Abstract
Caveolins mediate the formation of caveolae, which are small omega-shaped membrane invaginations involved in a variety of cellular processes. There are three caveolin isoforms, the third of which (Cav3) is expressed in smooth and skeletal muscles. Mutations in Cav3 cause a variety of human muscular diseases. In this work, we characterize the secondary structure, dynamics, and topology of the monomeric form of the full-length lipidated protein. Cav3 consists of a series of membrane-embedded or surface-associated helical elements connected by extramembrane connecting loops or disordered domains. Our results also reveal that the N-terminal domain undergoes a large scale pH-mediated topological rearrangement between soluble and membrane-anchored forms. Considering that roughly one-third of pathogenic mutations in Cav3 influence charged residues located in this domain, we hypothesize that this transition is likely to be relevant to the molecular basis of Cav3-linked diseases. These results provide insight into the structure of Cav3 and set the stage for mechanistic investigations of the effects of pathogenic mutations.
Introduction
Caveolae are present in the plasma membranes of a wide variety of mammalian cells. These small omega-shaped invaginations are enriched in glycosphingolipids, cholesterol, and lipid-anchored membrane proteins (1, 2), and support a wide range of cellular processes including endocytosis (3, 4), cellular signaling (5, 6), lipid metabolism (7, 8), and mechanosensation (9). The formation of caveolae is critically dependent on caveolin proteins, which include Caveolin 1 (Cav1), Caveolin 2 (Cav2), and Caveolin3 (Cav3) (10, 11, 12). Cav1 and Cav2 are broadly coexpressed in a variety of tissues including adipocytes, endothelia, and type-1 pneumocytes. On the other hand, Cav3 is the sole caveolin isoform expressed in skeletal and smooth muscle tissue (12, 13). Pathogenic mutations within the Cav3 gene are responsible for a variety of neuromuscular and cardiac disorders including limb-girdle muscular dystrophy 1-C, inherited rippling muscle disease, distal myopathy, idiopathic persistent elevation of serum creatine kinase, and familial hypertrophic cardiomyopathy (14, 15, 16, 17). Thus, insight into the structure and function of Cav3 is of direct relevance to human health and disease.
Although Cav1 is the most extensively characterized caveolin protein (18, 19, 20), the three isoforms are highly homologous (Cav3 is 65% identical and 85% similar to Cav1), indicating they are likely to adopt a similar topology and structure. Caveolin proteins are traditionally divided into four structural domains: a flexible N-terminal domain (residues 1–54), a caveolin scaffolding domain (55–74), a membrane embedded domain (75–106), and a C-terminal cytosolic domain (107–151) (21). The cytosolic N-terminal domain contains a highly conserved caveolin signature motif (residues 41–48, FEDVIAEP). The caveolin scaffolding domain is believed to mediate membrane association, cholesterol binding, oligomerization, and the formation of various protein-protein interactions (5, 22, 23, 24). The membrane-embedded domain consists of a helix-break-helix motif that enters and exits the inner membrane leaflet (25, 26). Finally, the cytosolic C-terminal domain of Cav1 was recently found to contain one long α-helix (27). This domain also contains three cysteine residues (residues 106, 116, and 129) known to undergo palmitoylation in Cav1 (28, 29). These cysteines are conserved in Cav3, which is also known to be palmitoylated (30), though the precise palmitoylation pattern has yet to be determined. Although such lipid modifications often mediate important biological or biophysical processes (31, 32, 33, 34, 35, 36, 37, 38), we recently found that lipid modifications have minimal influence on the structure of Cav3 in 1-palmitoyl-2-hydroxy-sn-glycero-3-phosphoglycerol (LPPG) micelles, conditions in which the protein appears to be monomeric (39), as described further below. Although various aspects of the secondary structure and topology of caveolin proteins have been previously characterized (19, 20, 26, 40, 41), a variety of questions regarding the structural properties of the monomer and the means by which they mediate the formation of oligomeric caveolae complexes remain unanswered.
Oligomerization of caveolin proteins is essential for the formation of caveolae membrane coats (10). However, the biologically relevant oligomeric states of caveolins remain the subject of some debate. Investigations involving a C-terminally truncated Cav1 construct provided evidence that caveolins form a heptameric oligomer (42). Alternative evidence has instead suggested Cav1 forms either a nonamer (43) or a 14–16 mer (24, 44). Though the precise nature of the oligomeric unit is unclear, the assembly oligomers is believed to result in the condensation of as many as 180 caveolin proteins in a single caveolae (45). The identity of the caveolin domain(s) mediating homooligomerization is also the subject of debate. Lateral interactions between caveolin scaffolding domains were initially believed to drive oligomerization (42, 46). However, a recent study of Cav3 has suggested that C-terminal interactions instead drive nonamerization (43). Regardless of the biochemically relevant oligomeric states, the structural properties of caveolin monomers must provide the initial driving force for the formation of oligomers. We previously identified favorable conditions for the characterization of full-length, monomeric Cav3 using solution NMR and described the influence of lipid modifications on its structural properties (39). In this study, we characterize the secondary structure, dynamics, and topology of a lipidated full-length human Cav3 construct. Furthermore, we demonstrate that the N-terminal domain undergoes a dramatic topological rearrangement in both micelles and vesicles that is reversibly mediated by pH. Given that a number of pathogenic mutations occur within this region, we suggest that the disruption of this conformational switch may be relevant to the molecular basis of disease.
Materials and Methods
Expression and purification of Cav3
Most of the experiments of this work employed a mutant form of Cav3 in which six of the native cysteine sites were mutated to Ala or Ser (C19S, C72A, C94A, C98A, C124S, and C140S). The fifth, sixth, and eighth Cys sites (C106, C116, and C129) were either lipidated (C116 and C129) or mutated to Phe (C106). This doubly lipidated mutant was previously found to yield a higher quality NMR spectra than the Cav3 mutant in which all three natively palmitoylated Cys sites were retained (39). Cloning, expression, purification of the doubly lipidated Cav3 variant into LPPG micelles, and lipidation were carried out as previously described (39). For the studies of this work lipidation involved attachment of a thiooctyl groups to the two Cys sites through a disulfide bond.
Solution NMR of Cav3 in POPG vesicles
For reconstitution of Cav3 into lipid vesicles, the protein was expressed, purified, and lipidated as previously described (39), except the protein was exchanged into sodium dodecyl sulfate (SDS) on the Ni-NTA column just before the final step of purification by equilibrating the Cav3 attached to the Ni-NTA resin with 12 column volumes of 25 mM sodium phosphate (pH 7.2) containing 0.5% SDS. The protein was then eluted with 25 mM sodium phosphate (pH 7.9) containing 250 mM imidazole and 0.5% SDS.
To make a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylglycerol (POPG) mixed micelle stock solution for use in the reconstitution protocol, 50 mM HEPES (pH 6.5) containing 300 mM NaCl, 1 mM EDTA, 50 mM POPG, and 250 mM SDS in the buffer was repeatedly flash-frozen in liquid nitrogen and thawed in a water bath sonicator until the solution clarified. To initiate reconstitution the POPG mixed micelle stock added to a solution of purified lipidated Cav3 to a final protein: lipid molar ratio of 1:250. The solution was then transferred to dialysis tubing and dialyzed against 4 L of 10 mM imidazole (pH 6.5) containing 0.5 mM EDTA, with stirring and multiple changes of buffer for 5 days. The vesicular sample was then concentrated and exchanged into 75 mM imidazole (pH 7.0) containing 25 mM sodium acetate and 0.5 mM EDTA. After collection of the first 1H 15N TROSY spectrum under this condition, the pH of the solution was subsequently adjusted to 4.5 with the addition of acetic acid. After acquisition of the second 1H 15N TROSY spectrum under this condition, the sample pH was adjusted back to pH 7.0 with NaOH before acquisition of the third 1H 15N TROSY spectrum.
Fluorescence measurements of Cav3 in POPG vesicles
Single-cysteine variants of 568F Cav3 (C19S, C72A, C94A, C98A, C124S, and C140S, with the three natively lipidated Cys sites mutated to Phe) were eluted in 250 mM imidazole, pH 7.8 containing 0.2% LPPG and concentrated to a final volume of 1200 μl. EDTA and Tris(2-carboxyethyl)phosphine were then added to final concentrations of 1 mM and 3 mM, respectively. Monobromobimane (mBBr) was then added to each protein sample to a final concentration of 10 mM, and the mixture was incubated at room temperature for 24 h. Excess mBBr and imidazole was removed using a 10 mL Econo-Pac 10DG buffer exchange column preequilibrated with 25 mM sodium phosphate buffer (pH 7.2) containing 0.2% LPPG. The protein was eluted within the first 4 ml of buffer, as judged by A280. The protein sample was then tumbled for 1 h with Ni-NTA resin that had been equilibrated in 25 mM sodium phosphate (pH 7.2). The protein was then eluted into SDS micelles and reconstituted into POPG vesicles as described previously. Proteoliposomes solutions were then diluted to a final protein concentration of 2 μM in a solution containing 15 mM acetic acid, 15 mM (2-(N-morpholino)ethanesulfonic acid) (MES), 30 mM Tris, 120 mM NaCl, and 0.5 mM EDTA at the indicated final pH (ranging from 3.5 to 9.0) (47). Fluorescence spectra were collected on a Jobin Yvon Fluoromax-3 (Horiba Scientific, Edison, NJ) at 25°C using an excitation wavelength of 384 nm.
Specific amino acid labeling of Cav3
Cav3 bearing specific isotopically labeled amino acids were produced to assist with the assignment of backbone amide 1H,15N NMR resonances. To produce these proteins, Cav3 was expressed in an auxotrophic Escherichia coli stain (CT19), which lacks the aspC, avtA, ilvE, trpB, and tyrB transaminase genes (48). Transformed CT19 cells were grown at 22°C with agitation (230 rpm) in 1 L of M9 media supplemented with 10 mg/ L ampicillin, 100 mg/L kanamycin, 20 mg/L tetracycline, glucose, minimum essential medium (MEM) vitamins, CaCl2, MgSO4, and 14N auxotrophic amino acids. Cells were harvested at OD600 = 0.6 by centrifugation at 3000 rpm for 15 min, and the cell pellet was resuspended in 1 L of M9 medium containing the same components except 0.2 g of the 15N-labeled target amino acid in place of the corresponding 14N amino acid. After 15 min of shaking at 22°C, Cav3 expression was then induced with the addition of 1 mM isopropyl-b-D-thiogalactoside to the culture media. Expression was allowed to proceed for 16 h, at which point the cells were harvested by centrifugation and the Cav3 was purified and prepared for NMR using the standard method (39). 14N-Cys/Ser/Thr-15N-Cav3 was also prepared using a similar protocol by inducing in M9 medium supplemented with 15N-NH4Cl and an excess of 14N-Cys/Ser/Thr (2 g/L M9).
Backbone resonance assignments and secondary structure prediction
Three-dimensional (3D) NMR data were recorded at 45°C on Bruker 800 or 900 NMR spectrometers equipped with cryoprobes. The sequential backbone resonance assignments for U-15N,13C-labeled Cav3 were carried out using TROSY-based triple resonance experiments (49, 50, 51), including HNCA, HNCACB, HN(CO)CA, HN(COCA)CB, HNCO, HN(CA)CO, HNCANNH, and 1H-15N NOESY-HSQC pulse sequences. NMR data were processed using NMRPipe (52) and analyzed using NMRView or Sparky software (53). Sequential assignments were established by assessing bond connectivities. α-Carbon connectivity was determined using a combination of HNCA and HN(CO)CA measurements. β-Carbon connectivity was determined using a combination of HN(CA)CB and HN(COCA)CB measurements. Finally, carbonyl carbon connectivity was determined using a combination of HN(CA)CO and HNCO measurements. To assist in the assignment of remaining unassigned peaks, a reference TROSY spectrum was compared with that of various single site Cav3 mutants including S61A, T67A, W70A, K108A, S109A, I114A, S118A, N122A, S124A, R126A, and T127A. The assigned Cα, Cβ, N, HN, and CO resonances of lipidated Cav3 were used to determine the secondary structure using chemical shift index (CSI) and Talos-N (54, 55).
Probe-induced paramagnetic peak reduction to probe membrane topology
The 900 MHz 1H 15N-TROSY spectrum of lipidated Cav3 was collected in the presence of either lipophilic 16-doxylstearic acid (16-DSA) or Gd(III)-DTPA at 45°C. To incorporate 16-DSA into NMR samples, a stock solution was first prepared by dissolving 16-DSA in methanol to a final concentration of 2.5 mg/ mL. An amount of the 16-DSA stock needed to achieve a final concentration of 4 mol % in the micelle phase was then dispensed into an empty NMR tube, followed by evaporation of the methanol. The micellar Cav3 sample was then transferred directly into the NMR tube to solubilize the 16-DSA. The pH of NMR samples was adjusted to 7.2 or 5.5 with the addition of NaOH before the acquisition of 1H 15N-TROSY spectra. NMR samples contained 180 uM 15N-lipidated Cav3 in a buffer containing 80 mM imidazole, 20 mM MES, 5% (w/v) LPPG, 1 mM EDTA, 10% D2O, and the indicated concentration of paramagnetic reagent. The ratios of peak heights in the spectrum from a paramagnetic sample to the heights of the corresponding peaks from a condition-matched diamagnetic sample were measured.
NMR relaxation analysis
To probe the dynamics of lipidated Cav3, the longitudinal relaxation time (T1) and transverse relaxation time (T2) of backbone 15N nuclei as well as the backbone heteronuclear NOEs (hetNOE) were assessed using established TROSY-based NMR experiments (56). Relaxation experiments were carried out at 800 MHz (18.7 Tesla). T1 values were measured from 1H-15N correlation spectra recorded with relaxation evolution delays of 10, 50, 100, 250, 500, 800, 1200, 1600, 2000, 4000, and 8000 ms. A 5 s delay was used between scans. T2 values were measured from 1H-15N correlation spectra recorded with relaxation evolution delays of 17, 34, 51, 68, 85, 102, 136, 170 and 204 ms. A delay of 4 s was used between scans. The steady-state 1H-15N heteronuclear NOE values were determined by calculating the differences in peak intensity between spectra collected with and without a 3 s presaturation of the proton dimension. To compare the dynamics of lipidated Cav3 at pH 5.5 and 6.5, T2 values of backbone of 15N nuclei were determined using an HSQC-based NMR experiment (57) at 900 MHz using relaxation evolution delays of 16, 32, 48, 64, 80, 105, 144, and 192 ms. A delay of 3.5 s was used between scans. The spectra were processed using NMRPipe and analyzed using Sparky or NMRView.
Far-ultraviolet circular dichroism
Circular dichroism (CD) spectra were acquired using a Jasco J-810 spectropolarimeter (Jasco, Easton, MD) equipped with a Peltier temperature control. Lipidated Cav3 was purified into LPPG micelles as described previously and exchanged into 25 mM sodium phosphate (pH 6.5) containing 0.2% (w/v) LPPG. The protein stock was then diluted to a final protein concentration of 10 μM in 25 mM sodium phosphate buffer with varying pH containing 0.2% (w/v) LPPG. CD spectra were acquired at 25°C using a 1 mm pathlength cuvette.
Results
Structural characterization of lipidated Cav3 by solution NMR
We recently described the initial characterization of a modified form of full length human Cav3 bearing three lipid modifications that mimic the putative palmitoylated form of the protein (39). These studies were carried out in LPPG micelles (see Fig. 1), because this medium was found to yield much better NMR spectra of the protein than other micelle and bicelle compositions tested. To enable specific lipidation of Cav3 at its putative palmitoylation sites (C106, C116, and C129), conservative mutations were made at the native cysteines predicted to fall within the soluble portion (C19S, C124S, and C140S) and membrane-embedded portion (C72A, C92A, and C98A) of Cav3. The lipidated form of this protein exhibited favorable properties for structural analysis by solution NMR spectroscopy (39). However, the yield of this construct was insufficient for the 3D NMR experiments needed to complete assignment of the 1H 15N TROSY spectrum. To circumvent this limitation, we mutated the first putative lipidation site to phenylalanine (C106F) while maintaining the other two lipidated cysteines (C116 and C129). This construct exhibited consistently improved yields relative to the original construct. Furthermore, the 1H dimension of the 1H 15N TROSY spectra were virtually identical regardless of whether Cav3 was modified with two or three acyl chains, which suggests that the structural properties of these constructs are quite similar. For these reasons, we used the doubly lipidated construct for these investigations, which will be referred to as Cav3 from this point forward.
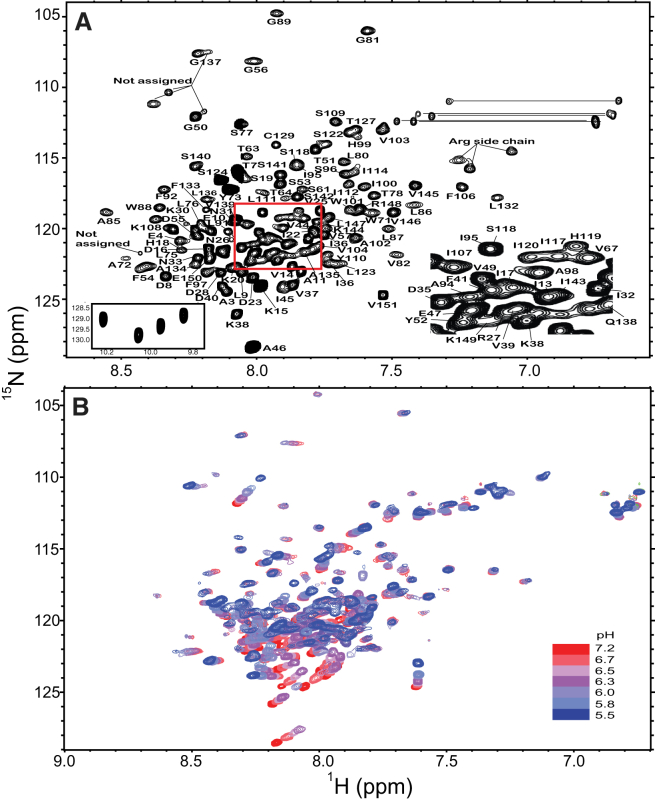
Figure 1.
Two-dimensional solution NMR spectra of full-length lipidated Cav3 in LPPG micelles. (A) The 1H 15N TROSY-HSQC spectrum of lipidated Cav3 in 100 mM imidazole (pH 6.5) containing 1 mM EDTA and 5% LPPG at 318 K and 800 MHz is shown. Peaks corresponding to Asn or Gln side-chain NH2 groups are connected with a line for reference. The red box highlights the region magnified in the right inset. The tryptophan side-chain peaks are shown in the left inset. The five amide peaks that could not be assigned are marked for reference. (B) The 1H 15N HSQC spectra of lipidated Cav3 in 75 mM imidazole containing 25 mM MES, 1 mM EDTA, and 5% LPPG are shown as a function of varying pH. To see this figure in color, go online.
The backbone amide resonances of lipidated Cav3 in the 1H 15N TROSY spectrum exhibit narrow dispersion (Fig. 1 A), suggesting that the protein is largely α-helical or disordered. Nevertheless, we succeeded in assigning 119 of 144 expected backbone resonances using 3D pulse sequences in conjunction with selective amino acid labeling (Fig. S1 in the Supporting Material) (Biological Magnetic Resonance Bank (BMRB):19903). Five peaks in the 1H 15N TROSY spectrum could not be assigned because the corresponding resonances were not observed in the 3D spectra. The remaining resonances could not be confidently identified or assigned due to line broadening. Based on these data, Talos-N and CSI analyses (54, 58) confirm that lipidated Cav3 contains both random coil domains (N-terminal domain and extreme C-terminus) and a series of α-helices: helix 1 (res. 55–80), helix 2 (84–98), helix 3 (106–113), helix 4 (117–120), helix5 (125–128), and helix 6 (132–146) (Fig. S2). Of importance, these predictions are consistent with the presence of a membrane-embedded helix-break-helix motif that is also present in Cav1 (25). Solution NMR dynamics measurements (T1, T2, and steady-state NOEs) reveal an enhanced mobility of the N-terminal domain (residues 1–55) relative to the micelle-associated portions of Cav3 (Fig. 2), which suggests this region may be dissociated from the micelle complex under this condition. Taken together, these results confirm the presence of a micelle-embedded helix-break-helix motif and suggest that the N-terminal domain is a structurally independent soluble domain.
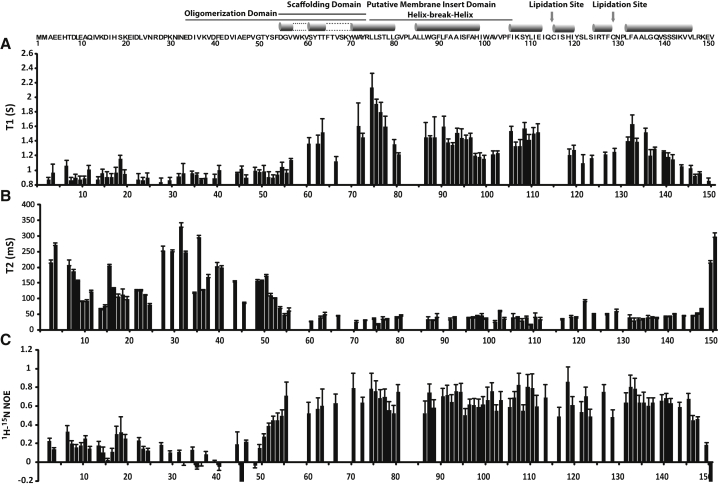
Figure 2.
Backbone dynamics of lipidated Cav3 in LPPG micelles. The dynamics of lipidated Cav3 in LPPG micelles at 45°C was probed by solution NMR. The longitudinal relaxation time (T1, A), transverse relaxation time (T2, B), and steady-state 1H 15N NOEs (C) were measured using an 800 MHz spectrometer at pH 6.5 and 318 K. Blank regions indicate residues that could not be assigned. Error bars for T1 and T2 reflect the uncertainties associated with the fitting of relaxation data. Error bars for 1H-15N NOE values reflect the spectral noise.
Relaxation measurements also provide insight into the oligomeric state of Cav3. Based on their low transverse relaxation times (T2) and high 1H 15N NOE values, a continuous stretch of the micelle-associated residues (72 to 112) appears to be uniformly immobile. We determined effective rotational correlation times for each of the observed peaks in this range from the T1/T2 ratio (57). Assuming a spherical aggregate, the average correlation times determined from these peaks (15.4 ± 2.7 ns) would imply an aggregate molecular mass in the range of 45 kDa. This is smaller than expected for the Cav3/ LPPG complex considering LPPG alone forms micelles in the range of 60–85 kDa (59, 60). This underestimation of the aggregate molecular mass could potentially be related to one or more of the following factors: 1) residual mobility in the micelle-associated portions of Cav3, 2) a nonspherical geometry of the protein-micelle complex, or 3) a potentially reduced LPPG aggregation number in the protein-micelle complex. Nevertheless, these results strongly suggest the observed signal is derived from a low-mass, monomeric form of Cav3. That Cav3 maintains a monomeric state is likely due to its effective dilution in the micelle phase (∼4 micelles per protein under these conditions) and/or the destabilization of oligomers by the anionic headgroup of the LPPG detergent.
Topological heterogeneity of lipidated Cav3 at pH 6.5
To probe the topological features of lipidated Cav3, we first characterized the accessibility of backbone amides to methylene hydrogens within the core of the LPPG micelle and/or to water using 15N-edited NOESY-HSQC. NOEs could be detected between detergent methylenes and many backbone amide hydrogens within each of the helical portions of Cav3 (Fig. 3 A), suggesting these regions of the molecule are either embedded within the micelle or associated with the micelle surface. The presence of detergent NOEs in the oligomerization domain, CSD, and membrane embedded domain are consistent with previous reports suggesting these residues are associated with the membrane (26, 61). Several of these residues within each helix are also in contact with water, indicating that the helices lie within the micelle interface or that water can penetrate the micelle to some extent. In contrast with the helical portions of Cav3, the amide hydrogens in the N-terminal domain only exhibited NOEs with water (due to exchange), which is consistent with its dissociation from the micelle complex under this condition. To confirm these topological features, we characterized the susceptibility of backbone amides to paramagnetic relaxation enhancement (PRE) from water-soluble (Gd-DTPA) or lipophilic (16-DSA) paramagnetic agents. Each of the helical portions of Cav3 were largely protected from Gd-DTPA (Fig. 3 B), which confirms that these segments of Cav3 are associated with the micelle surface or embedded within the micelle core. Indeed, PRE-induced peak broadening was most pronounced within helical regions in the presence of the detergent-soluble 16-DSA paramagnet (Fig. 3 B). Notably, the seemingly disordered N-terminal domain exhibits partial accessibility to both Gd-DTPA and 16-DSA (Fig. 3 B), which suggest this domain may exhibit heterogeneous topological properties under this condition. Taken together, these results suggest that Cav3 persists as a dynamic structural ensemble under these conditions in which the helical portions are associated with the micelle while the N-terminal domain populates both surface-associated and water-soluble states.
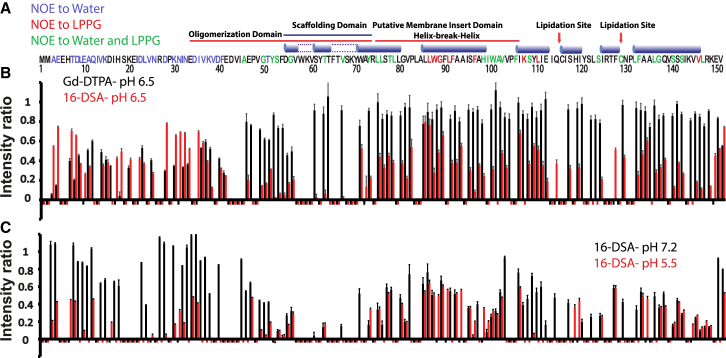
Figure 3.
Solvent NOEs and paramagnetic relaxation enhancement of backbone amide resonances by paramagnetic probes. (A) An 15N-edited NOESY-HSQC spectrum of lipidated Cav-3 was acquired and analyzed under conditions of 100 mM imidazole (pH 6.5) containing 1 mM EDTA and 5% LPPG at 45°C. Backbone amide residues that exhibited detectable NOEs with water (blue), LPPG methylene hydrogens (red), or both (green) are indicated in the amino acid sequence. Residues for which NOEs could not be clearly interpreted are shown in black. The 1H 15N HSQC spectrum of lipidated Cav3 was recorded in the presence and absence of water soluble (Gd-DTPA) or lipophilic (16-DSA) paramagnets under various conditions. (B) The intensity of backbone amide peaks in the presence of 10 mM Gd-DTPA (black) or 4 mol % 16-DSA (red) relative to those collected in the absence of paramagnetic reagents in 100 mM imidazole (pH 6.5), 1 mM EDTA, and 5% LPPG are shown for each residue. Negative peaks denote resonances for which the ratio could not be accurately determined. (C) The intensity of backbone amide peaks in the presence of 4 mol % 16-DSA relative to those collected in the absence of paramagnetic reagents are shown at pH 5.5 (red) and pH 7.2 (black). Negative bars denote residues for which the ratio could not be accurately determined. To see this figure in color, go online.
A pH-mediated conformational change in Cav3
There are 20 ionizable side chains within the N-terminal domain (pI = 4.2). We hypothesized that the ratio of the two conformational states observed at pH 6.5 may depend upon the charge state of the N-terminal domain. We therefore compared the 1H 15N HSQC spectrum of lipidated Cav3 under conditions with varying pH. Large and widespread chemical shift perturbations were apparent between pH 5.5 and 7.2 (Fig. 1 B), which confirms that the conformational ensemble of Cav3 is quite sensitive to pH. To determine whether this observation could potentially account for the topological heterogeneity observed at pH 6.5, we compared the susceptibility of backbone amides to paramagnetic relaxation enhancement by the membrane-permeable paramagnet 16-DSA at both pH 5.5 and pH 7.2. This proved feasible even though, as a consequence of peak broadening, only 73 residues could be confidently assigned in TROSY-HSQC spectrum at pH 5.5. PREs in the membrane-embedded domain characterized at pH 6.5 were quite similar to those under each of these conditions (Fig. 3 C). However, PREs from the lipophilic 16-DSA in the N-terminal domain were significantly enhanced at low pH (Fig. 3 C), suggesting this domain associates with the micelle surface under this condition. This interpretation is consistent with an observed reduction of the T2 values of N-terminal residues at pH 5.5 (Fig. S3). This reduction in backbone motions is consistent with the association of the N-terminal domain with the micelle interface at low pH.
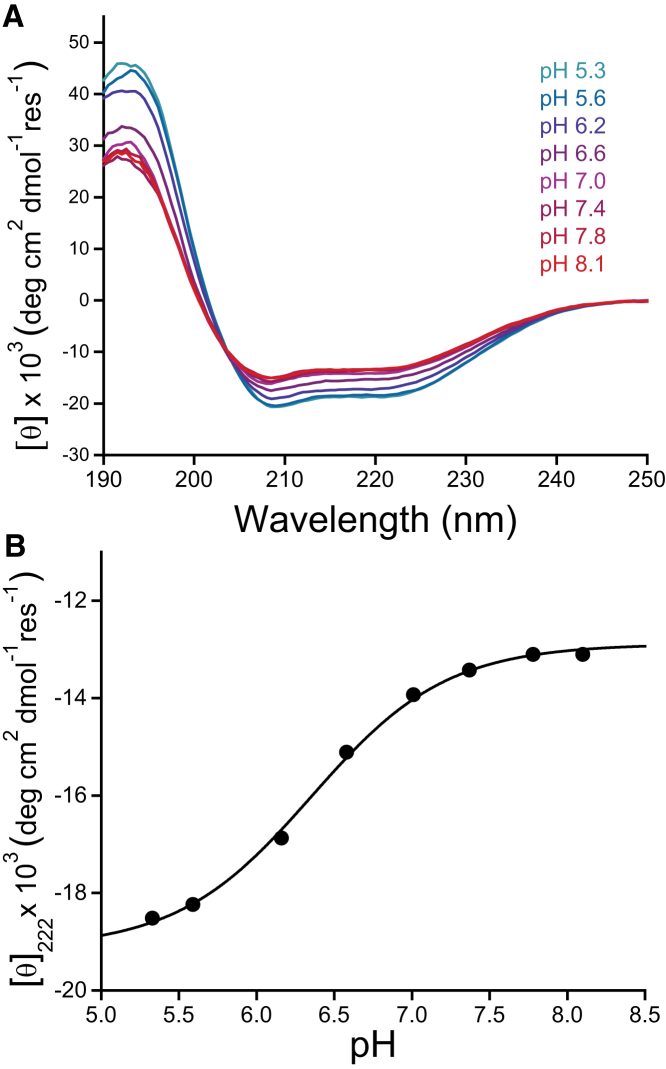
To assess the effect of pH on the secondary structure of Cav3, we collected the far-UV CD spectra under conditions with varying pH. Consistent with the NMR results and secondary structure predictions, the CD spectrum suggests Cav3 is significantly helical under all these conditions (Fig. 4 A). Furthermore, a prominent increase in helical content occurs at low pH (Fig. 4 A), which suggests this topological transition is coincident with the formation of additional α-helical secondary structure. The midpoint of this transition occurs at pH 6.35. Analysis of the CD spectra using K2D3 suggests that the helical content of lipidated Cav3 changes from 39% at pH 8.1 to 55% at pH 5.3 (62), which corresponds to an increase of roughly 26 additional helical residues at low pH. This is consistent with CSI analysis of the NMR data, which shows that at least 15 residues in the N-terminal domain (residues 10–24) contribute to this transition (Fig. S4; due to extensive line broadening at low pH, the remaining residues that participate in this transition could not be identified). Taken together, these results suggest that Cav3 undergoes a large-scale topological change in which the soluble N-terminal domain at neutral pH undergoes a transition to micelle-associated helix at low pH, with the midpoint of the transition occurring near pH 6.5.
Figure 4.
Influence of pH on the secondary structure of lipidated Cav3. The effect of pH on the secondary structure of lipidated Cav3 was investigated using CD spectroscopy at 25°C. (A) The far-UV CD spectrum of lipidated Cav3 in 25 mM sodium phosphate containing 0.2% LPPG was recorded with varying pH. The mean residue ellipticity is plotted against the wavelength. (B) The mean residue ellipticity at 222 nm is plotted against the pH. The data were fit with a model derived from the Henderson-Hasselbalch equation (black line), and the apparent pKa of the transition was determined to be 6.35 ± 0.04. To see this figure in color, go online.
Reversible association of the N-terminal domain with the membrane
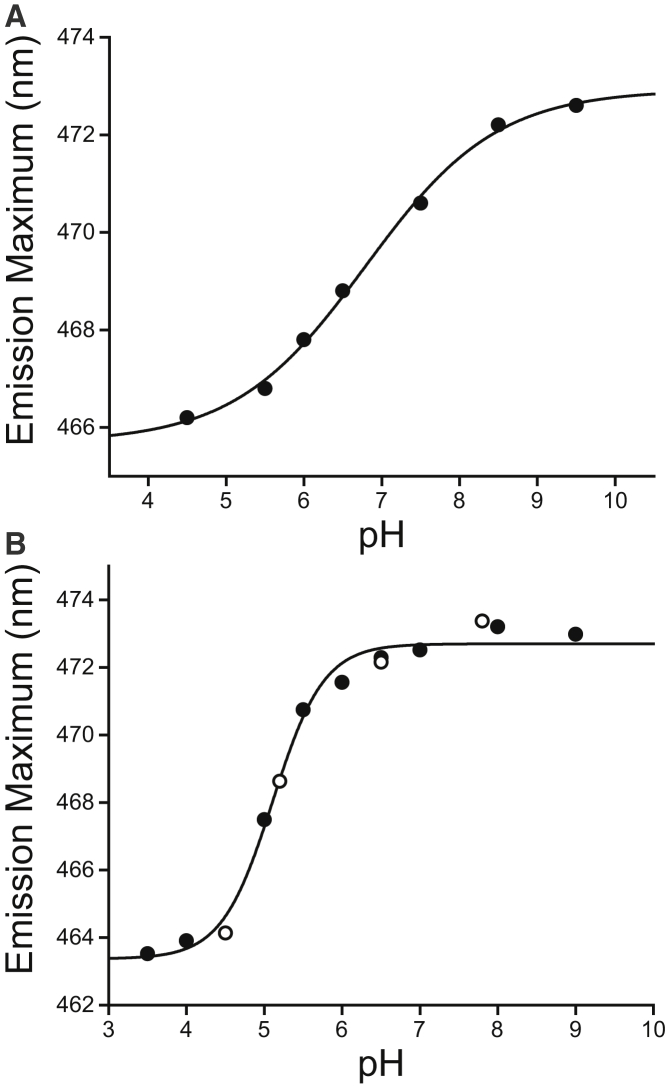
To probe the nature of the pH-mediated conformational change in Cav3, we produced a series of single-cysteine variants to facilitate the conjugation of fluorescent probes at various positions within the N-terminal domain (see Materials and Methods). For these studies the three putative palmitoylation sites of Cav3 were replaced with phenylalanines to serve as surrogate membrane anchors to facilitate specific modification of single cysteine residues with mBBr, a molecule that becomes fluorescent upon reaction with free thiols. The use of mBBr reduces the background fluorescence associated with unreacted fluorophores, which can be difficult to remove from these samples due to the association of excess fluorophore with detergent micelles. We then reconstituted the mBBR-labeled Cav3 variants into LPPG micelles and monitored the change in fluorescence emission spectrum under conditions with varying pH. Consistent with solution NMR measurements, reductions in pH were accompanied by a sigmoidal blue shift in the emission maximum of mBBr-labeled V25C Cav3 (Fig. 5 A), which likely reflects the association of the N-terminal domain with the interface. The fluorescence emission maximum of free mBBr-conjugated cysteine was found to be insensitive to pH, which confirms that the shift in the emission spectrum of mBBr-labeled Cav3 reflects a conformational change (data not shown). This conformational transition was observed to be similar when mBBr was conjugated to two other randomly chosen positions within the N-terminal domain (Fig. S5), which suggests that this conformational transition is independent to the labeling position. The apparent pKa of the transition occurs at 6.8 ± 0.1 (Fig. 5 A), which is similar to that observed by CD spectroscopy under these conditions (6.35 ± 0.04) (Fig. 4 B). These results suggest that a mixture of two conformational states persist at pH 6.5, which accounts for the topological heterogeneity detected by solution NMR under this condition, as described previously (Fig. 3). To confirm that this conformational transition also occurs in actual lipid bilayers, we carried out these experiments on mBBr-labeled Cav3 variants reconstituted in bilayered POPG vesicles. A similar conformation transition was observed in vesicles (Fig. 5 B), though the transition appears to be more cooperative and the apparent pKa (5.11 ± 0.06) is lower under these conditions. Again, the nature of the conformational transition in vesicles was found to be independent of the labeling position with the N-terminal domain (Fig. S5). Moreover, the forward and reverse titrations were quite similar (Fig. 5 B), suggesting that the conformational ensemble reaches equilibrium.
Figure 5.
pH-mediated conformational transition of the Cav3 N-terminal domain in LPPG micelles and in POPG vesicles. The influence of pH on the fluorescence spectrum of mBBr-labeled V25C Cav3 was assessed in micelles and vesicles. (A) The fluorescence emission spectrum of mBBr-labeled V25C Cav3 in 120 mM NaCl containing 15 mM acetic acid, 15 mM MES, 30mM Tris, 0.5 mM EDTA, and 5% LPPG was collected as a function of pH. The wavelength corresponding to the emission maximum is plotted against the pH. The data were fit with a model derived from the Henderson-Hasselbalch equation (black line), and the apparent pKa of the transition was determined to be 6.8 ± 0.1. (B) mBBr-labeled V25C Cav3 in POPG vesicles was equilibrated in 120 mM NaCl containing 15 mM acetic acid, 15 mM MES, 30 mM Tris, and 0.5 mM EDTA with varying pH before acquisition of the fluorescence emission spectrum. The wavelength corresponding to the emission maximum is plotted against the pH. Closed symbols reflect data for the titration from low to high pH. Open symbols reflect the data for the titration from high to low pH. The data were fit with a model derived from the Henderson-Hasselbalch equation (black line), and the apparent pKa of the transition was determined to be 5.11 ± 0.06.
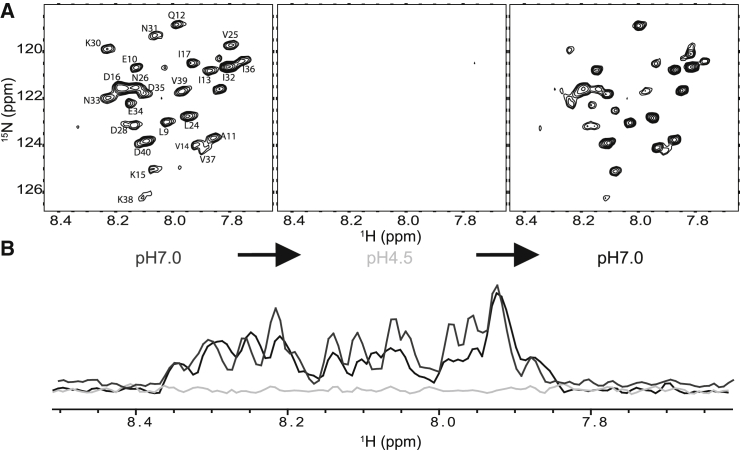
To confirm that this conformational transition primarily reflects the reversible association of the N-terminal domain with the membrane, we reconstituted 15N-labeled lipidated Cav3 into POPG lipid vesicles and compared the 1H 15N TROSY spectrum at high and low pH. POPG was employed as the lipid in experiments involving vesicles in this work to best match structure of the LPPG detergent used in micellar experiments. Due to the slow tumbling of the vesicles in solution, the majority of the backbone amide peaks were broadened beyond detection (Fig. 6). However, some peaks from the N-terminal domain were still detectable at pH 7.0 (Fig. 6), confirming that this domain is dynamic and occupies the solution phase at neutral pH. However, reduction of the pH to 4.5 resulted in the complete broadening of these peaks (Fig. 6), which occurs as a result of the association of the N-terminal domain with the membrane and a dramatic reduction in motion. Subsequent adjustment of the sample to pH 7.0 resulted in the recovery of the spectrum (Fig. 6), again confirming the reversibility of this conformational transition. To ensure that this transition did not arise as a result of the protonation of the N-terminal polyhistidine tag, we repeated this experiment using a construct featuring a C-terminal polyhistidine tag. The results were consistent regardless of the position of the tag (Fig. S6), which confirms that this transition is intrinsic to the native residues of the N-terminal domain. Taken together, these results demonstrate that the N-terminal domain of Cav3 reversibly associates with the membrane in a pH-dependent manner, as illustrated in the working model of Fig. 7.
Figure 6.
pH-mediated membrane association of the Cav3 N-terminal domain. (A) Lipidated Cav3 was reconstituted in POPG vesicles and equilibrated in 75 mM imidazole (pH 7.0) containing 25 mM sodium acetate and 0.5 mM EDTA before acquisition of the 1H 15N TROSY-HSQC spectrum (left). The pH of the sample was then lowered to pH 4.5 before a second acquisition of the 1H 15N TROSY-HSQC spectrum (center). Finally, the pH of the original sample was restored to 7.0 before acquisition of a third 1H 15N TROSY-HSQC spectrum (right). (B) The 1H spectrum is shown for the initial sample in lipid vesicles at pH 7.0 (dark gray), as well as those taken subsequently at pH 4.5 (light gray) and then again at 7.0 (black).
Figure 7.
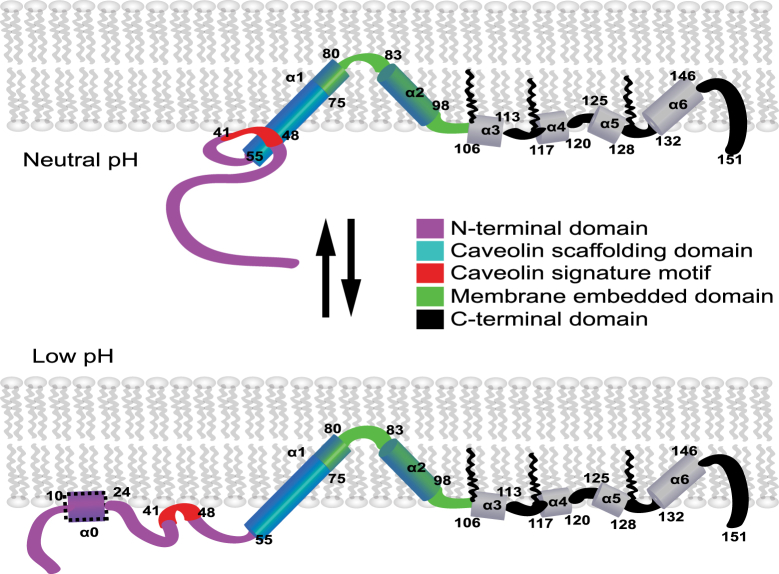
Working model for the organization of monomeric Cav3 in membranes and the nature of its pH-mediated conformational transition. A potential interaction between the acidic caveolin signature sequence and basic residues of the caveolin scaffolding domain is implied in the upper panel. Protonation of acidic residues at low pH may disrupt this interaction and drive association of the N-terminal domain with the membrane interface. It is emphasized that the exact palmitoylation pattern of native Cav3 is not known (the pattern shown here is based on Cav1) and that most of the experiments on which this working model are based were conducted with mutant forms of Cav3—usually chemically modified, and usually using LPPG micelles as the model membranes. To see this figure in color, go online.
Discussion
Although Cav3 is a critical mediator of caveolae biogenesis, the structural mechanisms that give rise to caveolin function remain unclear. In this work, we explored the structural properties of the monomeric form of lipidated Cav3. Our analysis of its secondary structure may help to resolve inconsistencies that have emerged from previous studies. For instance, previous characterizations of the caveolin scaffolding domain have drawn conflicting conclusions as to whether this portion of the molecule adopts α-helical or β-strand secondary structure (26, 40, 41). We cannot rule out the possibility that some of these differences might reflect complications due to the presence of a number of mutations in the Cav-3 construct used in this work combined with the fact that Cav-3 may have some different structural properties than Cav-1, despite their 65% sequence identity. Other possible contributing factors to these differences include the facts that all previous analyses were carried out using different truncated caveolin constructs and sometimes also using different membrane mimetics. Our analysis of the full-length lipidated form of Cav3 indicates that the scaffolding domain consists of a long membrane-associated α-helix that is contiguous with the helix-break-helix motif within the caveolin membrane domain (25, 26). Our results also demonstrate the presence of helical structure within the C-terminal domain, consistent with what has previously been observed for Cav1 (27). However, it should be noted that we observed four different helical segments within the C-terminal domain of Cav3 (helix 3, residues 106–113, helix 4, residues 117–120, helix 5, residues 125–128, and helix 6, residues 132–146) as opposed to one long C-terminal helix observed in nonlipidated Cav1 (27). The CSI values proximal to the lipidation sites in Cav3 (residues 114–116 and 129–131) are low compared to those values reported for the corresponding residues in Cav1 (Fig. S2) (27). Furthermore, a comparison of the CSI values of lipidated and nonlipidated Cav3 reveals differences proximal to the lipidation sites (Fig. S7). Taken together, these results suggest that the localized structural perturbations caused by lipid modifications likely reflect helical breaks within the C-terminal domain. In addition to altering the local secondary structure, lipidation of palmitoylated sites also reduced exchange-broadening, which ultimately facilitated assignments of amide peaks within the C-terminal domain.
Beyond providing insights into the secondary structure of Cav3, our results also reveal that the N-terminal domain undergoes a dramatic topological rearrangement that is mediated by pH under these conditions. This structural transition was accompanied by changes in the conformational dynamics (Fig. S3), secondary structure (Fig. 4), and topology (Fig. 3, B and C) of the N-terminal domain. Furthermore, this transition was observed in both micelles and in phospholipid vesicles (Figs. 5 and 6). Based on the fact that all of the peaks within the N-terminal domain are significantly broadened at low pH in vesicles (Fig. 6), it seems likely that these differences reflect a large-scale cooperative conformational change that involves the entire domain (see working model in Fig. 7). This likely stems from the protonation of conserved residues within the N-terminal domain in a manner that drives membrane association. The cluster of acidic residues in the caveolin signature sequence (residues 41–48, FEDVIAEP) could potentially serve as the pH sensor for this transition. This acidic motif has been previously suggested to interact with basic residues in the caveolin scaffolding domain (42). Thus, it is tempting to speculate that the protonation of the acidic residues may disrupt this electrostatic interaction and drive the membrane association of the signature motif at low pH, as is the rendered model in Fig. 7. We emphasize that this is a working model that is subject to revision based on future results. We acknowledge that the lyso-phosphatidylglycerol micelles and POPG vesicles employed in this work are not ideal mimics of caveolar membranes, such that future validation of this working model in more optimal media is merited. Furthermore, we note that validation of the hypothesized conformational switch is beyond the scope of this work, but remains an intriguing hypothesis for future investigations.
The biological relevance of this conformational change has yet to be explored. It has been shown that, during its degradation, Cav1 is delivered to lysosomes from multivesicular bodies/late endosomes, where it becomes exposed to the acidic lumen (63). Exposure of the protein to reduced pH promotes membrane association of the N-terminal domain in a manner that could disrupt its interactions with other proteins, interactions that originally formed at cytosolic pH. It is conceivable that mutations that compromise this process could potentially cause a buildup of Cav3 and/or the formation of toxic aggregates. Indeed, there are many examples of pathogenic mutations in integral membrane proteins that alter protein processing and/or degradation. It is also possible that the pH-induced conformational transition observed under our experimental conditions may be triggered by other environmental stimuli within the cell. The cooperativity and the apparent pKa of the transition are quite different in LPPG micelles versus POPG membranes, although LPPG and POPG each feature a glycerol backbone and phosphoglycerol headgroup (Fig. 5). This observation suggests that subtle differences in the membrane environment, such as the high degree of curvature occurring in caveolae or fluctuations in surface charge density, could potentially also modulate the ratio of these two conformational ensembles in vivo. Regardless of the precise physiochemical drivers of this conformational transition, it should be noted that nearly one-third of the currently identified pathogenic missense mutations in Cav3 alter native charged residues or introduce nonnative charged residues within the N-terminal domain (human gene mutation database, http://www.hgmd.cf.ac.uk). This observation strongly suggests that the charged residues in the N-terminal domain that mediate this conformational change are of critical importance for the native function of Cav3. Future studies are needed to determine the relevance of this conformational change to Cav3 function and the mechanistic effects of pathogenic mutations within the N-terminal domain.
Author Contributions
The experiments were designed by J.H.K. and C.R.S. The data were collected by J.H.K., J.P.S., Z.L., and K.C.R. The data were analyzed and interpreted by J.H.K., J.P.S., Z.L., D.P., and C.R.S. The article was written and edited by J.H.K., J.P.S., and C.R.S.
Acknowledgments
This work was supported by U.S. National Institutes of Health (NIH) grants U54 GM094608, RO1 HL122010, and RO1 GM106672. J.S. was supported by NIH F32 fellowship GM110929. The NMR instrumentation used in this work was supported by NIH grant S10 RR025677-01 and National Science Foundation Grant DBI-0922862.
Editor: Francesca Marassi.
Footnotes
Seven figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30280-6.
Supporting Material
References
- 1.Brown D.A., London E. Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- 2.Parat M.O. The biology of caveolae: achievements and perspectives. Int. Rev. Cell Mol. Biol. 2009;273:117–162. doi: 10.1016/S1937-6448(08)01804-2. [DOI] [PubMed] [Google Scholar]
- 3.Le P.U., Guay G., Nabi I.R. Caveolin-1 is a negative regulator of caveolae-mediated endocytosis to the endoplasmic reticulum. J. Biol. Chem. 2002;277:3371–3379. doi: 10.1074/jbc.M111240200. [DOI] [PubMed] [Google Scholar]
- 4.Sharma D.K., Brown J.C., Pagano R.E. Selective stimulation of caveolar endocytosis by glycosphingolipids and cholesterol. Mol. Biol. Cell. 2004;15:3114–3122. doi: 10.1091/mbc.E04-03-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.García-Cardeña G., Martasek P., Sessa W.C. Dissecting the interaction between nitric oxide synthase (NOS) and caveolin. Functional significance of the nos caveolin binding domain in vivo. J. Biol. Chem. 1997;272:25437–25440. doi: 10.1074/jbc.272.41.25437. [DOI] [PubMed] [Google Scholar]
- 6.Sowa G., Pypaert M., Sessa W.C. Distinction between signaling mechanisms in lipid rafts vs. caveolae. Proc. Natl. Acad. Sci. USA. 2001;98:14072–14077. doi: 10.1073/pnas.241409998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fielding C.J., Fielding P.E. Cellular cholesterol efflux. Biochim. Biophys. Acta. 2001;1533:175–189. doi: 10.1016/s1388-1981(01)00162-7. [DOI] [PubMed] [Google Scholar]
- 8.Le Lay S., Hajduch E., Dugail I. Cholesterol-induced caveolin targeting to lipid droplets in adipocytes: a role for caveolar endocytosis. Traffic. 2006;7:549–561. doi: 10.1111/j.1600-0854.2006.00406.x. [DOI] [PubMed] [Google Scholar]
- 9.Kozera L., White E., Calaghan S. Caveolae act as membrane reserves which limit mechanosensitive I(Cl,swell) channel activation during swelling in the rat ventricular myocyte. PLoS One. 2009;4:e8312. doi: 10.1371/journal.pone.0008312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rothberg K.G., Heuser J.E., Anderson R.G. Caveolin, a protein component of caveolae membrane coats. Cell. 1992;68:673–682. doi: 10.1016/0092-8674(92)90143-z. [DOI] [PubMed] [Google Scholar]
- 11.Scherer P.E., Okamoto T., Lisanti M.P. Identification, sequence, and expression of caveolin-2 defines a caveolin gene family. Proc. Natl. Acad. Sci. USA. 1996;93:131–135. doi: 10.1073/pnas.93.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang Z., Scherer P.E., Lisanti M.P. Molecular cloning of caveolin-3, a novel member of the caveolin gene family expressed predominantly in muscle. J. Biol. Chem. 1996;271:2255–2261. doi: 10.1074/jbc.271.4.2255. [DOI] [PubMed] [Google Scholar]
- 13.Way M., Parton R.G. M-caveolin, a muscle-specific caveolin-related protein. FEBS Lett. 1996;378:108–112. doi: 10.1016/0014-5793(96)82884-5. [DOI] [PubMed] [Google Scholar]
- 14.Galbiati F., Razani B., Lisanti M.P. Caveolae and caveolin-3 in muscular dystrophy. Trends Mol. Med. 2001;7:435–441. doi: 10.1016/s1471-4914(01)02105-0. [DOI] [PubMed] [Google Scholar]
- 15.Gazzerro E., Sotgia F., Minetti C. Caveolinopathies: from the biology of caveolin-3 to human diseases. Eur. J. Hum. Genet. 2010;18:137–145. doi: 10.1038/ejhg.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams T.M., Lisanti M.P. The Caveolin genes: from cell biology to medicine. Ann. Med. 2004;36:584–595. doi: 10.1080/07853890410018899. [DOI] [PubMed] [Google Scholar]
- 17.Woodman S.E., Sotgia F., Lisanti M.P. Caveolinopathies: mutations in caveolin-3 cause four distinct autosomal dominant muscle diseases. Neurology. 2004;62:538–543. doi: 10.1212/wnl.62.4.538. [DOI] [PubMed] [Google Scholar]
- 18.Dupree P., Parton R.G., Simons K. Caveolae and sorting in the trans-Golgi network of epithelial cells. EMBO J. 1993;12:1597–1605. doi: 10.1002/j.1460-2075.1993.tb05804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Lan C., Gallay J., Jamin N. Structural and dynamic properties of juxta-membrane segments of caveolin-1 and caveolin-2 at the membrane interface. Eur. Biophys. J. 2010;39:307–325. doi: 10.1007/s00249-009-0548-4. [DOI] [PubMed] [Google Scholar]
- 20.Rieth M.D., Lee J., Glover K.J. Probing the caveolin-1 P132L mutant: critical insights into its oligomeric behavior and structure. Biochemistry. 2012;51:3911–3918. doi: 10.1021/bi3001853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Root K.T., Plucinsky S.M., Glover K.J. Recent progress in the topology, structure, and oligomerization of caveolin: a building block of caveolae. Curr. Top. Membr. 2015;75:305–336. doi: 10.1016/bs.ctm.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Schlegel A., Schwab R.B., Lisanti M.P. A role for the caveolin scaffolding domain in mediating the membrane attachment of caveolin-1. The caveolin scaffolding domain is both necessary and sufficient for membrane binding in vitro. J. Biol. Chem. 1999;274:22660–22667. doi: 10.1074/jbc.274.32.22660. [DOI] [PubMed] [Google Scholar]
- 23.Wanaski S.P., Ng B.K., Glaser M. Caveolin scaffolding region and the membrane binding region of SRC form lateral membrane domains. Biochemistry. 2003;42:42–56. doi: 10.1021/bi012097n. [DOI] [PubMed] [Google Scholar]
- 24.Sargiacomo M., Scherer P.E., Lisanti M.P. Oligomeric structure of caveolin: implications for caveolae membrane organization. Proc. Natl. Acad. Sci. USA. 1995;92:9407–9411. doi: 10.1073/pnas.92.20.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee J., Glover K.J. The transmembrane domain of caveolin-1 exhibits a helix-break-helix structure. Biochim. Biophys. Acta. 2012;1818:1158–1164. doi: 10.1016/j.bbamem.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rui H., Root K.T., Im W. Probing the U-shaped conformation of caveolin-1 in a bilayer. Biophys. J. 2014;106:1371–1380. doi: 10.1016/j.bpj.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plucinsky S.M., Glover K.J. Secondary structure analysis of a functional construct of caveolin-1 reveals a long C-terminal helix. Biophys. J. 2015;109:1686–1688. doi: 10.1016/j.bpj.2015.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dietzen D.J., Hastings W.R., Lublin D.M. Caveolin is palmitoylated on multiple cysteine residues. Palmitoylation is not necessary for localization of caveolin to caveolae. J. Biol. Chem. 1995;270:6838–6842. doi: 10.1074/jbc.270.12.6838. [DOI] [PubMed] [Google Scholar]
- 29.Monier S., Dietzen D.J., Kurzchalia T.V. Oligomerization of VIP21-caveolin in vitro is stabilized by long chain fatty acylation or cholesterol. FEBS Lett. 1996;388:143–149. doi: 10.1016/0014-5793(96)00519-4. [DOI] [PubMed] [Google Scholar]
- 30.Galbiati F., Volonte D., Lisanti M.P. Phenotypic behavior of caveolin-3 mutations that cause autosomal dominant limb girdle muscular dystrophy (LGMD-1C). Retention of LGMD-1C caveolin-3 mutants within the golgi complex. J. Biol. Chem. 1999;274:25632–25641. doi: 10.1074/jbc.274.36.25632. [DOI] [PubMed] [Google Scholar]
- 31.Abrami L., Kunz B., van der Goot F.G. Palmitoylation and ubiquitination regulate exit of the Wnt signaling protein LRP6 from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2008;105:5384–5389. doi: 10.1073/pnas.0710389105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aicart-Ramos C., Valero R.A., Rodriguez-Crespo I. Protein palmitoylation and subcellular trafficking. Biochim. Biophys. Acta. 2011;1808:2981–2994. doi: 10.1016/j.bbamem.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 33.Blaskovic S., Blanc M., van der Goot F.G. What does S-palmitoylation do to membrane proteins? FEBS J. 2013;280:2766–2774. doi: 10.1111/febs.12263. [DOI] [PubMed] [Google Scholar]
- 34.Conibear E., Davis N.G. Palmitoylation and depalmitoylation dynamics at a glance. J. Cell Sci. 2010;123:4007–4010. doi: 10.1242/jcs.059287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joseph M., Nagaraj R. Conformations of peptides corresponding to fatty acylation sites in proteins. A circular dichroism study. J. Biol. Chem. 1995;270:19439–19445. doi: 10.1074/jbc.270.33.19439. [DOI] [PubMed] [Google Scholar]
- 36.Joseph M., Nagaraj R. Interaction of peptides corresponding to fatty acylation sites in proteins with model membranes. J. Biol. Chem. 1995;270:16749–16755. doi: 10.1074/jbc.270.28.16749. [DOI] [PubMed] [Google Scholar]
- 37.Linder M., Deschenes R. Protein palmitoylation. Methods. 2006;40:125–126. doi: 10.1016/j.ymeth.2006.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salaun C., Greaves J., Chamberlain L.H. The intracellular dynamic of protein palmitoylation. J. Cell Biol. 2010;191:1229–1238. doi: 10.1083/jcb.201008160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J.H., Peng D., Sanders C.R. Modest effects of lipid modifications on the structure of caveolin-3. Biochemistry. 2014;53:4320–4322. doi: 10.1021/bi5005238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoop C.L., Sivanandam V.N., van der Wel P.C. Structural characterization of the caveolin scaffolding domain in association with cholesterol-rich membranes. Biochemistry. 2012;51:90–99. doi: 10.1021/bi201356v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le Lan C., Neumann J.M., Jamin N. Role of the membrane interface on the conformation of the caveolin scaffolding domain: a CD and NMR study. FEBS Lett. 2006;580:5301–5305. doi: 10.1016/j.febslet.2006.08.075. [DOI] [PubMed] [Google Scholar]
- 42.Fernandez I., Ying Y., Anderson R.G. Mechanism of caveolin filament assembly. Proc. Natl. Acad. Sci. USA. 2002;99:11193–11198. doi: 10.1073/pnas.172196599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whiteley G., Collins R.F., Kitmitto A. Characterization of the molecular architecture of human caveolin-3 and interaction with the skeletal muscle ryanodine receptor. J. Biol. Chem. 2012;287:40302–40316. doi: 10.1074/jbc.M112.377085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monier S., Parton R.G., Kurzchalia T.V. VIP21-caveolin, a membrane protein constituent of the caveolar coat, oligomerizes in vivo and in vitro. Mol. Biol. Cell. 1995;6:911–927. doi: 10.1091/mbc.6.7.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gambin Y., Ariotti N., Parton R.G. Single-molecule analysis reveals self assembly and nanoscale segregation of two distinct cavin subcomplexes on caveolae. eLife. 2013;3:e01434. doi: 10.7554/eLife.01434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schlegel A., Lisanti M.P. A molecular dissection of caveolin-1 membrane attachment and oligomerization. Two separate regions of the caveolin-1 C-terminal domain mediate membrane binding and oligomer/oligomer interactions in vivo. J. Biol. Chem. 2000;275:21605–21617. doi: 10.1074/jbc.M002558200. [DOI] [PubMed] [Google Scholar]
- 47.Ellis K.J., Morrison J.F. Buffers of constant ionic strength for studying pH-dependent processes. Methods Enzymol. 1982;87:405–426. doi: 10.1016/s0076-6879(82)87025-0. [DOI] [PubMed] [Google Scholar]
- 48.Waugh D.S. Genetic tools for selective labeling of proteins with alpha-15N-amino acids. J. Biomol. NMR. 1996;8:184–192. doi: 10.1007/BF00211164. [DOI] [PubMed] [Google Scholar]
- 49.Salzmann M., Pervushin K., Wüthrich K. TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc. Natl. Acad. Sci. USA. 1998;95:13585–13590. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eletsky A., Kienhöfer A., Pervushin K. TROSY NMR with partially deuterated proteins. J. Biomol. NMR. 2001;20:177–180. doi: 10.1023/a:1011265430149. [DOI] [PubMed] [Google Scholar]
- 51.Schulte-Herbrüggen T., Sorensen O.W. Clean TROSY: compensation for relaxation-induced artifacts. J. Magn. Reson. 2000;144:123–128. doi: 10.1006/jmre.2000.2020. [DOI] [PubMed] [Google Scholar]
- 52.Delaglio F., Grzesiek S., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 53.Goddard T.D., Kneller D.G. University of California; San Francisco, CA: 2015. SPARKY 3. [Google Scholar]
- 54.Shen Y., Bax A. Protein backbone and side-chain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wishart D.S., Sykes B.D. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 56.Zhu G., Xia Y., Sze K.H. Protein dynamics measurements by TROSY-based NMR experiments. J. Magn. Reson. 2000;143:423–426. doi: 10.1006/jmre.2000.2022. [DOI] [PubMed] [Google Scholar]
- 57.Kay L.E., Torchia D.A., Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 58.Wishart D.S., Sykes B.D., Richards F.M. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 59.Chou J.J., Baber J.L., Bax A. Characterization of phospholipid mixed micelles by translational diffusion. J. Biomol. NMR. 2004;29:299–308. doi: 10.1023/B:JNMR.0000032560.43738.6a. [DOI] [PubMed] [Google Scholar]
- 60.Lipfert J., Columbus L., Doniach S. Size and shape of detergent micelles determined by small-angle x-ray scattering. J. Phys. Chem. B. 2007;111:12427–12438. doi: 10.1021/jp073016l. [DOI] [PubMed] [Google Scholar]
- 61.Ariotti N., Rae J., Parton R.G. Molecular characterization of caveolin-induced membrane curvature. J. Biol. Chem. 2015;290:24875–24890. doi: 10.1074/jbc.M115.644336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Louis-Jeune C., Andrade-Navarro M.A., Perez-Iratxeta C. Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins. 2012;80:374–381. doi: 10.1002/prot.23188. [DOI] [PubMed] [Google Scholar]
- 63.Hayer A., Stoeber M., Helenius A. Caveolin-1 is ubiquitinated and targeted to intralumenal vesicles in endolysosomes for degradation. J. Cell Biol. 2010;191:615–629. doi: 10.1083/jcb.201003086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.