Abstract
The enzyme N-myristoyltransferase (NMT) catalyzes the essential fatty acylation of substrate proteins with myristic acid in eukaryotes and is a validated drug target in the parasite Trypanosoma brucei, the causative agent of African trypanosomiasis (sleeping sickness). N-Myristoylation typically mediates membrane localization of proteins and is essential to the function of many. However, only a handful of proteins are experimentally validated as N-myristoylated in T. brucei. Here, we perform metabolic labeling with an alkyne-tagged myristic acid analogue, enabling the capture of lipidated proteins in insect and host life stages of T. brucei. We further compare this with a longer chain palmitate analogue to explore the chain length-specific incorporation of fatty acids into proteins. Finally, we combine the alkynyl-myristate analogue with NMT inhibitors and quantitative chemical proteomics to globally define N-myristoylated proteins in the clinically relevant bloodstream form parasites. This analysis reveals five ARF family small GTPases, calpain-like proteins, phosphatases, and many uncharacterized proteins as substrates of NMT in the parasite, providing a global view of the scope of this important protein modification and further evidence for the crucial and pleiotropic role of NMT in the cell.
Keywords: human African trypanosomiasis, N-myristoylation, chemical proteomics, click chemistry, protein lipidation, target validation
Human African trypanosomiasis (HAT), or African sleeping sickness, is a usually fatal tropical disease caused by unicellular eukaryotic parasites of the species Trypanosoma brucei and transmitted by an insect vector. Although the number of reported cases has dropped in recent years,1 an estimated 21 million people are at high to moderate risk of the disease.2 In addition, the analogous livestock disease, Nagana, causes an estimated 3 million cattle deaths per year with significant economic impact.3T. brucei gambiense, responsible for >98% of HAT cases, causes a chronic infection in which the early stage, lasting several months or years, is relatively asymptomatic; later in infection, parasites cross the blood–brain barrier and invade the central nervous system, ultimately leading to coma and death. There are few treatments currently available to treat late-stage HAT, and all suffer from high toxicity, high expense, or problematic delivery.4T. brucei is transmitted primarily by the bite of an infected tsetse fly, which injects the metacyclic trypomastigote form of the parasite into the mammalian host, although mother to fetus transmission can also occur. The parasite then transforms into the bloodstream form (BSF), which remains extracellular in the bloodstream and lymph. When a tsetse fly takes a blood meal from an infected host, parasites are taken up and transform into procyclic forms (PCFs) that multiply in the insect gut prior to transformation into epimastigotes, which travel to the insect salivary gland.5 The BSF is therefore of most interest for treatment of infection and progression of the disease, whereas the PCF is important for replication in the insect vector. The adaptive differences between BSF and PCF, and the process of differentiation, are important for druggability of BSF trypanosomes in the mammalian host.
The enzyme myristoyl-CoA:protein N-myristoyltransferase (NMT) is an essential eukaryotic enzyme that catalyzes attachment of the C14:0 fatty acid myristate from myristoyl-CoA to the N-terminal glycine residue of a subset of cellular proteins.6 N-Myristoylation mediates membrane localization, modulates stability, or regulates protein–protein interactions, and NMT has been investigated as a potential drug target in HAT,7 fungal infections,8 leishmaniases,9 malaria,10 nematodes,11 and cancer.12 Structure-based design and high-throughput screening have yielded multiple NMT inhibitor series, some with species selectivity.7,13 In T. brucei, RNAi knockdown of NMT results in abnormal morphology and defects in endocytic trafficking.14 Trafficking defects may in part be related to loss of myristoylation of members of the ADP-ribosylation factor (ARF) family of small GTPases involved in vesicular trafficking in eukaryotes. RNAi depletion of two N-myristoylated T. brucei ARFs showed that the proteins are essential for viability of BSF parasites and revealed defects in subcellular structures such as the flagellar pocket (the site of almost all endo- and exocytosis), vesicles, and the Golgi apparatus.15 In 2010, Frearson et al. reported a series of NMT inhibitors with high potency against the T. brucei enzyme, with the ability to cure trypanosomiasis in mice.7,13a,13b The phenotype of inhibitor treatment was distinct from RNAi knockdown of NMT, but the parasite did exhibit an enlarged flagellar pocket.7 Given the cotranslational nature of N-myristoylation, NMT inhibitors would be expected to affect viability of both BSF and PCF parasites, because both forms replicate; in terms of clinical treatment, however, targeting BSF parasites is of most interest.
Bioinformatic analyses suggest that more than 60 proteins may be N-myristoylated in T. brucei, resulting in the prediction that NMT inhibition will have pleiotropic effects on the parasite.16 There is some experimental evidence for N-myristoylation of a few parasite proteins: three ARFs,15a,15b,17 cytoskeleton-associated protein CAP5.5,18 Calflagin,19 phosphatase PPEF,16 flagellar-calcium binding protein FCaBP (in the related parasite T. cruzi),20 and virulence-associated metacaspase 4 (MCA4).21 However, global characterization of N-myristoylation by standard biochemical methods such as radiolabeling is hampered by low sensitivity, the need for specific antibodies for target proteins, and the frequent requirement for artificial overexpression to achieve detection. We and others have recently made use of the metabolic incorporation of bioorthogonally tagged fatty acids to globally profile myristoylation and other protein lipidations in diverse organisms,22 including the trypanosomatid parasite Leishmania donovani, which causes leishmaniasis.23 Here we apply this technology to study lipidation in T. brucei, comparing acylated proteins in the insect (PCF) and host (BSF) life stages using a tagged analogue of myristate and analyzing acylation patterns with a tagged palmitate analogue. Finally, we quantify changes in acylation levels in the presence of NMT inhibitors and demonstrate selective target engagement across the proteome through quantitative chemical proteomic analyses. Taken together, these data globally define NMT substrates in the key life stages of the parasite and provide insight into the mechanism of action of NMT inhibitors in T. brucei.
Results and Discussion
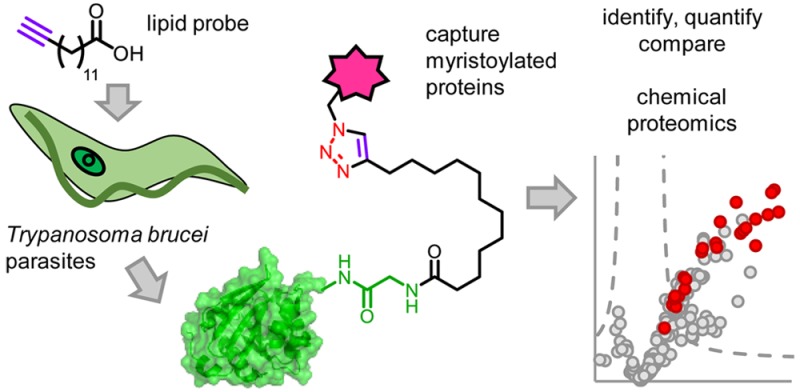
Fatty acids bearing terminal alkyne or azide modifications are known to be tolerated by the cellular machinery in diverse systems and incorporated into acylated proteins.22a Our approach uses myristic acid analogue YnMyr (Figure 1a), the coenzyme-A analogue of which is accepted as a substrate by NMTs and which we have previously shown is incorporated into N-myristoylated proteins in Plasmodium falciparum,10Leishmania donovani,23 human cancer cells,12 virus-infected cells,24 and zebrafish embryos.25 After cell lysis, alkyne-tagged proteins are captured by a click reaction, the copper-catalyzed cycloaddition of an alkyne and azide (CuAAC), appending a variety of groups such as a fluorophore for visualization and/or biotin for affinity pull-down.26 Enriched proteins are then subject to tryptic digest and shotgun LC-MS/MS analysis for proteomic identification (Figure 1a).
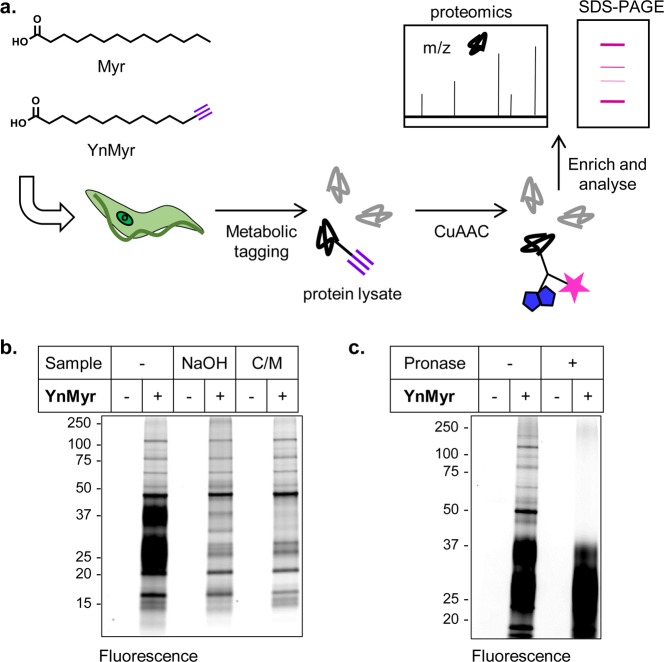
Figure 1.
YnMyr labels proteins in T. brucei PCF parasites. (a) Overview of tagging strategy. YnMyr or myristic acid (Myr) was added to T. brucei parasite cultures and incorporated metabolically into lipidated proteins. The alkyne tag was reacted by CuAAC with fluorophore and/or biotin-functionalized azide capture reagents (Supporting Information Figure S1) to allow downstream enrichment and analysis by proteomics and SDS-PAGE. (b) Labeling with YnMyr or Myr (−) in PCF parasites. After 18 h of incubation with probes at 100 μM, parasites were lysed, and proteins were reacted with AzTB and separated by gel for fluorescence scanning. Samples were treated with NaOH or precipitated with chloroform/methanol (C/M) as indicated. (c) Specific PCF labeling is resistant to treatment with Pronase. Coomassie gels are shown in Supporting Information Figure S2.
YnMyr Labels Proteins in T. brucei
To investigate whether YnMyr can be used to label proteins in T. brucei, cultures of PCF parasites were incubated for 18 h with 100 μM YnMyr or myristic acid (Myr) control. Following cell lysis, tagged proteins were ligated to biotin and TAMRA functionalized reagent AzTB (Supporting Information Figure S1) via CuAAC and visualized by in-gel fluorescence following separation by SDS-PAGE. In addition to multiple discrete bands, two diffuse bands were observed between 20 and 40 kDa (Figure 1b), which could be removed by chloroform–methanol precipitation or base treatment of proteins after CuAAC. Trypanosomatid parasites are abundant in complex glycolipids, and PCF T. brucei possesses a family of surface proteins, the procyclins, which bear a glycan-elaborated glycosylphosphatidylinositol (GPI)-anchor near their C-terminus and in some cases N-glycosylation in the N-terminal domain.27 When separated by SDS-PAGE, procyclins migrate as two polydisperse bands at ∼30 and 40 kDa and can be radiolabeled with [3H]-myristate.28 Treatment of YnMyr-labeled PCF samples with Pronase shifted a proportion of the diffuse bands to lower molecular weight and completely removed the majority of labeling (Figure 1c), consistent with the discrete bands corresponding to proteins and the diffuse bands corresponding to the partially protease-resistant procyclins. Alternatively, the diffuse bands may correspond to other glycolipid components, such as free poly-N-glycosylated GPIs that are also present on the surface of T. brucei PCF.28a
Having established that YnMyr could label proteins in the procyclic insect stage form of T. brucei, we focused on host stage parasites. Cultured BSF cells were incubated with 100 μM YnMyr or Myr for 4, 8, or 18 h and lysates processed as described above. Labeling intensity increased from 4 to 8 h but decreased again at 18 h, at which time point YnMyr-related toxicity was also observed (Figure 2a,b; Supporting Information Figure S3); parasites exhibited the so-called “Big Eye” phenotype, which is characterized by an enlarged flagellar pocket. This phenotype is the result of a block in receptor-mediated endocytosis and has previously been described following RNAi knockdown of clathrin heavy chain29 and the small GTPase ARF1.15b Consistent with our results, YnMyr was previously shown to be moderately toxic to T. brucei in a study seeking to identify inhibitors of variant surface glycoprotein (VSG) GPI myristoylation.30 The VSG coats the surface of BSF T. brucei and is unusual in incorporating specifically diacyl-myristate into its GPI anchor.31 This feature is unique to T. brucei BSF, and multiple cellular pathways have evolved to ensure that myristate alone is incorporated. We hypothesize that the observed YnMyr toxicity is related to disruption of the VSG myristate pathway, and an 8 h tagging step was therefore used in subsequent experiments to circumvent YnMyr-related toxicity and focus analysis on N-myristoylated proteins.
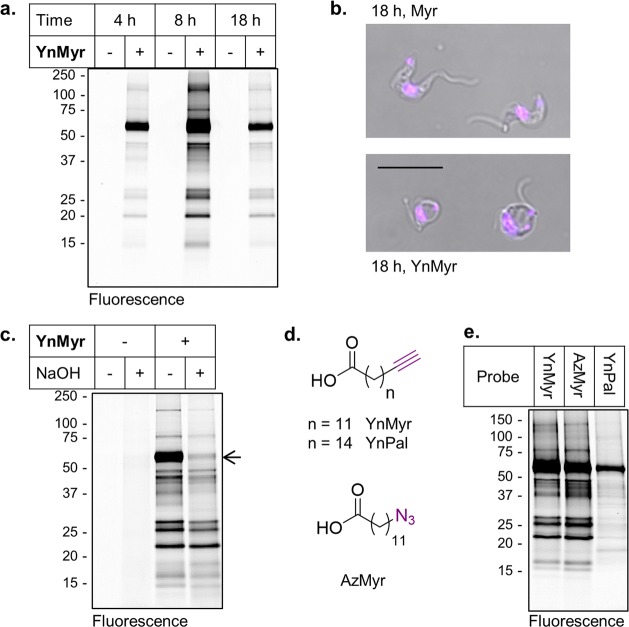
Figure 2.
YnMyr labels proteins in T. brucei BSF parasites. (a) Time-dependent metabolic incorporation of 100 μM YnMyr or Myr (−) in BSF parasites. (b) Phenotype of YnMyr treatment at 18 h in BSF. Scale bar = 10 μm. See also Supporting Information Figure S3. (b) NaOH treatment of lysates from BSF parasites incubated with YnMyr reveal labeling of a base sensitive band at ∼60 kDa. (d) Chemical structures of myristate analogues YnMyr and AzMyr and palmitate analogue YnPal. (e) Comparative labeling with different fatty acid analogues at 100 μM in BSF. Coomassie gels are shown in Supporting Information Figure S2. Additional data are shown in Supporting Information Figure S4.
A prominent band between 50 and 75 kDa showed sensitivity to treatment with strong base (NaOH; Figure 2c; Supporting Information Figure S4), indicating ester-linked YnMyr and consistent with incorporation of the probe into the GPI anchor of the VSG, as expected. We have previously observed significant incorporation of tagged fatty acids into GPI-anchored proteins in the malaria parasite P. falciparum.10 The majority of other bands were insensitive to base treatment, implying amide-linked YnMyr. Azido-myristate mimetic AzMyr gave very similar labeling to YnMyr, as expected, whereas longer chain palmitate analogue YnPal gave a distinct pattern (Figure 2d,e). These data are consistent with incorporation of tagged fatty acids into proteins by chain-length specific acyltransferases such as NMT and palmitoylacyltransferases (PATs).
Proteomic Identification of YnMyr-Labeled Proteins in T. brucei
We have previously shown that YnMyr is incorporated into putative N-myristoylated protein ARL6 via labeling of this protein after immunoprecipitation (IP).17 However, the IP method relies on the availability of an antibody to the protein of interest and has very low throughput. To carry out global identification of the proteins labeled in BSF and PCF parasites, tagged proteins were ligated to AzTB and enriched by pull-down onto NeutrAvidin-coated resin through the biotin affinity label (Supporting Information Figure S5). Bead-bound proteins were subject to tryptic digest and peptides analyzed by LC-MS/MS. Each sample set consisted of a YnMyr sample and Myr control, prepared in parallel. Raw data were searched using MaxQuant32 and a database of the T. brucei TREU 927 reference strain (TriTrypDB33). In-gel fluorescence analysis suggested that the VSG was also labeled with YnMyr. Since the VSG variant is strain-specific, initial BSF experimental data were therefore also searched against the T. brucei Lister strain 427, identifying VSG variant Tb427.BES40.22; this protein sequence was appended to the TREU 927 FASTA file for all subsequent searches. Data were analyzed using label-free quantification (LFQ), a technique that normalizes intensity measurements to enable comparison between different LC-MS/MS runs, in MaxQuant (“MaxLFQ”).34 After Log2 transformation of intensities, data were filtered to retain only proteins present in several replicates, missing values were imputed from a normal distribution to mimic values at the limit of detection, and permutation-corrected two-sample t tests were used to assess proteins significantly enriched in YnMyr samples over Myr controls (see Methods). A caveat to this analytical workflow is that proteins detected in multiple YnMyr samples but with low intensity will be assigned as nonsignificant even if they are absent from Myr controls because their intensity is too close to background; many of these proteins could be genuine hits but of low abundance.
In BSF experiments, proteins were filtered to retain only those present in at least three of four replicates and in biological duplicate, resulting in 101 significantly enriched proteins in YnMyr samples (Figure 3a; Supporting Information Table S1). Of these, 46 (46%) are likely to carry an N-terminal glycine implied by an MG motif at the N-terminus, which is thought to be a requirement for NMT-dependent myristoylation.35 In PCF experiments, proteins were filtered to retain only those present in at least four of the six technical replicates and in biological duplicate, identifying 91 proteins as significantly enriched in YnMyr samples (Figure 3b; Supporting Information Table S2); again, roughly half of these proteins have an N-terminal MG motif.
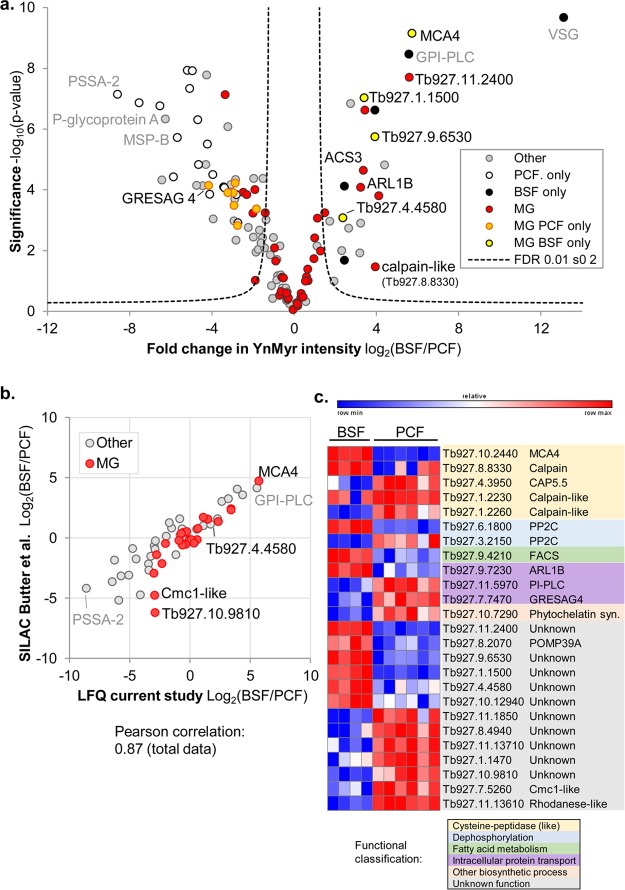
Figure 3.
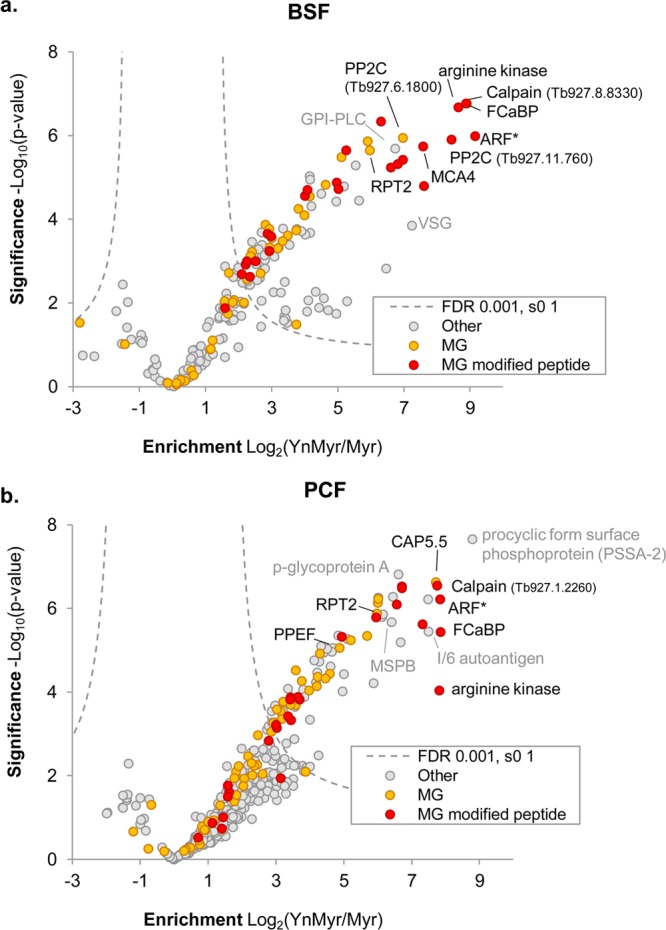
Identification of YnMyr-tagged proteins in (a) BSF and (b) PCF T. brucei. Volcano plots show significance of enrichment over background. Parasites were treated with Myr or YnMyr, lysates labeled by CuAAC, and proteins enriched by biotin–streptavidin interaction and digested by trypsin for LC-MS/MS. Proteins from four (BSF) or six (PCF) replicates (independent sample processing from the lysate stage; includes a biological replicate in each case) were quantified by label-free quantification (MaxLFQ). After filtering to retain only those proteins present in biological duplicate and in 3 of 4 (BSF) or 4 of 6 (PCF) samples, missing intensities were imputed from a normal distribution chosen to mimic noise level, and a modified t test with permutation-based FDR statistics was applied (250 permutations) to compare Myr and YnMyr groups. BSF and PCF: FDR = 0.001; s0 = 1. Proteins containing an N-terminal glycine (MG) and those for which the YnMyr-modified peptide was identified using reagents AzRB or AzRTB in the respective lifestage are indicated. ARF* indicates multiple proteins in this proteinGroup. See also Supporting Information Tables S1–S3.
Non-MG proteins highly enriched in YnMyr samples include glycosylphosphatidylinositol-specific phospholipase C (GPI-PLC), a protein that is S-myristoylated,36 in BSF samples, and MSP-B (a homologue of GPI-anchored Leishmania GP63 surface protease), procyclic form surface phosphoprotein (PSSA-2), and p-glycoprotein-A in PCF parasites. The GPI-anchored VSG was also detected in BSF samples and was enriched in YnMyr experiments compared to controls. Proteins previously shown to be N-myristoylated in T. brucei or related species included flagellar calcium binding protein (FCaBP, shown to be N-myristoylated in T. cruzi),20 cytoskeleton-associated protein CAP5.5,18 the proteasome regulatory ATPase subunit 2 (RPT2), which is known to be N-myristoylated in many eukaryotes,37 a phosphatase of the PPEF family,16 and a GRASP homologue (acylated in other protozoan parasites38). Metacaspase 4 (MCA4), previously shown to be palmitoylated at a cysteine residue proximal to a likely N-myristoylation site, was also identified, principally in BSF samples; this pseudopeptidase is an important virulence factor in T. brucei infection.21 Several ARF/ARLs, a well-studied family of N-myristoylated small GTPases, were also identified across the data sets: these included Tb927.7.6230, now called ARF3 (GeneDB) but previously known as ARL1, and shown to be both N-myristoylated and essential to survival of BSF T. brucei.15a,15c ARL6 (Tb927.8.5060), which we showed previously to be tagged with YnMyr in T. brucei using IP with an ARL6 specific antibody,17 was a significant hit identified in both BSF and PCF parasites.
In initial analyses using AzTB as capture reagent, no YnMyr-modified peptides were identified; this is unsurprising because the biotinylated peptide should remain partly anchored to the resin and the large TAMRA-containing label hinders detection by LC-MS/MS. We recently reported a series of related reagents incorporating a trypsin cleavage site between the TAMRA/biotin moieties and the azide capture group;25 these reagents enabled identification of YnMyr-tagged peptides in the malaria parasite P. falciparum,10 human cancer cells,12 zebrafish,25 and Leishmania parasites.23 In the current study we used AzRTB and AzRB (Supporting Information Figure S1), which both incorporate an arginine trypsin cleavage site and biotin, with AzRTB also featuring a TAMRA fluorophore. Searches were carried out with the additional mass fragment as a variable modification on any amino acid at a peptide N-terminus and data filtered to retain only those identifications meeting a stringent score threshold, as established in our previous studies25 (see Methods). Sixty-five identified modification sites across BSF and PCF samples were matched to 56 discrete proteins; peptides differently modified by methionine oxidation or with different lengths due to missed cleavages can derive from the same protein sequence (Supporting Information Table S3; see Supporting Information Figure S6 for examples of assigned modified peptide spectra and Supporting Information Data File for all spectra). Of these 65 peptides, only 3 lacked an N-terminal glycine and corresponded to proteins for which no other peptides were identified; therefore, these are likely to be false-positive identifications. When AzRTB was used to capture and enrich tagged proteins, 18 YnMyr-modified peptides (17 proteins) were detected in T. brucei PCF samples. AzRB, however, resulted in more identifications, with 30 modified N-terminal glycines detected in PCF and 26 in BSF samples. Sixteen YnMyr modified N-terminal glycines were detected independently with both AzRTB and AzRB; these reagents result in remnants on the modified peptide that are slightly different in mass (by one methylene unit), providing orthogonal evidence for modification of these peptides.
BSF and PCF parasites are adapted to very different environments, and the identification of stage-specific proteins expressed specifically in the insect vector or the host is an ongoing area of research.39 Comparison of YnMyr LFQ intensities of hits (defined as proteins identified as significantly enriched over Myr controls in one or both life stages) by t test revealed that a subset of non-MG and MG proteins were differentially detected in one life stage over the other (Figure 4; Supporting Information Table S4). The data are in good agreement with existing studies. The ratio of BSF/PCF YnMyr intensities for hits was plotted against the ratio from a published data set where stable isotope labeling by amino acids in cell culture (SILAC) quantitative proteomics was used to compare the two life stages:39 pleasingly, for the 59 proteins quantified by both studies (roughly half of the LFQ hits in our comparison), the correlation was high (Pearson 0.87; Figure 4b). Our data are also in similarly good agreement with an earlier SILAC-based study (Urbaniak et al.;40 Pearson correlation 0.83; Supporting Information Figure S7). These high correlations also show that, in most cases, YnMyr labeling tracks protein abundance.
Figure 4.
Comparison of acylated proteins in two life stages of T. brucei. (a) Volcano plot comparing YnMyr intensities of hits (defined as proteins significantly enriched over Myr controls in one or both life stages) by a two-sided two-sample permutation-corrected t test (250 permutations; FDR 0.01, s0 2). Those proteins identified in one life stage (BSF/PCF only) and N-terminal glycine-containing proteins (MG) are indicated. (b) Comparison of relative abundance of protein hits in BSF and PCF parasites in the current study (LFQ quantification; ratio of YnMyr intensities is plotted) with the study of Butter et al. (quantification via SILAC).39 Comparison with another previously reported data set is shown in Supporting Information Figure S7. See also Supporting Information Table S4. (c) Heatmap and functional classification of MG proteins found/enriched in one of the two stages. YnMyr intensities shown and color-coded within each row. The total data set is shown in Supporting Information Figure S8.
As expected, specific cell surface proteins such as GPI-PLC and VSG were prominent in BSF samples, whereas MSP-B and PSSA-2 were found in PCF samples, consistent with the large changes that occur on the parasite’s surface coat. Functional analysis of stage-enriched MG motif proteins revealed several hits in the calpain family of cysteine peptidases, two protein phosphatases with different stage specificities, and proteins involved in intracellular transport: phosphoinositide-specific phospholipase C and receptor adenylate cyclase (GRESAG4)41 in PCF parasites and ARL1B enriched in BSF parasites (Figure 4c). Calpains are calcium-dependent cysteine peptidases, and T. brucei possesses an expanded family of calpain-like proteins, some of which have apparently no proteolytic activity but are targeted by lipidation or other signals to different subcellular locations.42 Although the functions of these proteins are largely unknown, they have been hypothesized to play regulatory roles, for example, in cytoskeletal remodeling in the case of CAP5.5,18 or in virulence in the mammalian host in the case of MCA4.21 Given the widespread differential phosphorylation of BSF and PCF T. brucei,43 the identified stage-enriched lipidated phosphatases could also be particularly interesting for future studies; for example, a recent analysis of two N-myristoylated phosphatases demonstrated their importance at specific stages of development in the malaria parasite.44 Thirteen MG proteins currently of unknown function were also enriched in one of the two stages. Interestingly, 12 lipidated proteins significantly enriched in our PCF data set were found to be up-regulated early in differentiation from BSF to PCF parasites in a recent proteomic study by Dejung et al.45 (Supporting Information Figure S7). Other interesting hits are ARF protein ARLB and calpain-like protein Tb927.1.2230, both enriched in our BSF data and identified as transiently up-regulated during differentiation by Dejung et al., suggestive of roles in the differentiation process. Overall, our data show that lipidated proteins involved in carbohydrate metabolism, phosphorylation processes, small-molecule, ion, and protein transport, and signal transduction vary between BSF and PCF parasites (Supporting Information Figure S8), consistent with the significant changes to cell structure and metabolism that accompany adaptation of the cell to its different host environments.
Proteomic Identification of YnPal-Labeled Proteins in T. brucei
As discussed above, GPI-anchored proteins and glycoconjugates are prevalent in T. brucei, and these are known to incorporate fatty acyl chains of various lengths. In addition, fatty acids can be incorporated into proteins on cysteine side chains; S-acylation with the 16-carbon fatty acid palmitate is a common modification across eukaryotes, including protozoan parasites.46 In contrast to the high specificity of NMT for myristoylation,6 S-acylation enzymes and pathways have been shown to be promiscuous in accepting fatty acid analogues with a variety of chain lengths—in mammalian cells at least.47 In an additional layer of complexity, some proteins are dually acylated, being N-myristoylated at the N-terminal glycine and S-acylated with palmitate on a nearby cysteine residue.46 Furthermore, T. brucei is known to metabolize long-chain fatty acids to meet its needs in different life stages and culture conditions,48 although whether alkyne-tagged analogues would also be substrates for these metabolic enzymes is not known.
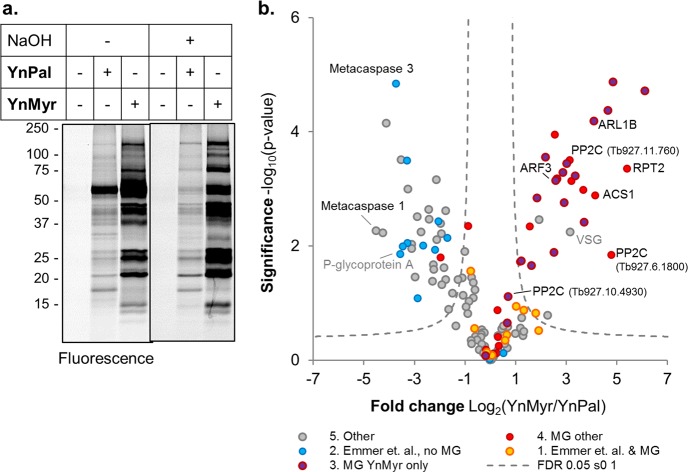
To provide additional insight into whether YnMyr was also incorporated into S-palmitoylation sites and to establish a broader picture of lipidation in T. brucei, longer chain palmitate analogue YnPal (Figure 2d) was incubated with BSF parasites at 100 μM for 4, 8, or 18 h, and samples were processed as before. In-gel fluorescence analysis revealed a distinct band pattern for the two analogues (Figure 5a). Interestingly, in contrast to YnMyr, YnPal was not visibly toxic to parasites, and labeling intensity continued to increase slightly up to 18 h (Supporting Information Figure S4). Similar to YnMyr, YnPal was also incorporated into a base-sensitive band at ∼60 kDa, although to a much lesser extent (Figure 5a). YnPal proteins were enriched and analyzed by LC-MS/MS as described above, and palmitic acid (Pal) controls were run in parallel. Results were filtered to retain only those in at least two of the three replicates and data analyzed by LFQ (Supporting Information Figure S9). This YnPal data set was compared to the YnMyr BSF data set and also cross-compared with a data set of potential palmitoylated proteins identified by Emmer et al.49 in T. brucei PCF using acyl-biotin exchange chemistry (ABE) (Supporting Information Table S5). ABE is an approach complementary to click chemistry for palmitoyl-protein discovery and identifies proteins that bear hydroxylamine-labile linkages at cysteine residues. Protein hits were then categorized in the following way: (1) N-terminal glycine (MG) motif plus detection in the experiments of Emmer et al.; (2) detection by Emmer et al. but no MG motif; (3) MG proteins found in YnMyr data sets only; (4) other MG motif proteins; (5) others (Figure 5b). YnMyr and YnPal intensities of hits, defined as proteins significant in one or both data sets and/or with direct detection of the N-terminal YnMyr modified peptide, were compared by t test (Figure 5b). A high degree of overlap was observed between YnMyr and YnPal data sets: of 134 protein hits, only 3 were exclusive to YnPal samples and 15 to YnMyr samples. Most MG hits were identified either in YnMyr alone (15 proteins) or in both data sets (32 proteins), whereas non-MG hits also identified by Emmer et al. were mostly enriched in YnPal samples compared to YnMyr, as expected (Figure 5b). MG proteins detected as S-acylated by Emmer et al. also identified in the current study include FCaBP, PPEF, and two calpain-like proteins with homology to Leishmania small myristoylated protein 1 (SMP-1), Tb927.1.2230, and Tb927.1.2260; there is published evidence for the S-acylation of these proteins on cysteines proximal to the N-terminal myristoylation site.16,20,50 LFQ analyses also revealed enrichment of known S-palmitoylated protein MCA4.21 Interestingly, other members of this family (MCA3 and MCA1) were also YnPal-enriched hits, suggesting that these interesting pseudoproteases may be S-acylated. Around 70 proteins lacking an MG motif and identified as YnPal hits here were not found by Emmer et al., likely due to stage-specific differences in the proteome (insect stage PCF parasites in Emmer et al. and human stage BSF cultures in the present study), and may also reflect the complementarity of ABE and CuAAC-based chemical proteomics for palmitoylated proteome analysis.
Figure 5.
Comparison of myristate and palmitate tagging in T. brucei BSF. (a) BSF parasites were incubated with YnMyr or YnPal (Figure 2d) probes for 8 h and tagged proteins visualized by in-gel fluorescence after CuAAC. The gel was subject to treatment with NaOH and reimaged to identify base labile bands. (b) Volcano plot comparing YnMyr and YnPal intensity for protein hits (significantly enriched over Myr or Pal controls). Two-sided two-sample permutation-corrected t test (250 permutations; FDR 0.05, s0 1). Proteins are categorized and color coded to indicate if they were identified as palmitoylated by Emmer et al. (Tb PCF palmitoylome) or contain an N-terminal glycine (MG). “YnMyr only” indicates protein not a hit in YnPal analyses. See also Supporting Information Figure S9 and Table S5.
Proteins in the ARF/ARL family are not generally dually acylated, and the large majority were indeed identified only in YnMyr analyses. An interesting exception is the ARF protein Tb927.9.13650 (and Tb927.9.13680, differing by just one amino acid), which was detected in YnPal analyses and contains a cysteine strongly predicted to be palmitoylated (CSS-Palm; Supporting Information Table S5).51 Consistent with gel-based results (Figure 5a), the VSG was also enriched in YnPal samples (Supporting Information Table S5), albeit to a lesser extent than with YnMyr. Although previous data indicate that myristate is specifically incorporated into the final GPI anchor of this protein, intermediates in the GPI anchor remodeling process contain longer chain fatty acids, including potentially stearate (C18:0).31,52 Alternatively, there are other examples of variable lipid labeling in T. brucei, with evidence for promiscuous incorporation and metabolism prior to incorporation of lipids. For example, the S-acylated protein glycosylphosphatidylinositol-specific phospholipase C (GPI-PLC), which is responsible for processing the GPI anchor of VSG and other substrates, can be radiolabeled with myristate, palmitate, or stearate;36 consistent with this, GPI-PLC was detected with almost equal enrichment in our YnMyr and YnPal analyses. In addition, labeling with either [3H]-myristate or [3H]-palmitate results in both myristate and palmitate being present on GPI-PLC, suggesting that trypanosomes can interconvert these fatty acids. Indeed, at least in some culture conditions, myristate is readily chain elongated in T. brucei to palmitate and stearate.53 It is conceivable that YnMyr and YnPal are processed by chain elongation/reduction by the trypanosome fatty acid biosynthetic machinery and incorporated into the VSG and other proteins; because processing occurs at the carboxyl end of the lipid, this would be expected to conserve the alkyne tag. Further work will be required to explore this possibility.
In summary, approximately 100 YnPal tagged targets have been identified here, including both S-acylated and GPI-anchored proteins.
Chemical Knockdown of NMT
NMT has been validated preclinically as a drug target in T. brucei BSF via RNAi knockdown and chemical inhibition of the enzyme, and target engagement of inhibitor with NMT inside parasites was demonstrated through the reduction of protein radiolabeling with [3H]-myristic acid following treatment with a T. brucei NMT inhibitor (NMTi).7 Azido-myristate has also been used to demonstrate that NMTi reduces tagging in the related organism T. cruzi in a dose-dependent manner at the level of in-gel fluorescence,54 although tagged proteins were not identified. Following a strategy similar to our recent studies in human cells12 and L. donovani,23 we aimed to use quantitative proteomics in combination with YnMyr and a TbNMT inhibitor to provide orthogonal evidence for the identity of NMT substrates while simultaneously defining those substrates that may mediate the phenotypes observed on inhibition.
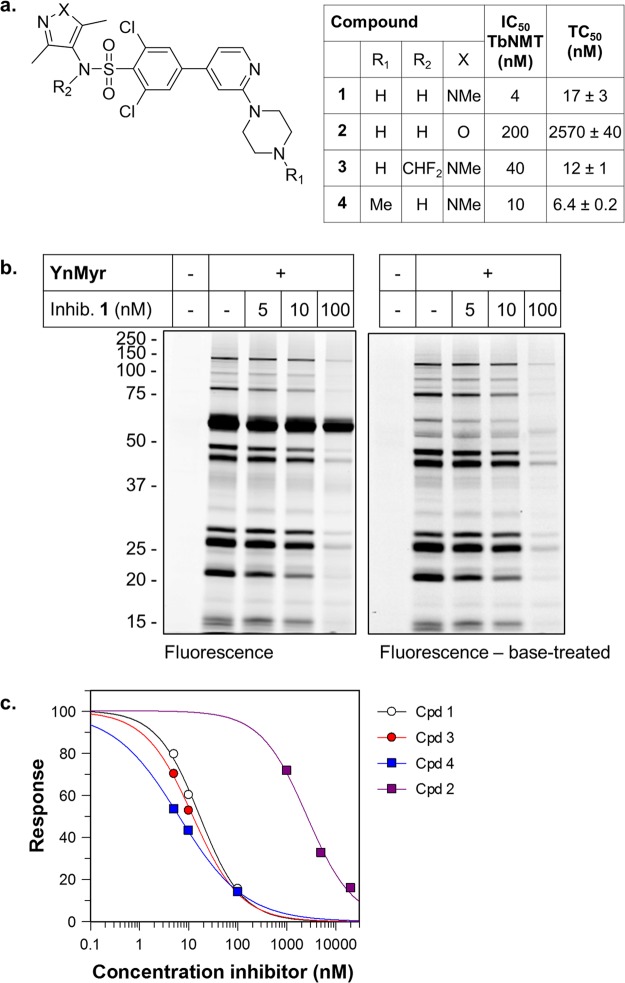
TbNMT inhibitor 1(7) (Figure 6a) was co-incubated with YnMyr in BSF parasites at concentrations ranging from 5 to 100 nM. A dose-dependent drop in labeling of most bands was observed, with the notable exception of the VSG, consistent with target engagement in the parasite (Figure 6b). Inhibitor 1 is reported to have an EC50 of 2 nM on BSF parasites and sub-nanomolar IC50 against TbNMT and is effective in eliminating parasites in a mouse model of trypanosomiasis.7 A small panel of analogues of 1,13a,55 with varying potency against TbNMT (compounds 2–4, Figure 6a), was analyzed using YnMyr tagging. In all cases the same trend toward decreased labeling was observed, with the VSG band remaining largely unaffected by NMT inhibition (Supporting Information Figure S10). Following base treatment, quantification of the decrease in fluorescence intensity suggested intracellular inhibition of N-myristoylation with a TC50 (concentration of compound resulting in a 50% decrease in tagging) in the low nanomolar range for 1, 3, and 4 and in the low micromolar range for 2, in line with the measured enzyme potencies for these compounds (Figure 6a,c). These data are consistent with the on-target action of these inhibitors in parasites and provide further evidence that the majority of base treatment-insensitive YnMyr labeling in T. brucei BSF is NMT-dependent.
Figure 6.
TbNMT inhibitors dose-dependently knock down YnMyr labeling. (a) Structures of previously reported TbNMT inhibitors 1–4, their IC50 values against TbNMT, and tagging-IC50 (concentration of compound required for 50% inhibition of tagging, TC50) calculated based on fluorescent gels. (b) In-gel fluorescence analysis of samples from parasites treated with indicated concentrations of 1 during YnMyr tagging. (c) Quantification of in-gel fluorescence signal from NaOH-treated gels (n = 2–3) of samples from parasites co-incubated with inhibitors 1–4 and YnMyr. See Supporting Information Figure S10 for example gels.
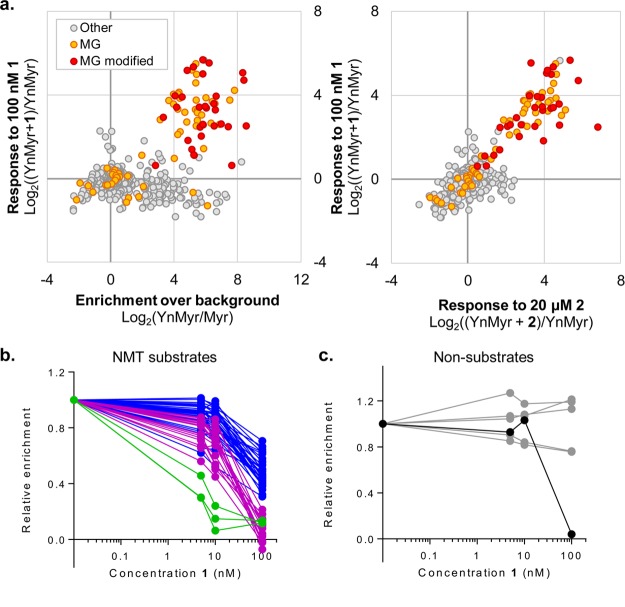
To identify proteins for which acylation levels were selectively affected by NMT inhibition, YnMyr-tagged BSF parasites were treated with 1 at 5, 10, or 100 nM or with 2 at 1, 5, or 20 μM, concentrations designed to probe the range of tagging reduction observed in gels for more and less potent examples of TbNMT inhibitors. The resulting samples (16 total, including replicates) were subject to CuAAC, base treatment to remove GPI-anchor tagging, enrichment, and LC-MS/MS analysis; Myr controls were processed in parallel (Supporting Information Figure S11). Data were analyzed by MaxLFQ,34 and proteins quantified in both no-inhibitor replicates were selected for analysis of enrichment levels over background (YnMyr/Myr) and response to inhibitor (Supporting Information Table S6). A subset of MG proteins, including almost all proteins identified as having a YnMyr-modified N-terminus, were enriched over Myr controls, and this enrichment reduced in response to the highest concentrations of inhibitor (100 nM 1 and 20 μM 2) (Figure 7a). Examination of enrichment ratios (enrichment over Myr controls, normalized to samples with no inhibitor; see Methods for detailed description of data processing and analysis) revealed 54 proteins that responded robustly to the highest concentrations of both inhibitors 1 and 2. Of this subset, only one did not contain an N-terminal glycine. Hierarchical clustering was performed on the 54 putative hits, and 4 clusters of response were defined (Supporting Information Figure S12); protein responses were further examined by plotting enrichment relative to concentration of inhibitor (Table 1; Figure 7b; Supporting Information Figure S13). The sole non-MG protein (Tb927.8.2250, annotated as a putative tRNA ligase) responded only to the highest concentration of NMT inhibitor (Figure 7c) and may be a downstream or low-affinity off-target of the inhibitor. Interestingly, this protein was identified by Emmer et al. as S-acylated in PCF parasites.49 The remaining 53 hits, classed as “high confidence”, showed a variety of robust dose–responses; a range of substrate sensitivity toward NMT inhibition was also observed in human cells and Leishmania parasites.12,23
Figure 7.
Chemical proteomic analysis of YnMyr tagging in the presence of NMT inhibitors. BSF parasites were labeled with YnMyr in the presence of inhibitors 1 (5, 10, 100 nM) and 2 (1, 5, 20 μM). Proteins were subject to CuAAC, base treatment, enrichment, and on-bead digest in technical duplicate and analyzed by LC-MS/MS with quantification by LFQ. (a) Global data visualization after filtering to retain only those proteins quantified in both YnMyr replicates and imputation of missing values (see Methods for details). (Left) Response to 100 nM inhibitor 1 plotted against enrichment over myristic acid (Myr) controls. (Right) Response to inhibitors 1 (100 nM) versus 2 (20 μM). Proteins containing an N-terminal glycine (MG) and those for which the YnMyr-modified peptide was identified using reagent AzRB or AzRTB are indicated. (b) Dose–response plots (treatment with 1) for protein hits. Curves are color-coded based on clustering (degree of response; see Supporting Information Figure S12). (c) Dose–response plots (treatment with 1) for other MG proteins not assigned as hits (gray) and for outlier non-MG protein (Tb927.8.2250, black) that decreases only at high concentrations of inhibitors. See also Supporting Information Table S6 and Figure S13 (dose–response curves for treatment with 2 and for “medium confidence” hits).
Table 1. High- and Medium-Confidence N-Myristoylated Protein Hitsa.

Proteins were identified by quantitative proteomics with YnMyr in combination with NMT inhibitors. Mean normalized enrichment ratios (YnMyr/Myr) are given, and cells are color-coded based on value (blue = 0, red = 1, yellow = 50 percentile). Proteins for which the YnMyr-modified N-terminus was detected (Mod. Pept.) and those identified as hits in PCF samples are indicated. Some protein groups contained multiple proteins (“&others”). See also Supporting Information Table S6.
In addition to the 53 high-confidence hits, a further 10 proteins showed a weaker dose–response to NMT inhibitors, but this group included 7 proteins in which the YnMyr-modified peptide was identified on the N-terminal glycine. The 10 proteins were thus assigned as “medium-confidence” hits and include FCaBP (previously identified as N-myristoylated)20 and putative N-myristoylated protein MCA4.21 The remaining MG proteins, which neither responded in a dose-responsive manner to NMT inhibitors nor had an identified N-terminal modified glycine, were classed as nonsubstrates and included ribosomal proteins and others not expected to be myristoylated. Notably, two widely used bioinformatics tools35,56 for prediction of whether a protein is a likely NMT substrate disagreed for 18 of the high-confidence hits identified here and predicted no myristoylation for an additional 7 (Supporting Information Figure S14 and Table S7). This is not necessarily surprising given that these tools were trained on data sets from other organisms and highlights the value of experimentally identifying NMT substrates.
Proteins for which the level of YnMyr tagging responded in a dose-dependent manner to both NMT inhibitors are highly likely to be NMT substrates (i.e., high-confidence hits). These included Golgi reassembly stacking protein (GRASP), proteasome regulatory ATPase subunit 2 (RPT2), five members of the ADP-ribosylation factor (ARF) family of GTPases, four proteins involved in fatty acyl CoA synthesis, four phosphatases (including PPEF), calpain-like proteins, and many proteins of unknown function. Fifty percent of high-confidence NMT substrate proteins identified here are associated with a loss of fitness in RNAi knockdown experiments in different life stages or conditions (Supporting Information Table S6; Alsford et al.;57 and data extracted from TriTrypDB33) and merit further investigation as substrates with the potential to mediate the antiparasitic effects of TbNMT inhibitors.
Conclusions
Here we have used chemical proteomics and a suite of chemical tools—bioorthogonally tagged fatty acid analogues, CuAAC capture reagents with cleavable moieties for myristoylation site identification, and N-myristoyltransferase inhibitors—to explore protein lipidation in the protozoan pathogen T. brucei. We identify lipidated proteins in bloodstream and procyclic form parasites and report the first comparative quantitative analysis of stage-enriched lipidated proteins. We also show that alkyne palmitate analogue YnPal can be used to tag S-acylated proteins in T. brucei; this analogue should prove a useful tool for further exploring palmitoylacyltransferase enzymes as potential drug targets. Furthermore, as we have previously shown in other organisms,12,23 quantitative chemical proteomics combined with well-characterized enzyme inhibitors proved to be a powerful combination for clearly defining NMT substrates amid the complexity of metabolic tagging. The N-myristoylated proteins identified here are involved in many important cellular processes, and our data sets provide a rich resource for future investigation of the complex and pleiotropic impact of NMT inhibition in T. brucei. Data are available via ProteomeXchange with identifier PXD004053. Finally, tagging of proteins by other mechanisms, for example, incorporation of YnMyr into the GPI anchor of the VSG as shown here, suggests that such analogues may also be useful tools for probing the GPI anchor and lipid metabolism pathways in trypanosomes.
Methods
Chemical Tools
The following chemical tools were synthesized as described previously: YnMyr, YnPal, and AzTB;58 AzMyr and YnTB;26 inhibitors 1 and 2;12 and AzRB and AzRTB.25 Myristic and palmitic acids and all other chemicals were purchased.
Parasite Culture
The T. brucei brucei BSF strain Lister 427 was maintained in vitro at 37 °C with 5% CO2 in HMI-9 medium containing 2 μg/mL geneticin (Invitrogen).59 The Lister 427 strain is monomorphic and has lost the ability to differentiate from long-slender trypomastigotes into the short-stumpy form. Cells were routinely maintained at a density <1 × 106/mL. The T. brucei brucei procyclic strain 449 was maintained in vitro at 26 °C in SDM-79 medium containing 25 μg/mL phleomycin.59 Cells were routinely maintained at a density <1.5 × 107/mL. All culture media contained 10% tetracycline-free fetal bovine serum (Autogen Bioclear).
Metabolic Tagging Experiments
Parasites were metabolically labeled by the addition of 100 μM myristic acid or YnMyr to T. brucei BSF (set up at 2.5 × 105/mL in HMI-9 medium the previous day) or PCF (5 × 106/mL in SDM-79). Cells were then grown for 8 h at 37 °C with 5% CO2 (BSF) or for 18 h at 26 °C (PCF) before harvesting. Parasites were lysed in ice-cold RIPA buffer (50 mM Tris, pH 7.4, 1% NP-40, 1% sodium deoxycholate, 150 mM NaCl, 0.5% SDS, and 1× Complete EDTA-free protease inhibitor cocktail (Roche)), sonicated 3 × 10 s at amplitude 45 with 1 min intervals on ice, and then centrifuged at 16000g for 30 min at 4 °C. For inhibition experiments, parasites were pretreated for 1 h with inhibitors 1–4 at indicated concentrations, and then Myr or YnMyr probe was added for the remaining labeling time (8 h for BSF, 18 h for PCF).
CuAAC Labeling, Pull-down, and Gel-Based Analysis
CuAAC chemistry, pull-down, and gel-based analysis were performed as described previously.23 In brief, proteins were precipitated and resuspended, and CuAAC was performed as described.26 Proteins were enriched on streptavidin or neutravidin-coated beads. Visualization was carried out using an Ettan DIGE Imager (Amersham Biosciences)-Cy3 channel to detect TAMRA-labeled proteins.
Proteomic Sample Preparation and Analysis
Sample preparation for proteomics analysis and LC-MS/MS was carried out as described previously (also detailed in the Supporting Information).12
Data Processing: General Comments
The data were processed with MaxQuant version 1.5.3.8, and the peptides were identified from the MS/MS spectra searched against the TriTrypDB-25 T. brucei TREU927 database using the Andromeda search engine. The VSG protein for the 427 strain was not present in this database, and so initial experiments B1 and B2 (see below) were searched against the T. brucei Lister strain 427. The TriTrypDB sequence for the identified VSG variant Tb427.BES40.22 was appended to the TREU927 FASTA file. Cysteine carbamidomethylation was used as a fixed modification, and methionine oxidation and N-terminal acetylation were used as variable modifications. The false-discovery rate was set to 0.01 for peptides, proteins, and sites. Other parameters were used as preset in the software. “Unique and razor peptides” mode was selected to allow for protein grouping; this calculates ratios from unique and razor peptides (razor peptides are uniquely assigned to protein groups and not to individual proteins). LFQ experiments in MaxQuant were performed using the built-in label-free quantification algorithm (MaxLFQ).34 Data were elaborated using Perseus version 1.5.0.31, Excel, and Graphpad Prism. The data have been deposited to the ProteomeXchange with identifier PXD004053.
Data Analysis
Enrichment LC-MS/MS experimental design for YnMyr samples (for each experiment, both Myr and YnMyr samples were processed; Table 2).
Table 2. Enrichment LC-MS/MS Experimental Design for YnMyr Samples.
| expt | biological sample | reagent | expt | biological sample | reagent | |
|---|---|---|---|---|---|---|
| B1 | BSF1 | AzTB | P1 | PCF1 | AzTB | |
| B2 | BSF2 | AzTB | P2 | PCF2 | AzTB | |
| B3 | BSF2 | AzTB | P3 | PCF1 | AzTB | |
| B4 | BSF2 | AzRB | P4 | PCF2 | AzTB | |
| P5 | PCF2 | AzRTB | ||||
| P6 | PCF1 | AzRB |
BSF Analysis. Data Set: Experiments B1–B4
For the search, “match between runs” was enabled within parameter groups but not between them (parameter groups: Myr, YnMyr). Replicates were grouped together (groups: Myr, YnMyr). The YnMyr protein group was filtered to require three valid values across the four replicates and then filtered to retain only those proteins present in biological duplicate (present in both experiments B1 and B2/3/4). Label-free intensities were logarithmized (base 2), and empty values were imputed with random numbers from a normal distribution, the mean and standard deviation of which were chosen to simulate low-abundance values close to noise level (impute criteria: width 0.1 and down shift 1.8; imputation for each sample individually). A modified t test with permutation-based FDR statistics was applied (250 permutations; FDR 0.001; s0 1) to compare Myr and YnMyr groups.
PCF Analysis. Data Set: Experiments P1–P6
For the search, “match between runs” was enabled within parameter groups but not between them (parameter groups: Myr, YnMyr). Replicates were grouped together (groups: Myr, YnMyr). The YnMyr protein group was filtered to require three valid values across the four replicates and then filtered to retain only those proteins present in biological duplicate (present in both experiments P1/3/6 and P2/4/5). Label-free intensities were logarithmized (base 2) and empty values were imputed with random numbers from a normal distribution (impute criteria: width 0.1 and down shift 1.8; imputation for each sample individually). A modified two-sample two-sided t test with permutation-based FDR statistics was applied (250 permutations; FDR 0.001; s0 1) to compare Myr and YnMyr groups.
BSF and PCF Comparisons. Data Set: Experiments B1–B4 and P1–P6
Data sets were searched together with MaxLFQ. Data were filtered for at least three valid values in at least one group (groups: BSF_Myr, BSF_YnMyr, PCF_Myr, PCF_YnMyr). The total data set was filtered to require three BSF_YnMyr or four PCF_YnMyr valid values and then cross-referenced with individual analyses (described above) to retain hits only. Missing values were imputed (impute criteria: width 0.1 and down shift 1.8) and two-sample t tests used to compare YnMyr intensities across BSF and PCF (250 permutations; FDR 0.01, s0 2). Data were elaborated in Excel and compared to literature data sets.39,40,45
Modified Peptide Analyses. Data Sets: Experiments B4, P5, and P6
MaxQuant searches were carried out as above with the following modifications: the minimum peptide length was reduced to 5. This is because the N-terminus of NMT substrates often contains a lysine residue,35 resulting in short N-terminal tryptic peptides. Modification with YnMyr and the expected portion of AzRB or AzRTB was specified as a variable modification. YnMyr-modified peptide matches were filtered to retain only those with a score (the Andromeda score for the best identified among the MS/MS spectra) >40 and delta score (the score difference to the second best peptide identification) >20.
YnPal and YnMyr Comparisons (BSF). Data Set: YnMyr Experiments B2–B4. YnPal: Three Technical Repeats (A–C, Processing from the Lysate Stage) of One Biological Sample (YnPal and Pal Samples)
Data were searched together by MaxLFQ. Data were filtered for at least two valid values in at least one group (groups: Myr, YnMyr, Pal, YnPal). Label-free intensities were logarithmized (base 2). YnMyr and YnPal data sets were then analyzed separately: filtered (at least two valid values in either Myr or YnMyr; at least two valid values in either Pal or YnPal), missing values were imputed (impute criteria: width 0.1 and down shift 1.8) and two-sample t tests used to define potential hits in each case (250 permutations; FDR 0.05, s0 1). The total data set was then cross-referenced with individual analyses to retain hits only (t test significant proteins and those where modified peptide was identified). Missing values were imputed (impute criteria: width 0.1 and down shift 1.8, across total data set) and two-sample t tests used to compare YnMyr and YnPal intensities (250 permutations; FDR 0.05, s0 1). Data were elaborated in Excel and compared to a literature data set.49
YnMyr Tagging in the Presence of NMT Inhibitors (BSF). Groups (Conditions): YnMyr, Myr, 5 nM 1, 10 nM 1, 100 nM 1, 1 μM 2, 5 μM 2, and 20 μM 2
. All samples were prepared and processed in duplicate (technical replicates A and B from the lysate stage). The “match between runs” option (time window 0.7 min, alignment time window 20 min) in MaxQuant was enabled during the searches. LFQ data were grouped and filtered to retain only those proteins present in both YnMyr replicates. The mean of the technical replicates was calculated, and then missing values were imputed from a normal distribution (width 0.1, downshift 1.8; separately for each data set). Response ratios (YnMyr/(YnMyr + inhibitor)) and enrichment ratios ((YnMyr + inhibitor)/Myr) were calculated, and enrichment ratios were subsequently normalized to YnMyr only (no inhibitor) to compare between proteins and across conditions. Proteins with −Log2((YnMyr + inhibitor)/Myr) > 2 (response ratio > 2) for 100 nM 1 and 20 μM 2 were selected as potential hits. These hits were further analyzed by plotting the dose–response curves (normalized enrichment ratios).
Acknowledgments
This work was supported by grants from the U.K. Engineering and Physical Sciences Research Council (studentship and Doctoral Prize Fellowship awards to M.H.W.), Wellcome Trust (087792), and U.K. Biotechnology and Biological Sciences Research Council (BB/D02014X/1). We thank L. Haigh for assistance with mass spectrometry and W. P. Heal for assistance with the synthesis of inhibitors 1 and 2, and we acknowledge M. Broncel and R. Serwa for AzRB/AzRTB design.
Glossary
Abbreviations
- ARF
ADP ribosylation factor
- BSF
bloodstream form
- CoA
coenzyme A
- CuAAC
copper catalyzed azide–alkyne cycloaddition
- GPI
glycosylphosphatidylinositol
- HAT
human African trypanosomiasis
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- LFQ
label-free quantification (intensity)
- Myr
myristic acid
- NMT
myristoyl-CoA:protein N-myristoyltransferase
- PCF
procyclic form
- RNAi
RNA interference
- VSG
variant surface glycoprotein
- YnMyr
alkynyl-myristate (13-tetradecynoic acid)
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfecdis.6b00034.
Accession Codes
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository60 with the data set identifier PXD004053.
Author Present Address
§ (M.H.W.) Department of Chemistry, Technische Universität München, Garching 85478, Germany.
Author Present Address
⊗ (D.P.) Wellcome Trust Centre for Molecular Parasitology, Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK.
Author Present Address
Δ (H.P.P.) Centre for Applied Entomology and Parasitology, School of Life Sciences, Keele University, Staffordshire ST5 5BG, UK.
Author Contributions
M.H.W. performed sample handling and analysis downstream of parasite culture. D.P. and H.P.P. performed parasite culture and metabolic tagging. D.F.S. and E.W.T. conceived the study. M.H.W. wrote the manuscript with input from all other authors.
The authors declare no competing financial interest.
Supplementary Material
References
- Simarro P. P.; Diarra A.; Ruiz Postigo J. A.; Franco J. R.; Jannin J. G. (2011) The human African trypanosomiasis control and surveillance programme of the World Health Organization 2000–2009: the way forward. PLoS Neglected Trop. Dis. 5 (2), e1007. 10.1371/journal.pntd.0001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simarro P. P.; Cecchi G.; Franco J. R.; Paone M.; Diarra A.; Ruiz-Postigo J. A.; Fevre E. M.; Mattioli R. C.; Jannin J. G. (2012) Estimating and mapping the population at risk of sleeping sickness. PLoS Neglected Trop. Dis. 6 (10), e1859. 10.1371/journal.pntd.0001859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchi G., and Mattioli R. C. (2009) Global Geospatial Datasets for African Trypanosomiasis Management: A Review, pp 1–39, FAO, Rome, Italy. [Google Scholar]
- Nagle A. S.; Khare S.; Kumar A. B.; Supek F.; Buchynskyy A.; Mathison C. J.; Chennamaneni N. K.; Pendem N.; Buckner F. S.; Gelb M. H.; Molteni V. (2014) Recent developments in drug discovery for leishmaniasis and human African trypanosomiasis. Chem. Rev. 114 (22), 11305–11347. 10.1021/cr500365f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H.; Stephens J. L.; Englund P. T. (2007) A fatty-acid synthesis mechanism specialized for parasitism. Nat. Rev. Microbiol. 5 (4), 287–297. 10.1038/nrmicro1617. [DOI] [PubMed] [Google Scholar]
- Wright M. H.; Heal W. P.; Mann D. J.; Tate E. W. (2010) Protein myristoylation in health and disease. J. Chem. Biol. 3 (1), 19–35. 10.1007/s12154-009-0032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frearson J. A.; Brand S.; McElroy S. P.; Cleghorn L. A.; Smid O.; Stojanovski L.; Price H. P.; Guther M. L.; Torrie L. S.; Robinson D. A.; Hallyburton I.; Mpamhanga C. P.; Brannigan J. A.; Wilkinson A. J.; Hodgkinson M.; Hui R.; Qiu W.; Raimi O. G.; van Aalten D. M.; Brenk R.; Gilbert I. H.; Read K. D.; Fairlamb A. H.; Ferguson M. A.; Smith D. F.; Wyatt P. G. (2010) N-myristoyltransferase inhibitors as new leads to treat sleeping sickness. Nature 464 (7289), 728–732. 10.1038/nature08893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopapadakou N. H. (2002) Antifungals targeted to protein modification: focus on protein N-myristoyltransferase. Expert Opin. Invest. Drugs 11 (8), 1117–1125. 10.1517/13543784.11.8.1117. [DOI] [PubMed] [Google Scholar]
- a Brannigan J. A.; Smith B. A.; Yu Z.; Brzozowski A. M.; Hodgkinson M. R.; Maroof A.; Price H. P.; Meier F.; Leatherbarrow R. J.; Tate E. W.; Smith D. F.; Wilkinson A. J. (2010) N-Myristoyltransferase from Leishmania donovani: structural and functional characterisation of a potential drug target for visceral leishmaniasis. J. Mol. Biol. 396 (4), 985–999. 10.1016/j.jmb.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Tate E. W.; Bell A. S.; Rackham M. D.; Wright M. H. (2014) N-Myristoyltransferase as a potential drug target in malaria and leishmaniasis. Parasitology 141 (1), 37–49. 10.1017/S0031182013000450. [DOI] [PubMed] [Google Scholar]
- Wright M. H.; Clough B.; Rackham M. D.; Rangachari K.; Brannigan J. A.; Grainger M.; Moss D. K.; Bottrill A. R.; Heal W. P.; Broncel M.; Serwa R. A.; Brady D.; Mann D. J.; Leatherbarrow R. J.; Tewari R.; Wilkinson A. J.; Holder A. A.; Tate E. W. (2014) Validation of N-myristoyltransferase as an antimalarial drug target using an integrated chemical biology approach. Nat. Chem. 6 (2), 112–121. 10.1038/nchem.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvin B. D.; Li Z.; Villemaine E.; Poole C. B.; Chapman M. S.; Pollastri M. P.; Wyatt P. G.; Carlow C. K. (2014) A target repurposing approach identifies N-myristoyltransferase as a new candidate drug target in filarial nematodes. PLoS Neglected Trop. Dis. 8 (9), e3145. 10.1371/journal.pntd.0003145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thinon E.; Serwa R. A.; Broncel M.; Brannigan J. A.; Brassat U.; Wright M. H.; Heal W. P.; Wilkinson A. J.; Mann D. J.; Tate E. W. (2014) Global profiling of co- and post-translationally N-myristoylated proteomes in human cells. Nat. Commun. 5, 4919. 10.1038/ncomms5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Brand S.; Cleghorn L. A.; McElroy S. P.; Robinson D. A.; Smith V. C.; Hallyburton I.; Harrison J. R.; Norcross N. R.; Spinks D.; Bayliss T.; Norval S.; Stojanovski L.; Torrie L. S.; Frearson J. A.; Brenk R.; Fairlamb A. H.; Ferguson M. A.; Read K. D.; Wyatt P. G.; Gilbert I. H. (2012) Discovery of a novel class of orally active trypanocidal N-myristoyltransferase inhibitors. J. Med. Chem. 55 (1), 140–152. 10.1021/jm201091t. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Brand S.; Norcross N. R.; Thompson S.; Harrison J. R.; Smith V. C.; Robinson D. A.; Torrie L. S.; McElroy S. P.; Hallyburton I.; Norval S.; Scullion P.; Stojanovski L.; Simeons F. R.; van Aalten D.; Frearson J. A.; Brenk R.; Fairlamb A. H.; Ferguson M. A.; Wyatt P. G.; Gilbert I. H.; Read K. D. (2014) Lead optimization of a pyrazole sulfonamide series of Trypanosoma brucei N-myristoyltransferase inhibitors: identification and evaluation of CNS penetrant compounds as potential treatments for stage 2 human African trypanosomiasis. J. Med. Chem. 57 (23), 9855–9869. 10.1021/jm500809c. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Spinks D.; Smith V.; Thompson S.; Robinson D. A.; Luksch T.; Smith A.; Torrie L. S.; McElroy S.; Stojanovski L.; Norval S.; Collie I. T.; Hallyburton I.; Rao B.; Brand S.; Brenk R.; Frearson J. A.; Read K. D.; Wyatt P. G.; Gilbert I. H. (2015) Development of Small-Molecule Trypanosoma brucei N-Myristoyltransferase Inhibitors: Discovery and Optimisation of a Novel Binding Mode. ChemMedChem 10 (11), 1821–1836. 10.1002/cmdc.201500301. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Bell A. S.; Mills J. E.; Williams G. P.; Brannigan J. A.; Wilkinson A. J.; Parkinson T.; Leatherbarrow R. J.; Tate E. W.; Holder A. A.; Smith D. F. (2012) Selective Inhibitors of Protozoan Protein N-myristoyltransferases as Starting Points for Tropical Disease Medicinal Chemistry Programs. PLoS Neglected Trop. Dis. 6 (4), e1625. 10.1371/journal.pntd.0001625. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Hutton J. A.; Goncalves V.; Brannigan J. A.; Paape D.; Wright M. H.; Waugh T. M.; Roberts S. M.; Bell A. S.; Wilkinson A. J.; Smith D. F.; Leatherbarrow R. J.; Tate E. W. (2014) Structure-Based Design of Potent and Selective Leishmania N-Myristoyltransferase Inhibitors. J. Med. Chem. 57 (20), 8664–8670. 10.1021/jm5011397. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Rackham M. D.; Brannigan J. A.; Moss D. K.; Yu Z.; Wilkinson A. J.; Holder A. A.; Tate E. W.; Leatherbarrow R. J. (2013) Discovery of novel and ligand-efficient inhibitors of Plasmodium falciparum and Plasmodium vivax N-myristoyltransferase. J. Med. Chem. 56 (1), 371–375. 10.1021/jm301474t. [DOI] [PMC free article] [PubMed] [Google Scholar]; g Olaleye T. O.; Brannigan J. A.; Roberts S. M.; Leatherbarrow R. J.; Wilkinson A. J.; Tate E. W. (2014) Peptidomimetic inhibitors of N-myristoyltransferase from human malaria and leishmaniasis parasites. Org. Biomol. Chem. 12 (41), 8132–8137. 10.1039/C4OB01669F. [DOI] [PMC free article] [PubMed] [Google Scholar]; h Rackham M. D.; Brannigan J. A.; Rangachari K.; Meister S.; Wilkinson A. J.; Holder A. A.; Leatherbarrow R. J.; Tate E. W. (2014) Design and synthesis of high affinity inhibitors of Plasmodium falciparum and Plasmodium vivax N-myristoyltransferases directed by ligand efficiency dependent lipophilicity (LELP). J. Med. Chem. 57 (6), 2773–2788. 10.1021/jm500066b. [DOI] [PMC free article] [PubMed] [Google Scholar]; i Rackham M. D.; Yu Z.; Brannigan J. A.; Heal W. P.; Paape D.; Barker K. V.; Wilkinson A. J.; Smith D. F.; Leatherbarrow R. J.; Tate E. W. (2015) Discovery of high affinity inhibitors of Leishmania donovani N-myristoyltransferase. MedChemComm 6 (10), 1761–1766. 10.1039/C5MD00241A. [DOI] [PMC free article] [PubMed] [Google Scholar]; j Yu Z.; Brannigan J. A.; Rangachari K.; Heal W. P.; Wilkinson A. J.; Holder A. A.; Leatherbarrow R. J.; Tate E. W. (2015) Discovery of pyridyl-based inhibitors of Plasmodium falciparum N-myristoyltransferase. MedChemComm 6 (10), 1767–1772. 10.1039/C5MD00242G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Price H. P.; Guther M. L.; Ferguson M. A.; Smith D. F. (2010) Myristoyl-CoA:protein N-myristoyltransferase depletion in trypanosomes causes avirulence and endocytic defects. Mol. Biochem. Parasitol. 169 (1), 55–58. 10.1016/j.molbiopara.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Price H. P.; Menon M. R.; Panethymitaki C.; Goulding D.; McKean P. G.; Smith D. F. (2003) Myristoyl-CoA:protein N-myristoyltransferase, an essential enzyme and potential drug target in kinetoplastid parasites. J. Biol. Chem. 278 (9), 7206–7214. 10.1074/jbc.M211391200. [DOI] [PubMed] [Google Scholar]
- a Price H. P.; Goulding D.; Smith D. F. (2005) ARL1 has an essential role in Trypanosoma brucei. Biochem. Soc. Trans. 33 (4), 643–645. 10.1042/BST0330643. [DOI] [PubMed] [Google Scholar]; b Price H. P.; Stark M.; Smith D. F. (2007) Trypanosoma brucei ARF1 plays a central role in endocytosis and golgi-lysosome trafficking. Mol. Biol. Cell 18 (3), 864–873. 10.1091/mbc.E06-08-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Price H. P.; Panethymitaki C.; Goulding D.; Smith D. F. (2005) Functional analysis of TbARL1, an N-myristoylated Golgi protein essential for viability in bloodstream trypanosomes. J. Cell Sci. 118 (4), 831–841. 10.1242/jcs.01624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills E.; Price H. P.; Johner A.; Emerson J. E.; Smith D. F. (2007) Kinetoplastid PPEF phosphatases: dual acylated proteins expressed in the endomembrane system of Leishmania. Mol. Biochem. Parasitol. 152 (1), 22–34. 10.1016/j.molbiopara.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price H. P.; Hodgkinson M. R.; Wright M. H.; Tate E. W.; Smith B. A.; Carrington M.; Stark M.; Smith D. F. (2012) A role for the vesicle-associated tubulin binding protein ARL6 (BBS3) in flagellum extension in Trypanosoma brucei. Biochim. Biophys. Acta, Mol. Cell Res. 1823 (7), 1178–1191. 10.1016/j.bbamcr.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz-Fowler C.; Ersfeld K.; Gull K. (2001) CAP5.5, a life-cycle-regulated, cytoskeleton-associated protein is a member of a novel family of calpain-related proteins in Trypanosoma brucei. Mol. Biochem. Parasitol. 116 (1), 25–34. 10.1016/S0166-6851(01)00296-1. [DOI] [PubMed] [Google Scholar]
- Emmer B. T.; Souther C.; Toriello K. M.; Olson C. L.; Epting C. L.; Engman D. M. (2009) Identification of a palmitoyl acyltransferase required for protein sorting to the flagellar membrane. J. Cell Sci. 122 (6), 867–874. 10.1242/jcs.041764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godsel L. M.; Engman D. M. (1999) Flagellar protein localization mediated by a calcium-myristoyl/palmitoyl switch mechanism. EMBO J. 18 (8), 2057–2065. 10.1093/emboj/18.8.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proto W. R.; Castanys-Munoz E.; Black A.; Tetley L.; Moss C. X.; Juliano L.; Coombs G. H.; Mottram J. C. (2011) Trypanosoma brucei metacaspase 4 is a pseudopeptidase and a virulence factor. J. Biol. Chem. 286 (46), 39914–39925. 10.1074/jbc.M111.292334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Tate E. W.; Kalesh K. A.; Lanyon-Hogg T.; Storck E. M.; Thinon E. (2015) Global profiling of protein lipidation using chemical proteomic technologies. Curr. Opin. Chem. Biol. 24, 48–57. 10.1016/j.cbpa.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Peng T.; Thinon E.; Hang H. C. (2016) Proteomic analysis of fatty-acylated proteins. Curr. Opin. Chem. Biol. 30, 77–86. 10.1016/j.cbpa.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright M. H.; Paape D.; Storck E. M.; Serwa R. A.; Smith D. F.; Tate E. W. (2015) Global analysis of protein N-myristoylation and exploration of N-myristoyltransferase as a drug target in the neglected human pathogen Leishmania donovani. Chem. Biol. 22 (3), 342–354. 10.1016/j.chembiol.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwa R. A.; Abaitua F.; Krause E.; Tate E. W.; O’Hare P. (2015) Systems Analysis of Protein Fatty Acylation in Herpes Simplex Virus-Infected Cells Using Chemical Proteomics. Chem. Biol. 22 (8), 1008–1017. 10.1016/j.chembiol.2015.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broncel M.; Serwa R. A.; Ciepla P.; Krause E.; Dallman M. J.; Magee A. I.; Tate E. W. (2015) Multifunctional reagents for quantitative proteome-wide analysis of protein modification in human cells and dynamic profiling of protein lipidation during vertebrate development. Angew. Chem., Int. Ed. 54 (20), 5948–5951. 10.1002/anie.201500342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heal W. P.; Wright M. H.; Thinon E.; Tate E. W. (2012) Multifunctional protein labeling via enzymatic N-terminal tagging and elaboration by click chemistry. Nat. Protoc. 7 (1), 105–117. 10.1038/nprot.2011.425. [DOI] [PubMed] [Google Scholar]
- Treumann A.; Zitzmann N.; Hulsmeier A.; Prescott A. R.; Almond A.; Sheehan J.; Ferguson M. A. (1997) Structural characterisation of two forms of procyclic acidic repetitive protein expressed by procyclic forms of Trypanosoma brucei. J. Mol. Biol. 269 (4), 529–547. 10.1006/jmbi.1997.1066. [DOI] [PubMed] [Google Scholar]
- a Vassella E.; Butikofer P.; Engstler M.; Jelk J.; Roditi I. (2003) Procyclin null mutants of Trypanosoma brucei express free glycosylphosphatidylinositols on their surface. Mol. Biol. Cell 14 (4), 1308–1318. 10.1091/mbc.E02-10-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Butikofer P.; Ruepp S.; Boschung M.; Roditi I. (1997) ‘GPEET’ procyclin is the major surface protein of procyclic culture forms of Trypanosoma brucei brucei strain 427. Biochem. J. 326 (2), 415–423. 10.1042/bj3260415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen C. L.; Goulding D.; Field M. C. (2003) Clathrin-mediated endocytosis is essential in Trypanosoma brucei. EMBO J. 22 (19), 4991–5002. 10.1093/emboj/cdg481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doering T. L.; Lu T.; Werbovetz K. A.; Gokel G. W.; Hart G. W.; Gordon J. I.; Englund P. T. (1994) Toxicity of myristic acid analogs toward African trypanosomes. Proc. Natl. Acad. Sci. U.S.A. 91 (21), 9735–9739. 10.1073/pnas.91.21.9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson M. A.; Low M. G.; Cross G. A. (1985) Glycosyl-sn-1,2-dimyristylphosphatidylinositol is covalently linked to Trypanosoma brucei variant surface glycoprotein. J. Biol. Chem. 260 (27), 14547–14555. [PubMed] [Google Scholar]
- Cox J.; Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26 (12), 1367–1372. 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Aslett M.; Aurrecoechea C.; Berriman M.; Brestelli J.; Brunk B. P.; Carrington M.; Depledge D. P.; Fischer S.; Gajria B.; Gao X.; Gardner M. J.; Gingle A.; Grant G.; Harb O. S.; Heiges M.; Hertz-Fowler C.; Houston R.; Innamorato F.; Iodice J.; Kissinger J. C.; Kraemer E.; Li W.; Logan F. J.; Miller J. A.; Mitra S.; Myler P. J.; Nayak V.; Pennington C.; Phan I.; Pinney D. F.; Ramasamy G.; Rogers M. B.; Roos D. S.; Ross C.; Sivam D.; Smith D. F.; Srinivasamoorthy G.; Stoeckert C. J. Jr.; Subramanian S.; Thibodeau R.; Tivey A.; Treatman C.; Velarde G.; Wang H. (2010) TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 38 (database issue), D457–D462. 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J.; Hein M. Y.; Luber C. A.; Paron I.; Nagaraj N.; Mann M. (2014) Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell. Proteomics 13 (9), 2513–2526. 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S.; Eisenhaber B.; Eisenhaber F. (2002) N-terminal N-myristoylation of proteins: prediction of substrate proteins from amino acid sequence. J. Mol. Biol. 317 (4), 541–557. 10.1006/jmbi.2002.5426. [DOI] [PubMed] [Google Scholar]
- Armah D. A.; Mensa-Wilmot K. (1999) S-Myristoylation of a glycosylphosphatidylinositol-specific phospholipase C in Trypanosoma brucei. J. Biol. Chem. 274 (9), 5931–5938. 10.1074/jbc.274.9.5931. [DOI] [PubMed] [Google Scholar]
- Kimura A.; Kato Y.; Hirano H. (2012) N-myristoylation of the Rpt2 subunit regulates intracellular localization of the yeast 26S proteasome. Biochemistry 51 (44), 8856–8866. 10.1021/bi3007862. [DOI] [PubMed] [Google Scholar]
- Struck N. S.; de Souza Dias S.; Langer C.; Marti M.; Pearce J. A.; Cowman A. F.; Gilberger T. W. (2005) Re-defining the Golgi complex in Plasmodium falciparum using the novel Golgi marker PfGRASP. J. Cell Sci. 118 (23), 5603–5613. 10.1242/jcs.02673. [DOI] [PubMed] [Google Scholar]
- Butter F.; Bucerius F.; Michel M.; Cicova Z.; Mann M.; Janzen C. J. (2013) Comparative proteomics of two life cycle stages of stable isotope-labeled Trypanosoma brucei reveals novel components of the parasite’s host adaptation machinery. Mol. Cell. Proteomics 12 (1), 172–179. 10.1074/mcp.M112.019224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak M. D.; Guther M. L.; Ferguson M. A. (2012) Comparative SILAC proteomic analysis of Trypanosoma brucei bloodstream and procyclic lifecycle stages. PLoS One 7 (5), e36619. 10.1371/journal.pone.0036619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saada E. A.; Kabututu Z. P.; Lopez M.; Shimogawa M. M.; Langousis G.; Oberholzer M.; Riestra A.; Jonsson Z. O.; Wohlschlegel J. A.; Hill K. L. (2014) Insect stage-specific receptor adenylate cyclases are localized to distinct subdomains of the Trypanosoma brucei flagellar membrane. Eukaryotic Cell 13 (8), 1064–1076. 10.1128/EC.00019-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.; Apagyi K.; McLeavy L.; Ersfeld K. (2010) Expression and cellular localisation of calpain-like proteins in Trypanosoma brucei. Mol. Biochem. Parasitol. 169 (1), 20–26. 10.1016/j.molbiopara.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Urbaniak M. D.; Martin D. M.; Ferguson M. A. (2013) Global quantitative SILAC phosphoproteomics reveals differential phosphorylation is widespread between the procyclic and bloodstream form lifecycle stages of Trypanosoma brucei. J. Proteome Res. 12 (5), 2233–2244. 10.1021/pr400086y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttery D. S.; Poulin B.; Ramaprasad A.; Wall R. J.; Ferguson D. J.; Brady D.; Patzewitz E. M.; Whipple S.; Straschil U.; Wright M. H.; Mohamed A. M.; Radhakrishnan A.; Arold S. T.; Tate E. W.; Holder A. A.; Wickstead B.; Pain A.; Tewari R. (2014) Genome-wide functional analysis of Plasmodium protein phosphatases reveals key regulators of parasite development and differentiation. Cell Host Microbe 16 (1), 128–140. 10.1016/j.chom.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejung M.; Subota I.; Bucerius F.; Dindar G.; Freiwald A.; Engstler M.; Boshart M.; Butter F.; Janzen C. J. (2016) Quantitative Proteomics Uncovers Novel Factors Involved in Developmental Differentiation of Trypanosoma brucei. PLoS Pathog. 12 (2), e1005439. 10.1371/journal.ppat.1005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldston A. M.; Sharma A. I.; Paul K. S.; Engman D. M. (2014) Acylation in trypanosomatids: an essential process and potential drug target. Trends Parasitol. 30 (7), 350–360. 10.1016/j.pt.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron G.; Zhang M. M.; Yount J. S.; Wilson J.; Raghavan A. S.; Shamir E.; Hang H. C. (2009) Robust fluorescent detection of protein fatty-acylation with chemical reporters. J. Am. Chem. Soc. 131 (13), 4967–4975. 10.1021/ja810122f. [DOI] [PubMed] [Google Scholar]
- Lee S. H.; Stephens J. L.; Paul K. S.; Englund P. T. (2006) Fatty acid synthesis by elongases in trypanosomes. Cell 126 (4), 691–699. 10.1016/j.cell.2006.06.045. [DOI] [PubMed] [Google Scholar]
- Emmer B. T.; Nakayasu E. S.; Souther C.; Choi H.; Sobreira T. J.; Epting C. L.; Nesvizhskii A. I.; Almeida I. C.; Engman D. M. (2011) Global analysis of protein palmitoylation in African trypanosomes. Eukaryotic Cell 10 (3), 455–463. 10.1128/EC.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tull D.; Vince J. E.; Callaghan J. M.; Naderer T.; Spurck T.; McFadden G. I.; Currie G.; Ferguson K.; Bacic A.; McConville M. J. (2004) SMP-1, a member of a new family of small myristoylated proteins in kinetoplastid parasites, is targeted to the flagellum membrane in Leishmania. Mol. Biol. Cell 15 (11), 4775–4786. 10.1091/mbc.E04-06-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J.; Wen L.; Gao X.; Jin C.; Xue Y.; Yao X. (2008) CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng., Des. Sel. 21 (11), 639–644. 10.1093/protein/gzn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterson W. J.; Raper J.; Doering T. L.; Hart G. W.; Englund P. T. (1990) Fatty acid remodeling: a novel reaction sequence in the biosynthesis of trypanosome glycosyl phosphatidylinositol membrane anchors. Cell 62 (1), 73–80. 10.1016/0092-8674(90)90241-6. [DOI] [PubMed] [Google Scholar]
- Doering T. L.; Pessin M. S.; Hoff E. F.; Hart G. W.; Raben D. M.; Englund P. T. (1993) Trypanosome metabolism of myristate, the fatty acid required for the variant surface glycoprotein membrane anchor. J. Biol. Chem. 268 (13), 9215–9222. [PubMed] [Google Scholar]
- Roberts A. J.; Torrie L. S.; Wyllie S.; Fairlamb A. H. (2014) Biochemical and genetic characterization of Trypanosoma cruzi N-myristoyltransferase. Biochem. J. 459 (2), 323–332. 10.1042/BJ20131033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand S., and Wyatt P. (2010) N-Myristoyl transferase inhibitors. WO2010026365 A1.
- Bologna G.; Yvon C.; Duvaud S.; Veuthey A. L. (2004) N-Terminal myristoylation predictions by ensembles of neural networks. Proteomics 4 (6), 1626–1632. 10.1002/pmic.200300783. [DOI] [PubMed] [Google Scholar]
- Alsford S.; Turner D. J.; Obado S. O.; Sanchez-Flores A.; Glover L.; Berriman M.; Hertz-Fowler C.; Horn D. (2011) High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 21 (6), 915–924. 10.1101/gr.115089.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heal W. P.; Jovanovic B.; Bessin S.; Wright M. H.; Magee A. I.; Tate E. W. (2011) Bioorthogonal chemical tagging of protein cholesterylation in living cells. Chem. Commun. (Cambridge, U. K.) 47 (14), 4081–4083. 10.1039/c0cc04710d. [DOI] [PubMed] [Google Scholar]
- Wirtz E.; Leal S.; Ochatt C.; Cross G. A. (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99 (1), 89–101. 10.1016/S0166-6851(99)00002-X. [DOI] [PubMed] [Google Scholar]
- Vizcaino J. A.; Cote R. G.; Csordas A.; Dianes J. A.; Fabregat A.; Foster J. M.; Griss J.; Alpi E.; Birim M.; Contell J.; O’Kelly G.; Schoenegger A.; Ovelleiro D.; Perez-Riverol Y.; Reisinger F.; Rios D.; Wang R.; Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41 (database issue), D1063–D1069. 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.