Abstract
Tetraspanins are master organizers in the plasma membrane, forming tetraspanin-enriched microdomains with one another and other surface molecules. Their rod-shaped structure includes a large extracellular loop (LEL) that plays a pivotal role in tetraspanin network formation. We performed comparative atomistic and coarse-grain molecular-dynamics simulations of the LEL in isolation and full-length CD81, and reproduced LEL flexibility patterns known from wet-lab experiments in which the LEL δ-loop region showed a pronounced flexibility. In a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine lipid bilayer and a plasma membrane environment, the conformational flexibility of the δ-loop initiates CD81-CD81 contacts for oligomerization. Furthermore, in the plasma membrane, CD81-ganglioside bridges arising from preformed glycolipid patches cross-link the complexes. The data suggest that exposing a flexible domain enables binding to interaction partners by circumventing the restriction of orientation and conformational freedom of membrane proteins.
Introduction
Tetraspanins are ubiquitously expressed plasma membrane proteins in animals, plants, and fungi. They are involved in an astonishing variety of processes, including cell proliferation, adhesion, spreading, and migration, as well as signal transduction, intracellular vesicle trafficking, cell-cell fusion, pathogen entry, cancer, and a number of other diseases (1, 2, 3, 4, 5). Apparently, individual tetraspanins are not assigned to one specific function, but rather act as master organizers or scaffolding proteins. Most importantly, they oligomerize to a high degree, forming functional sites called tetraspanin-enriched microdomains (TEMs) by associating laterally with one another and other interaction partners (6, 7, 8) such as integrins (9), immunoglobulin superfamily proteins (10), cellular receptors (11), and glycolipids (12, 13, 14, 15). To date, we are just beginning to understand the molecular mechanisms underlying the formation of these large molecular networks.
Tetraspanins are named according to their common structural feature, which comprises four transmembrane (TM) domains that form a tetrahelical bundle (16). They possess two extracellular domains, a small extracellular loop (SEL) between TM helix 1 (TM1) and TM2, and a large extracellular loop (LEL) between TM3 and TM4. Based on a comparison of segmental lengths and amino acid variability among ≈100 different tetraspanins, the LEL was grouped into a conserved domain and a variable domain (VD) (17). It has been speculated that the variance of the VD determines which interaction partner(s) is selected for network formation (18, 19).
The structurally best-studied tetraspanin is CD81. Three available x-ray structures of the water-soluble, human LEL (PDB: 1G8Q, 1IV5, and 3X0E at a resolution of 1.6, 2.6, and 1.84 Å, respectively) resolve five largely helical segments (α, β, γ, δ, and ε). Disulfide bridges form loops comprising the γ and γ/δ sections, respectively. Although α, β, and ε show little conformational variation, the structures differ in their VD (γ and δ), which is poorly resolved, suggesting conformational fluctuations even in the packed crystal lattices (20, 21, 22). In addition, a solution NMR study of the LEL showed pronounced conformational flexibility in this region (23). However, to date, it remains to be determined whether the conformational flexibility of the LEL is of functional importance.
Apart from the VD, other tetraspanin regions have also been shown to mediate interactions. For instance, segments in proximity to TM1 and TM2 are required for dimerization (24), and CD81 association with its primary interaction partner, EWI-2, involves TM3 and TM4 (25).
To date, conformational variability has been observed only experimentally in the isolated LEL (20, 21, 22, 23), without membrane anchoring and other putatively stabilizing regions such as the SEL. This raises the question as to whether the observed flexibility is maintained in the membrane-embedded, full-length protein. Furthermore, it remains to be elucidated whether the flexibility of a defined protein domain has any influence on the protein’s ability to form networks or oligomeric states. Here, we sought to address these questions by conducting atomistic and coarse-grain (CG) molecular-dynamics (MD) simulations.
Materials and Methods
Atomistic simulations of CD81LEL
The MD model of the water-soluble CD81 LEL domain is based on chain A of the dimeric CD81 crystal structure (PDB: 1G8Q) (20), which was isolated for further preparation. Hydrogen atoms were added to the protein residues according to the standard protonation states of the GROMOS96 54A7 united atom force field (26), mimicking neutral pH conditions. Only the imidazole ring of His151 was considered in the fully protonated state, based on its orientation and the predicted hydrogen-bond configuration in the crystal structure. Both pairs of the disulfide bridge-forming LEL cysteine residues (Cys156/190 and Cys157/175) were linked by definition of the disulfide bridges. The mushroom-shaped CD81 LEL structure was placed in the center of a rhombic dodecahedron simulation box, with a minimum distance of 1 nm to the box edges. A steepest-descent energy minimization in vacuo was performed before CD81LEL was solvated in an explicit 150 mM NaCl/SPC water (27) environment. Two additional Na+ counterions were added to neutralize the overall system charge. After a second energy minimization step, the solvated system was relaxed using a protocol of six consecutive protein relaxation steps, with harmonically (position) restrained protein heavy atoms. First, a 500 ps MD simulation in the NVT ensemble (constant volume and temperature) was performed, with a position restraints force constant kpr of 1000 kJ⋅mol−1⋅nm−2, followed by five 2 ns NPT (constant pressure and temperature) simulations with kpr = 1000, 500, 100, 50, and 10 kJ⋅mol−1⋅nm−2, respectively. This relaxation scheme, followed by the unbiased 1 μs MD simulation, was applied to four parallel simulation runs (CD81LEL,A to CD81LEL,D), each with a different initial velocity vector distribution (multicopy approach (28, 29)) at T = 310 K. During relaxation, a temperature of 310 K was maintained every 200 fs (NVT and first NPT run) or 400 fs (other NPT runs) for the protein and solvent separately, and a pressure of 1 bar was adjusted isotropically every 4 ps (NPT runs) using the Berendsen methodology (30). For the 1 μs MD simulations, the Nosé-Hoover thermostat (31, 32) was used, maintaining a temperature of 310 K every 1.6 ps for the protein and the aqueous phase separately. A pressure of 1 bar was regulated each 4 ps using the Parrinello-Rahman barostat (33, 34, 35) with isotropic coupling. Periodic boundary conditions were applied in all three dimensions. Electrostatic interactions were handled using the smooth particle mesh Ewald (PME) methodology (36, 37). The Verlet cutoff scheme (38) was used, and neighbor lists were updated every 40th step of the 2 fs integration steps. All bonds were constrained using the LINCS algorithm (39, 40), and the angle of the SPC water molecules was constrained using the SETTLE methodology (41). All atomistic CD81LEL simulations were performed using GROMACS 4.6.5 (42, 43) on a CPU/GPU computing node.
Atomistic simulations of CD81FL
The atomistic MD model of the full-length CD81 is based on the CD81 homology model (PDB: 2AVZ) (16). Cross-linked cysteine residues, as well as the protonated state of the central histidine residue in the LEL, were defined as in the CD81LEL model. To incorporate CD81FL into a preequilibrated 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipid bilayer patch (44, 45), the programs LAMBADA and InflateGRO2 (46) were used to automatically orient, align, and embed the membrane protein system, resulting in a rectangular simulation box of x = y = 6.487 nm (according to the dimensions of the bilayer plane) and z = 12.208 nm (based on the protein dimensions along the membrane-normal vector). During the embedding procedure, seven lipids in the upper (representing the extracellular) leaflet, and eight lipids in the lower leaflet were deleted. As with the CD81LEL system, the simulation box was filled up with a 150 mM NaCl/SPC water (27) solution before it was energy minimized. Using a multicopy approach (28, 29), the same relaxation protocol used for CD81LEL was performed for the four CD81FL MD systems before the protein was released into the unbiased 1 μs MD simulations. To allow an extensive equilibration of the bilayer lipids around the embedded full-length protein, for system relaxation we used a longer first NPT simulation, with kpr = 1000 kJ⋅mol−1⋅nm−2 (20 instead of 2 ns). All atomistic CD81FL simulations were performed using GROMACS 4.6.5 (42, 43) on a CPU/GPU computing node.
CG simulations of CD81 in a POPC lipid bilayer and the plasma membrane
To study CD81’s oligomerization behavior, we used two MARTINI CG models with different lipid bilayer environments: a homogeneous POPC lipid bilayer patch and a heterogeneous membrane model, mimicking an idealized mammalian plasma membrane (47). To generate the preequilibrated POPC membrane patch, the CHARMM-GUI Membrane Builder (48, 49) was used for the initial placement of 1) MARTINI POPC lipids on a 36 × 36 nm bilayer grid along the xy plane, and 2) solvent particles (150 mM NaCl/MARTINI water solution). The assembled system was then copied along both lateral membrane vectors x and y (yielding xy system dimensions of 72 × 72 nm) and relaxed following a protocol of six consecutive simulations as described previously (48). According to the model of the plasma membrane (47), the effect of a cytoskeleton network is imitated by using weak position restraints along the membrane normal vector z on ≈15.5% of the POPC lipids in the upper (outer) leaflet, to limit large-scale bilayer undulations. The relaxed 70 × 70 nm POPC bilayer patch was then energy minimized using the steepest-descent methodology and equilibrated for 100 ns. For the plasma membrane system, consisting of 63 different lipid types (grouped into phosphatidic acids, phosphatidylcholines (PCs), phosphatidylethanolamines, phosphatidylinositols, phosphatidylserines, lyso-PCs, diacylglycerols, gangliosides, sphingomyelins, ceramides, phosphatidylinositol (mono/bis/tris)-phosphate (PIP), and cholesterol), the configuration after a 40 μs unbiased MD simulation was used (47). For further system preparation steps, the aqueous phase of the preequilibrated membrane systems was removed.
The CG structure and topology of the full-length protein (CD81FL) were generated based on the previously used homology model (16) (PDB: 2AVZ) using the martinize.py program (50). The MARTINI 2.2 force field was used (50). Both cysteine pairs that formed the disulfide bridges in the LEL region (Cys156/190 and Cys157/175) were automatically detected and linked via 0.24 nm bond constraints (instead of the published 0.39 nm with a force constant of 5000 kJ⋅mol−1⋅nm−2, as recommended by the authors of the martinize.py program: D. de Jong, J.J. Uusitalo, and T.A. Wassenaar). After an in vacuo steepest-descent energy minimization, 49 copies of the CG proteins were placed on a 7 × 7 grid along the xy plane, randomly oriented around a central axis parallel to the (membrane normal) vector z. Thus, all proteins have the same general direction, with their extracellular domain oriented toward the positive z direction, which corresponds to the upper or extracellular leaflet of the preequilibrated POPC or plasma membrane patch, respectively. The overall xy box dimensions of the 7 × 7 grid are equal to the corresponding system dimensions of the respective membrane model (68 × 68 nm for POPC, 71 × 71 nm for the plasma membrane). LAMBADA (46) was used to automatically align each membrane patch according to the hydrophobic belts of the proteins placed on the 7 × 7 grid and to combine both components, respectively. Protein incorporation into the preequilibrated membrane patches was done using the InflateGRO2 membrane protein-embedding routine (46), which was executed iteratively for the individual proteins. As the lipids in the original plasma membrane model are organized in two sets (corresponding to their respective leaflet), an additional resorting step was necessary to execute InflateGRO2 with its internal bookkeeping routine for lipid deletions. For this purpose, a program was developed that sorts the lipids in both the structure and the corresponding topology file into a unique group with respect to their first occurrence. The lipid composition after protein embedding/lipid deletion still reflects the composition of the preequilibrated plasma membrane patch (Fig. S3 a in the Supporting Material). Retaining the z dimension of the preequilibrated plasma membrane model (47), the simulation box of both systems was filled up with MARTINI water beads. For the POPC membrane system, NaCl ions yielding a concentration of 150 mM NaCl were added along with a total of 245 Na+ counterions to neutralize the overall system charge. For the plasma membrane system, the original numbers of Na+ and Cl− ions plus one additional Cl− counterion were added. The resulting simulation systems contained a total of 493,711 (POPC membrane) and 513,228 (plasma membrane) CG particles.
Using the same system setup protocol previously described for the plasma membrane model (47), a steepest-descent energy minimization was performed for both systems, followed by a 500 ps NVT and 25 ns NPT simulation. During the relaxation steps, the protein backbone particles were restrained to their initial positions using a harmonic potential with a force constant kpr of 1000 kJ⋅mol−1⋅nm−2. The simulation parameters for the respective relaxation steps were set as described previously (47) except for 1) the (long-range) electrostatics, for which the smooth PME methodology was used, and 2) the pressure coupling, for which the Berendsen barostat was used. A second 25 ns equilibration step with Berendsen weak (pressure) coupling was performed in which the proteins were able to diffuse freely before the unbiased simulation (using the Parrinello-Rahman barostat (33, 34, 35)) was started. For the 2.5 μs (POPC membrane) and 7.5 μs (plasma membrane) MD production runs, all parameters were set as described previously (47) except for the electrostatics, which were handled using the smooth PME method (36, 37). For each simulation system, both trajectories in which the proteins could diffuse freely were concatenated for analyses. All CG simulations were performed using GROMACS 4.6.5 (42, 43) on a CPU cluster (16 nodes, each with one eight-core Intel Xeon E5-2670 CPU (2.60 GHz, two threads per core)).
Molecular visualization and segment definitions for analysis
Molecular illustrations were generated using the program VMD (version 1.9.2) including the internal Tachyon rendering engine (51, 52). Visualization of secondary structural elements was based on a VMD-internal STRIDE analysis (53), which was used to determine helical elements in the segment definitions as follows: 12–38 (TM1), 56–83 (TM2), 89–136 (TM3/helix α), 141–154 (helix β), 158–174 (γ-loop), 176–189 (δ-loop), and 190–228 (helix ε/TM4). The SEL region was defined as residues 39–55 and the LEL region was defined as residues 113–202, based on the CD81 LEL crystal structure. For illustration of the full-length protein, helices TM3/α and ε/TM4 were split into residues 89–112 (TM3) and 113–136 (α-helix), as well as 190–202 (ε-helix) and 203–228 (TM4), respectively.
Conformational drift
Conformational deviations with respect to the MD initial structure were monitored by calculating the root mean-square deviation (RMSD) of the protein α-carbon (Cα) atoms for each trajectory snapshot using the GROMACS tool gmx rms (after least-square fitting the Cα atoms to the MD initial structure; Figs. S1 c and S2 c).
Helical structure occurrence and protein flexibility
To monitor helical structure elements (Figs. 1 b, top, and 2 b, top), the program DSSP (54) was used to determine the secondary-structure distribution along the protein chain for each 5 ns step of the four concatenated MD simulation trajectories. The first 20 ns of each MD trajectory were excluded to reduce the bias of the identical initial configurations. We chose to exclude the first 20 ns from the analyses because ≈50% of the average conformational deviations from the starting configuration took place within that time range (Figs. S1 c and S2 c). The helical (α-, π-, and 310-helices) structure elements of the concatenated MD trajectories were summarized, counted, and normalized according to the total number of analyzed frames.
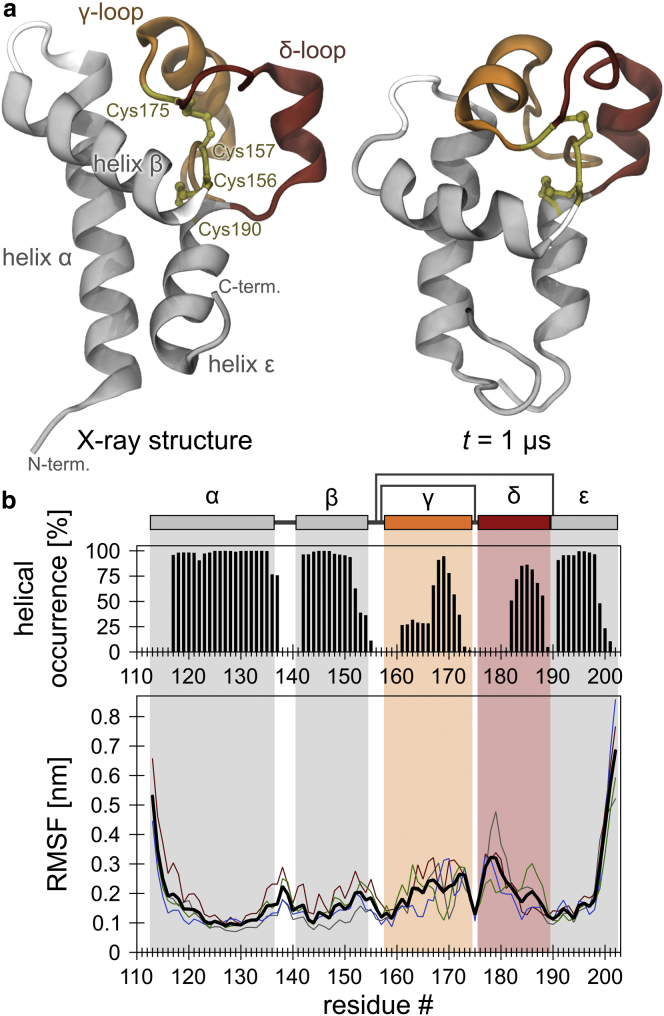
Figure 1.
Helical stability and backbone flexibility of the isolated LEL of CD81. (a) Left: CD81LEL initial conformation (chain A of the x-ray structure (20) (PDB: 1G8Q)). Right: after a 1 μs unbiased, atomistic MD simulation (run B; see also Fig. S1b). (b) Top: occurrence of helical structure per residue (based on the ϕ/ψ dihedral angles) over time, averaged over four runs. Bottom: Cα RMSF as a measure of the local protein backbone flexibility. The highest flexibility of the fluctuating termini is due to the lack of anchoring to the TM segments present in the full-length protein (compare with Fig. 2b, which shows the flexibility of this region in the full-length protein). Colored lines show the flexibility profiles from the individual MD runs; average (n = 4) is shown in black. The pictogram shows the CD81 LEL domain structure, including cysteine disulfide bridges indicated by additional black lines connecting residues 157 with 175, and 156 with 190. To see this figure in color, go online.
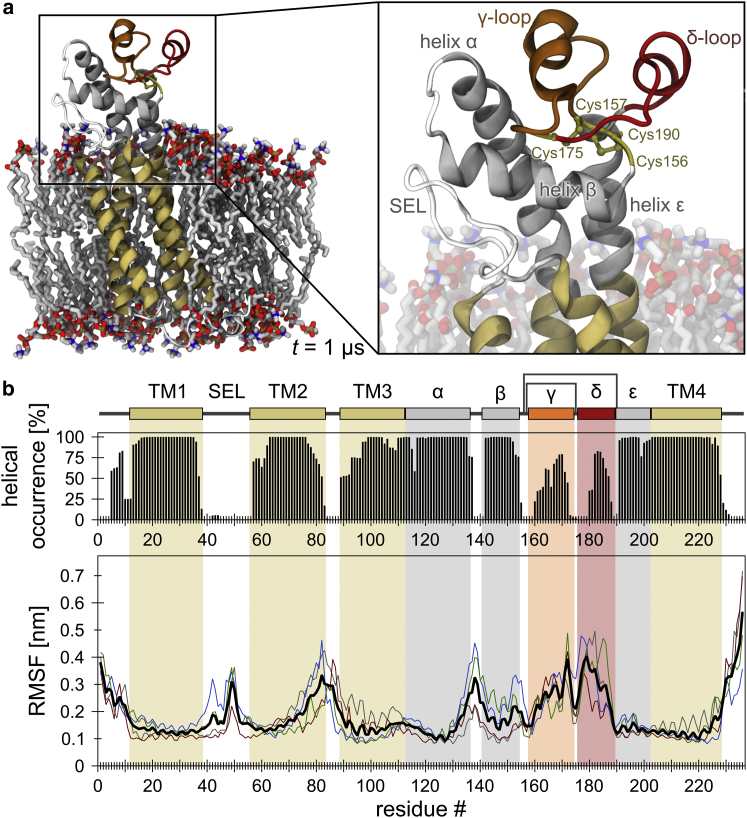
Figure 2.
Helical stability and backbone flexibility of CD81FL. (a) Left: conformation of the full-length protein embedded in a POPC bilayer after a 1 μs unbiased, atomistic MD simulation (run D; see also Fig. S2b). TM1–4 in dark yellow; helices α, β, and ε in gray; loops γ and δ in orange and red, respectively. Red, blue, and tan atoms of the lipid bilayer represent oxygen, nitrogen, and phosphate, respectively. Right: enlarged view of the SEL and LEL. (b) Helical structure per residue (top) and Cα RMSF (bottom). Note that a direct comparison of the average RMSFs in (b) (shown in black; n = 4) and Fig. 1b suggests that the LEL domain flexibility increases in CD81FL; however, this is a result of least-square fitting to all Cα atoms of the full-length protein (for comparison of exclusively LEL segments, see Fig. 3a). To see this figure in color, go online.
The local flexibility along the protein chain (Figs. 1 b, bottom, 2 b, bottom, 3 a, 4 c, and S3 c) was measured by computing the RMS fluctuations (RMSFs) of the Cα atoms (atomistic models) and backbone particles (CG model) per CD81 molecule, respectively. Before computing the RMSF, structures were superimposed on the initial structure using a least-square fit based on the respective atoms. To avoid the influence of large fluctuations resulting from the transition of the MD initial configuration (crystal/predicted/CG structure) to the dynamic structure in solution, we excluded the first 20 ns for the atomistic models (20–1000 ns trajectory, 4901 frames) and the first 500 ns for the CG systems (POPC: 500–2500 ns trajectory, 2501 frames; plasma membrane: 500–7500 ns trajectory, 7001 frames).
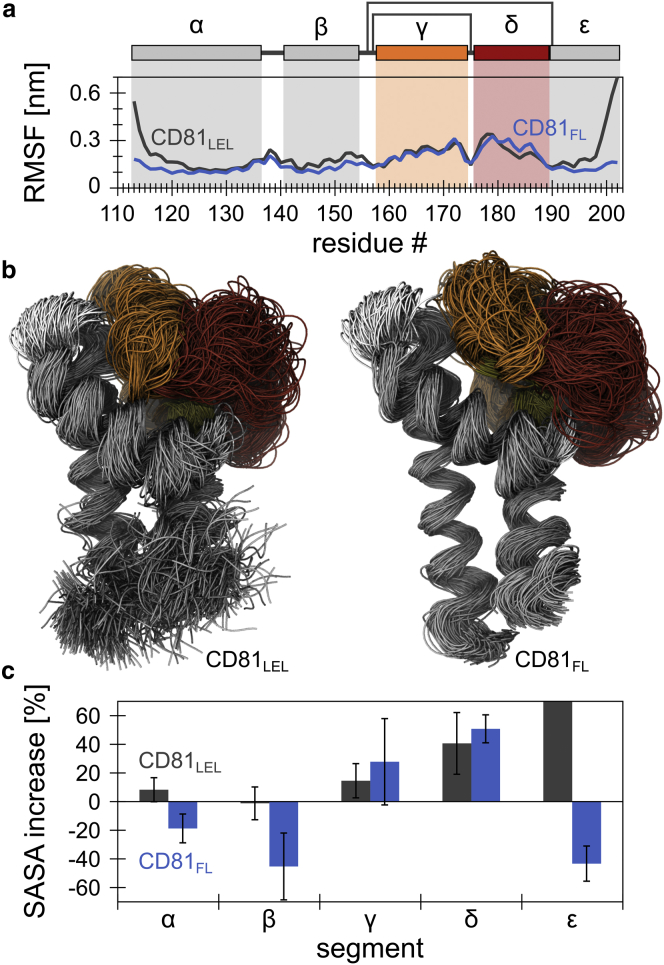
Figure 3.
LEL segmental flexibility enables the space exploration activity of the δ-loop. (a) Comparison of the average Cα RMSFs of CD81LEL (n = 4) (same as in Fig. 1b) and CD81FL (n = 4), with only its LEL section included for analysis. The shape of the blue trace is slightly different from the respective section in Fig. 2b (in particular at the termini) due to the protein segment considered for fitting. (b) Left: superimposed protein backbone traces obtained from the four MD trajectories (shown are 4 × 98 snapshots from 10 ns intervals, excluding the first 20 ns) of the isolated LEL. Right: the LEL in CD81FL. For superimposition, Cα atoms that exhibited an average RMSF value smaller than 0.15 nm were used for least-square fitting. Protein segments are colored as in the pictogram in (a). (c) Increase of the SASA of the individual LEL segments, referring to the respective MD initial structure (gray bars, CD81LEL; blue bars, CD81FL) in percent (averaged over the four MD runs; n = 4, values are given as means ± SD). For clarity, the extreme value obtained for the ε-helix in CD81LEL is not fully illustrated because it does not result from relevant dynamics (see text). To see this figure in color, go online.
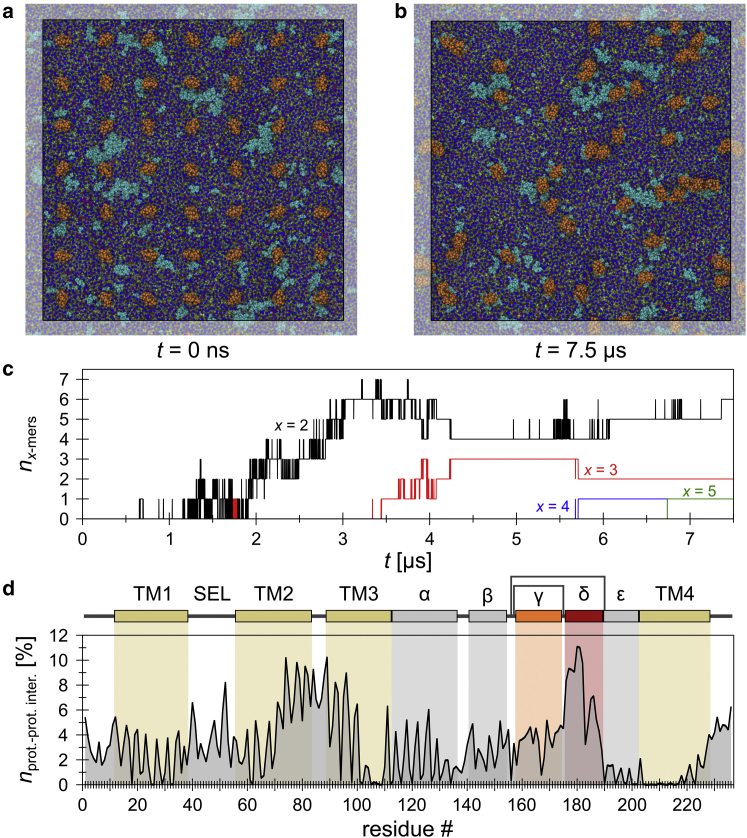
Figure 4.
Structural flexibility and interactions of CD81 in a CG POPC bilayer system. (a) Left: mapping of the atomistic structure of the full-length CD81 onto the CG scale (gray, backbone particles; orange, side-chain particles). Right: top and side views of the MD initial configuration, containing 49 copies of CG CD81 embedded into a preequilibrated POPC lipid bilayer model (≈70 × 70 nm along the bilayer plane). The aqueous phase is shown as a semitransparent volume surface (for clarity, ions are not shown). (b) System configuration after a 2.5 μs unbiased MD simulation. (c) The black trace indicates the mean Cα RMSF, including all individual CD81 chains (values are given as means ± SD; n = 49, the range of the SD is indicated in gray). (d) Protein-protein RDF. The red area indicates the range used to define direct CD81-CD81 interactions (distance threshold = 0.6 nm). (e) CD81 oligomerization over time. (f) Illustration of a CD81 dimer (the protein surface is semitransparent and the CD81 domain is colored according to the topology diagram in c). (g) CD81-CD81 contacts normalized per residue (the sum exceeds 100%, as one residue can be involved in more than one contact). To see this figure in color, go online.
Solvent-accessible surface area of the superimposed protein structures
The solvent-accessible surface area (SASA) of the superimposed protein structures for each MD simulation run was computed using the GROMACS program gmx sasa (55) (van der Waals radii were taken from (56)). Superimposition of the protein structures (Fig. 3 b) was based on a least-square fit of the Cα atoms exhibiting an average Cα RMSF value of <0.15 nm (Fig. 3 a). The Cα atoms used for fitting were from residues 119–134, 140, 143–147, 150, 156–159, 175, and 189–194 (CD81LEL), or 115–135, 137, 140–151, 153–159, 175, and 189–202 (CD81FL). Separate groups for the individual LEL segments were defined to extract the SASAs of the respective segments from the entire surface.
Time evolution of oligomerization and lipid interactions
Monitoring oligomerization over time (Figs. 4 e and 5 c), we counted the number of directly interacting proteins, forming x-mers (x = 2, 3, … 9) for each trajectory frame. Direct CD81-CD81 interactions were defined to occur within a distance of 0.6 nm, as revealed by the radial distribution function (RDF) of each CG particle of CD81 to each particle of other CD81 proteins (Fig. 4 d). Since oligomerization started after 500 ns (POPC membrane) and 1.5 μs (plasma membrane), we used time ranges of 700–2.5 μs and 2–7.5 μs, respectively, to measure the protein-protein RDF.
Figure 5.
CD81 oligomerization in a CG model of the plasma membrane. (a) Top view of the MD initial configuration, containing 49 copies of CG CD81 embedded into a preequilibrated plasma membrane model (≈70 × 70 nm along the bilayer plane). (b) System configuration after a 7.5 μs unbiased MD simulation. (c and d) CD81 oligomerization over time (c) and CD81-CD81 contacts per residue (d) analyzed as in Fig. 4. To see this figure in color, go online.
To identify preferred lipid interactions (Fig. S4), we counted the total number of contacts between the CG CD81 molecules and the individual lipid species of the plasma membrane bilayer for each trajectory step. As with the identification of contacts between CG CD81 molecules, preferred lipid-binding partners were defined as occurring within a distance of 0.6 nm (Fig. 6 c). The total number of contacts was then normalized to the concentration of the respective lipid species (Fig. S4). Highlighting the preferential interactions with gangliosides (GM) versus nonpreferential interactions with PC headgroup lipids, Fig. 6 d illustrates the sum of the individual, not normalized, curves of the respective lipid species per time step.
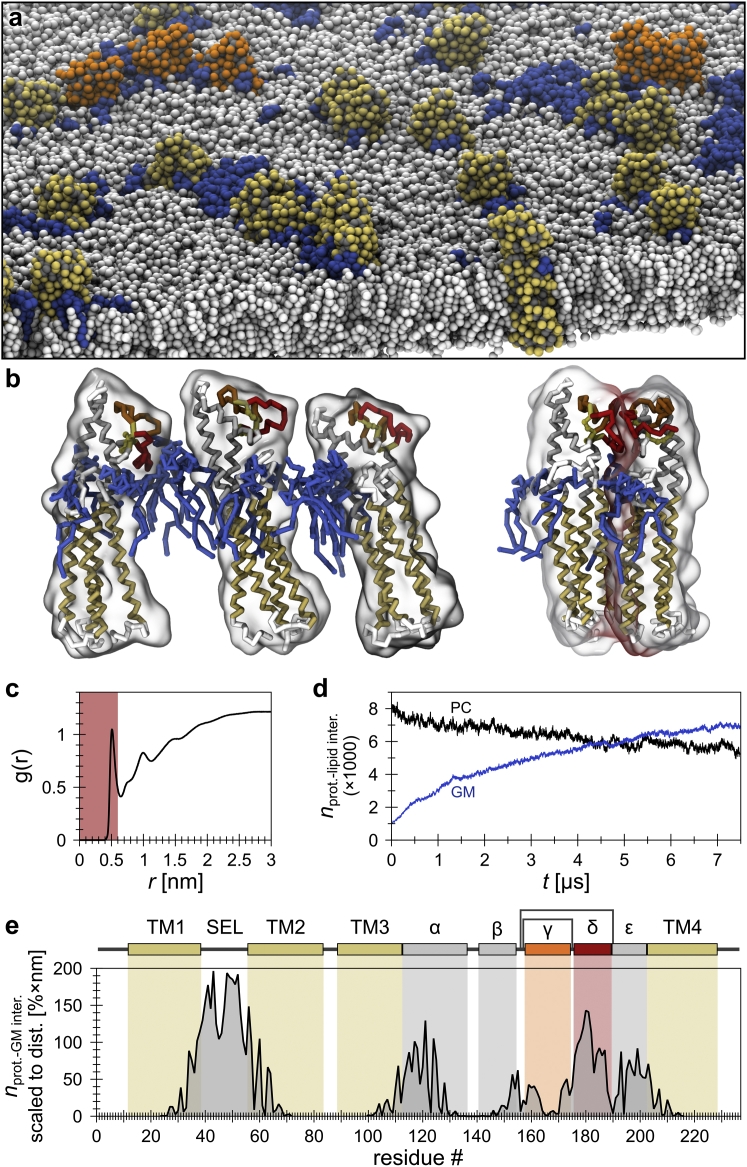
Figure 6.
CD81-GM contacts over time and interaction sites. (a) Snapshot of the simulation system after a 7.5 μs MD simulation. Gangliosides are colored in blue and proteins are in yellow/gray (side-chain/backbone particles) or, in the case of the two example states in (b), in orange/gray. (b) Left: configuration in which gangliosides form glycolipid bridges among three nondirectly interacting CD81 molecules. Right: direct CD81-CD81 interaction. Protein surfaces are shown as semitransparent. A gray-to-red color gradient indicates protein interactions per residue within 0.6 nm. (c) Protein-lipid RDF. The red area indicates the range of CD81-lipid distances used for contact definition (distance threshold = 0.6 nm). (d) Number of protein-lipid interactions as a function of time (black, CD81-PC interactions; blue, CD81-GM interactions). (e) CD81-GM contacts per residue scaled to the residue distance to the hydrophobic belt center of the protein (see Fig. S6 for original data and residue distances). To see this figure in color, go online.
CD81-CD81 and CD81-lipid contacts per residue
We monitored the occurrence of direct CD81 interactions per protein residue (Figs. 4 g and 5 d) by measuring the minimum distance between each residue of the individual protein chains to each chain of the respective other proteins per trajectory frame. For the POPC membrane system the first 700 ns were excluded for analysis, and for the plasma membrane system the first 2 μs were not considered. Measured distances within 0.6 nm per residue were counted and normalized (divided by the product of the number of analyzed trajectory frames (1801 for the POPC membrane and 5501 for the plasma membrane) and the number of potentially interacting proteins (n = 48)).
For the plasma membrane system, the number of direct CD81-GM interactions per residue (Fig. S6, top) was monitored using a similar protocol: distances within 0.6 nm between each residue of the individual protein chains with all GM lipids were counted and normalized (divided by the product of the number of analyzed trajectory frames (see above) and the number of proteins (n = 49)).
Mean-square displacement as a function of time
To compare the lateral diffusion of CD81 in the different CG membrane environments (POPC and plasma membrane), we computed the mean-square displacement of the individual proteins along the xy plane as a function of time (Fig. S3 b) using the GROMACS program gmx msd (42).
Degradation of the glycocalyx from Jurkat T cells and cluster grade analysis
Jurkat T-cells were cultured and transfected essentially as previously described (57). In brief, 105 Jurkat T-cells transfected with a plasmid encoding for CD81-GFP were plated onto poly-lysine-coated coverslips in 1 mL prewarmed Ringer solution (130 mM NaCl, 4 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 48 mM D(+)glucose, 10 mM HEPES, pH 7.4) for control samples or prewarmed Ringer solution supplemented with 0.5 unit/mL neuraminidase (#N2876; Sigma-Aldrich, St. Louis, MO) and 4 units/mL β 1-4 galactosidase (#P0730; New England Biolabs, Ipswich, MA) for treated samples. Cell suspensions were plated on poly-L-lysine-coated coverslips in six-well plates and incubated for 1 h in a cell incubator. Subsequently, membrane sheets were generated, fixed, and washed as described previously (57). The membrane sheets were further stained for sialic acid and N-acetyl-glucosaminyl using wheat germ agglutinin (WGA) conjugated to Alexa Fluor 594 (#W11262; Invitrogen, Waltham, MA) at a concentration of 5 μg/mL in PBS (137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, pH 7.4) for 1 h at room temperature. After washing, the membrane sheets were imaged using an epifluorescence microscope as described previously (57). The lipid dye 1-(4-trimethyl-ammonium-phenyl)-6-phenyl-1,3,5-hexatriene p-toluene-sulfonate was added to visualize the membrane, and imaging was performed in the blue (membrane), green (CD81-GFP), and red (WGA-Alexa Fluor 594) channels.
For analysis, images were exported as tiff files and analyzed using the program ImageJ. Regions of interest were placed on individual membrane sheets selected in the blue channel, avoiding damaged membrane areas. The standard deviation (SD) of the mean intensity was determined within the selected regions of interest in the green channel and then related to the background-corrected intensity to determine the relative SD, which quantitatively estimates the degree of clustering (58). From individual membrane sheets, the relative SD was plotted against the background-corrected mean intensity. To quantify the degree of glycocalyx degradation, in the red channel we measured the background-corrected mean intensities on membrane sheets from transfected and untransfected cells.
Results
The δ-loop exhibits the highest flexibility in the isolated LEL and in the membrane-embedded full-length CD81
First, we asked whether the conformational flexibility pattern revealed by NMR spectroscopy (23) could be reproduced in silico. Atomistic MD simulations of the isolated soluble LEL region of human CD81 (CD81LEL) were performed in a 150 mM NaCl solution (Fig. S1 a) in four independent, unbiased 1 μs simulation runs. The continuous chain (monomer A) of the highest-resolution (1.6 Å) crystal structure of human CD81LEL (20) was used as the initial configuration. During the simulations, the LEL regions essentially maintained their compact conformations (Figs. 1 a and S1 b). Two parameters were measured to characterize the basic structural properties. First, we monitored the occurrence of helical secondary-structure elements. Compared with the crystal structure, helices α, β, and ε mainly preserved their helical configuration, whereas the γ- and δ-loop regions showed a diminished amount of helical content (Fig. 1 b, top). Then, to assess the flexibility along the protein chain, we analyzed the RMSFs of the backbone Cα atoms. Inside the protein chain, maximal flexibility occurred within the δ-loop (Fig. 1 b, bottom). Additionally, some flexibility was observed in the γ-loop, whereas helices α, β, and ε remained stable. Local flexibility minima were observed at cysteine-disulfide bridges flanking the flexible loops. The overall flexibility profile is well in line with the previously reported conformational variance pattern of isolated CD81LEL observed by NMR spectroscopy (23).
It is unclear whether anchoring the fluctuating N- and C-termini also influences the flexibility of other LEL segments. To address this issue, we performed atomistic MD simulations as described above with membrane-embedded, full-length CD81 (CD81FL), incorporating its predicted structure (16) into a homogeneous POPC bilayer (Fig. S2 a). In the simulations, the CD81FL structure remained mainly stable, with the exception of segments within the LEL (Figs. 2 a and S2 b). As for CD81LEL, we quantified changes in helical structure and flexibility along the protein backbone chain (Fig. 2 b). The LEL flexibility profile was the same within the full-length CD81 (Fig. 3 a) except for the flexible termini observed in the isolated LEL, which now experienced conformational constraints due to anchoring. This shows that when the LEL is an integral part of the rod-shaped structure, it does not experience any restraints that diminish its intrinsic flexibility as observed in solution.
CD81 establishes contacts to interaction partners via its flexible δ-loop
We next analyzed the relationship between local flexibility and the gaining of access to the environmental space. From each of the four MD runs, we superimposed 98 structures (time range 20–1000 ns) by fitting only the Cα atoms that exhibited the highest stability (Fig. 3 b). It is clear that although the δ-loop constitutes the smallest segment, it screens the largest environmental volume. Quantifying this effect, we determined per simulation run the SASA of the superimposed structures (Fig. 3 c) from which the SASAs of the individual LEL segments were extracted. With respect to the MD initial structures, in CD81FL, helices α, β, and ε minimize the explored space, whereas in CD81LEL, helices α and β hardly change and ε shows an exceedingly high increase in the SASA (>200%). However, this is a result of not anchoring the C-terminus, in conjunction with the compact conformation of the ε-helix in the initial structure. More importantly, the flexibility of the δ-loop leads to an increased average SASA of 41% in the isolated CD81LEL and 51% in CD81FL, whereas the respective γ-loops increase by just 15% and 28%.
To elucidate whether there is any relationship between this increase in explored environmental space and interaction-partner recruitment, we studied CD81 interactions in two large membrane environments incorporating many CD81 molecules. We turned to a MARTINI CG model (59, 60) to gain insights on extended timescales and length scales. To study CD81-CD81 interactions, we incorporated 49 full-length CD81 proteins into a preequilibrated ≈70 × 70 nm POPC lipid bilayer patch (Fig. 4 a). After a short initial relaxation, the system was simulated in an unbiased MD run over 2.5 μs (see Fig. 4 b for the final snapshot). To verify that CG CD81 molecules exhibit the same dynamical properties as CD81 in the atomistic model (Fig. 2 b, bottom), we determined the RMSFs of the protein backbone particles and calculated the average from the 49 molecules (Fig. 4 c). A comparison of both RMSF profiles (atomistic and CG) reveals a high similarity between the flexible and nonflexible regions. Again, the CG CD81 molecules exhibit the highest flexibility in the γ- and δ-loop regions.
To analyze CD81-CD81 interactions, we determined the RDF between each CG atom (of one CD81 molecule) to all CG atoms of the other CD81s (Fig. 4 d). The function peaked at a radius of ≈0.5 nm. Considering a narrow distribution of contact distances around this peak, a radius of 0.6 nm was defined as the CD81-CD81 contact threshold for monitoring oligomerization over time. The first contact between two CD81 proteins was detected after 78 ns (Fig. 4 e). Within the first 2 μs, more than half of the molecules formed dimers (for example, see Fig. 4 f), and from then on the dimers diminished as they became incorporated into higher oligomers (Fig. 4 e). To ascertain whether there is any relation between segmental flexibility and contact establishment, we determined CD81 interaction partner contacts per protein residue. We found one major interaction segment, the δ-loop (Fig. 4 g). Hence, the δ-loop plays a pivotal role in the establishment of direct CD81-CD81 contacts.
We next performed a simulation in a complex lipid environment resembling the lipid composition of a mammalian plasma membrane (47). This membrane is composed of 63 different lipid species, including glycolipids, which formed patchy platforms during the simulation (indicated by light-blue molecule crowds in Fig. 5 a). In addition to facilitating studies of CD81-CD81 interactions, this system also allows the identification of preferred lipid-binding partners. After a short initial relaxation, we performed an unbiased MD run over 7.5 μs (see Fig. 5 b for the final snapshot). Here, a longer simulation time was required because the diffusional mobility of CD81 was decreased when compared with that of the POPC system (Fig. S3 b).
CD81-CD81 interactions were analyzed in a manner similar to that employed for the POPC simulation (the RDF also yielded a threshold of 0.6 nm; for the RMSF, see Fig. S3 c). The first transient contact between two CD81 proteins was detected after 650 ns (Fig. 5 c; please note that simulation times cannot be directly compared between the POPC and plasma membrane system due to different CD81 mobilities, as illustrated in Fig. S3 b). At the end of the simulation, about half of the CD81 molecules were present in dimers and several higher oligomers (Fig. 5 c; see also Fig. 6, a and b). Interestingly, in the plasma membrane, the initial rise in dimers was only half as high as in the POPC system because they were recruited more efficiently into higher oligomers. Another difference is that we identified a second major interaction site comprising the intracellular leaflet-embedded TM2-TM3 region (Fig. 5 d).
To identify preferred lipid-binding partners, we analyzed the probabilities for direct interactions between the CD81 proteins and each of the 63 available lipid species. To this end, we determined the RDF between all CG atoms of CD81 and lipids (Fig. 6 c). As illustrated in Fig. 6 d, the absolute number of contacts with the eight charged GM species increased over time, whereas direct interactions with, e.g., phosphatidylcholine (PC) species, surrounding the proteins initially, decreased. The replacement of PC lipids by gangliosides rendered the negatively charged extracellular glycolipids a preferred interaction partner, yielding CD81-GM complexes that remained until the end of the simulation. This shows that gangliosides are preferred interaction partners, whereas the 10-fold higher concentrated PC lipids essentially do not bind. Notably, charge alone is not sufficient for interactions, as other charged lipids in the intracellular leaflet (PIP1, PIP2, and PIP3) showed no preferred binding tendency (Fig. S4). These observations are in line with previous experimental studies showing that CD81 forms complexes with gangliosides (13). Moreover, the simulation shows that CD81-GM interactions lead to domains in which multiple, not directly interacting CD81 molecules concentrate locally through their association with ganglioside patches that have formed in the preequilibrated plasma membrane (47). Therefore, oligomeric platforms are formed by both types of interactions (CD81-CD81 and CD81-GM; for examples, see Fig. 6 b, right and left, respectively), which reflects the tendency of tetraspanins to be engaged in a molecular network.
Although direct interactions between CD81 molecules in network formation have been described (6, 7, 8), experimental evidence for a similar role of gangliosides is lacking. When we degraded the glycocalyx using α-neuraminidase and β-galactosidase, we found a small effect on the lateral distribution of CD81 domains, which became more dispersed and uniformly distributed (Fig. S5). This supports the finding from the CG simulation, suggesting that gangliosides promote further tetraspanin network growth of small CD81 oligomers.
When determining CD81-GM contacts per protein residue, we found that most interactions occurred at the membrane proximal regions of CD81, including the TM helices and the SEL (Fig. S6). Surprisingly, relative to the initial residue distance to the plasma membrane, the δ-loop exhibited a clear involvement in direct interactions (Fig. 6 e). Thus, although the δ-loop does not play a major role in GM contacts, it may be able to exert stabilizing effects.
Discussion
As was previously pointed out (61), disordered segments in proteins are often functional, although the underlying mechanisms are unclear. This functionality might be related to disordered segments binding to a target or comprising a flexible linker. Here, we used MD simulations to gain insights into the role of local conformational flexibility in nanodomain formation in the plasma membrane, using the tetraspanin molecule CD81 as a model.
First, we verified whether the flexibility pattern revealed by wet-lab experiments could be reproduced in silico. In atomistic simulations, CD81LEL generally retained its compact structure, with the helices of the conserved region (α, β, and ε) exhibiting a high stability, whereas the helical components of the variable region showed a tendency for structural dissolution. This is in agreement with the conformational variance of CD81LEL observed by NMR spectroscopy (23). In full-length CD81, residues forming the highly flexible terminal ends of the isolated CD81LEL necessarily became conformationally stable. However, the flexibility patterns of the remaining LEL domains were indistinguishable, suggesting that insights obtained from the isolated CD81LEL can be directly transferred to full-length CD81. Moreover, the flexibility of the LEL loop regions is an intrinsic feature of the LEL itself and is neither restricted nor promoted by other domains. Furthermore, the simulations reveal that flexibility around the exposed δ-loop increases the volume that the molecule can probe for interaction partners. Notably, the overall rigid scaffold of the protein, acting like a molecular flagpole, is a prerequisite for extended sampling of the environmental space. This suggests that although the rigidity of membrane-embedded proteins may slow down contact establishment, such a restriction can be circumvented by a locally exposed domain that features pronounced conformational flexibility.
Using a CG model of multiple CD81 molecules in a POPC and a plasma membrane (47) environment, we found that the LEL flexibility profile matched that obtained from atomistic simulations, rationalizing its use for studying the relationship between flexibility and interaction-partner recruitment. We determined the protein residues involved in physical contacts to find out whether the exposed δ-loop displays a higher activity in establishing contacts. In the POPC membrane, the δ-loop plays a prominent role in contact establishment, whereas in the plasma membrane this activity is diminished. Here also the TM2-TM3-region makes a significant contribution. Since the δ-loop binds to GM lipids in the plasma membrane (for example, see Fig. 6 b), we suggest that this mechanism diminishes the fraction of δ-loop-mediated CD81-CD81 contacts.
CD81 oligomeric states are produced by both CD81-CD81 and CD81-GM contacts. In this scenario, the propensity of gangliosides to form platforms promotes CD81 crowding via indirect glycolipid-bridging effects. This explains how tetraspanins can form large, two-dimensionally growing molecular networks by using specifically forming binary complexes as building blocks: patches of lipids act as a molecular glue, gathering together different CD81-binary complexes (e.g., CD81-CD81 and CD81-EWI-2). A similar mechanism may explain why tetraspanin palmitoylation, which eventually facilitates association with lipid phases, stabilizes TEMs (6, 7, 8).
Importantly, although the interactions were stable until the end of the simulation, we did not observe a defined structural interface between two CD81 molecules. Anyway, this is not expected, as simulations even in the range of several microseconds are too short to form equilibrated protein-binding modes or defined quaternary structures. Therefore, the observed interactions reflect rather promiscuous initial contacts, and it is difficult to predict which of the two preferred interactions leads to stable complexes. However, since deletion of the δ-loop (deleted region: 176–186) is sufficient to destabilize CD81 dimers or higher oligomers (57), we argue that the observed role of the flexible δ-loop in silico is meaningful and provides a mechanistic explanation for cellular experiments. Surprisingly, this flexibility allows the δ-loop to interact even with the distally located GM molecules. However, other segments of CD81 are also strongly involved, and therefore the contribution of the δ-loop to GM bridge formation is likely of a supportive nature.
In conclusion, the MD simulation data reproduce the conformational flexibility observed in experimental studies. More importantly, they reveal the functional significance of flexibility in contact establishment and how large molecular platforms can be built from binary complexes. In general, the segmental flexibility of membrane proteins is likely to facilitate screening of the environmental space and thus play a pivotal role in mediating binding to interaction partners.
Author Contributions
T.H.S. performed MD simulations and analyses. Y.H. performed experiments and data analysis involving cells. T.H.S. and T.L. conceived the project, designed the experiments, and wrote the manuscript.
Acknowledgments
We thank Dr. Luise Florin for comments on the manuscript, Dr. Christian Kandt for proofreading and valuable advice, and the Paderborn Center for Parallel Computing (PC2) for providing computational facilities as part of the Ressourcenverbund NRW.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (LA1272/6-1 to T.L.).
Editor: Bert de Groot.
Footnotes
Six figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30279-X.
Contributor Information
Thomas H. Schmidt, Email: thomas.schmidt@uni-bonn.de.
Thorsten Lang, Email: thorsten.lang@uni-bonn.de.
Supporting Material
References
- 1.van Spriel A.B., Figdor C.G. The role of tetraspanins in the pathogenesis of infectious diseases. Microbes Infect. 2010;12:106–112. doi: 10.1016/j.micinf.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Veenbergen S., van Spriel A.B. Tetraspanins in the immune response against cancer. Immunol. Lett. 2011;138:129–136. doi: 10.1016/j.imlet.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 3.Zhang X.A., Huang C. Tetraspanins and cell membrane tubular structures. Cell. Mol. Life Sci. 2012;69:2843–2852. doi: 10.1007/s00018-012-0954-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monk P.N., Partridge L.J. Tetraspanins: gateways for infection. Infect. Disord. Drug Targets. 2012;12:4–17. doi: 10.2174/187152612798994957. [DOI] [PubMed] [Google Scholar]
- 5.Hemler M.E. Tetraspanin proteins promote multiple cancer stages. Nat. Rev. Cancer. 2014;14:49–60. doi: 10.1038/nrc3640. [DOI] [PubMed] [Google Scholar]
- 6.Charrin S., Manié S., Rubinstein E. Multiple levels of interactions within the tetraspanin web. Biochem. Biophys. Res. Commun. 2003;304:107–112. doi: 10.1016/s0006-291x(03)00545-x. [DOI] [PubMed] [Google Scholar]
- 7.Hemler M.E. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 8.Levy S., Shoham T. The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol. 2005;5:136–148. doi: 10.1038/nri1548. [DOI] [PubMed] [Google Scholar]
- 9.Bassani S., Cingolani L.A. Tetraspanins: interactions and interplay with integrins. Int. J. Biochem. Cell Biol. 2012;44:703–708. doi: 10.1016/j.biocel.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 10.Claas C., Stipp C.S., Hemler M.E. Evaluation of prototype transmembrane 4 superfamily protein complexes and their relation to lipid rafts. J. Biol. Chem. 2001;276:7974–7984. doi: 10.1074/jbc.M008650200. [DOI] [PubMed] [Google Scholar]
- 11.Tarrant J.M., Robb L., Wright M.D. Tetraspanins: molecular organisers of the leukocyte surface. Trends Immunol. 2003;24:610–617. doi: 10.1016/j.it.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 12.Kawakami Y., Kawakami K., Hakomori S. Tetraspanin CD9 is a “proteolipid,” and its interaction with α 3 integrin in microdomain is promoted by GM3 ganglioside, leading to inhibition of laminin-5-dependent cell motility. J. Biol. Chem. 2002;277:34349–34358. doi: 10.1074/jbc.M200771200. [DOI] [PubMed] [Google Scholar]
- 13.Toledo M.S., Suzuki E., Hakomori S. Effect of ganglioside and tetraspanins in microdomains on interaction of integrins with fibroblast growth factor receptor. J. Biol. Chem. 2005;280:16227–16234. doi: 10.1074/jbc.M413713200. [DOI] [PubMed] [Google Scholar]
- 14.Todeschini A.R., Dos Santos J.N., Hakomori S.I. Ganglioside GM2-tetraspanin CD82 complex inhibits met and its cross-talk with integrins, providing a basis for control of cell motility through glycosynapse. J. Biol. Chem. 2007;282:8123–8133. doi: 10.1074/jbc.M611407200. [DOI] [PubMed] [Google Scholar]
- 15.Todeschini A.R., Hakomori S. Functional role of glycosphingolipids and gangliosides in control of cell adhesion, motility, and growth, through glycosynaptic microdomains. Biochim. Biophys. Acta. 2008;1780:421–433. doi: 10.1016/j.bbagen.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seigneuret M. Complete predicted three-dimensional structure of the facilitator transmembrane protein and hepatitis C virus receptor CD81: conserved and variable structural domains in the tetraspanin superfamily. Biophys. J. 2006;90:212–227. doi: 10.1529/biophysj.105.069666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seigneuret M., Delaguillaumie A., Conjeaud H. Structure of the tetraspanin main extracellular domain. A partially conserved fold with a structurally variable domain insertion. J. Biol. Chem. 2001;276:40055–40064. doi: 10.1074/jbc.M105557200. [DOI] [PubMed] [Google Scholar]
- 18.Stipp C.S., Kolesnikova T.V., Hemler M.E. Functional domains in tetraspanin proteins. Trends Biochem. Sci. 2003;28:106–112. doi: 10.1016/S0968-0004(02)00014-2. [DOI] [PubMed] [Google Scholar]
- 19.DeSalle R., Mares R., Garcia-España A. Evolution of cysteine patterns in the large extracellular loop of tetraspanins from animals, fungi, plants and single-celled eukaryotes. Mol. Phylogenet. Evol. 2010;56:486–491. doi: 10.1016/j.ympev.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 20.Kitadokoro K., Bordo D., Bolognesi M. CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs. EMBO J. 2001;20:12–18. doi: 10.1093/emboj/20.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitadokoro K., Ponassi M., Bolognesi M. Subunit association and conformational flexibility in the head subdomain of human CD81 large extracellular loop. Biol. Chem. 2002;383:1447–1452. doi: 10.1515/BC.2002.164. [DOI] [PubMed] [Google Scholar]
- 22.Yang W., Zhang M., Cui S. An intramolecular bond at cluster of differentiation 81 ectodomain is important for hepatitis C virus entry. FASEB J. 2015;29:4214–4226. doi: 10.1096/fj.15-272880. [DOI] [PubMed] [Google Scholar]
- 23.Rajesh S., Sridhar P., Overduin M. Structural basis of ligand interactions of the large extracellular domain of tetraspanin CD81. J. Virol. 2012;86:9606–9616. doi: 10.1128/JVI.00559-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kovalenko O.V., Metcalf D.G., Hemler M.E. Structural organization and interactions of transmembrane domains in tetraspanin proteins. BMC Struct. Biol. 2005;5:11. doi: 10.1186/1472-6807-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montpellier C., Tews B.A., Cocquerel L. Interacting regions of CD81 and two of its partners, EWI-2 and EWI-2wint, and their effect on hepatitis C virus infection. J. Biol. Chem. 2011;286:13954–13965. doi: 10.1074/jbc.M111.220103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmid N., Eichenberger A.P., van Gunsteren W.F. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur. Biophys. J. 2011;40:843–856. doi: 10.1007/s00249-011-0700-9. [DOI] [PubMed] [Google Scholar]
- 27.Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., Hermans J. Interaction models for water in relation to protein hydration. In: Pullman B., editor. Intermolecular Forces. Reidel; Dordrecht: 1981. pp. 331–342. [Google Scholar]
- 28.Caves L.S.D., Evanseck J.D., Karplus M. Locally accessible conformations of proteins: multiple molecular dynamics simulations of crambin. Protein Sci. 1998;7:649–666. doi: 10.1002/pro.5560070314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koch D.C., Schmidt T.H., Kandt C. Structural dynamics of the cell wall precursor lipid II in the presence and absence of the lantibiotic nisin. Biochim. Biophys. Acta. 2014;1838:3061–3068. doi: 10.1016/j.bbamem.2014.07.024. [DOI] [PubMed] [Google Scholar]
- 30.Berendsen H.J.C., Postma J.P.M., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 31.Nose S. A unified formulation of the constant temperature molecular-dynamics methods. J. Chem. Phys. 1984;81:511–519. [Google Scholar]
- 32.Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 33.Parrinello M., Rahman A. Crystal structure and pair potentials: a molecular-dynamics study. Phys. Rev. Lett. 1980;45:1196–1199. [Google Scholar]
- 34.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 35.Parrinello M., Rahman A. Strain fluctuations and elastic constants. J. Chem. Phys. 1982;76:2662. [Google Scholar]
- 36.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 37.Essmann U., Perera L., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 38.Páll S., Hess B. A flexible algorithm for calculating pair interactions on SIMD architectures. Comput. Phys. Commun. 2013;184:2641–2650. [Google Scholar]
- 39.Hess B., Bekker H., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 40.Hess B. P-LINCS: a parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 2008;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 41.Miyamoto S., Kollman P.A. SETTLE: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 42.Van Der Spoel D., Lindahl E., Berendsen H.J.C. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 43.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 44.Poger D., Van Gunsteren W.F., Mark A.E. A new force field for simulating phosphatidylcholine bilayers. J. Comput. Chem. 2010;31:1117–1125. doi: 10.1002/jcc.21396. [DOI] [PubMed] [Google Scholar]
- 45.Poger D., Mark A.E. On the validation of molecular dynamics simulations of saturated and cis-monounsaturated phosphatidylcholine lipid bilayers: a comparison with experiment. J. Chem. Theory Comput. 2010;6:325–336. doi: 10.1021/ct900487a. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt T.H., Kandt C. LAMBADA and InflateGRO2: efficient membrane alignment and insertion of membrane proteins for molecular dynamics simulations. J. Chem. Inf. Model. 2012;52:2657–2669. doi: 10.1021/ci3000453. [DOI] [PubMed] [Google Scholar]
- 47.Ingólfsson H.I., Melo M.N., Marrink S.J. Lipid organization of the plasma membrane. J. Am. Chem. Soc. 2014;136:14554–14559. doi: 10.1021/ja507832e. [DOI] [PubMed] [Google Scholar]
- 48.Jo S., Kim T., Im W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One. 2007;2:e880. doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jo S., Kim T., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 50.de Jong D.H., Singh G., Marrink S.J. Improved parameters for the Martini coarse-grained protein force field. J. Chem. Theory Comput. 2013;9:687–697. doi: 10.1021/ct300646g. [DOI] [PubMed] [Google Scholar]
- 51.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 52.Hsin J., Arkhipov A., Schulten K. Using VMD: an introductory tutorial. Curr. Protoc. Bioinformatics. 2008;Chapter 5 doi: 10.1002/0471250953.bi0507s24. Unit 5.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frishman D., Argos P. Knowledge-based protein secondary structure assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 54.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 55.Eisenhaber F., Lijnzaad P., Scharf M. The double cubic lattice method: efficient approaches to numerical integration of surface area and volume and to dot surface contouring of molecular assemblies. J. Comput. Chem. 1995;16:273–284. [Google Scholar]
- 56.Bondi A. van der Waals volumes and radii. J. Phys. Chem. 1964;68:441–451. [Google Scholar]
- 57.Homsi Y., Schloetel J.-G., Lang T. The extracellular δ-domain is essential for the formation of CD81 tetraspanin webs. Biophys. J. 2014;107:100–113. doi: 10.1016/j.bpj.2014.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zilly F.E., Halemani N.D., Lang T. Ca2+ induces clustering of membrane proteins in the plasma membrane via electrostatic interactions. EMBO J. 2011;30:1209–1220. doi: 10.1038/emboj.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marrink S.J., Risselada H.J., de Vries A.H. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 60.Marrink S.J., Tieleman D.P. Perspective on the Martini model. Chem. Soc. Rev. 2013;42:6801–6822. doi: 10.1039/c3cs60093a. [DOI] [PubMed] [Google Scholar]
- 61.Dyson H.J., Wright P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.