Abstract
Eukaryotic cells condense their genetic material in the nucleus in the form of chromatin, a macromolecular complex made of DNA and multiple proteins. The structure of chromatin is intimately connected to the regulation of all eukaryotic organisms, from amoebas to humans, but its organization remains largely unknown. The nucleosome repeat length (NRL) and the concentration of linker histones (ρLH) are two structural parameters that vary among cell types and cell cycles; the NRL is the number of DNA basepairs wound around each nucleosome core plus the number of basepairs linking successive nucleosomes. Recent studies have found a linear empirical relationship between the variation of these two properties for different cells, but its underlying mechanism remains elusive. Here we apply our established mesoscale chromatin model to explore the mechanisms responsible for this relationship, by investigating chromatin fibers as a function of NRL and ρLH combinations. We find that a threshold of linker histone concentration triggers the compaction of chromatin into well-formed 30-nm fibers; this critical value increases linearly with NRL, except for long NRLs, where the fibers remain disorganized. Remarkably, the interaction patterns between core histone tails and chromatin elements are highly sensitive to the NRL and ρLH combination, suggesting a molecular mechanism that could have a key role in regulating the structural state of the fibers in the cell. An estimate of the minimized work and volume associated with storage of chromatin fibers in the nucleus further suggests factors that could spontaneously regulate the NRL as a function of linker histone concentration. Both the tail interaction map and DNA packing considerations support the empirical NRL/ρLH relationship and offer a framework to interpret experiments for different chromatin conditions in the cell.
Introduction
Chromatin is a macromolecular complex made of DNA and proteins that condenses the genomic DNA in the nucleus of eukaryotic cells. The fundamental structural unit of chromatin is the nucleosome. This cylindrical, nanometric bead is made of 1.75 turns of DNA wrapped around the nucleosome core, an octamer that contains two copies each of the core histones H2A, H2B, H3, and H4 (1). In each chromosome, a single DNA molecule connects consecutive nucleosomes through linker DNAs, which determine the nucleosome repeat length (NRL), a crucial structural variable of chromatin defined by the sum of DNA basepairs (bp) of nucleosome-wound and linker DNA (2, 3, 4).
Physiological salt conditions and the presence of linker histones H1/H5 compact this beads-on-a-string polymer of nucleosomes and linker DNAs, leading to a thicker or interdigitated fiber organization that keeps hierarchically folding until shaping a chromosome in the nucleus (5, 6, 7). The structure of chromatin is well characterized at the nucleosome level (8, 9), and modern experimental techniques have made great improvements regarding visualizing chromatin at the chromosomal level (10, 11). Although the organization of chromatin at intermediate levels has remained controversial for more than three decades (7, 12), a combination of experiments, new imaging techniques, and computational models is producing significant progress (13, 14). See a recent perspective focusing on multiscale aspects of chromatin and the merging of theory and experiment (15). In particular, recent experimental and computational work indicates that the NRL impacts chromatin organization strongly and thus affects biological regulation in different cells. In fact, in vivo biochemical experiments have found that linker histone (LH) concentration linearly increases the NRL (16), but the underlying mechanism of this phenomenon remains unexplored. Our systematic computational examination of the NRL and LH concentrations aims to explain this experimental observation and shed further light on the molecular mechanisms responsible.
The NRL is made up of two parts: the DNA wrapped around the nucleosome (∼147 bp), and the linker DNA length connecting consecutive nucleosomes. The NRL varies among organisms, from ∼160 bp in yeast to ∼230 bp in sea urchin sperm cells (2, 3). The NRL can also vary among cell types, from ∼160 bp in principal neurons to ∼200 bp in glia cells in the nervous system of mammals (17). The NRL also changes across cell stages: for example, mouse cells in the embryonic stem stage have NRL ∼185 bp, but ∼195 bp when they differentiate into thymus and liver cells (18).
Recent structural studies have shown that the NRL crucially impacts the organization of chromatin: short NRLs favor the formation of fibers rich in zigzag motifs, while long NRLs lead to more heteromorphic structures (3, 19). However, although long NRLs expose more linker DNA to the cellular machinery, transcriptionally active chromatin usually adopts a shorter NRL than silenced chromatin, as noted above in the differentiation of mouse cells (16). What controls the NRL in the nucleus, and why do short NRLs favor transcription?
Woodcock et al. (16) have recently found in vivo a remarkable empirical relationship that could help us in understanding the variation of NRL in cells. Their analysis revealed a direct linear dependence between the NRL and the concentration of LHs in the nucleus, which is surprisingly robust over several organisms and cell types. Additionally, experiments controlling the LH concentration in mouse cells show that this NRL/LH concentration relationship is not a mere correlation: higher LH concentrations directly modulate the NRL in the nucleus (20). These observations suggest that a general cellular mechanism might be controlling internal factors in the nucleus to impact chromatin organization. But what is this mechanism, and, more fundamentally, how does the cell sense that these internal factors require modification at each cell stage?
To begin to address these questions, we systematically investigate the structure of chromatin fibers at different NRL and LH concentrations using our established chromatin mesoscopic model (21, 22, 23). This equilibrium approach represents a first-order approximation to a complex, dynamic system operating in cells, which can involve nucleosome sliding, DNA unwrapping, and other processes. Our analyses reveal a critical NRL linearly dependent LH concentration that compacts fibers; for long NRL, compaction is limited by the exposed areas of the linker DNA. We also discover that the NRL/LH concentration relationship is intimately connected with the pattern of the core histone tail interactions, and that it optimizes the volume and energy per basepair of chromatin. These findings suggest underlying structural and thermodynamic mechanisms for the relationship between the NRL and LH concentration, which act to regulate chromatin structure in the cell.
Materials and Methods
Mesocale chromatin model
Our computational model represents chromatin at the mesoscale level, coarse-graining each relevant molecular element at an appropriate resolution. This strategy has shed light on many properties of chromatin fibers, like the role of the flexible and positively charged core histone tails in the compaction of fibers (21) or the impact of uniform and nonuniform NRLs in chromatin architecture (3, 22). The importance of divalent ions in stabilizing heteromorphic fibers has been confirmed by electron-microscopy assisted nucleosome interaction capture experiments (19), and the mechanisms associated with LHs and Mg2+ in the unfolding of fibers (24) are in agreement with single-molecule experiments as reviewed in Collepardo-Guevara and Schlick (25). Additional experimental verifications are summarized in the literature (15, 21, 23, 26). Cryo-EM three-dimensional reconstruction of chromatin fibers (14) have confirmed our prediction that short-NRL fibers adopt 30-nm zigzag structures (3, 27). Our refined LH model also captured the spontaneous condensation of the LH C-terminal domain upon nucleosome binding and characterized the associated electrostatic mechanism that regulates the local and global organization of chromatin (23). The observed asymmetry in LH binding could also be captured and related to the asymmetry of the CTD itself. Our recent multiscale study also reproduced the effects of a common epigenetic modification on global chromatin architecture (28). Most recently, in agreement with cross-linking data, our model has reproduced internucleosome interaction patterns for interphase and metaphase chromatin to suggest a hierarchical looping mechanism for condensed chromatin (29).
Our mesoscale model of chromatin fibers has been described in detail in Perišić et al. (3) and Arya and Schlick (21). Here we summarize important aspects of the model. The nucleosome core is a relatively rigid structure made of eight core histones and 147 bp of wrapped DNA; we coarse-grain the nucleosome crystal structure (8, 9) as a rigid surface made of 300 pseudo-charges, which are adjusted at different monovalent salt concentrations using discrete surface charge optimization, a technique developed in our group to reproduce full-atom electrostatic field of protein/DNA complexes by coarse-grained models (30). The surface of the nucleosome contains 10 core histone tails that are flexible and positively charged; accordingly, we model them as elastic and charged peptides with a resolution of 5 amino acids per spherical bead and adjust the parameters using the Warshel-Levitt united-atom protein model (21). Linker DNAs connect consecutive nucleosomes and are coarse-grained using an elastic wormlike chain of nb spherical beads associated to each NRL. The number of linker DNA beads associated with each NRL value are 2, 3, 4, 5, 6, 7, and 8 for NRL = 173, 182, 191, 200, 209, 218, and 226 bps, respectively (3). The negative charge of DNA’s phosphate backbone is approximated at different salt concentrations using the Stigter’s procedure (31) (as detailed by Schlick et al. (32)). These three elements—nucleosome core, histone tails, and linker DNA—constitute each nucleosome in the model.
Additionally, we incorporate LH using our refined coarse-grained model of linker histone H1.4 (23). We discard the short N-terminal domain because it does not significantly impact chromatin structure (33). We coarse-grain the globular head using six rigid beads by applying the shaped-based coarse-graining method developed in the Schulten lab (34, 35), and adjusting the charges using discrete surface charge optimization (3, 30). We assume a symmetric positioning of the globular head as observed in Meyer et al. (36). Because the unstructured C-terminal domain shares elastic and electrostatic properties as the core histone tails, we model this 111-residue domain using 22 beads with 5-amino-acid resolution, electrostatic scaling factors, and elastic force field as in the core histone tail model. The combination of this refined LH model with the mesocale chromatin model revealed the synergy between local and global structural mechanisms in the condensation of fibers and provided a framework to interpret the role of LH posttranslational modifications (23).
All these molecular components (nucleosome complex with wrapped DNA, linker histone, and linker DNA) interact through electrostatic and excluded volume potentials as detailed in Perišić et al. (3) and Luque et al. (23). We adjust Debye-Hückel charges at different concentrations of NaCl to approximate the properties of the atomistic electric field of each element (21). However, within this framework, our model cannot treat very high monovalent salt concentrations (>200 mM), including observed depletion of nucleosomes (37). Our study focuses on physiological conditions below this ionic strength.
Monte Carlo simulations
We apply the well-known Monte Carlo sampling method to equilibrate and explore the conformational ensamble of chromatin fibers as a function of NRL and LH concentration. We use five Monte Carlo moves that efficiently explore the mesoscale properties of our model: 1) local translation and 2) local rotation of DNA beads and nucleosome cores, 3) global pivot of a fiber segment, 4) frequent regrowth of core histone tails, and 5) translation of LH CTD beads (see details in Luque et al. (23)).
We simulate fibers made of 12 nucleosomes (12-unit oligonucleosomes) at 150 mM NaCl and temperature of 293 K with different uniform NRLs: 173, 182, 191, 200, 209, 218, and 226 bp. For each case we simulate fibers containing 0, 3, 6, 9, and 12 LHs, that is, per nucleosome. We assume that LHs are randomly distributed in the chromatin fibers. Fibers saturated with LHs and without LHs are simulated starting from a zigzag configuration, dominant for the intermediate salt concentration used here (19). Fibers with intermediate concentrations of LHs are reequilibrated from saturated fibers. For each set of conditions, we simulate eight independent trajectories, applying 40–90 million Monte Carlo steps, as needed to guarantee the convergence of the energy as well as global and local quantities (3, 27). The last 10 million steps are used for statistical analysis. We store the fiber properties every 10,000 steps to decorrelate structures in the average analysis.
Structural analysis
We compute structural properties as described in Perišić et al. (3) and Luque et al. (23), namely internucleosome interactions (I(k)), dimer (dd), and tripled (td) nucleosome distances; triplet nucleosome angle (ta), fiber length , and width , sedimentation coefficient (S20,w), and packing ratio (pr). The value I(k) represents the average number of nucleosome-nucleosome interactions per nucleosome with their k neighbors, mimicking measured cross-linking contacts (19, 29). Additionally, we compute the sedimentation coefficient as a function of the LH concentration by extending our method as follows. Our protocol is grounded on the standard relation (38, 39), where nm is the radius of a nucleosome, is the sedimentation coefficient of a single nucleosome, is the number of nucleosomes in the array, and is the distance between the centers of two different nucleosomes. The sedimentation of a nucleosome without LH is (Svedberg) (40) and with LH is S (41). To incorporate a variable concentration of LHs in a fiber, we assume an effective value of that averages the contribution of nucleosomes with and without LHs, leading to the linear relationship .
Results
Threshold LH concentration as a function of the NRL
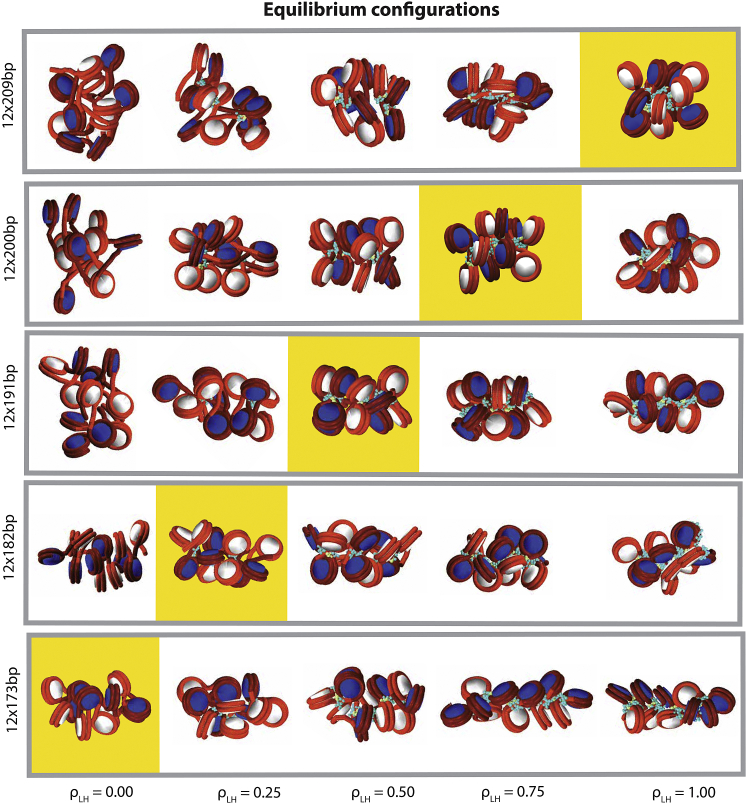
We start by analyzing the equilibrium conformations of 12-unit oligonucleosomes as shown in Fig. 1 and Fig. S1 in the Supporting Material for combinations of NRLs (rows) and LH concentrations (columns). For short NRLs, like 173 and 182 bp, well-formed fibers emerge at low LH concentrations (Fig. 1, A and 1 B). At high LH concentrations, fibers instead adopt a more stretched (173 bp) or irregularly shaped (182 bp) conformation; fibers are so tight that some LH C-terminal domains cannot condense in the nucleosome dyad region and explore extended configurations around the nucleosome (see, for instance, Fig. 1 B at ). For medium NRLs, like 191, 200, and 209 bp, fibers are disordered at low LH concentrations but well formed at medium and full LH saturation (Fig. 1, C–E). For larger NRLs, like 218 and 226 bp, fibers are highly disorganized at low LH concentrations and remain polymorphic at saturated LH conditions (Fig. S1, F and G). This analysis indicates that there is a threshold LH concentration responsible for the formation of well-defined fibers. This concentration increases linearly with the NRL, until saturating the fiber at NRL ∼209 bp (one LH per nucleosome, ). For larger NRLs, the LH C-terminal domain—condensed in the nucleosome—is too short to impose order in the fiber (3, 23).
Figure 1.
Equilibrium configurations for 12-unit oligonucleosomes at 150 mM NaCl for different LH concentrations, NRLs (rows), and (columns). Yellow boxes highlight the critical LH concentration associated to the formation of zigzag fibers for short and medium NRLs. In Fig. S1, we include equilibrium confirgurations for fibers with larger NRLs. To see this figure in color, go online.
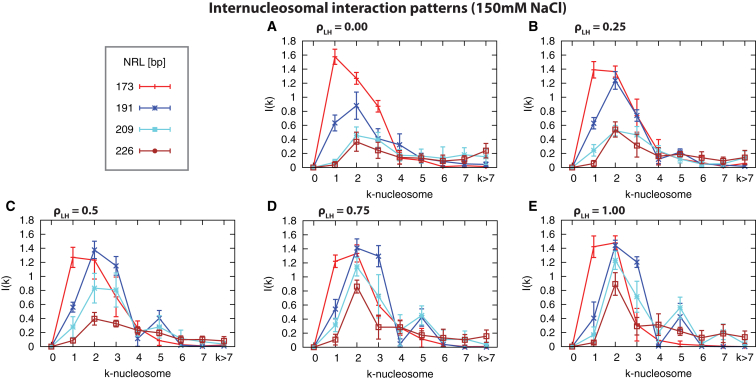
To understand better the organization of the fibers, Figs. 2 and S2 show the core-core interaction patterns associated to equilibrium chromatin structures at different conditions (Figs. 1 and S1). For short NRLs and low LH concentrations, nucleosomes are very close to each other, and first-neighbor interactions are dominant (Fig. 2 A); the increase of LH concentration, however, favors the emergence of second-neighbor interactions (Figs. 2, B–E, and S2, A and B). For medium NRLs, second-neighbor interactions dominate even in the absence of LHs, although the overall interaction pattern is weaker than for short NRLs (Fig. 2 A). The presence of LHs increases the overall interaction between nucleosomes and favors the formation of secondary peaks between fifth and seventh neighbors (Figs. 2, B–E, and S2, C–E). For larger NRLs, internucleosome interactions decrease at all concentrations and the secondary peaks smear out (Figs. 2, A–E, and S2, F and G). Thus, in general, the increase of LH concentration favors the formation of zigzag motifs, but, for large-NRL fibers, zigzag patterns are less pronounced even when they are saturated with LHs.
Figure 2.
Internucleosome interaction patterns for 12-unit oligonucleosomes at 150 mM NaCl at different NRLs (173, 191, 209, and 226 bp) grouped at different LH concentrations: (A) , (B) , (C) , (D) , and (E) . In the Supporting Material, we compare the effect of LH concentration on each NRL for additional NRLs (Fig. S2). To see this figure in color, go online.
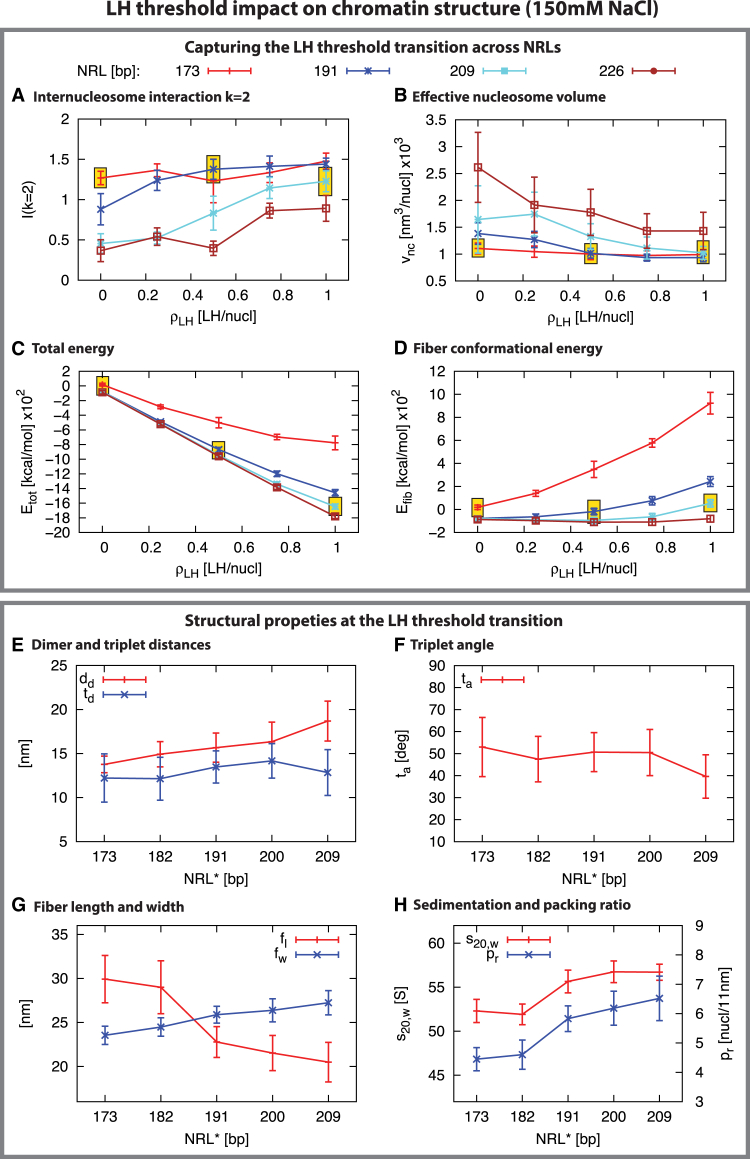
Interestingly, as highlighted in Fig. 3, the well-formed fibers that emerge at the threshold LH concentration (short and medium NRLs) share similar structural and mechanical properties; for instance, the intensity of the zigzag motifs (Fig. 3 A). This is due to the similar compaction adopted by the fibers at this transition point, as illustrated by the effective volume per nucleosome in each fiber (Fig. 3 B).
Figure 3.
LH threshold analysis for 12-unit oligonucleosomes at 150 mM NaCl. The first box compares structural properties at different LH concentrations and NRLs: (A) second neighbor core-core frequency interaction (k = 2), (B) effective volume per nucleosome , (C) internal energy, and (D) fiber conformational effective energy, , where and −11.80 kcal/mol. Yellow boxes highlight the threshold LH concentration that triggers the formation of well-defined fibers (see Fig. 1). The second box compares structural properties for different NRL fibers at the LH theshold concentration (NRL∗ = NRL at ): (E) dimer and triplet distances; (F) triplet angle; (G) sedimentation coefficient (left axis) and packing ratio (right axis); and (H) fiber length and width. In the Supporting Material we include additional structural analyses of the chromatin fibers (Figs. S3 and S4). To see this figure in color, go online.
Structural properties of chromatin are also affected by the NRL and LH concentration values. Fig. S4 shows associated sedimentation value, packing ratio, dimer distance, triplet distance, triplet angle, fiber length, fiber width, and length/width. We see that an increase of LH concentration raises the sedimentation value of fibers for all NRLs, although values saturate at high LH concentrations for medium and short NRL fibers (Fig. S4 A). Medium NRL structures reach the highest values, consistent with our work that emphasized a moderate NRL for optimal chromatin compaction (3). Long NRLs, however, are more loose (Fig. 1, F and G) and adopt the lowest values, as also noted in Perišić et al. (3). The packing ratio also tends to increase with the concentration of LHs (Fig. S4 B). Medium NRL fibers reach highest values, ∼6 nucleosomes/11 nm, while fibers with the shortest NRL, 173 bp, adopt a relatively constant value, ∼4 nucleosomes/11 nm. This was observed to result from their ladderlike organization (3).
The change in compaction is associated with internal structural changes related to the enhanced electrostatic screening of the LH C-terminal domain condensation between the entry and exit linker DNAs (23). Increasing reduces dimer distances in the fiber for all NRLs (Fig. S4 C); these distances naturally increase linearly with the NRL. The triplet nucleosome distance shows a more heterogeneous behavior (Fig. S4 D): the shorter the NRL, the less pronounced the dependence on . Reaching small triplet distances is crucial to form well-defined zigzag structures, explaining why lower LH concentrations for shorter NRLs provide optimal compaction. The trinucleosome angle follows a similar trend (Fig. S4 E). Again, medium NRLs adopt the smallest values. As expected, an inverse correlation between fiber length and packing ratio is seen (Fig. S4, F and B). The fiber width remains relatively constant for all NRLs, although for the shortest NRL fiber, 173 bp, it decreases significantly (Fig. S4 G). This indicates that the overall fiber width remains relatively stable due to excluded volume effects of the nucleosomes.
Histone tail sensitivity to NRL and LH concentration
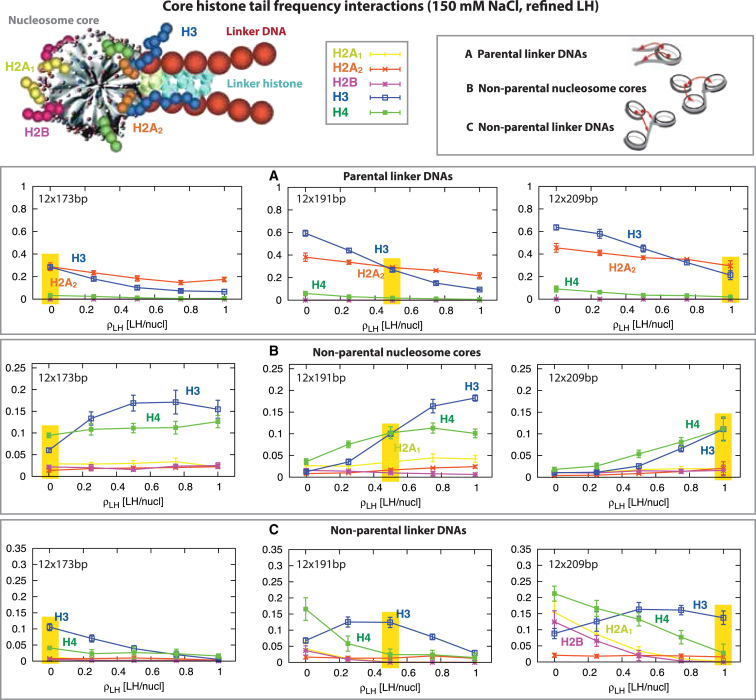
Besides LH concentrations and linker lengths, core histone tail interactions with the chromatin elements of the fiber (26) also define the chromatin folded state. In particular, we observe that the tail interaction patterns can differentiate between different NRL and LH concentration states of the chromatin fiber and produce similar patterns overall for each NRL at the threshold LH concentration that defines compact zigzag states (yellow highlights in Fig. 4, A–C). This sensitivity suggests a biophysical histone code working in analogy to a charge-modulated histone code.
Figure 4.
Core histone tail frequency interactions for 12-oligonucleosomes at 150 mM NaCl at different concentrations and NRLs (173, 191, and 209 bp). At the top we show the main elements of the nucleosome core and the color code for the histone tails: H2A1 (yellow), H2A2 (orange), H2B (purple), H3 (blue), and H4 (green). We plot the interaction of the tails with (A) parental linker DNAs, (B) nonparental nucleosome cores, and (C) nonparental linker DNAs. Yellow boxes highlight the LH concentration that triggers compact fibers at each NRL (see equilibrium structures in Fig. 1). Fig. 4 includes additional tail interactions and NRLs. To see this figure in color, go online.
Figs. 4 and S5 display the interaction patterns between core histone tails and both nucleosomes and linker DNAs as functions of NRL and LH concentration. We observe a general trend that an increase in enhances tail interactions with parent and neighboring nucleosomes (Figs. 4 B and S5, A and C) but decreases tail/linker DNA interactions (Fig. 4, A and C). The details, however, are tail-specific, and NRL- and -dependent.
For the interactions with parental nucleosome-wound DNA (Fig. S5 A), intensity generally increases with . H3 and H2A2 show the highest frequencies for parental linker DNA, followed by H4 (Fig. 4 A). For medium and long NRLs, H3 dominates over H2A2 at lower LH concentration, but H2A2 takes the lead at higher LH concentrations. The threshold associated with this crossover is higher for longer NRLs and corresponds to the onset of well-formed zigzag fibers (Fig. 1).
Tail interactions with nonparental nucleosome-wound and linker DNA, on the other hand, show different characteristics. The H4 and H3 tails are notable for their interactions with nonparental nucleosomes (Fig. 4 B), which agrees with our prior findings (26). The interaction between core histone tails and nonparental linker DNAs show the most complex pattern (Fig. 4 C). For short NRLs, H3 leads the interaction at low LH concentrations, but the intensity fades with increasing . For medium NRLs, H3’s domination over H4 occurs around the threshold associated to optimally compact chromatin fibers.
Remarkably, when we compare the histone tail interactions at the threshold for different NRL fibers (highlighted yellow regions in Fig. 4, A–C), we observe that the patterns are similar. This is due to the similar structural configuration of the fibers at the threshold despite the difference in NRL. Thus, in general, histone tails are sensitive to the variation of NRL and , except at the threshold . As we discuss below, this sensitivity offers a molecular map that could help regulate the NRL and LH concentration relationship in the cell.
Thermodynamic mechanism relating NRL and LH concentration
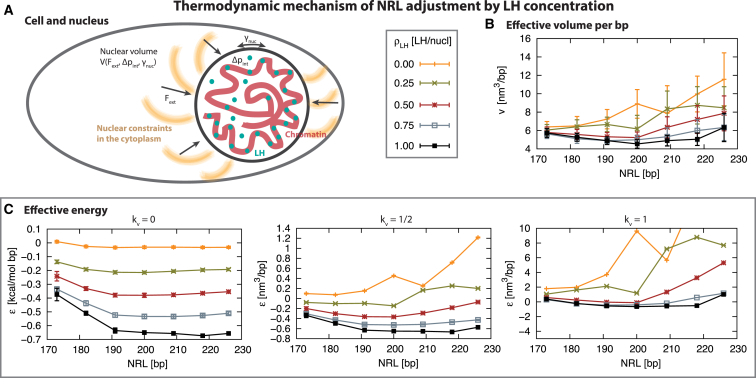
We now analyze our results with a simple, first-order approximation in the context of the cell, to investigate how the NRL and may be affected by cellular physical constraints (see Fig. 5 A). In the nucleus, the presence of salt at physiological concentration, core histones, and linker histones condenses bps of genetic material in the nuclear volume, , at a density , which also defines the volume per basepair . Using our mesoscale model we can estimate how the NRL and LH concentration impacts the effective volume per basepair in chromatin fibers, . Here is the effective fiber volume (calculated from our converged fibers as the cylindrical volume, defined by the length and width of the fiber, ) and is the number of basepairs in the fiber ( number of nucleosomes).
Figure 5.
Thermodynamic properties for 12-unit oligonucleosomes at 150 mM NaCl. (A) Representation of the nucleus constraints in a cell. The nuclear membrane surface tension is subject to an external force from the cytoplasm, and an internal pressure from the contained chromatin. The tradeoff between these forces defines the nuclear volume . (B) Effective volume per basepair (, see text) as a function of NRL for various LH concentrations, . (C) Effective energy as a function of NRL for various LH concentrations. The effective energy is the energy per basepair plus the volume-imposed energy term: . Here, E is the potential energy of the fiber, is the total number of basepairs in the fiber, represents compressibility of the nuclear membrane, v is the effective volume, and is the optimal effective volume associated with NRL = 200 bp and . We plot effective energies for a negligible , comparable , and dominant volume term. To see this figure in color, go online.
Fig. 5 B plots the effective volume per basepair, , for different NRLs and LH concentrations. Without LHs, the volume is smaller for short NRLs and rapidly increases for larger NRLs. Increasing reduces the effective volume of DNA in larger NRLs, shifting the minimum volume around medium NRLs, which reaches the smallest effective volume at full LH saturation for NRL ≈ 200 bp with ≈4.5 nm3/bp. Thus, the NRL that optimizes the packing density of chromatin in the nucleus increases with , supporting the empirical linear relationship observed experimentally.
From a thermodynamic standpoint, storing the genome in the nucleus requires a work per basepair associated with the compaction energy of chromatin and the tension in the nuclear membrane (see Fig. 5 A). We approximate the energy per basepair of a chromatin fiber using the potential energy obtained from our converged trajectories, , where E is the total internal energy of the fiber. The cell organelles and the nuclear membrane impose a reference nuclear volume, that defines the volume per basepair . If chromatin exceeds this volume, the resistance of the nucleus to deform will impose an energetic penalty, approximated here as a quadratic function around the reference volume , , where is the effective stiffness of the elastic nucleus. In this context, the total work per basepair is given by , which is compared in Fig. 5 for different NRLs and LH concentrations. For an unconstrained nucleus (i.e., ), as shown in Fig. 5 C, the internal energy dominates, and a shallow global minimum occurs at full LH saturation for long NRL (, NRL ≈ 218 bp). For a semicompressible nucleus (i.e., ), there is a tradeoff between the chromatin energy term and the volume energy term. At low LH concentrations, short NRL minimizes the energy, but, for medium NRLs, the mininum (or threshold LH concentration) increases. At full LH saturation, there is a shallow global minimum at NRL ∼ 209 bp. For a relatively incompressible nucleus (i.e., ), the volume term dominates, and the work dependence approaches the effective volume per basepair (Fig. 5 B). Thus, the presence of energy minima for different NRLs and LH concentration suggests a spontaneous thermodynamic mechanism controlling the cellular response of NRL as a function of LH concentration, which for constrained and relatively stiff nuclei supports the empirical linear relationship between these internal factors.
Discussion
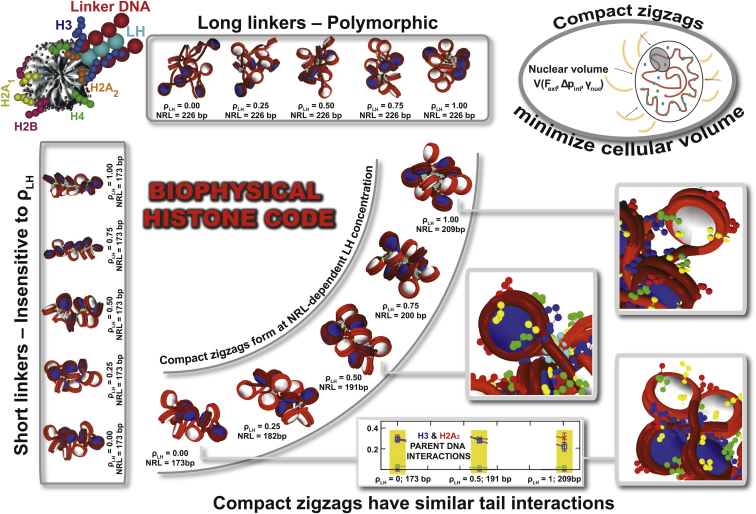
The interplay between NRL and the LH density as well as the effect of these variables on higher-order chromatin organization have been investigated experimentally. Although the H1 and H5 variants of LH exhibit some differences regarding their effects on nucleosome spacing (42) and chromatin condensation (43), the overall patterns are robust. In general, LH binding to nucleosomes is associated with larger NRLs (44). Thus, an increase in H1 results in a gradual but saturable increase of NRLs (45). Conversely, knockdown of H1 during chromatin assembly reduces linker lengths (46). Experiments also show that as linker length increases, the structure of chromatin fibers become more sensitive to LH; fibers with short linkers are insensitive to LH due to their ladderlike shape (46). Our computational work aims to shed further light into this sensitivity via a systematic study of chromatin fibers with various NRLs and in the presence of varying LH concentrations. Our results in terms of the chromatin geometry and dynamics and internucleosome interaction patterns reveal an onset of critical, NRL-dependent, LH concentration that triggers the formation of well-formed, compact chromatin fibers. This threshold of LHs increases linearly with the NRLs of the fiber from for 173 bp to for 209 bp. For larger NRLs, the length of the LH CTD condensed in the nucleosome is too short to fully form nucleosome stems to stabilize the zigzag fiber, and the resulting polymorphic topologies are less than optimally compact (3, 23, 47). Significantly, this relation between the LH concentration and NRL correlates with the linear dependence observed experimentally, for instance, in mouse tissue cells, glia cells, and chicken erythrocytes (16, 18, 20).
The qualitative framework presented here to interpret the state of chromatin and understand the regulatory role of core histone tails underscores how the organization of chromatin across organisms and cell states is related to physical parameters such as the nuclear volume, average NRL, LH concentration, and LH binding efficiency. Specifically, we suggest that cells may employ a general thermodynamic mechanism that relates these three internal variables to control the NRL of the fibers by modifying the LH concentration. Thus, the free energy for the system depends on the cellular volume constraints (Fig. 5). This free energy depends on NRL/LH combinations and imposes a penalty for larger NRLs. Thus, short NRLs have low threshold compared to medium NRL at large . This analysis is again in line with the empirical relationship observed experimentally (16). The shallow energy minimum observed for the critical at medium and high shifts from NRL ≈ 190–200 bp to NRL ≈ 200–210 bp. This shift echoes the NRL pattern observed for proliferating cells, like HeLa cells, i.e., NRL ≈ 188 bp with bulk concentration of 0.8 LHs per nucleosome (29) to differentiated cells, like chicken erythrocytes, i.e., NRL ≈ 212 bp with bulk concentration of 1.3 LHs per nucleosome (48). That NRL in different organisms has a peak at ∼200 bp (2) supports our general operative mechanism as outlined.
From a molecular perspective, the concentration of linker histones can be modulated by the nucleosome sliding and DNA unwrapping (49) to obtain the optimal NRL-dependent value. Because nucleosome unwrapping and sliding are relatively slow (50, 51), molecular remodelers must play a crucial role by speeding the adjustment of linker lengths. Indeed, observations that nucleosomal DNA is more highly methylated than flanking DNA (52) suggest that the binding of LHs can promote DNA methylation (53, 54), thereby providing a positive feedback mechanism that facilitates the removal of nucleosomes. Additionally, LHs can also methylate core histone tails, like H3 (53) to recruit chromatin remodelers such as ISWI/ACF, which interact with core histone tails and regulate the linker DNA length (55). As we discuss below, a sensitive dependence of the core histone tail interactions with linker DNA length and LH concentrations could also be interpreted in the context of a biophysical histone code that can sense the structural state of chromatin (see Fig. 6 and its caption).
Figure 6.
A possible biophysical code for the interplay between the NRL and LH in chromatin fibers. At a critical LH density that is NRL-dependent, chromatin fibers with different NRLs form compact zigzags (see fiber images inside arc). This occurs for moderate NRLs; fibers with short linkers are insensitive to LH because they adopt ladderlike forms, and fibers with long linkers bend easily because the LH-induced stem is short compared to the linker DNA. These compact zigzag forms also minimize cell volume, therefore adding a thermodynamic component. Intriguingly, we also find that the dominant tail interaction patterns are similar for different NRLs at these critical zigzag-inducing LH concentrations (as shown above for tail/parent linker DNA interactions, see Fig. 4 also), suggesting that the histone tails can act as sensors. Together, these structural and thermodynamic components suggest a biophysical code by which the NRL and LH can be adjusted as needed for cellular compaction of chromatin fibers. To see this figure in color, go online.
Our thermodynamic analysis offers a framework to explain the empirical linear relationship observed between the NRL and the LH concentration (16). Our analysis also suggests alternative scenarios for the relationship between these two structural factors. If the nuclear volume would not be a constraint, for instance, Fig. S4 C , longer NRLs could be favored thermodynamically. This could explain the unusual large NRL of echinoderm sperm cells (∼230–240 bp) (2), which have nuclei similar to human cells in size, ∼5 μm, but store an order-of-magnitude less DNA in length. Thus, relatively shorter DNA in the sea urchin sperm cells favor the formation of chromatin fibers with longer NRLs compared to compact chromatin in human cells. Additionally, the behavior of cells with nearly constant NRL over different LH concentrations, like yeast (∼165 bp) and principal rat neurons (∼162 bp) (16), could also be explained by the two mechanisms presented here. First, the presence of nucleosome positioning sequences could limit the length of linker DNAs in a chromatin fiber. This could be the main mechanism for yeast, because its genome is rich in overlapping positioning clusters (56). Second, phosphorylation reduces the binding affinity of LHs in the nucleosomes (54). This might cause a high percentage of the available LH to favor unbound or loosely bound states. Thus, despite the abundance of linker histones in high LH concentration, neuronal chromatin might bind to a limited number of those, explaining why NRLs of chromatin in principal neurons fall outside of the linear relation between NRL and LH.
Besides the thermodynamic relationship among NRLs, , and cellular volume, a sensitive relationship exists among the core histone tail interaction pattern, the NRL, and the LH concentration. That is, at the threshold where each fiber becomes a compact zigzag, the overall histone tail interactions are very similar for different NRLs (yellow highlights in Fig. 4, A–C). Such a biophysical code could regulate the cell access to DNA by the structural state of chromatin fibers in the nucleus, as required for biological processes. The predictable frequencies of interactions between core histone tails and the different chromatin elements could facilitate the binding of chromatin remodelers to adjust NRL and values at each cell state. Such a biophysical histone code (Fig. 6) could complement the chemical histone code, or epigenetic implications, characterized by posttranslational modifications (57). The combination of both signaling mechanisms could provide a communication channel between the physical and chemical states of chromatin, facilitating the active regulation of the cell nucleus. Further modeling, in particular, multiscale simulations that can treat megabase of DNA, could ultimately lead to a quantitative framework to explain and predict the dynamic regulation of chromatin structure in eukaryotic cells.
Author Contributions
A.L. designed research, performed research, analyzed data, and wrote the article; G.O. analyzed data and wrote the article; and T.S. designed research, analyzed data, and wrote the article.
Acknowledgments
Computing support from New York University, High Performance Computing at the University of South Queensland, the Bowery and Cardiac clusters, and Blue Gene at the Center for Comparative Neuroimaging is acknowledged.
This work was supported by National Institutes of Health grant No. R01 GM55164 (to T.S.). Acknowledgment is also made to Philip Morris USA and Philip Morris International for partial support of this research to T.S.
Editor: Michele Vendruscolo.
Footnotes
Five figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30219-3.
Supporting Material
References
- 1.Alberts B., Johnson A., Walter P. Garland Science/Taylor & Francis Group; Milton Park, Abingdon-on-Thames, UK: 2007. Molecular Biology of the Cell. [Google Scholar]
- 2.van Holde K. Springer; Heidelberg, Germany: 1989. Chromatin. [Google Scholar]
- 3.Perišić O., Collepardo-Guevara R., Schlick T. Modeling studies of chromatin fiber structure as a function of DNA linker length. J. Mol. Biol. 2010;403:777–802. doi: 10.1016/j.jmb.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grigoryev S.A. Nucleosome spacing and chromatin higher-order folding. Nucleus. 2012;3:493–499. doi: 10.4161/nucl.22168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 6.Maeshima K., Hihara S., Eltsov M. Chromatin structure: does the 30-nm fibre exist in vivo? Curr. Opin. Cell Biol. 2010;22:291–297. doi: 10.1016/j.ceb.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Grigoryev S.A., Woodcock C.L. Chromatin organization—the 30 nm fiber. Exp. Cell Res. 2012;318:1448–1455. doi: 10.1016/j.yexcr.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 8.Luger K., Mäder A.W., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 9.Davey C.A., Sargent D.F., Richmond T.J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 10.Dekker J. Mapping in vivo chromatin interactions in yeast suggests an extended chromatin fiber with regional variation in compaction. J. Biol. Chem. 2008;283:34532–34540. doi: 10.1074/jbc.M806479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naumova N., Imakaev M., Dekker J. Organization of the mitotic chromosome. Science. 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quénet D., McNally J.G., Dalal Y. Through thick and thin: the conundrum of chromatin fibre folding in vivo. EMBO Rep. 2012;13:943–944. doi: 10.1038/embor.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlick T., Hayes J., Grigoryev S. Toward convergence of experimental studies and theoretical modeling of the chromatin fiber. J. Biol. Chem. 2012;287:5183–5191. doi: 10.1074/jbc.R111.305763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song F., Chen P., Li G. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344:376–380. doi: 10.1126/science.1251413. [DOI] [PubMed] [Google Scholar]
- 15.Ozer G., Luque A., Schlick T. The chromatin fiber: multiscale problems and approaches. Curr. Opin. Struct. Biol. 2015;31:124–139. doi: 10.1016/j.sbi.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woodcock C.L., Skoultchi A.I., Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 17.Pearson E.C., Bates D.L., Thomas J.O. Neuronal nuclei and glial nuclei from mammalian cerebral cortex. Nucleosome repeat lengths, DNA contents and H1 contents. Eur. J. Biochem. 1984;144:353–360. doi: 10.1111/j.1432-1033.1984.tb08471.x. [DOI] [PubMed] [Google Scholar]
- 18.Fan Y., Nikitina T., Skoultchi A.I. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol. Cell. Biol. 2003;23:4559–4572. doi: 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grigoryev S.A., Arya G., Schlick T. Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proc. Natl. Acad. Sci. USA. 2009;106:13317–13322. doi: 10.1073/pnas.0903280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan Y., Nikitina T., Skoultchi A. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 21.Arya G., Schlick T. A tale of tails: how histone tails mediate chromatin compaction in different salt and linker histone environments. J. Phys. Chem. A. 2009;113:4045–4059. doi: 10.1021/jp810375d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collepardo-Guevara R., Schlick T. Chromatin fiber polymorphism triggered by variations of DNA linker lengths. Proc. Natl. Acad. Sci. USA. 2014;111:8061–8066. doi: 10.1073/pnas.1315872111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luque A., Collepardo-Guevara R., Schlick T. Dynamic condensation of linker histone C-terminal domain regulates chromatin structure. Nucleic Acids Res. 2014;42:7553. doi: 10.1093/nar/gku491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collepardo-Guevara R., Schlick T. Crucial role of dynamic linker histone binding and divalent ions for DNA accessibility and gene regulation revealed by mesoscale modeling of oligonucleosomes. Nucleic Acids Res. 2012;40:8803–8817. doi: 10.1093/nar/gks600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collepardo-Guevara R., Schlick T. Insights into chromatin fibre structure by in vitro and in silico single-molecule stretching experiments. Biochem. Soc. Trans. 2013;41:494–500. doi: 10.1042/BST20120349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arya G., Schlick T. Role of histone tails in chromatin folding revealed by a mesoscopic oligonucleosome model. Proc. Natl. Acad. Sci. USA. 2006;103:16236–16241. doi: 10.1073/pnas.0604817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlick T., Perisić O. Mesoscale simulations of two nucleosome-repeat length oligonucleosomes. Phys. Chem. Chem. Phys. 2009;11:10729–10737. doi: 10.1039/b918629h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collepardo-Guevara R., Portella G., Orozco M. Chromatin unfolding by epigenetic modifications explained by dramatic impairment of internucleosome interactions: a multiscale computational study. J. Am. Chem. Soc. 2015;137:10205–10215. doi: 10.1021/jacs.5b04086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grigoryev S.A., Bascom G., Schlick T. Hierarchical looping of zigzag nucleosome chains in metaphase chromosomes. Proc. Natl. Acad. Sci. USA. 2016;113:1238–1243. doi: 10.1073/pnas.1518280113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Q., Beard D.A., Schlick T. Constructing irregular surfaces to enclose macromolecular complexes for mesoscale modeling using the discrete surface charge optimization (DiSCO) algorithm. J. Comput. Chem. 2003;24:2063–2074. doi: 10.1002/jcc.10337. [DOI] [PubMed] [Google Scholar]
- 31.Stigter D. Interactions of highly charged colloidal cylinders with applications to double-stranded. Biopolymers. 1977;16:1435–1448. doi: 10.1002/bip.1977.360160705. [DOI] [PubMed] [Google Scholar]
- 32.Schlick T., Li B., Olson W.K. The influence of salt on the structure and energetics of supercoiled DNA. Biophys. J. 1994;67:2146–2166. doi: 10.1016/S0006-3495(94)80732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allan J., Mitchell T., Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. J. Mol. Biol. 1986;187:591–601. doi: 10.1016/0022-2836(86)90337-2. [DOI] [PubMed] [Google Scholar]
- 34.Yin Y., Arkhipov A., Schulten K. Simulations of membrane tubulation by lattices of amphiphysin N-BAR domains. Structure. 2009;17:882–892. doi: 10.1016/j.str.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 36.Meyer S., Becker N.B., Everaers R. From crystal and NMR structures, footprints and cryo-electron-micrographs to large and soft structures: nanoscale modeling of the nucleosomal stem. Nucleic Acids Res. 2011;39:9139–9154. doi: 10.1093/nar/gkr573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao B., Freedman B.S., Marko J.F. Histone H1 compacts DNA under force and during chromatin assembly. Mol. Biol. Cell. 2012;23:4864–4871. doi: 10.1091/mbc.E12-07-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bloomfield V., Dalton W.O., van Holde K.E. Frictional coefficients of multisubunit structures. I. Theory. Biopolymers. 1967;5:135–148. doi: 10.1002/bip.1967.360050202. [DOI] [PubMed] [Google Scholar]
- 39.Kirkwood J. The general theory of irreversible processes in solutions of macromolecules. J. Polym. Sci. Polym. Phys. 1954;12:1–14. [Google Scholar]
- 40.Garcia-Ramirez M., Dong F., Ausio J. Role of the histone “tails” in the folding of oligonucleosomes depleted of histone H1. J. Biol. Chem. 1992;267:19587–19595. [PubMed] [Google Scholar]
- 41.Butler P.J., Thomas J.O. Dinucleosomes show compaction by ionic strength, consistent with bending of linker DNA. J. Mol. Biol. 1998;281:401–407. doi: 10.1006/jmbi.1998.1954. [DOI] [PubMed] [Google Scholar]
- 42.Sancho M., Diani E., Jordan A. Depletion of human histone H1 variants uncovers specific roles in gene expression and cell growth. PLoS Genet. 2008;4:e1000227. doi: 10.1371/journal.pgen.1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clausell J., Happel N., Beato M. Histone H1 subtypes differentially modulate chromatin condensation without preventing ATP-dependent remodeling by SWI/SNF or NURF. PLoS One. 2009;4:e0007243. doi: 10.1371/journal.pone.0007243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Öberg C., Izzo A., Belikov S. Linker histone subtypes differ in their effect on nucleosomal spacing in vivo. J. Mol. Biol. 2012;419:183–197. doi: 10.1016/j.jmb.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 45.Belikov S., Åstrand C., Wrange O. Mechanism of histone H1-stimulated glucocorticoid receptor DNA binding in vivo. Mol. Cell. Biol. 2007;27:2398–2410. doi: 10.1128/MCB.01509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Routh A., Sandin S., Rhodes D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc. Natl. Acad. Sci. USA. 2008;105:8872–8877. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang H., Clark D.J., Hayes J.J. DNA and nucleosomes direct distinct folding of a linker histone H1 C-terminal domain. Nucleic Acids Res. 2012;40:1475–1484. doi: 10.1093/nar/gkr866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bates D.L., Thomas J.O. Histones H1 and H5: one or two molecules per nucleosome? Nucleic Acids Res. 1981;9:5883–5894. doi: 10.1093/nar/9.22.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schiessel H. The physics of chromatin. J. Phys. Condens. Matter. 2003;15:R699–R774. doi: 10.1088/0953-8984/27/6/060301. [DOI] [PubMed] [Google Scholar]
- 50.Blossey R., Schiessel H. The dynamics of the nucleosome: thermal effects, external forces and ATP. FEBS J. 2011;278:3619–3632. doi: 10.1111/j.1742-4658.2011.08283.x. [DOI] [PubMed] [Google Scholar]
- 51.Pennings S., Meersseman G., Bradbury E.M. Linker histones H1 and H5 prevent the mobility of positioned nucleosomes. Proc. Natl. Acad. Sci. USA. 1994;91:10275–10279. doi: 10.1073/pnas.91.22.10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chodavarapu R.K., Feng S., Pellegrini M. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang S.M., Kim B.J., Skoultchi A.I. H1 linker histone promotes epigenetic silencing by regulating both DNA methylation and histone H3 methylation. Proc. Natl. Acad. Sci. USA. 2013;110:1708–1713. doi: 10.1073/pnas.1213266110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Over R.S., Michaels S.D. Open and closed: the roles of linker histones in plants and animals. Mol. Plant. 2014;7:481–491. doi: 10.1093/mp/sst164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Florescu A.-M., Schiessel H., Blossey R. Kinetic control of nucleosome displacement by ISWI/ACF chromatin remodelers. Phys. Rev. Lett. 2012;109:118103. doi: 10.1103/PhysRevLett.109.118103. [DOI] [PubMed] [Google Scholar]
- 56.Cole H.A., Nagarajavel V., Clark D.J. Perfect and imperfect nucleosome positioning in yeast. Biochim. Biophys. Acta. 2012;1819:639–643. doi: 10.1016/j.bbagrm.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.