sci. U. S. A. 2012;109:20537–20542. doi: 10.1073/pnas.1209119109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nelson-Sathi S. Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature. 2015;517:77–80. doi: 10.1038/nature13805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nitschke W., Russell M.J. Beating the acetyl coenzyme A-pathway to the origin of life. Philos. Trans. R. Soc. B. 2013;368 doi: 10.1098/rstb.2012.0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ohmoto H., Watanabe Y., Ikemi H., Poulson S.R., Taylor B.E. Sulphur isotope evidence for an oxic Archaean atmosphere. Nature. 2006;442:908–911. doi: 10.1038/nature05044. [DOI] [PubMed] [Google Scholar]
- 80.Ouchane S., Steunou A.-S., Picaud M., Astier C. Aerobic and anaerobic Mg-protoporphyrin monomethyl ester cyclases in purple bacteria: a strategy adopted to bypass the repressive oxygen control system. J. Biol. Chem. 2004;279:6385–6394. doi: 10.1074/jbc.M309851200. [DOI] [PubMed] [Google Scholar]
- 81.Ouzounis C.A., Kunin V., Darzentas N., Goldovsky L. A minimal estimate for the gene content of the last universal common ancestor — exobiology from a terrestrial perspective. Res. Microbiol. 2006;157:57–68. doi: 10.1016/j.resmic.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 82.Pereira M.M., Santana M., Teixeira M. A novel scenario for the evolution of haem–copper oxygen reductases. BBA Bioenergetics. 2001;1505:185–208. doi: 10.1016/s0005-2728(01)00169-4. [DOI] [PubMed] [Google Scholar]
- 83.Proskurowski G. Abiogenic hydrocarbon production at lost city hydrothermal field. Science. 2008;319:604–607. doi: 10.1126/science.1151194. [DOI] [PubMed] [Google Scholar]
- 84.Puigbò P., Wolf Y.I., Koonin E.V. Search for a “Tree of Life” in the thicket of the phylogenetic forest. J. Biol. 2009;8:59. doi: 10.1186/jbiol159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ragsdale S.W. Nickel-based enzyme systems. J. Biol. Chem. 2009;284:18571–18575. doi: 10.1074/jbc.R900020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ragsdale S.W. Life with carbon monoxide. Crit. Rev. Biochem. Mol. Biol. 2010;39:165–195. doi: 10.1080/10409230490496577. [DOI] [PubMed] [Google Scholar]
- 87.Raymann K., Brochier-Armanet C., Gribaldo S. The two-domain tree of life is linked to a new root for the Archaea. Proc. Natl. Acad. Sci. 2015;112:6670–6675. doi: 10.1073/pnas.1420858112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Raymond J., Segre D. The effect of oxygen on biochemical networks and the evolution of complex life. Science. 2006;311:1764–1767. doi: 10.1126/science.1118439. [DOI] [PubMed] [Google Scholar]
- 89.Rhodes M.E., Spear J.R., Oren A., House C.H. Differences in lateral gene transfer in hypersaline versus thermal environments. BMC Evol. Biol. 2011;11:199. doi: 10.1186/1471-2148-11-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rivera M.C., Lake J.A. The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature. 2004;431:152–155. doi: 10.1038/nature02848. [DOI] [PubMed] [Google Scholar]
- 91.Rodionov D.A., Vitreschak A.G., Mironov A.A., Gelfand M.S. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem. 2003;278:41148–41159. doi: 10.1074/jbc.M305837200. [DOI] [PubMed] [Google Scholar]
- 92.Roessner C.A., Santander P.J., Scott A.I. Multiple biosynthetic pathways for vitamin B12: variations on a central theme. Vitam. Horm. 2001;61:267–297. doi: 10.1016/s0083-6729(01)61009-4. [DOI] [PubMed] [Google Scholar]
- 93.Roldan A. Bio-inspired CO2 conversion by iron sulfide catalysts under sustainable conditions. Chem. Commun. 2015;51:7501–7504. doi: 10.1039/c5cc02078f. [DOI] [PubMed] [Google Scholar]
- 94.Schoepp-Cothenet B. On the universal core of bioenergetics. BBA Bioenergetics. 2013;1827:79–93. doi: 10.1016/j.bbabio.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 95.Schomburg I. BRENDA: a resource for enzyme data and metabolic information. Trends Biochem. Sci. 2002;27:54–56. doi: 10.1016/s0968-0004(01)02027-8. [DOI] [PubMed] [Google Scholar]
- 96.Schönheit P., Buckel W., Martin W.F. On the origin of heterotrophy. Trends Microbiol. 2015;24:12–25. doi: 10.1016/j.tim.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 97.Schrenk M.O., Brazelton W.J., Lang S.Q. Serpentinization, carbon, and deep life. Rev. Mineral. Geochem. 2013;75:575–606. [Google Scholar]
- 98.Schuchmann K., Müller V. Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria. Nat. Rev. Microbiol. 2014;12:809–821. doi: 10.1038/nrmicro3365. [DOI] [PubMed] [Google Scholar]
- 99.Sessions A.L., Doughty D.M., Welander P.V., Summons R.E., Newman D.K. The continuing puzzle of the great oxidation event. Curr. Biol. 2009;19:R567–R574. doi: 10.1016/j.cub.2009.05.054. [DOI] [PubMed] [Google Scholar]
- 100.Snel B., Bork P., Huynen M.A. Genomes in flux: the evolution of archaeal and proteobacterial gene content. Genome Res. 2002;12:17–25. doi: 10.1101/gr.176501. [DOI] [PubMed] [Google Scholar]
- 101.Sofia H.J., Chen G., Hetzler B.G., Reyes-Spindola J.F., Miller N.E. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sojo V, Herschy B, Whicher A, Camprubí E, Lane N. 2016. The origin of life in alkaline hydrothermal vents. Astrobiology (⋯:⋯–⋯ in press). [DOI] [PubMed]
- 103.Sojo V., Pomiankowski A., Lane N. A bioenergetic basis for membrane divergence in Archaea and Bacteria. PLoS Biol. 2014;12 doi: 10.1371/journal.pbio.1001926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sorokin D.Y. Elemental sulfur and acetate can support life of a novel strictly anaerobic haloarchaeon. ISME J. 2015 doi: 10.1038/ismej.2015.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sousa F.L., Alves R.J., Pereira-Leal J.B., Teixeira M., Pereira M.M. A bioinformatics classifier and database for heme-copper oxygen reductases. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sousa F.L., Martin W.F. Biochemical fossils of the ancient transition from geoenergetics to bioenergetics in prokaryotic one carbon compound metabolism. BBA Bioenergetics. 2014;1837:964–981. doi: 10.1016/j.bbabio.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 107.Sousa F.L. Early bioenergetic evolution. Philos. Trans. R. Soc. Lond. B. 2013;368:20130088. doi: 10.1098/rstb.2013.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Spang A. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015;521:173–179. doi: 10.1038/nature14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sperling E.A. Statistical analysis of iron geochemical data suggests limited late Proterozoic oxygenation. Nature. 2015;523:451–454. doi: 10.1038/nature14589. [DOI] [PubMed] [Google Scholar]
- 110.Tatusov R.L., Koonin E.V., Lipman D.J. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 111.Thauer R.K., Jungermann K.K., Decker K. Energy-conservation in chemotropic anaerobic bacteria. Bacteriol. Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tuller T., Birin H., Gophna U., Kupiec M., Ruppin E. Reconstructing ancestral gene content by coevolution. Genome Res. 2010;20:122–132. doi: 10.1101/gr.096115.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Williams R., Fraústo Da Silva J. Evolution Was Chemically Constrained. J. Theor. Biol. 2003;220:323–343. doi: 10.1006/jtbi.2003.3152. [DOI] [PubMed] [Google Scholar]
- 114.Williams T.A., Foster P.G., Cox C.J., Embley T.M. An archaeal origin of eukaryotes supports only two primary domains of life. Nature. 2013;504:231–236. doi: 10.1038/nature12779. [DOI] [PubMed] [Google Scholar]
- 115.Woese C.R., Kandler O., Wheelis M.L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Woese C.R., Olsen G.J., Ibba M., Söll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wolf Y.I., Aravind L., Grishin N.V., Koonin E.V. Evolution of aminoacyl-tRNA synthetases — analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 1999;9:689–710. [PubMed] [Google Scholar]
- 118.Wolf Y.I., Koonin E.V. Genome reduction as the dominant mode of evolution. BioEssays. 2013;35:829–837. doi: 10.1002/bies.201300037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang Q., van der Donk W.A., Liu W. Radical-mediated enzymatic methylation: a tale of two SAMs. Acc. Chem. Res. 2011;45:555–564. doi: 10.1021/ar200202c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Müller M., Mentel M., van Hellemond J.J., Henze K., Woehle C., Gould S.B., Yu R.Y., van der Giezen M., Tielens A.G.M., Martin W.F. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol. Mol. Biol. Rev. 2012;76:444–495. doi: 10.1128/MMBR.05024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fujiwara T., Fukumori Y., Yamanaka T. aa3-type cytochrome c oxidase occurs in Halobacterium halobium and its activity is inhibited by higher concentrations of salts. Plant Cell Physiol. 1987;28:29–36. [Google Scholar]
- 122.Tanaka M., Ogawa N., Ihara K., Sugiyama Y., Mukohata Y. Cytochrome aa3 in Haloferax volcanii. J. Bacteriol. 2002;184:840–845. doi: 10.1128/JB.184.3.840-845.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oxygen requirements of the organisms present in the 1045 protein families.
Functional annotation.
Transparency document.