Abstract
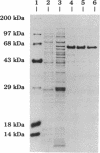
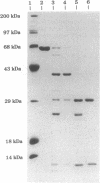
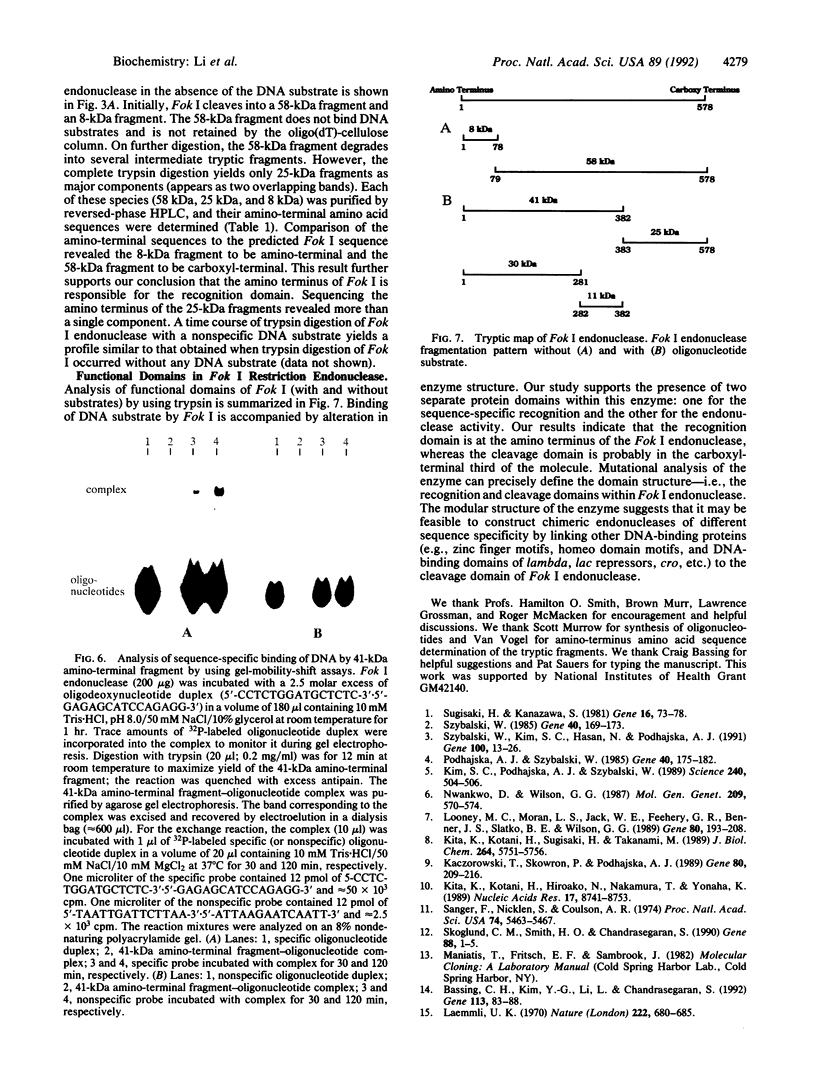
The PCR was used to alter transcriptional and translational signals surrounding the Flavobacterium okeanokoites restriction endonuclease (fokIR) gene, so as to achieve high expression in Escherichia coli. By changing the ribosome-binding site sequence preceding the fokIR gene to match the consensus E. coli signal and by placing a positive retroregulator stem-loop sequence downstream of the gene, Fok I yield was increased to 5-8% of total cellular protein. Fok I was purified to homogeneity with phosphocellulose, DEAE-Sephadex, and gel chromatography, yielding 50 mg of pure Fok I endonuclease per liter of culture medium. The recognition and cleavage domains of Fok I were analyzed by trypsin digestion. Fok I in the absence of a DNA substrate cleaves into a 58-kDa carboxyl-terminal and 8-kDa amino-terminal fragment. The 58-kDa fragment does not bind the DNA substrate. Fok I in the presence of a DNA substrate cleaves into a 41-kDa amino-terminal fragment and a 25-kDa carboxyl-terminal fragment. On further digestion, the 41-kDa fragment degrades into 30-kDa amino-terminal and 11-kDa carboxyl-terminal fragments. The cleaved fragments both bind DNA substrates, as does the 41-kDa fragment. Gel-mobility-shift assays indicate that all the protein contacts necessary for the sequence-specific recognition of DNA substrates are encoded within the 41-kDa fragment. Thus, the 41-kDa amino-terminal fragment constitutes the Fok I recognition domain. The 25-kDa fragment, purified by using a DEAE-Sephadex column, cleaves nonspecifically both methylated (pACYCfokIM) and nonmethylated (pTZ19R) DNA substrates in the presence of MgCl2. Thus, the 25-kDa carboxyl-terminal fragment constitutes the Fok I cleavage domain.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bassing C. H., Kim Y. G., Li L., Chandrasegaran S. Overproduction, purification and characterization of M.HinfI methyltransferase and its deletion mutant. Gene. 1992 Apr 1;113(1):83–88. doi: 10.1016/0378-1119(92)90672-c. [DOI] [PubMed] [Google Scholar]
- Kaczorowski T., Skowron P., Podhajska A. J. Purification and characterization of the FokI restriction endonuclease. Gene. 1989 Aug 15;80(2):209–216. doi: 10.1016/0378-1119(89)90285-0. [DOI] [PubMed] [Google Scholar]
- Kim S. C., Podhajska A. J., Szybalski W. Cleaving DNA at any predetermined site with adapter-primers and class-IIS restriction enzymes. Science. 1988 Apr 22;240(4851):504–506. doi: 10.1126/science.2833816. [DOI] [PubMed] [Google Scholar]
- Kita K., Kotani H., Hiraoka N., Nakamura T., Yonaha K. Overproduction and crystallization of FokI restriction endonuclease. Nucleic Acids Res. 1989 Nov 11;17(21):8741–8753. doi: 10.1093/nar/17.21.8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita K., Kotani H., Sugisaki H., Takanami M. The fokI restriction-modification system. I. Organization and nucleotide sequences of the restriction and modification genes. J Biol Chem. 1989 Apr 5;264(10):5751–5756. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Looney M. C., Moran L. S., Jack W. E., Feehery G. R., Benner J. S., Slatko B. E., Wilson G. G. Nucleotide sequence of the FokI restriction-modification system: separate strand-specificity domains in the methyltransferase. Gene. 1989 Aug 15;80(2):193–208. doi: 10.1016/0378-1119(89)90284-9. [DOI] [PubMed] [Google Scholar]
- Nwankwo D., Wilson G. Cloning of two type II methylase genes that recognise asymmetric nucleotide sequences: FokI and HgaI. Mol Gen Genet. 1987 Oct;209(3):570–574. doi: 10.1007/BF00331164. [DOI] [PubMed] [Google Scholar]
- Podhajska A. J., Szybalski W. Conversion of the FokI endonuclease to a universal restriction enzyme: cleavage of phage M13mp7 DNA at predetermined sites. Gene. 1985;40(2-3):175–182. doi: 10.1016/0378-1119(85)90040-x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund C. M., Smith H. O., Chandrasegaran S. Construction of an efficient overproducer clone of HinfI restriction endonuclease using the polymerase chain reaction. Gene. 1990 Mar 30;88(1):1–5. doi: 10.1016/0378-1119(90)90052-s. [DOI] [PubMed] [Google Scholar]
- Sugisaki H., Kanazawa S. New restriction endonucleases from Flavobacterium okeanokoites (FokI) and Micrococcus luteus (MluI). Gene. 1981 Dec;16(1-3):73–78. doi: 10.1016/0378-1119(81)90062-7. [DOI] [PubMed] [Google Scholar]
- Szybalski W., Kim S. C., Hasan N., Podhajska A. J. Class-IIS restriction enzymes--a review. Gene. 1991 Apr;100:13–26. doi: 10.1016/0378-1119(91)90345-c. [DOI] [PubMed] [Google Scholar]
- Szybalski W. Universal restriction endonucleases: designing novel cleavage specificities by combining adapter oligodeoxynucleotide and enzyme moieties. Gene. 1985;40(2-3):169–173. doi: 10.1016/0378-1119(85)90039-3. [DOI] [PubMed] [Google Scholar]