Abstract
The mechanical properties of the microenvironment and direct contact-mediated cell-cell interactions are two variables known to be important in the determination of stem cell differentiation fate, but little is known about the interplay of these cues. Here, we use a micropatterning approach on polyacrylamide gels of tunable stiffnesses to study how homotypic cell-cell contacts and mechanical stiffness affect different stages of osteogenesis of mesenchymal stem cells (MSCs). Nuclear localization of transcription factors associated with osteogenesis depended on substrate stiffness and was independent of the degree of cell-cell contact. However, expression of alkaline phosphatase, an early protein marker for osteogenesis, increased only in cells with both direct contact with neighboring cells and adhesion to stiffer substrates. Finally, mature osteogenesis, as assessed by calcium deposition, was low in micropatterned cells, even on stiff substrates and in multicellular clusters. These results indicate that substrate stiffness and the presence of neighboring cells regulate osteogenesis in MSCs.
Keywords: micropatterning, mesenchymal stem cells, differentiation, osteogenesis, mechanical properties
Introduction
Mesenchymal stem cells (MSCs) are a promising adult stem cell source with regenerative and immunomodulatory properties [1]. Understanding the factors influencing their differentiation in vitro is of major therapeutic interest. Cell-cell contact and mechanical properties of the substrate have been shown to be two important factors affecting MSC differentiation, but their interplay has received little attention [2]. In a cytoskeleton and integrin-mediated mechanism, mesenchymal stem cells (MSCs) will respond to substrate stiffnesses by differentiating down specific lineages [3–6]. For example, stiff substrates abet osteogenesis, while softer substrates enhance adipogenesis and lead to the expression of genes associated with transdifferentiation into neurons. Stiffness also influences other biological activity, such as proliferation, as MSCs on soft substrates show decreased proliferative activity compared to cells grown on stiffer surfaces [7]. Recent studies have shown that mechanical cues can impact differentiation state even after weeks of culture, by which time differentiation programs have already been executed [8].
Separate studies have shown the importance of cell-cell contact on MSC behavior. Direct co-culture with osteoblasts increases osteogenesis in MSCs but via pathways different from indirect co-culture conditions, in which cell-cell contact is precluded [9]. Direct contact with cells from a different lineage, such as endothelial cells, can also influence MSC phenotype [10]. Homotypic cell-cell contact is also important, as limiting the degree of cell-cell contacts between MSCs inhibits the adipogenic, osteogenic, and chondrogenic arms of differentiation, and vice versa [11–13]. MSC seeding density has been shown to affect response to mechanical cues, with cells seeded at high densities upregulating osteogenic genes regardless of substrate stiffness [14].
We hypothesized that both direct cell-cell contact and appropriate substrate mechanical properties would be required for MSC osteogenic differentiation. To test this hypothesis, we utilized a micropatterning approach, as previous studies have exploited micropatterning platforms to stringently control cell spreading and cell-cell contact, among other behaviors [15]. Specifically, we patterned circular 20 and 30 micrometer-diameter islands of fibronectin, an adhesion molecule, at a uniform spacing onto polyacrylamide (PA) gels of varying Young’s moduli. Uniform spacing between patterned islands normalizes cell exposure to paracrine molecules, and pattern dimension controls cell shape and the degree of cell-cell contact, all factors that have been shown to impact differentiation [11,12], [17–23]. Because the number of cells adhering to a single micropatterned fibronectin island falls in a distribution [24], we were able to study both isolated single cells as well as small groups of cells that were in contact with each other on a single fibronectin island.
Synthetic hydrogels, such as PA gels, can be fabricated to convey specific mechanical cues to adherent cells [25]. By varying monomer and crosslinker ratios in the PA gel, we could precisely tune the substrates’ Young’s moduli without changing adhesion ligand density, which has been shown to affect MSC differentiation [26]. We chose three stiffnesses to reflect the range over which MSCs respond to the mechanical cues of their environment. In three dimensions, matrices with an elastic modulus of 20 kPa were found to most effective at inducing osteogenesis, whereas elastic moduli of 2 kPa and lower result in far less osteogenesis [5]. We chose the clonally-derived murine MSC D1 line to culture on micropatterned islands [27–29], as clonal populations provide greater cell-to-cell homogeneity [5] compared to primary cells. To assess their differentiation, MSCs were fixed at different times points and stained for a variety of osteogenic markers, including transcription factors Yes-associated protein (YAP), Runt-related transcription factor 2, and osterix; the early osteogenic protein marker alkaline phosphatase (ALP); and Alizarin Red for calcium deposition. Concurrently, the numbers of D1 cells per island of fibronectin was quantified to correlate osteogenesis with direct cell-cell contact.
Materials and Methods
Substrate preparation and stiffness measurement
PA gels were fabricated on glass cover slips and functionalized with hydrazine [30]. 18-mm glass coverslips were cleaned by sonicating in 70% ethanol and then functionalized with 3-aminopropyltrimethoxysilane, followed by 1% gluteraldehyde. Various mixtures of acrylamide and bis-acrylamide were cast between a Sigmacote(R)-treated glass slide and functionalized coverslips to form PA gels. Ammonium persulfate and N,N,N′,N′-tetramethylethylenediamine were used to polymerize polyacrylamide. The PA gel surface was then functionalized with 100% hydrazine for 20 hours, quenched with 5% acetic acid in deionized water, washed with deionized water, and air dried before micropatterning. The Young’s modulus of bulk PA gels were measured by casting gels into a 10-mm diameter mold and compressing without confinement using an Instron 3342 mechanical apparatus at 1 mm min−1. To perform atomic force microscopy (AFM) measurements, a MFP-3D system (Asylum Research) was used with silicon nitride cantilevers (MLCT, Bruker AFM Probes). The stiffness was calibrated by determining a spring constant of the cantilever from the thermal fluctuations at room temperature, ranging from 20~50 mN/m. The cantilever was moved towards the stage at a rate of 1 μm s−1 for indentations.
Micropatterning
Standard soft lithography techniques were used to create PDMS stamps. In short, negative photoresist SU-8 2025 (MicroChem, Newton, MA) was spun onto clean silicon wafers to a thickness of 25 um, and patterned by exposure of UV light through a transparency photomask (CAD/Art Services, Bandon, OR). After the photoresist was developed, poly(dimethylsiloxane) (PDMS) mixed with crosslinker at a 10:1 ratio was poured on, crosslinked at 65 °C, and cut into stamps. Stamps were cleaned by sonication in 70% ethanol. After drying, 20 ug/mL of human fibronectin in deionized water was incubated on stamp surfaces for an hour. After the fibronectin was aspirated, the stamp was air-dried and stamped gently onto the PA gel for two minutes. The stamp was removed, and both the PA gel and the tissue culture well harboring the gel were treated with 1% Pluronic-F127 for five minutes and washed three times with Dulbecco’s phosphate buffered saline before culture. Transfer of fibronectin was assessed using immunofluorescence. Substrates were blocked with 1% bovine serum albumin. Primary antibody labeling was performed in 1% BSA with rabbit anti-fibronectin (Sigma, St. Louis, MO), and secondary antibody labeling was performed using the same procedure with DyLight 488 conjugated anti-rabbit goat IgG (Rockland, Limerick, PA). A uniform intensity threshold was used to identify immunostained fibronectin micropatterns on all substrates, and area-average intensities were compared. Fluorescence images for fibronectin immunofluorescence were acquired using an Olympus IX81 inverted microscope (BD Biosciences, San Jose, CA) and a Coolsnap HQ2 camera (Prior Scientific, Rockland, MA).
Cell culture
Prior to seeding on micropatterns, clonally derived mouse MSCs (D1s) purchased from American Type Cell Culture were expanded subconfluently in high-glucose, Dulbecco’s Modified Eagle media supplemented with 10% fetal bovine serum (complete DMEM). MSCs were trypsinized, seeded onto micropatterned islands, and allowed to adhere for 3 hours, after which non-adhered cells were washed off via three changes of media. Twenty-four hours after seeding, osteogenic differentiation was induced and maintained by supplementing complete DMEM with 10 mM β-glycerophosphate and 250 μM L-ascorbic acid, cycling every two days. Cells were fixed at seven or 24 hours after seeding for transcription factor analysis, six days after osteogenic induction for ALP analysis, and two weeks after osteogenic induction for calcium deposition.
Analysis of cell proliferation
To assess cell populations on fibronectin islands at 3 hours, cells were fixed, as described below, and stained with DAPI. To analyze cell proliferation, cells were incubated with EdU for 24 hours at three days after seeding onto micropatterns. Immediately after incubation with EdU, cells were fixed and EdU was detected with a Click-IT EdU AlexaFluor 555 imaging kit (Life Technologies, Eugene, OR). Images were taken on a Nikon E800 upright microscope and scored as either EdU positive or negative.
Analysis of transcription factor localization, alkaline phosphatase (ALP) production, calcium deposition, and other lineage markers
Nuclear localization of YAP and Runx2 was assessed using immunofluorescence. For YAP staining, samples were fixed, permeabilized with Triton X-100, blocked with 10% goat serum, incubated with a rabbit YAP-only primary antibody overnight (Cell Signaling 4912S, Danvers, MA), and then incubated with an anti-rabbit FITC secondary antibody (Sigma, St. Louis, MO). Osx (AbCam) and sclerostin (R&D Systems, Minneapolis, MN) staining was done following the same procedure. Runx2 staining was performed in a similar manner (Novus Biologicals, San Diego, CA), except the primary was conjugated with a fluorescent tag. ImageJ was used to quantify intensity of immunostain and nuclear localizations of transcription factors. Nuclei were counterstained with DAPI and imaged using confocal microscopy (Upright Zeiss LSM 710). Slices with the brightest nuclear signal were used for analysis. For ALP analysis, D1s were fixed and stained with elf-97, following the manufacturer’s instructions. Staining was stopped through washing with excess of PBS after 90 seconds. Fixed cells were further stained with the red membrane linker dye PKH26, and cell nuclei were stained with DAPI. An Olympus IX81 inverted microscope with a Carv II Nipkow-type Spinning Disc Confocal Attachment (BD Biosciences, San Jose, CA) and a Coolsnap HQ2 camera (Prior Scientific, Rockland, MA) were used to acquire elf-97 staining and day 6 DAPI images. PKH26-stained regions were thresholded in ImageJ to demarcate outlines of micropatterned cells. To ensure there was no false positive from DAPI fluorescence, DAPI images were substracted from elf-97 before quantification. The area-average fluorescence of cells stained with elf-97 within PKH26-stained outlines was quantified with ImageJ. The number of nuclei per micropattern was counted by eye. Calcium deposition was assessed with Alizarin Red staining. Cells were equilibrated in distilled water following fixation, incubated with Alizarin Red stain for 5 minutes, and washed until clear. For neutral lipid production, samples were equilibriated into 60% 2-propanol and incubated with Oil Red O (Sigma) before being washed until clear. For proteoglycan production, samples were fixed and incubated with a 1% Alcian Blue 8 GX (Sigma) solution at pH 2.5 before washed until clear. All samples were counterstained with DAPI and imaged with an Nikon Eclipse E800.
Statistical testing
Analysis of a continuous variable against one factor was performed using a 1-way ANOVA, followed by pairwise comparisons with Tukey-Kramer compensation. Analysis of a continuous variable against two factors, for example substrate stiffness and island cellularity, was performed using an n-way ANOVA, followed by pairwise comparisons with Tukey-Kramer compensation, in MATLAB. Comparisons of EdU staining on different substrate stiffnesses was analyzed using Chi-square test for trend in GraphPad Prism. Correlation coefficients and best fit lines were computed using the least squares method in MATLAB. A p value of less than 0.05 was considered significant.
Results
Substrate preparation and cell proliferation
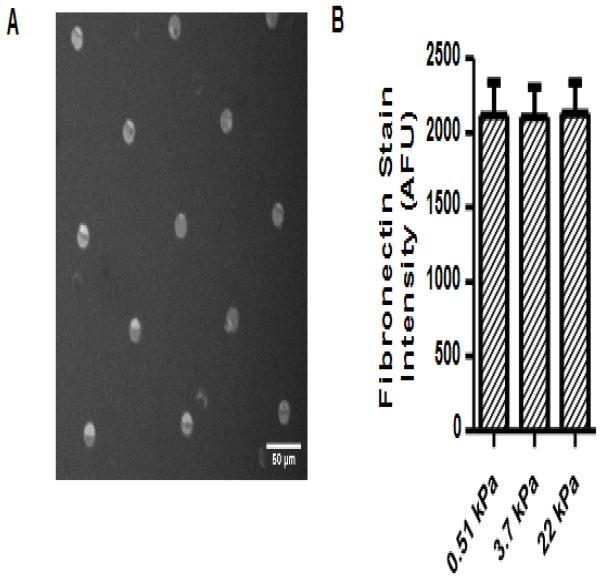
Altering the ratio of acrylamide to bis-acrylamide produced hydrogel substrates of varying Young’s moduli. Air drying after hydrogel fabrication, followed by rehydration, did not induce local stiffness increases, as determined by AFM (Supp Fig. 1). Microcontact printing generated fibronectin islands that, as verified with immunohistochemistry (Fig. 1A), did not differ in fibronectin intensity across stiffnesses, indicating that the amounts of conjugated protein on the different surfaces were consistent (Fig. 1B).
Figure 1.
Fibronectin micropatterning on substrates of varying Young’s moduli. Fibronectin micropatterns were visualized with immunohistochemistry (A), and the fluorescent intensity of fibronectin across substrates of different moduli was not significantly different (B). Experimental repeats: 2.
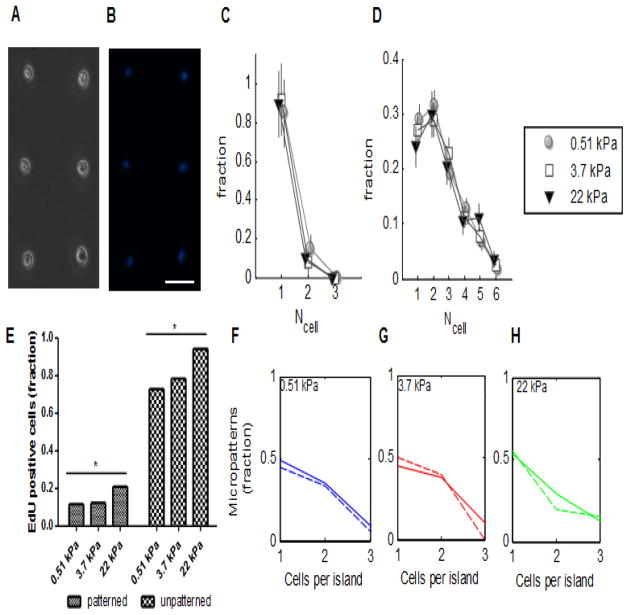
The distribution of MSC numbers on 20-micrometer diameter micropatterned fibronectin islands was assessed at 3 hours after seeding (Fig. 2A, 2B), a time span short enough to preclude cell division as a contributor to cell number. Most fibronectin islands harbored only one cell, with a few doublets. The distribution was similar on micropatterned surfaces of all three stiffnesses (Fig. 2C). After six days of culture, MSC numbers on fibronectin islands were again assessed. The cell number distributions had broadened and shifted rightwards (Fig. 2D), presumably reflecting rounds of cell division, as excess cells had been washed away immediately after seeding. Even after six days, islands with only single cells were observed, suggesting that the fibronectin pattern size was sufficiently small to retain some cells that had not undergone division, allowing cells without a neighbor to be studied. The finding that adjacent cell islands did not join after six days of culture indicates that the Pluronic-treated PA gel surface between patterned islands adequately repelled cell adhesion. The number of cells per island was therefore limited by the size of the fibronectin pattern. The cell distributions on gels of different moduli were not significantly different from each other, as assessed by the chi-squared test.
Figure 2.
Cell adhesion and proliferation over time on patterned and unpatterned substrates. MSCs adhered to fibronectin on micropatterned islands on day 0, brightfield (A) and DAPI-stained (B). (C) Normalized frequency of numbers of cells per island on substrates of different stiffness on day 0. Error bars show standard deviation; n > 26 per stiffness. (D) Normalized frequency of numbers of cells per island on substrates of different stiffness on day 6. Error bars show standard deviation; n = >180 per stiffness. (E) Fraction of EdU positive cells on patterned and unpatterned substrates of different elastic moduli. Increased stiffness was associated with increased EdU staining in both patterned and unpatterned substrates. * = p < 0.05, Chi-squared test for trend. (F–H) Normalized frequency of cell number on micropatterned islands that were EdU positive cells (solid line), compared to the frequency of cell number on micropatterned islands overall (dotted line). Scale bar = 50 microns; error bars denote standard deviation. For (E) – (H), n = 262 micropatterned and 581 unpatterned cells. Experimental repeats: 2.
Using incorporation of a thymidine analog, 5-ethynyl-2′-deoxyuridine (EdU), the proliferation of D1s was assessed at 3 days, an intermediate time point between cell seeding and differentiation analysis with ALP, on both patterned and unpatterned substrates. A 24 hour incubation period was chosen because it allowed EdU incorporation on both patterned and unpatterned substrates, without any one condition resulting in a saturation of EdU incorporation in all cells. The fraction of MSCs that incorporated EdU on unpatterned substrates increased significantly with stiffness, as analyzed by a Chi-squared test (Fig. 2E). A smaller fraction of the total number of cells cultured on micropatterns incorporated EdU, likely because proliferation was reduced by the growth area restrictions of the micropatterns. As on unpatterned substrates, proliferation increased significantly with substrate stiffness (Fig. 2E). In order to assess whether cellularity impacted proliferation, the distribution of cell number per EdU-positive micropatterned island was compared to the cell number distribution of micropatterned islands in general, on substrates of each of the three moduli (Fig. 2F–H). The fraction of EdU positive micropatterns decreased with increased cellularity, presumably because the micropatterned island size had constrained cell growth, thereby decreasing EdU incorporation. The cell number distributions of EdU-positive micropatterned island was found not to differ significantly from the cell number distribution of micropatterned islands in general, as assessed by a Kolmogorov-Smirnov test, suggesting that proliferation was unaffected by cellularity of micropatterned islands.
Osteogenic differentiation
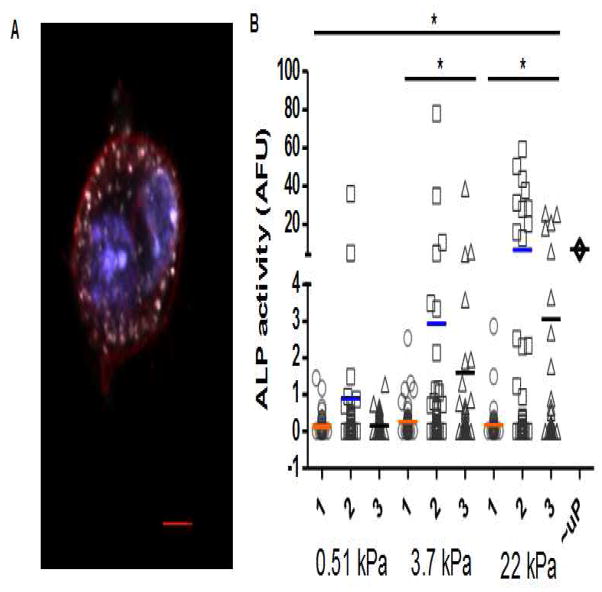
D1 MSCs seeded onto 20-micrometer diameter fibronectin micropatterns were cultured in osteogenic differentiation media for 6 days and stained for ALP, an early marker for osteogenesis (Fig 3A). Cells on fibronectin-coated, unpatterned 22 kPa substrates were cultured and stained in parallel (Fig 3D). Cells on both patterned and unpatterned substrates in basal media without osteogenic supplements were cultured and stained in parallel as negative controls (Supp Fig. 2). Naive D1s were found to stain negatively for ALP as well as for the neutral lipid stain Oil Red O and the proteoglycan stain Alcian Blue, dyes for assessing adipogenic and chondrogenic differentiation, respectively. Cells at an earlier time point stained negative for Alcian Blue, but were positive at a later time point, indicating that cells with chondrogenic characteristics may have developed as well (Supp Fig. 3), as has been observed in some MSC cultures [31]. Under media conditioned with osteogenic supplements, single cells, doublets, and triplets showed no significant differences in ALP expression when differentiated on 0.51 kPa and on 3.7 kPa substrates, and all exhibited low levels of ALP. Conversely, doublets and triplets produced significantly higher amounts of ALP on the 22 kPa substrate, while single cells on the 22 kPa substrate still exhibited low ALP expression (Fig. 3B, Supp Fig. 4). Among micropatterned islands with the same number of MSCs, doublet and triplet cells cultured on the 22 kPa substrate expressed higher levels of ALP than their counterparts on 0.51 kPa and 3.7 kPa substrates; this effect was not observed in single cells. Besides cell number, we looked for correlations between projected cell area and differentiation (Supp Fig. 5), since spread area has been linked to differentiation [18]. Cells were not observed on the unpatterned areas of PA gel surfaces, and cells remained confined to the fibronectin islands as noted above. However, except for on 0.51 kPa substrates, no significant correlation between area and differentiation was observed, and this correlation vanished when queried within sets of islands harboring the same number of cells.
Figure 3.
ALP expression at day 6 in micropatterns with different numbers of cells, and on different substrate stiffnesses. (A) Confocal image of a micropatterned cell positively stained for ALP. Blue, nucleus; red, membrane; white, ALP. (B) Quantification of the level of ALP (area average) as a function of cell number on substrates with E of 0.51 kPa, 3.7 kPa, and 22 kPa, respectively. ALP expression on unpatterned 22 kPa substrates (uP) is also provided for comparison. ALP expression varies significantly as a function of substrate stiffness and cellularity (n-way ANOVA, p < 0.01). While 2- and 3-cell islands expressed significantly more ALP on stiff than on soft substrates (p < 0.05; 1-way ANOVA with Tukey compensation), this was not the case for 1-cell islands. Per stiffness, between 170 and 220 micropatterned islands were analyzed; experimental repeats: 2.
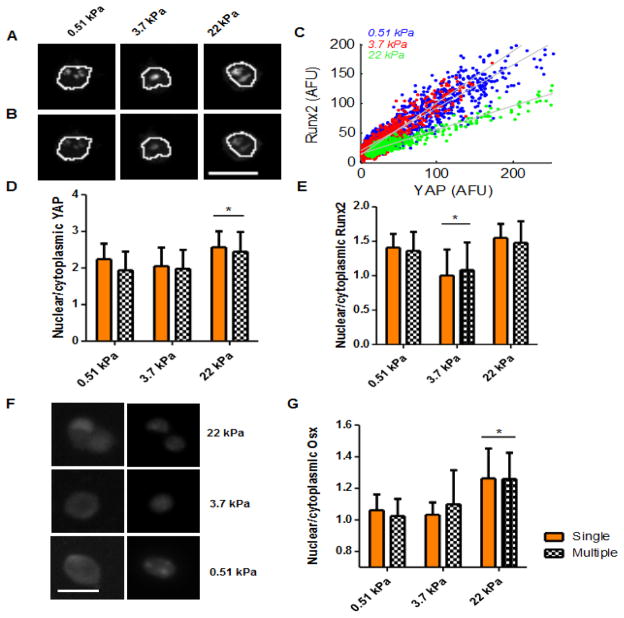
MSCs seeded on fibronectin micropatterns were then assessed for nuclear localization of YAP and Runx2. Runx2 has been shown to be crucial to osteogenic commitment of MSCs, serving as a master regulator of the osteoblast differentiation process, while YAP has been implicated as having a central role in mediating mechanotransduction [32,33]. Since most 20-um diameter micropattern harbored single cells, a combination of 20-um and 30-um fibronectin islands were patterned, enabling both single cell and multiple cell clusters to be assessed at early time points. Both YAP and Runx2 localized to the nucleus to some extent on all substrate stiffnesses (Fig 4A, 4B, Supp Fig 6). Moreover, the two transcription factors appeared to localize to the same regions within the nucleus, an observation confirmed by pixel intensity analysis, wherein pixels with a high intensity of one transcription factor correlated with high intensity pixels of the other (Fig 4C). Analysis of variance of the slope of best fit lines determined significant differences, indicating varying relative intensities of YAP and Runx2 per area as a function of stiffness. The ratios of nuclear to cytoplasmic localization of both transcription factors on varying E was subsequently quantified. Similar levels of YAP nuclear localization were present in cells grown on 0.51 and 3.7 kPa substrates, whereas cells cultured on 22 kPa substrates exhibited a higher ratio of nuclear to cytoplasmic YAP localization (Fig 4D). Interestingly, whereas similar levels of Runx2 nuclear localization were observed in cells grown on 0.51 and 22 kPa substrates, cells grown on 3.7 kPa substrates exhibited the lowest nuclear to cytoplasmic ratio of this transcription factor (Fig 4E). For both transcription factors and across varying E, no significant differences in nuclear localization were observed in single versus multiple cell clusters. To determine downstream effects of YAP and Runx2 nuclear localization, the same analysis at a later time point was performed on osterix (Osx), a transcription factor necessary for osteoblastic differentiation after Runx2 activation of the osteogenesis program [33]. In the stiffest substrates, Osx localized to and occupied the nucleus more homogeneously than Runx2 and YAP, which appeared punctile (Fig. 4F). As with these earlier transcription factors, increased modulus of substrates correlated with increased nuclear localization of Osx, independent of cellularity (Fig. 4G). Sclerostin, a transcription factor associated with the inhibition of osteogenesis [34], was stained for as a negative control (Supp Fig. 6C).
Figure 4.
Transcription factor localization in micropatterned cells at 7 hours of culture. Representative image of YAP stain (A) and runx2 stain (B) in a micropatterned cell grown on substrates of varying E. White lines outline nucleus. (C) Scatter plots of the pixel intensities of nuclear YAP and runx2 stains within the nucleus, in three micropatterned cells per stiffness condition. Gray lines represent least squares fit. Quantification of nuclear localization of YAP (D) and runx2 (E) on micropatterns on different E for islands with single or multiple cells, n > 25 cells per stiffness. * = p <0.05, n-way ANOVA. (F) Representative images of osterix (left) and nuclear (right) stained cells on different stiffnesses, n > 25 cells per stiffness. (G) Quantification of nuclear localization of osterix, n between 23 and 36 cells. * = p <0.05, n-way ANOVA. Scale bar, 20 um. YAP and Runx2 experimental repeats: 3; osterix experiment was performed once.
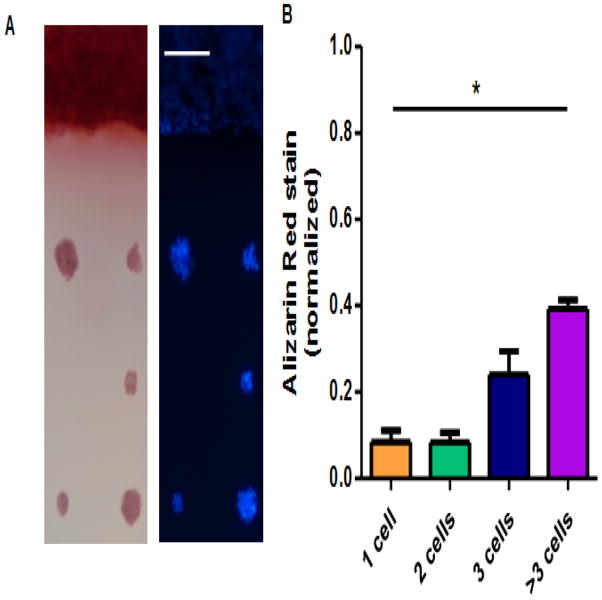
Finally, progression to mature osteogenesis in micropatterned cells was assessed by staining for calcium deposition in 2 week cultures. The spacing between micropatterns was increased to 150 micrometers for this study, in order to decrease potential cell growth bridging adjacent micropatterns. A large area of fibronectin, 1 mm wide on its narrowest dimension, was also patterned alongside islands in order to verify that cells could produce late osteogenesis markers on substrates in the absence of growth area restraints. Robust calcium deposition was observed in cells grown on the large area of patterned fibronectin at 22 kPa, with much less calcium staining occurring in patterned islands, in a cell-number dependent manner (Fig 5A–B). Basal media controls showed very little calcium deposition (Supp Fig. 2).
Figure 5.
Osteogenic differentiation at 2 weeks. (A) Alizarin red stain (left) and DAPI stain (right) of cells grown on 22 kPa PA gels, either on unpatterned regions (top of picture) or on 30-micron diameter islands. (B) Quantification of Alizarin red stain, normalized to cells that stained positive grown on unpatterned region on the same PA gel. * = p < 0.05, 1-way ANOVA, n = 28 micropatterns, experimental repeats: 2. Scale bar, 100 um.
Discussion
The osteogenic response of MSCs was found to depend on both the modulus of the adhesion substrate and the number of immediate neighbors, although to varying degrees depending on the stage of differentiation analyzed. The interplay of mechanical and cell-cell contact cues was most apparent in ALP production, which is an early protein marker for osteogenesis. Single MSCs produced little ALP, regardless of the modulus. However, on substrates with sufficiently high E, only cells experiencing direct cell-cell contact produced significant amounts of ALP, as shown by cell doublets and triplets cultured on 22 kPa PA gels robustly expressing ALP compared to single cells on the same stiffness. The higher ALP production on 22 kPa gels and lower ALP production on softer substrates agrees with the effects of substrate stiffness on osteogenesis that have been observed in previous studies [3,5]. This is also in line with studies that have linked greater cell-cell contact in MSCs with higher levels of ALP production [11,12], as well as with research demonstrating that cell density in general impacts various biology phenomena, including adipogenic and neuronal differentiation [35,36] in MSCs, expansion of hematopoietic stem cells [37], and induction of pluripotent stem cells [38]. Interestingly, production of ALP in doublet and triplet cells was greatly reduced on softer substrates, indicating that cell-cell contact is insufficient for osteogenesis and that differentiation requires appropriate mechanical cues, at least for cells seeded on micropatterns at this density. The similarity in immunostaining intensity of fibronectin on different stiffnesses implies that any phenotypic changes that were observed were due to mechanical properties alone. Since spread area has been well established to be linked to differentiation [8,18], the relationship between cell or cell cluster area with differentiation was also assessed. However, as cell spread area was generally confined by fibronectin patterning of the same size and as some cells in multicellular clusters appeared to overlap (Supp Fig. 3a), quantifying the volume of a cell or cell cluster may be a more revealing metric than total spread area for cell fate in this system. Indeed, only cells grown on 0.51 kPa substrates were found to exhibit any correlation between area and differentiation (Supp Fig. 3b). The specific mechanisms prompting osteogenesis as a result of increased cell number per micropatterned island are unknown, although previous studies suggest that gap junctions [11] and cadherins [9] may be implicated. Moreover, as increased cell-cell contact occurred without increase in island size, the changes here may have also been driven by increased local cell density [39]. As traction forces generated by cell clusters differ from single cells, differences in ligand clustering and subsequent activation of integrin-mediated pathways may play a role in increased differentiation in cell clusters [40–42].
Markers for osteogenesis at time points earlier and later than ALP production were found to respond differently to cellularity. Transcription factor activity, which was assessed at an earlier time point than ALP production by immunofluorescence of nuclear localization, exhibited a dependency on substrate stiffness but not on micropattern cellularity. This suggests that the effects of increased cell-cell contact on osteogenic differentiation resulted from biological processes downstream of the activity of these transcription factors. While YAP is generally considered to be a key transcription factor mediating mechanotransduction [32], it is important to note that it may have multiple roles. For example, YAP colocalized with Runx2 in the nucleus has been implicated in the inhibition of osteocalcin expression [45]. Morever, although YAP nuclear localization has been observed on compliant substrates and on confined areas comparable to those used here, the D1 MSCs used in this study are generally smaller and have less spread area than human MSCs, potentially requiring a more confined area to recapitulate the cytoplasmic localization seen in previous work [32]. Moreover, the morphology of YAP localized to the nucleus in this study was punctile, differing from the more homogeneous distribution seen in prior work. Although the implications of differing morphologies of nuclear localized transcriptions factor are unclear, it may point to a different mechanism of transcription control. Because the MSCs used here are a clonal cell line, variability in phenotype are likely stochastic, rather than genetic or epigenetic, in origin. This variability may be amplified by the proliferation inhibition imposed by growth area restrictions of micropatterned islands [46].
Most micropatterning studies to date use proliferation inhibitors in order to prevent cell division, presumably in order to control spread area and to prevent cell attachment to regions between micropatterned islands. No proliferation inhibitor was employed in these experiments, in order to increase long term cell viability and enable assessment of mature osteogenesis markers. The 20 um diameter size was chosen for longer term experiments because a range of cell numbers per island, including single cells, was maintained in culture, enabling comparison of osteogenesis in single and multiple cell clusters at different time points. Adherent cells have a minimum area required for attachment and survival [24], and the continued, albeit reduced, proliferative capacity of micropatterned MSCs suggests that the chosen micropattern size was sufficient for supporting cell viability while at the same time retarding division to produce a spectrum of cells per island. The proliferation of micropatterned MSCs was unaffected by the number of neighboring cells per island, but increased, though modestly, with increasing E. After six days of culture, the distributions of cell numbers on the three moduli had similar shapes, and the distributions themselves were found not to be significantly different from each other. However, it is possible that if insufficient area for cell growth is available, a newly formed daughter cell resulting from cell divisions may detach from the micropatterned island, and this could confound the interpretation.
Although micropatterning studies have investigated early stage osteogenesis of MSCs, late stage osteogenesis in the context of micropatterning has received little attention. While MSCs grown on a large area of fibronectin were capable of robust calcium deposition over the entirety of their growth area, MSCs on micropatterns had a much more modest degree of calcium deposition, even on a substrate stiffness and in cell clusters sufficient to produce high levels of ALP. Indeed, whereas some micropatterns harboring doublets and triplets of cells produced ALP comparable to and exceeding ALP production in cells cultured on an unpatterned substrate coated with fibronectin (Fig. 3B), micropatterned islands containing even more than three cells failed to deposit calcium at the levels seen in a large area of cell growth. This suggests that substrate mechanical properties and cell-cell interaction cues that are sufficient for inducing expression of early stage osteogenesis markers are insufficient for inducing late stage osteogenesis. This could relate to the ability of a culture system comprised of only a few cells to allow for assembly of a collagen secreted by the cells into a crosslinked matrix, which is a key step in osteoblast differentiation and maturation [47,48].
Conclusions
The present study shows that substrate modulus and cell-cell interactions both regulate osteogenic differentiation of MSCs. The production of ALP was affected by both substrate stiffness and by the number of adjacent cells, with high expression requiring both a cell cluster and stiffer substrate. However, matrix mechanics alone impacted transcription factor localization to the nucleus. Conversely, none of the micropatterned cells led to significant deposition of calcium, even on substrates that were capable of supporting this late osteogenesis marker in cells cultured without growth constraints. This study has clear implications for MSC biology generally, and for high-throughput single-cell analysis platforms and delivery of stem cells at micrometer scale constructs specifically.
Supplementary Material
Acknowledgments
This work was funded by National Institute of Health (R01 EB014703). A.S.M. acknowledges funding from the National Science Foundation Graduate Research Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Krampera M, Pasini A, Pizzolo G, Cosmi L, Romagnani S, Annunziato F. Regenerative and immunomodulatory potential of mesenchymal stem cells. Curr Opin Pharm. 2006;6(4):435–41. doi: 10.1016/j.coph.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Ye K, Cao L, Li S, Yu L, Ding J. Interplay of matrix stiffness and cell-cell contact in regulating differentiation of stem cells. ACS Appl Mater Interfaces. doi: 10.1021/acsami.5b09746. [DOI] [PubMed] [Google Scholar]
- 3.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 4.Ross TD, Coon BG, Yun S, Baeyens N, Tanaka K, Ouyang M, et al. Integrins in mechanotransduction. Current Opinion in Cell Biology. 2013;25(5):613–18. doi: 10.1016/j.ceb.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali OA, Bencherif SA, et al. Harnessing traction-mediated manipulation of the cell/matrix interfrace to control stem-cell fate. Nat Mater. 2010;9(6):518–26. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Li S, Yan C, Liu P, Ding J. Fabrication of RGD mcro/nanopattern and corresponding study of stem cell differentiation. Nano Lett. 2015;15:1457–1467. doi: 10.1021/nl5049862. [DOI] [PubMed] [Google Scholar]
- 7.Winer JP, Janmey PA, McCormick ME, Funaki M. Bone marrow-derived human mesenchymal stem cells become quiescent on soft substrates but remain responsive to chemical or mechanical stimuli. Tissue Engineering: Part A. 2009;15(1):147–54. doi: 10.1089/ten.tea.2007.0388. [DOI] [PubMed] [Google Scholar]
- 8.Lee J, Abdeen AA, Kilian KA. Rewiring mesenchymal stem cell lineage specification by switching the biophysical microenvironment. Sci Rep. 2014;5(4):5188. doi: 10.1038/srep05188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Volloch V, Pindrus MA, Blasioli DJ, Cheng J, Kaplan DL. Murine osteoblasts regulate mesenchymal stem cells via WNT and cadherin pathways: mechanism depends on cell-cell contact mode. J Tissue Eng Regen Med. 2007;1(1):39–50. doi: 10.1002/term.6. [DOI] [PubMed] [Google Scholar]
- 10.Ball SG, Shuttleworth AC, Kielty CM. Direct cell contact influences bone marrow mesenchymal stem cell fate. Int J Biochem Cell Biol. 2004;36(4):714–27. doi: 10.1016/j.biocel.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 11.Tang J, Peng R, Ding J. The regulation of stem cell differentiation by cell-cell contact on micropatterned material surfaces. Biomaterials. 2010;31(9):2470–76. doi: 10.1016/j.biomaterials.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Song W, Kawazoe N, Chen G. The osteogenic differentiation of mesenchymal stem cells by controlled cell–cell interaction on micropatterned surfaces. J Biomed Mater Res Part A. 2013;101A(12):3388–95. doi: 10.1002/jbm.a.34645. [DOI] [PubMed] [Google Scholar]
- 13.Cao B, Li Z, Peng R, Ding J. Effects of cell-cell contact and oxygen tension on chondrogenic differentiation of stem cells. Biomaterials. 2015;64:21–32. doi: 10.1016/j.biomaterials.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 14.Xue R, Li JY, Yeh Y, Chien S. Effects of matrix elasticity and cell density on human mesenchymal stem cells differentiation. J Orthop Res. 2013;31(9):1360–65. doi: 10.1002/jor.22374. [DOI] [PubMed] [Google Scholar]
- 15.Yao X, Peng R, Ding J. Cell-material interactions revealed via material techniques of surface patterning. Adv Mater. 2013;25:5257–5286. doi: 10.1002/adma.201301762. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Li S, Yan C, Liu P, Ding J. Fabrication of RGD mcro/nanopattern and corresponding study of stem cell differentiation. Nano Lett. 2015;15:1457–1467. doi: 10.1021/nl5049862. [DOI] [PubMed] [Google Scholar]
- 17.Domenech M, Yu H, Warrick J, Badders NM, Meyvantsson I, Alexander CM, et al. Cellular observations enabled by microculture: paracrine signaling and population demographics. Integr Biol. 2009;1(3):267–74. doi: 10.1039/b823059e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and rhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–95. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 19.Kilian KA, Bugarija B, Lahn BT, Mrksich M. Geometric cues for directing the differentiation of mesenchymal stem cells. PNAS. 2010;107(11):4872–4877. doi: 10.1073/pnas.0903269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charest JL, Jennings JM, King WP, Kowalczyk AP, Garcia AJ. Cadherin-mediated cell-cell contact regulates keratinocyte differentiation. J Invest Dermatol. 2009;129(3):564–72. doi: 10.1038/jid.2008.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng R, Yao X, Ding J. Effect of cell anisotropy on differentiation of stem cells on micropatterned surfaces through the controlled single cell adhesion. Biomaterials. 2011;32(32):8048–57. doi: 10.1016/j.biomaterials.2011.07.035. [DOI] [PubMed] [Google Scholar]
- 22.Yao X, Peng R, Ding J. Effects of aspect ratios of stem cells on lineage commitments with and without induction media. Biomaterials. 2013;34:930–939. doi: 10.1016/j.biomaterials.2012.10.052. [DOI] [PubMed] [Google Scholar]
- 23.Cao B, Peng R, Li Z, Ding J. Effects of spreading areas and aspect ratios of single cells on dedifferentiation of chondrocytes. Biomaterials. 2014;35:6871–6881. doi: 10.1016/j.biomaterials.2014.04.107. [DOI] [PubMed] [Google Scholar]
- 24.Yan C, Sun J, Ding J. Critical areas of cell adhesion on micropatterned surfaces. Biomaterials. 2011;32(16):3931–38. doi: 10.1016/j.biomaterials.2011.01.078. [DOI] [PubMed] [Google Scholar]
- 25.Tseng Q, Wang I, Duchemin-Pelletier E, Azioune A, Carpi N, Gao J, et al. A new micropatterning method of soft substrates reveals that different tumorigenic signals can promote or reduce cell contraction levels. LoC. 2011;11(13):2231–40. doi: 10.1039/c0lc00641f. [DOI] [PubMed] [Google Scholar]
- 26.Lee KY, Alsberg E, Hsiong S, Comisar W, Linderman J, Ziff R, et al. Nanoscale adhesion ligand organization regulates osteoblast proliferation and differentiation. Nano Letters. 2004;4(8):1501–06. doi: 10.1021/nl0493592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diduch DR, Coe MR, Joyner C, Owen ME, Balian G. Two cell lines from bone marrow that differ in terms of collagen synthesis, osteogenic characteristics, and matrix mineralization. J Bone Joint Surg Am. 1993;75-A:92–105. doi: 10.2106/00004623-199301000-00012. [DOI] [PubMed] [Google Scholar]
- 28.Cui Q, Wang GJ, Balian G. Steroid-induced adipogenesis in a pluripotent cell line from bone marrow. J Bone Joint Surg Am. 1997;79-A:1054–1063. doi: 10.2106/00004623-199707000-00012. [DOI] [PubMed] [Google Scholar]
- 29.Dahir GA, Cui Q, Anderson P, Simon C, Joyner C, Triffitt JT, Balian G. Pluripotent mesenchymal cells repopulate bone marrow and retain osteogenic properties. Clin Orthop Relat Res. 2000;279:S134–S145. doi: 10.1097/00003086-200010001-00018. [DOI] [PubMed] [Google Scholar]
- 30.Damljanovic V, Lagerholm C, Jacobson K. Bulk and micropatterned conjugation of extracellular matrix proteins to characterized polyacrylamide substrates for cell mechanotransduction assays. BioTechniques. 2005;39(6):847–51. doi: 10.2144/000112026. [DOI] [PubMed] [Google Scholar]
- 31.Sasaki J-I, Matsumoto T, Egusa H, Matsusaki M, Nishiguchi A, Nakano T, Akashi M, Imazato S, Yatani H. In vitro reproduction of endochondral ossification using a 3D mesenchymal stem cell construct. Integr Biol. 2012;4:1207–1214. doi: 10.1039/c2ib20027a. [DOI] [PubMed] [Google Scholar]
- 32.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–83. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 33.Bruderer M, Richards RG, Alini M, Stoddart MJ. Role and regulation of RUNX2 in osteogenesis. Eur Cell Mater. 2014;28:269–85. doi: 10.22203/ecm.v028a19. [DOI] [PubMed] [Google Scholar]
- 34.Thouverey C, Caverzasio J. Sclerostin inhibits osteoblast differentiation without affecting BMP2/SMAD1/5 or Wnt3a/b-catenin signaling but through activation of platelet-derived growth factor receptor signaling in vitro. Bonekey Rep. 2012;4:757. doi: 10.1038/bonekey.2015.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gobaa S, Hoehnel S, Roccio M, Negro A, Kobel S, Lutolf MP. Artificial niche microarrays for probing single stem cell fate in high throughput. Nat Methods. 2011;8(11):949–55. doi: 10.1038/nmeth.1732. [DOI] [PubMed] [Google Scholar]
- 36.Tsai RY, McKay RD. Cell contact regulates fate choice by cortical stem cells. J Neurosci. 2000;20(10):3725–35. doi: 10.1523/JNEUROSCI.20-10-03725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khosrowshahi YB, Khoshfetrat AB, Shamsasenjan K. Ex vivo expansion of hematopoietic stem cells in a proliferation chamber with external stirred conditioning tank: sequential optimization of growth factors. Eng Life Sci. doi: 10.1002/elsc.201500053. Advance online publication. [DOI] [Google Scholar]
- 38.Chen T, Yuan D, Wei B, Jiang J, Kang J, Ling K, et al. E-cadherin-mediated cell-cell contact is critical for induced pluripotent stem cell generation. Stem Cells. 2010;28:1315–25. doi: 10.1002/stem.456. [DOI] [PubMed] [Google Scholar]
- 39.Peng R, Yao X, Cao B, Tang J, Ding J. The effect of culture conditions on the adipogenic and osteogenic inductions of mesenchymal stem cells on micropatterned surfaces. Biomaterials. 2012;33:6008–6019. doi: 10.1016/j.biomaterials.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 40.Pankov R, Yamada KM. Fibronectin at a glance. J Cell Sci. 115:3861–63. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- 41.Tang X, Tofangchi A, Anand SV, Saif TA. A novel cell traction force microscopy to study multicellular system. PloS Comput Biol. 2014;10(6):e1003631. doi: 10.1371/journal.pcbi.1003631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slater JH, Frey W. Nanopatterning of fibronectin and the influence of integrin clustering on endothelial cell spreading proliferation. J Biomed Mater Res A. 2008;87:176–95. doi: 10.1002/jbm.a.31725. [DOI] [PubMed] [Google Scholar]
- 43.Ueda HR, Hayashi S, Matsuyama S, Yomo T, Hashimoto S, Kay SA, et al. Universality and flexibility in gene expression from bacteria to human. PNAS. 2004;101(11):3765–69. doi: 10.1073/pnas.0306244101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kucera JP, Hueschkel MO, Renaud P, Rohr S. Power-law behavior of beat-rate variability in monolayer cultures of neonatal rat ventricular myocytes. Circ Res. 2000;86:1140–45. doi: 10.1161/01.res.86.11.1140. [DOI] [PubMed] [Google Scholar]
- 45.Zaidi SK, Sullivan AJ, Medina R, Ito Y, van Wijnen AJ, Stein JL, et al. Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. EMBO J. 2004;23:790–99. doi: 10.1038/sj.emboj.7600073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neildez-Nguyen TM, Parisot A, Vignal C, Rameau P, Stockholm D, Picot J, Allo V, et al. Epigenetic gene expression noise and phenotype diversification of clonal cell populations. Differentiation. 2008;76:33–40. doi: 10.1111/j.1432-0436.2007.00219.x. [DOI] [PubMed] [Google Scholar]
- 47.Aubin JE, Liu F, Malaval L, Gupta AK. Osteoblast and chondroblast differentiation. Bone. 1995;17(2):77S–83S. doi: 10.1016/8756-3282(95)00183-e. [DOI] [PubMed] [Google Scholar]
- 48.Chau JF, Leong WF, Li B. Signaling pathways governing osteoblast proliferation, differentiation and function. Histol Histopathol. 2009;24:1593–1606. doi: 10.14670/HH-24.1593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.