Abstract
Fibroblast growth factors (FGFs) have long been attributed to influence morphogenesis in embryonic development. Signaling by FGF morphogen encodes positional identity of tissues by creating a concentration gradient over the developing embryo. Various mechanisms that influence the development of such gradient have been elucidated in the recent past. These mechanisms of FGF gradient formation present either as an extracellular control over FGF ligand diffusion or as a subcellular control of FGF propagation and signaling. In this review, we describe our current understanding of FGF as a morphogen, the extracellular control of FGF gradient formation by heparan sulfate proteoglycans (HSPGs) and mechanisms of intracellular regulation of FGF signaling that influence gradient formation.
Keywords: Heparan sulfate proteoglycans, Morphogen, Fibroblast growth factors
1. Introduction
Early embryonic development comprises of several crucial processes such as cell proliferation and differentiation, cell survival, patterning of the embryonic axis, maintenance of cell lineages, cell migration and morphogenesis. These processes are tightly regulated by various factors. A group of signaling molecules – the secreted growth factors such as fibroblast growth factors (FGFs), Wnt pathway factors, Hedgehog family factors (Hhs), transforming growth factors (TGFs), bone morphogenetic proteins (BMPs) play crucial roles in the patterning and morphogenesis of the early developing embryo. The potency and importance of morphogens in early development were demonstrated by a series of elegant transplantation studies by Spemann and Mangold in the early 1900s. More recent studies have discovered that morphogens secreted by cells diffuse extracellularly to their target cells and bind their cell surface receptors to influence intracellular signaling cascades. The extracellular diffusion of morphogens determines a concentration gradient in the developing embryo and is crucial for establishing positional identities of differentiating cells. It is thus pertinent to understand the various mechanisms governing the establishment of a morphogen concentration gradient. Recent discoveries elucidate the extracellular role of HSPGs in creating, maintaining and regulating morphogen concentration gradients. Morphogen concentration gradients are also impacted by subcellular processes that control morphogen signal propagation. In this review, we focus specifically on the mechanisms of extracellular control of FGF morphogen gradient formation by HSPGs and also discuss the latest advances in the knowledge of intracellular control on FGF signaling.
2. FGF as a morphogen during embryonic development
FGFs are a family of polypeptide growth factors that have been evolutionarily conserved across metazoan species. A member of the family was first identified in pituitary extracts and described for its role in stimulating cell growth and proliferation of mouse fibroblasts [1]. Since their discovery, FGFs have been implicated in regulating a plethora of cell processes ranging from cell proliferation and differentiation to cell migration, survival and apoptosis (reviewed in [2–4]). Various members of the FGF family are actively involved in regulating crucial developmental events [5,6], including but not restricted to, coordinating cell migration and formation of primitive streak [7–9], mesoderm induction and patterning [9], neural induction [10,11] and endoderm formation and patterning [12,13]. The diversity in function exhibited by the FGF family derives directly from the diversity in FGF ligands and FGF receptors. In the mouse and human genomes, 22 FGF genes have been described and can be phylogenetically classified into distinct subfamilies: intracellular FGF subfamily, hormone-like or endocrine FGF subfamily and canonical or paracrine FGF subfamily which further contains five subfamilies [14–16]. Intracellular FGFs are localized to the cytoplasm and are not secreted. They interact with voltage-gated sodium channels in a manner that is independent of FGF receptors [17]. Hormone-like or endocrine FGFs are able to enter the bloodstream and can hence influence target tissues located farther away from the source of their secretion. Endocrine FGFs employ the Klotho gene family of transmembrane proteins as co-receptors to bind and signal via FGF receptors [18]. Canonical or paracrine FGFs are secreted and localized in the extracellular matrix (ECM) and the cell surface close to the site of their secretion. They rely primarily on diffusion to influence target tissues and, like endocrine FGFs, signal through FGF receptors. Here, we focus primarily on paracrine FGFs and the mechanisms through which their concentration gradient is created.
The status of FGFs as morphogens has been well established. Pioneering studies in the Xenopus embryos have demonstrated that secreted FGFs are diffusible and present a concentration gradient at the target tissue leading to expression of different marker genes at varying concentrations [19–21]. Early studies in mouse embryos have also shown that FGF morphogen released from the rostral midline specifies the rostral identity of the developing neocortex [22,23]. Furthermore, studies on the spinal cord in the developing chick embryo have demonstrated that the positional identity of thoracic and branchial motor neurons is established by an FGF gradient that directly influences Hox-c expression profiles [24–26]. Taken together, FGFs are secreted signaling molecules that diffuse to form a concentration gradient at the target tissue, thereby influencing the specification of cell fates in a dose-dependent manner. FGF therefore complies with the classical definition of a morphogen.
FGF morphogen ligands bind with a high degree of specificity to cell surface FGF receptor (FGFR) tyrosine kinases. In vertebrates, four highly related FGFRs (FGFR1-R4) are capable of binding FGF ligands with varying degrees of specificity [27,28]. FGF receptors themselves also exist in alternative splicing isoforms. This generates diversity in FGF ligand–receptor binding that translates into a functional diversity [3,29]. FGFRs consist of an extracellular ligand-binding domain, a transmembrane domain and a cytoplasmic tyrosine kinase domain [30]. Dimerization of FGFR by FGF ligand binding leads to tyrosine kinase activation that can further direct three independent signal transduction pathways, Ras/MAPK, PI3k/Akt and PLCγ/Ca2+ – that contribute to the functional diversity elicited by FGFs. Binding of FGF to FGFR and downstream activation of FGF signaling is regulated by heparin/heparan sulfate (HS) [5,31–36]. Details on mechanisms of HS modulation of FGF gradient formation and signaling are presented in the following section.
3. Extracellular control of FGF gradient formation and signaling by HSPGs
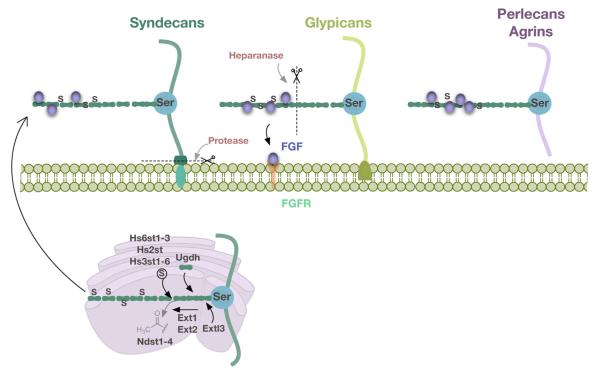
Heparan sulfate proteoglycans (HSPGs) are glycoproteins consisting of a core protein to which heparan sulfate glycosaminoglycan polymer chains are covalently linked (Fig. 1). HSPGs are typically categorized based on the location of their core protein – transmembrane type HSPGs (syndecans) and glycerophosphatidylinositide (GPI) anchored HSPGs (glypicans) that are confined to the cell surface, and extracellular matrix HSPGs that are directly secreted and localized in the extracellular matrix (ECM) and basal lamina (perlecans, agrin) [37–42]. HSPG core proteins are evolutionarily conserved across species and are capable of binding ligands and matrix components to regulate biological processes of various growth factors [43,44]. HS side-chains exhibit a great degree of structural heterogeneity based on their length, composition and the microenvironment during development [45]. A concerted effort by both HS side chains and the HSPG core protein effectively regulates growth factor diffusion and gradient formation. A review detailing the importance of HS side chains and HSPG core protein interactions with different morphogens is available [41]. While HSPGs interact with a wide plethora of factors, of particular interest to this review are the FGF ligands that bind HSPGs. By binding FGF with moderate affinity, HSPGs serve many purposes such as restricting diffusion of ligands, preventing their enzymatic degradation, functioning as reservoirs for ligands, transporting ligand to neighboring cells, physically approximating ligands to receptors and promoting stability of ligand–receptor complex by acting as co-receptors [37,39]. HSPGs hence play a pivotal role in eliciting FGF signaling.
Fig. 1.
Biosynthesis and structural diversity of HSPGs: biosynthesis of HSPGs begins in the Golgi apparatus within the cytoplasm. Specific serine residues on the core protein undergo xylosylation that allows for the incorporation of repeating units of GlcA and GlcNAc residues. Ugdh is an enzyme early in the biosynthesis cascade that is required for the synthesis of GlcA and GlcNAc residues. Extl3 is an exostosin-like enzyme that attaches the first GlcNAc to the linkage tetrasaccharide on the core protein. GlcA and GlcNAc residues are added to the side chain by exostoses Ext1 and Ext2. Enzymes Ndst1-4 add N-sulfation groups following the removal of acetyl groups. Additional O-sulfation groups at various positions are added to the chain by enzymes Hs2st, Hs3st1-6, Hs6st1-3. The precursor molecule is then transported to the cell surface or ECM where it undergoes further processing. Heparanase cleaves HS side chains while specific serine proteases cause shedding of HSPGs. Shown here for three kinds of HSPGs: transmembrane type (Syndecans), GPI anchored type (Glypicans) and secreted type (Perlecans, Agrins).
Biosynthesis of HSPGs begins in the Golgi apparatus wherein a precursor polymer is synthesized, which is then transported to the cell surface for further modifications. For the interested reader, details of HSPG biosynthesis have been extensively described in several reviews [37,38,41,46]. Briefly, the HSPG biosynthesis cascade begins with a xylosylation at specific serine residues on the core protein (Fig. 1). This is followed by the incorporation of repeating units of glucuronic acid (GlcA) and N-acetyl-d-glucosamine (GlcNAc) residues that constitute the HS side chain. Ugdh (UDP-6-glucose-dehydrogenase) is required for the synthesis of GlcA and GlcNAc residues. Extl3 (exostosin-like glycosyltransferase) attaches the first GlcNAc molecule to a linkage tetrasaccharide composed of glucuronic acid-galactose-galactosexylose. GlcA and GlcNAc residues are then alternately added to the HS side chain by exostosin glycosyltransferase Ext1 and Ext2 [47]. The extending side chain undergoes further processing such as removal of acetyl groups followed by addition of sulfate groups by N-deacetylase-N-sulfotransferases (Ndst1-4) [48] and addition of sulfate groups by uronyl-sulfotransferases (Hs2st) and glucosaminyl-sulfotransferases (Hs3st1-6, Hs6st1-3) [49]. Structural and hence functional heterogeneity exhibited by HSPGs are in part attributable to these enzymes since modifications only occur in some of the GlcA and GlcNAc units, which varies considerably depending on the type of HSPGs and developmental processes. This HS side chain precursor is then transported to the cell surface or ECM where it undergoes further processing by 6-O-Endosulfatases (Sulf1 and Sulf2) which remove specific 6-O sulfate groups [50]. HSPGs and HS chains can also be cleaved extracellularly by proteases and heparanase, causing their release into the extracellular matrix [51]. The specificity of each of these enzymatic modifications translates to a differential regulation of FGF morphogen gradient and signaling.
3.1. HSPGs control FGF morphogen gradient formation by regulating their diffusion
One of the mechanisms by which a morphogen gradient can be established in the developing embryo is simple diffusion. Several mathematical models and biological theories have supported the existence of simple diffusion to explain gradient formation of various morphogens [52,53]. However, the speed and extent of such diffusion must be controlled carefully to ensure proper morphogenesis. HSPGs are abundantly expressed in the extracellular matrix, which creates ample opportunities for them to interact with signaling molecules. The strength of such interaction, however, varies greatly due to the enormous structural heterogeneity of HSPGs, making HSPGs ideally suited to shape the morphogen gradient.
Experimental evidence first suggested that HSPGs function as ‘reservoirs’ for FGF ligands. Germinal in vitro studies established that HSPGs bind FGF ligands to prevent their degradation and proteolysis, thereby increasing their radius of diffusion [54–57]. More recently, analysis of FGF gradient spanning the embryonic mouse midbrain, however, suggests that FGF protein is immobilized by its interaction with the cell surface HSPGs [58]. Live imaging of single-molecule dynamics in gastrulating Zebrafish embryos show that the majority of FGF8 exists as freely diffusible single molecules, but a small fraction of FGF8 are HSPG-bound that move significantly slower than predicted for Brownian motion [59]. Enzymatic degradation of HSPGs extended the range of FGF signaling domain, mimicking the gain-of-function FGF phenotype. Further single cell imaging shows that the movement of FGF2 in the extracellular matrix can be slow or fast, short or long range, depending on the translocation of FGF2 molecules from one HSPG to another [60]. The importance of FGF–HSPG interaction is also revealed by the dynamics of FGF dimerization. Only homodimers of FGF9 and FGF20 are effective in binding to HSPGs, which restrict their diffusion [61]. A natural occurring mouse mutation disrupting FGF9 dimerization allows FGF9 to diffuse long distances, leading to ectopic FGF9 signaling [62]. Finally, explant studies on lacrimal gland and submandibular gland branching morphogenesis showed that FGF10 and FGF7 promotes bud elongation or branching respectively, which correlates with their ranges of diffusion in the presence of HSPGs [63]. A single amino acid change in FGF10 that reduces its affinity to HSPGs converts FGF10 into a FGF7-like ligand, resulting in extensive branching of the glands. But what happens when all HSPGs are removed to allow unfettered diffusion of FGF? This question is addressed in a study of lacrimal gland development where the gene encoding a key biosynthetic enzyme Ugdh is knocked out to ablate the synthesis of all glycosaminoglycans including HSPGs [64]. Ugdh null embryos display excessive dispersion of FGF10 in mesenchyme and failure of lacrimal gland budding, resembling FGF10 loss-of-function phenotype. Therefore, whereas modest reduction in FGF–HSPG interaction extends the signaling range of FGF, a complete loss of HSPGs abrogates the morphogen gradient of FGF.
More recently, details of the specific core protein and HS side chain of HSPGs that regulates the binding and subsequent release of FGF ligand are being elucidated. A recent study on the collective cell migration in Zebrafish lateral line noted that the loss of HS side chains of Syndecans and Glypicans arising from mutations in Extl3 and Ext2 resulted in increased diffusion of FGF ligands into the surrounding tissue [65]. Similarly, local retention of FGF4 and FGF8 ligands by HS side chains was shown to be important for the activation of FGF signaling in mouse extraembryonic ectoderm [66]. Moreover, this study showed that FGF ligands are more likely to bind and interact with trans-membrane type of cell-surface tethered HSPGs such as Syndecans than with secreted type HSPGs such as Perlecans. It was previously noted in lung development that HS low in sulfation are expressed in the mesenchyme surrounding the prospective bud while the highly sulfated HS are present in basement membranes of the epithelial tubules, suggesting that the dynamic pattern of HS modification may be important for branching morphogenesis [67]. Interestingly, lacrimal gland development is normal when HS 2-O and 6-O sulfation are genetically ablated in the mesenchyme, but it is abolished by deleting Ndst genes, which not only affects HS N-sulfation but also the overall level of HS sulfation [64,68]. Hence at least for FGF10, the density instead of the position of sulfation groups in HS is likely to play a more dominant role in restraining FGF10 diffusion.
3.2. Shedding of HSPGs and HS side chains regulate distribution of FGF ligands
Proteases and endoglycosidases cleave HSPGs and/or HS side chains respectively from the cell membrane or the extracellular matrix to facilitate the distribution of FGF ligands. HtrA1 is a serine protease that cleaves membrane bound HSPGs such as syndecans and glypicans, which facilitate the long-range dispersal of FGF ligands to affect mesoderm formation and neuronal differentiation in developing Xenopus embryos [69]. Similarly in mouse, proteolytic cleavage of FGF-bound HS side chains has been shown to facilitate and extend the non-cell autonomous FGF signaling range in the extra-embryonic ectoderm [66]. Heparanase cleaves ECM HSPG perlecan to release FGF10, which is required for submandibular gland branching morphogenesis [70]. Heparanase is also required for the ectodomain shedding of syndecan-1 to release HS fragments that can bind and activate FGF2 signaling during tissue repair [71]. While HSPGs maintain local FGF gradient by binding FGF ligands, these results show that the cleavage of HS side chains or core proteins by endoglycosidases and proteases can also aid in the release of FGF ligands.
3.3. HSPGs act as co-receptors to modulate FGF signaling gradient
Pioneering in vitro studies first suggested the role of HSPGs as co-receptors in FGF signaling [33–36,72,73]. In this role, HSPGs function by physically approximating FGF ligands to FGF receptors and by simultaneously binding FGF ligands and receptors to induce a conformational change required for FGF signal activation. Genetic manipulation of HS biosynthesis enzymes provides evidence that there exists heterogeneity in the manner by which HSPGs influence FGF downstream signaling [74–79]. In addition, the action of 6-O-Endosulfatases Sulf1 and Sulf2 on HSPGs lead to negative regulation of FGF signaling [80,81]. The differential distribution of HSPGs in tissues could plausibly establish a gradient of FGF co-receptors and in turn elicit a graded FGF signaling. This has been observed in mammalian lens development, where HSPGs are expressed in an anteriorhigh–posterlow pattern in the lens [82]. Ligand and carbohydrate engagement (LACE) assay reveals that the in situ assembly of FGF/FGFR complexes in the lens becomes increasingly stronger from the anterior to the posterior side. This corresponds to the spatial gradient of FGF signaling necessary to promote proliferation of the anterior epithelial cells and differentiation of the posterior fiber cells [83]. Indeed, genetic ablation of HSPGs by deleting Ndst1 and Ndst2 disrupted FGF signaling in the lens, leading to the cell proliferation and differentiation defects [82]. Thus HSPGs can utilize their co-receptor function to actively shape the FGF gradient in the signal receiving cells.
4. FGF gradient is controlled by the secretion and internalization of FGF ligands and receptors
The secretion of FGF ligand from the signaling cells is also a crucial step in establishing FGF morphogen gradient. Studies have described the intracellular trafficking of FGF ligands and function of HSPG to act as a ‘molecular trap’ for the translocation of secreted FGF2 across the cell membrane [84,85]. It has been recently discovered that subcellular trafficking of Drosophila FGF branchless in flight muscle can be redirected from surface to T-tubules, which is necessary to promote tracheal invasion [86]. In Zebrafish embryos, the migrating lateral line primordium forms a microluminal structure, which locally traps and concentrate freely diffusible FGF ligands [87]. These results demonstrate the importance of tissue cavities in regulating FGF diffusion.
According to a source-sink model, FGF gradient also relies on the process of endocytosis for the removal of FGF ligands and receptors [59]. Evidence for the role of HSPGs in internalizing FGF initially came from in vitro studies [88–90]. However, following internalization, presumably by endocytosis, the fate of FGF ligands in the cell is widely debated [91]. While some studies indicate that FGF follows a lysosomal degradative pathway after HSPG-mediated endocytosis[92,93], others argue that the internalization of HSPG-bound FGF is likely to be protected from lysosomal proteases [88]. While there is some evidence that internalization of FGF2 by HSPGs act as a shuttle for nuclear transport [94], a more recent study found that syndecan-4 internalized FGF2 and FGFR1 by the process of macropinocytosis to form functional cytoplasmic endosomes [95].
The availability of FGF receptor on the cell surface is important for sensing FGF gradient. Studies show that Src and Esp8 mediate FGF receptor endocytosis by recruiting activated FGFRs into clathrin-coated pits in a dynamin-dependent pathway [96,97]. Ubiquitination of FGFRs is required for lysosomal sorting of FGFRs, causing them to be degraded or recycled to affect FGF signaling [98–102]. Extended-synaptotagmin related membrane protein ESyt2 has been shown to act as an FGFR adaptor for clathrin-mediated endocytosis [103–105]. Alternatively, FGFRs can also be endocytosed in a clathrin- and dynamin-independent pathway depending on the type of FGFR [106]. While endocytosis of FGFR mostly follows a degradative pathway, sometimes FGFRs are subjected to intracellular trafficking. Signaling pathways and ligands that promote the recycling of endosomal FGFRs to the cell surface during crucial developmental processes have been described[107,108]. Taken together, the internalization and trafficking of FGFs and FGFRs modulate morphogen gradient formation during development.
5. Other mechanisms that influence FGF morphogen gradient formation
A gradual decay of FGF mRNA is yet another mechanism of gradient formation [109]. In the developing mouse embryo, Fgf8 mRNA is produced in the posterior tip of the developing embryo. As the axial elongation of differentiated tissue proceeds toward the anterior portion of the embryo, Fgf8 mRNA decays progressively in a similar fashion. This leads to the formation of an Fgf8 mRNA gradient along the anterior-posterior axis of the developing embryo, which translates into FGF8 protein gradient that alters downstream FGF signaling in a dose-dependent manner.
FGF signal interpretation also depends on several modulators of the FGF signaling cascade. Sprouty was identified in Drosophila as an inhibitor of FGF pathway via its suppression of Ras-MAPK pathway [110,111]. Spred is a Sprouty related tyrosine kinase substrate that also acts as an inhibitor of FGF signaling [112]. Actions of both Sprouty and Spred are essential for FGF signal interpretation during the development of Xenopus mesoderm [113]. Another modulator of FGF signaling – Sef is positively regulated by FGF in Xenopus and Zebrafish models. It regulates FGF signaling via a feedback mechanism by binding FGFR and affects various aspects of vertebrate development [114–118]. Recently, serpin protease nexin-1 (PN1) has been identified in developing Xenopus embryos as another feedback regulator of FGF signaling. Activated by FGF signaling, PN1 binds to and inhibits serine protease Htra1, thereby preventing the degradation of Syndecan 4 leading to an altered FGF signaling [119].
6. Perspectives
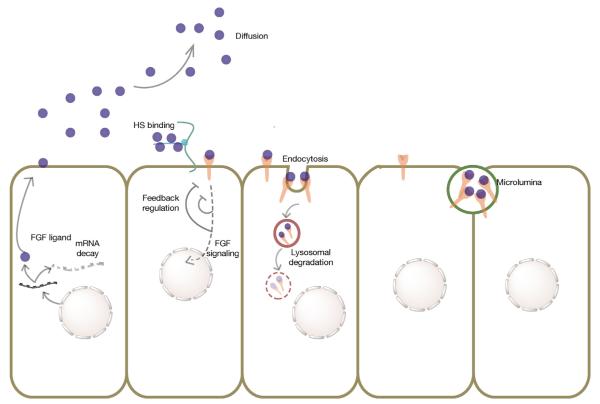
In summary, the role of FGF signaling molecules as morphogen has been established in various systems. Largely regulated extracellularly by HSPGs, FGF morphogen concentration gradient across the developing embryo induces sub-cellular signaling in a dose-dependent manner. FGF gradient is also controlled by other mechanisms such as mRNA decay, ligand trafficking and secretion, endocytosis of ligand and receptor, tissue cavity and feedback mechanisms by inhibitors of FGF pathway (summarized in Fig. 2).
Fig. 2.
Summary of mechanisms regulating FGF gradient formation: FGF ligands from the source cell are bound by HSPG side chains to prevent their diffusion farther away from the source tissue. FGF ligands bound to the receptors initiate FGF signaling cascade that is subject to feedback regulation at various levels, mainly by FGF inhibitors. Alternatively, FGF mRNA can undergo decay as cells move away from the local expression domain. FGF ligands and receptors can also be endocytosed to follow a mostly degradative pathway. FGF ligands can also be locally sequestered by the formation of microluminal structures.
While various mechanisms influencing FGF morphogen gradient have been described, the recent discovery of microluminal structures to confine FGF ligand movement [87] suggests new paradigms of gradient formation are still being revealed. Further studies are needed to determine if microluminal structures are formed during mammalian embryogenesis as well. The availability and ease of genetic manipulations in various model systems have enabled us to delve further into the specifics of HSPGs in creating a FGF concentration gradient. The molecular details of FGF movement within the extracellular matrix itself still remains to be elucidated, which will likely be addressed by technical advances in super resolution microscopy and synthesis of heparan sulfates with defined structure. Resolving how FGF morphogen gradient is controlled may provide important insights for future development of pharmacological interventions.
Acknowledgements
The authors thank members of the Zhang lab for discussions. The work was supported by grants from NIH (EY017061 and EY018868 to XZ). XZ is supported by Jules and Doris Stein Research to Prevent Blindness Professorship.
References
- [1].Armelin HA. Pituitary extracts and steroid hormones in the control of 3T3 cell growth. Proc Natl Acad Sci U S A. 1973;70:2702–6. doi: 10.1073/pnas.70.9.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wilson R, Leptin M. Fibroblast growth factor receptor-dependent morphogenesis of the Drosophila mesoderm. Philos Trans R Soc Lond Ser B, Biol Sci. 2000;355:891–5. doi: 10.1098/rstb.2000.0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-3-reviews3005. REVIEWS 3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Borland CZ, Schutzman JL, Stern MJ. Fibroblast growth factor signaling in Caenorhabditis elegans. BioEssays. 2001;23:1120–30. doi: 10.1002/bies.10007. [DOI] [PubMed] [Google Scholar]
- [5].Bottcher RT, Niehrs C. Fibroblast growth factor signaling during early vertebrate development. Endocr Rev. 2005;26:63–77. doi: 10.1210/er.2003-0040. [DOI] [PubMed] [Google Scholar]
- [6].Dorey K, Amaya E. FGF signalling: diverse roles during early vertebrate embryogenesis. Development. 2010;137:3731–42. doi: 10.1242/dev.037689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Deng CX, Wynshaw-Boris A, Shen MM, Daugherty C, Ornitz DM, Leder P. Murine FGFR-1 is required for early postimplantation growth and axial organization. Genes Dev. 1994;8:3045–57. doi: 10.1101/gad.8.24.3045. [DOI] [PubMed] [Google Scholar]
- [8].Yamaguchi TP, Harpal K, Henkemeyer M, Rossant J. fgfr-1 is required for embryonic growth and mesodermal patterning during mouse gastrulation. Genes Dev. 1994;8:3032–44. doi: 10.1101/gad.8.24.3032. [DOI] [PubMed] [Google Scholar]
- [9].Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- [10].Wilson SI, Graziano E, Harland R, Jessell TM, Edlund T. An early requirement for FGF signalling in the acquisition of neural cell fate in the chick embryo. Curr Biol. 2000;10:421–9. doi: 10.1016/s0960-9822(00)00431-0. [DOI] [PubMed] [Google Scholar]
- [11].Streit A, Berliner AJ, Papanayotou C, Sirulnik A, Stern CD. Initiation of neural induction by FGF signalling before gastrulation. Nature. 2000;406:74–8. doi: 10.1038/35017617. [DOI] [PubMed] [Google Scholar]
- [12].Arman E, Haffner-Krausz R, Chen Y, Heath JK, Lonai P. Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc Natl Acad Sci U S A. 1998;95:5082–7. doi: 10.1073/pnas.95.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wells JM, Melton DA. Early mouse endoderm is patterned by soluble factors from adjacent germ layers. Development. 2000;127:1563–72. doi: 10.1242/dev.127.8.1563. [DOI] [PubMed] [Google Scholar]
- [14].Popovici C, Roubin R, Coulier F, Birnbaum D. An evolutionary history of the FGF superfamily. BioEssays. 2005;27:849–57. doi: 10.1002/bies.20261. [DOI] [PubMed] [Google Scholar]
- [15].Itoh N, Ornitz DM. Functional evolutionary history of the mouse Fgf gene family. Dev Dyn. 2008;237:18–27. doi: 10.1002/dvdy.21388. [DOI] [PubMed] [Google Scholar]
- [16].Itoh N, Ornitz DM. Fibroblast growth factors: from molecular evolution to roles in development, metabolism and disease. J Biochem. 2011;149:121–30. doi: 10.1093/jb/mvq121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Goldfarb M. Voltage-gated sodium channel-associated proteins and alternative mechanisms of inactivation and block. Cell Mol Life Sci. 2012;69:1067–76. doi: 10.1007/s00018-011-0832-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kuro-o M. Endocrine FGFs and Klothos: emerging concepts. Trends Endocrinol Metab. 2008;19:239–45. doi: 10.1016/j.tem.2008.06.002. [DOI] [PubMed] [Google Scholar]
- [19].Green JB, New HV, Smith JC. Responses of embryonic Xenopus cells to activin and FGF are separated by multiple dose thresholds and correspond to distinct axes of the mesoderm. Cell. 1992;71:731–9. doi: 10.1016/0092-8674(92)90550-v. [DOI] [PubMed] [Google Scholar]
- [20].Kengaku M, Okamoto H. bFGF as a possible morphogen for the antero-posterior axis of the central nervous system in Xenopus. Development. 1995;121:3121–30. doi: 10.1242/dev.121.9.3121. [DOI] [PubMed] [Google Scholar]
- [21].Christen B, Slack JM. Spatial response to fibroblast growth factor signalling in Xenopus embryos. Development. 1999;126:119–25. doi: 10.1242/dev.126.1.119. [DOI] [PubMed] [Google Scholar]
- [22].Fukuchi-Shimogori T, Grove EA. Neocortex patterning by the secreted signaling molecule FGF8. Science. 2001;294:1071–4. doi: 10.1126/science.1064252. [DOI] [PubMed] [Google Scholar]
- [23].Sansom SN, Livesey FJ. Gradients in the brain: the control of the development of form and function in the cerebral cortex. Cold Spring Harbor Perspect Biol. 2009;1:a002519. doi: 10.1101/cshperspect.a002519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Dasen JS, Liu JP, Jessell TM. Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature. 2003;425:926–33. doi: 10.1038/nature02051. [DOI] [PubMed] [Google Scholar]
- [25].Studer M, Lumsden A, Ariza-McNaughton L, Bradley A, Krumlauf R. Altered segmental identity and abnormal migration of motor neurons in mice lacking Hoxb-1. Nature. 1996;384:630–4. doi: 10.1038/384630a0. [DOI] [PubMed] [Google Scholar]
- [26].Bel-Vialar S, Itasaki N, Krumlauf R. Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development. 2002;129:5103–15. doi: 10.1242/dev.129.22.5103. [DOI] [PubMed] [Google Scholar]
- [27].Johnson DE, Williams LT. Structural and functional diversity in the FGF receptor multigene family. Adv Cancer Res. 1993;60:1–41. doi: 10.1016/s0065-230x(08)60821-0. [DOI] [PubMed] [Google Scholar]
- [28].Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem. 2006;281:15694–700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Itoh N, Ornitz DM. Evolution of the Fgf and Fgfr gene families. Trends Genet. 2004;20:563–9. doi: 10.1016/j.tig.2004.08.007. [DOI] [PubMed] [Google Scholar]
- [30].Mohammadi M, McMahon G, Sun L, Tang C, Hirth P, Yeh BK, et al. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science. 1997;276:955–60. doi: 10.1126/science.276.5314.955. [DOI] [PubMed] [Google Scholar]
- [31].Pellegrini L, Burke DF, von Delft F, Mulloy B, Blundell TL. Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature. 2000;407:1029–34. doi: 10.1038/35039551. [DOI] [PubMed] [Google Scholar]
- [32].Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A, et al. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell. 2000;6:743–50. doi: 10.1016/s1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- [33].Yayon A, Klagsbrun M, Esko JD, Leder P, Ornitz DM. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell. 1991;64:841–8. doi: 10.1016/0092-8674(91)90512-w. [DOI] [PubMed] [Google Scholar]
- [34].Rapraeger AC, Krufka A, Olwin BB. Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science. 1991;252:1705–8. doi: 10.1126/science.1646484. [DOI] [PubMed] [Google Scholar]
- [35].Ornitz DM, Yayon A, Flanagan JG, Svahn CM, Levi E, Leder P. Heparin is required for cell-free binding of basic fibroblast growth factor to a soluble receptor and for mitogenesis in whole cells. Mol Cell Biol. 1992;12:240–7. doi: 10.1128/mcb.12.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ornitz DM, Leder P. Ligand specificity and heparin dependence of fibroblast growth factor receptors 1 and 3. J Biol Chem. 1992;267:16305–11. [PubMed] [Google Scholar]
- [37].Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–7. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- [38].Hacker U, Nybakken K, Perrimon N. Heparan sulphate proteoglycans: the sweet side of development. Nat Rev Mol Cell Biol. 2005;6:530–41. doi: 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- [39].Matsuo I, Kimura-Yoshida C. Extracellular modulation of fibroblast growth factor signaling through heparan sulfate proteoglycans in mammalian development. Curr Opin Genet Dev. 2013;23:399–407. doi: 10.1016/j.gde.2013.02.004. [DOI] [PubMed] [Google Scholar]
- [40].Matsuo I, Kimura-Yoshida C. Extracellular distribution of diffusible growth factors controlled by heparan sulfate proteoglycans during mammalian embryogenesis. Philos Trans R Soc Lond Ser B, Biol Sci. 2014;369 doi: 10.1098/rstb.2013.0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sarrazin S, Lamanna WC, Esko JD. Heparan sulfate proteoglycans. Cold Spring Harbor Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bulow HE, Hobert O. The molecular diversity of glycosaminoglycans shapes animal development. Annu Rev Cell Dev Biol. 2006;22:375–407. doi: 10.1146/annurev.cellbio.22.010605.093433. [DOI] [PubMed] [Google Scholar]
- [43].Rogalski TM, Williams BD, Mullen GP, Moerman DG. Products of the unc-52 gene in Caenorhabditis elegans are homologous to the core protein of the mammalian basement membrane heparan sulfate proteoglycan. Genes Dev. 1993;7:1471–84. doi: 10.1101/gad.7.8.1471. [DOI] [PubMed] [Google Scholar]
- [44].Shworak NW, Kojima T, Rosenberg RD. Isolation and characterization of ryudocan and syndecan heparan sulfate proteoglycans, core proteins, and cDNAs from a rat endothelial cell line. Haemostasis. 1993;23(Suppl 1):161–76. doi: 10.1159/000216925. [DOI] [PubMed] [Google Scholar]
- [45].Kato M, Wang H, Bernfield M, Gallagher JT, Turnbull JE. Cell surface syndecan-1 on distinct cell types differs in fine structure and ligand binding of its heparan sulfate chains. J Biol Chem. 1994;269:18881–90. [PubMed] [Google Scholar]
- [46].Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–71. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- [47].Zak BM, Crawford BE, Esko JD. Hereditary multiple exostoses and heparan sulfate polymerization. Biochim Biophys Acta. 2002;1573:346–55. doi: 10.1016/s0304-4165(02)00402-6. [DOI] [PubMed] [Google Scholar]
- [48].Grobe K, Ledin J, Ringvall M, Holmborn K, Forsberg E, Esko JD, et al. Heparan sulfate and development: differential roles of the N-acetylglucosamine N-deacetylase/N-sulfotransferase isozymes. Biochim Biophys Acta. 2002;1573:209–15. doi: 10.1016/s0304-4165(02)00386-0. [DOI] [PubMed] [Google Scholar]
- [49].Thacker BE, Xu D, Lawrence R, Esko JD. Heparan sulfate 3-O-sulfation: a rare modification in search of a function. Matrix Biol. 2014;35:60–72. doi: 10.1016/j.matbio.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lamanna WC, Kalus I, Padva M, Baldwin RJ, Merry CL, Dierks T. The heparanome – the enigma of encoding and decoding heparan sulfate sulfation. J Biotechnol. 2007;129:290–307. doi: 10.1016/j.jbiotec.2007.01.022. [DOI] [PubMed] [Google Scholar]
- [51].Manon-Jensen T, Itoh Y, Couchman JR. Proteoglycans in health and disease: the multiple roles of syndecan shedding. FEBS J. 2010;277:3876–89. doi: 10.1111/j.1742-4658.2010.07798.x. [DOI] [PubMed] [Google Scholar]
- [52].Lander AD, Nie Q, Wan FY. Do morphogen gradients arise by diffusion? Dev Cell. 2002;2:785–96. doi: 10.1016/s1534-5807(02)00179-x. [DOI] [PubMed] [Google Scholar]
- [53].Wartlick O, Kicheva A, Gonzalez-Gaitan M. Morphogen gradient formation. Cold Spring Harbor Perspect Biol. 2009;1:a001255. doi: 10.1101/cshperspect.a001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Gospodarowicz D, Cheng J. Heparin protects basic and acidic FGF from inactivation. J Cell Physiol. 1986;128:475–84. doi: 10.1002/jcp.1041280317. [DOI] [PubMed] [Google Scholar]
- [55].Saksela O, Rifkin DB. Release of basic fibroblast growth factor-heparan sulfate complexes from endothelial cells by plasminogen activator-mediated proteolytic activity. J Cell Biol. 1990;110:767–75. doi: 10.1083/jcb.110.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Sommer A, Rifkin DB. Interaction of heparin with human basic fibroblast growth factor: protection of the angiogenic protein from proteolytic degradation by a glycosaminoglycan. J Cell Physiol. 1989;138:215–20. doi: 10.1002/jcp.1041380129. [DOI] [PubMed] [Google Scholar]
- [57].Flaumenhaft R, Moscatelli D, Rifkin DB. Heparin and heparan sulfate increase the radius of diffusion and action of basic fibroblast growth factor. J Cell Biol. 1990;111:1651–9. doi: 10.1083/jcb.111.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Chen Y, Mohammadi M, Flanagan JG. Graded levels of FGF protein span the midbrain and can instruct graded induction and repression of neural mapping labels. Neuron. 2009;62:773–80. doi: 10.1016/j.neuron.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Yu SR, Burkhardt M, Nowak M, Ries J, Petrasek Z, Scholpp S, et al. Fgf8 morphogen gradient forms by a source-sink mechanism with freely diffusing molecules. Nature. 2009;461:533–6. doi: 10.1038/nature08391. [DOI] [PubMed] [Google Scholar]
- [60].Duchesne L, Octeau V, Bearon RN, Beckett A, Prior IA, Lounis B, et al. Transport of fibroblast growth factor 2 in the pericellular matrix is controlled by the spatial distribution of its binding sites in heparan sulfate. PLoS Biol. 2012;10:e1001361. doi: 10.1371/journal.pbio.1001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kalinina J, Byron SA, Makarenkova HP, Olsen SK, Eliseenkova AV, Larochelle WJ, et al. Homodimerization controls the fibroblast growth factor 9 subfamily’s receptor binding and heparan sulfate-dependent diffusion in the extracellular matrix. Mol Cell Biol. 2009;29:4663–78. doi: 10.1128/MCB.01780-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Harada M, Murakami H, Okawa A, Okimoto N, Hiraoka S, Nakahara T, et al. FGF9 monomer-dimer equilibrium regulates extracellular matrix affinity and tissue diffusion. Nat Genet. 2009;41:289–98. doi: 10.1038/ng.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Makarenkova HP, Hoffman MP, Beenken A, Eliseenkova AV, Meech R, Tsau C, et al. Differential interactions of FGFs with heparan sulfate control gradient formation and branching morphogenesis. Sci Signal. 2009;2:ra55. doi: 10.1126/scisignal.2000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Qu X, Pan Y, Carbe C, Powers A, Grobe K, Zhang X. Glycosaminoglycan-dependent restriction of FGF diffusion is necessary for lacrimal gland development. Development. 2012;139:2730–9. doi: 10.1242/dev.079236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Venero Galanternik M, Kramer KL, Piotrowski T. Heparan sulfate proteoglycans regulate Fgf signaling and cell polarity during collective cell migration. Cell Rep. 2015 doi: 10.1016/j.celrep.2014.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Shimokawa K, Kimura-Yoshida C, Nagai N, Mukai K, Matsubara K, Watanabe H, et al. Cell surface heparan sulfate chains regulate local reception of FGF signaling in the mouse embryo. Dev Cell. 2011;21:257–72. doi: 10.1016/j.devcel.2011.06.027. [DOI] [PubMed] [Google Scholar]
- [67].Izvolsky KI, Shoykhet D, Yang Y, Yu Q, Nugent MA, Cardoso WV. Heparan sulfate-FGF10 interactions during lung morphogenesis. Dev Biol. 2003;258:185–200. doi: 10.1016/s0012-1606(03)00114-3. [DOI] [PubMed] [Google Scholar]
- [68].Qu X, Carbe C, Tao C, Powers A, Lawrence R, van Kuppevelt TH, et al. Lacrimal gland development and Fgf10-Fgfr2b signaling are controlled by 2-O- and 6-O-sulfated heparan sulfate. J Biol Chem. 2011;286:14435–44. doi: 10.1074/jbc.M111.225003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Hou S, Maccarana M, Min TH, Strate I, Pera EM. The secreted serine protease xHtrA1 stimulates long-range FGF signaling in the early Xenopus embryo. Dev Cell. 2007;13:226–41. doi: 10.1016/j.devcel.2007.07.001. [DOI] [PubMed] [Google Scholar]
- [70].Patel VN, Knox SM, Likar KM, Lathrop CA, Hossain R, Eftekhari S, et al. Heparanase cleavage of perlecan heparan sulfate modulates FGF10 activity during ex vivo submandibular gland branching morphogenesis. Development. 2007;134:4177–86. doi: 10.1242/dev.011171. [DOI] [PubMed] [Google Scholar]
- [71].Kato M, Wang H, Kainulainen V, Fitzgerald ML, Ledbetter S, Ornitz DM, et al. Physiological degradation converts the soluble syndecan-1 ectodomain from an inhibitor to a potent activator of FGF-2. Nat Med. 1998;4:691–7. doi: 10.1038/nm0698-691. [DOI] [PubMed] [Google Scholar]
- [72].Nurcombe V, Ford MD, Wildschut JA, Bartlett PF. Developmental regulation of neural response to FGF-1 and FGF-2 by heparan sulfate proteoglycan. Science. 1993;260:103–6. doi: 10.1126/science.7682010. [DOI] [PubMed] [Google Scholar]
- [73].Heath WF, Cantrell AS, Mayne NG, Jaskunas SR. Mutations in the heparin-binding domains of human basic fibroblast growth factor alter its biological activity. Biochemistry. 1991;30:5608–15. doi: 10.1021/bi00236a039. [DOI] [PubMed] [Google Scholar]
- [74].Pan Y, Carbe C, Powers A, Zhang EE, Esko JD, Grobe K, et al. Bud specific N-sulfation of heparan sulfate regulates Shp2-dependent FGF signaling during lacrimal gland induction. Development. 2008;135:301–10. doi: 10.1242/dev.014829. [DOI] [PubMed] [Google Scholar]
- [75].Pan Y, Woodbury A, Esko JD, Grobe K, Zhang X. Heparan sulfate biosynthetic gene Ndst1 is required for FGF signaling in early lens development. Development. 2006;133:4933–44. doi: 10.1242/dev.02679. [DOI] [PubMed] [Google Scholar]
- [76].Patel VN, Likar KM, Zisman-Rozen S, Cowherd SN, Lassiter KS, Sher I, et al. Specific heparan sulfate structures modulate FGF10-mediated submandibular gland epithelial morphogenesis and differentiation. J Biol Chem. 2008;283:9308–17. doi: 10.1074/jbc.M709995200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Lin X, Buff EM, Perrimon N, Michelson AM. Heparan sulfate proteoglycans are essential for FGF receptor signaling during Drosophila embryonic development. Development. 1999;126:3715–23. doi: 10.1242/dev.126.17.3715. [DOI] [PubMed] [Google Scholar]
- [78].Kamimura K, Koyama T, Habuchi H, Ueda R, Masu M, Kimata K, et al. Specific and flexible roles of heparan sulfate modifications in Drosophila FGF signaling. J Cell Biol. 2006;174:773–8. doi: 10.1083/jcb.200603129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Norton WH, Ledin J, Grandel H, Neumann CJ. HSPG synthesis by zebrafish Ext2 and Extl3 is required for Fgf10 signalling during limb development. Development. 2005;132:4963–73. doi: 10.1242/dev.02084. [DOI] [PubMed] [Google Scholar]
- [80].Otsuki S, Hanson SR, Miyaki S, Grogan SP, Kinoshita M, Asahara H, et al. Extracellular sulfatases support cartilage homeostasis by regulating BMP and FGF signaling pathways. Proc Natl Acad Sci U S A. 2010;107:10202–7. doi: 10.1073/pnas.0913897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Wang S, Ai X, Freeman SD, Pownall ME, Lu Q, Kessler DS, et al. QSulf1, a heparan sulfate 6-O-endosulfatase, inhibits fibroblast growth factor signaling in mesoderm induction and angiogenesis. Proc Natl Acad Sci U S A. 2004;101:4833–8. doi: 10.1073/pnas.0401028101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Qu X, Hertzler K, Pan Y, Grobe K, Robinson ML, Zhang X. Genetic epistasis between heparan sulfate and FGF-Ras signaling controls lens development. Dev Biol. 2011;355:12–20. doi: 10.1016/j.ydbio.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Lovicu FJ, McAvoy JW. Growth factor regulation of lens development. Dev Biol. 2005;280:1–14. doi: 10.1016/j.ydbio.2005.01.020. [DOI] [PubMed] [Google Scholar]
- [84].Nickel W. Unconventional secretion: an extracellular trap for export of fibroblast growth factor 2. J Cell Sci. 2007;120:2295–9. doi: 10.1242/jcs.011080. [DOI] [PubMed] [Google Scholar]
- [85].Taverna S, Rigogliuso S, Salamone M, Vittorelli ML. Intracellular trafficking of endogenous fibroblast growth factor-2. FEBS J. 2008;275:1579–92. doi: 10.1111/j.1742-4658.2008.06316.x. [DOI] [PubMed] [Google Scholar]
- [86].Peterson SJ, Krasnow MA. Subcellular trafficking of FGF controls tracheal invasion of Drosophila flight muscle. Cell. 2015;160:313–23. doi: 10.1016/j.cell.2014.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Durdu S, Iskar M, Revenu C, Schieber N, Kunze A, Bork P, et al. Luminal signalling links cell communication to tissue architecture during organogenesis. Nature. 2014;515:120–4. doi: 10.1038/nature13852. [DOI] [PubMed] [Google Scholar]
- [88].Roghani M, Moscatelli D. Basic fibroblast growth factor is internalized through both receptor-mediated and heparan sulfate-mediated mechanisms. J Biol Chem. 1992;267:22156–62. [PubMed] [Google Scholar]
- [89].Rusnati M, Urbinati C, Presta M. Internalization of basic fibroblast growth factor (bFGF) in cultured endothelial cells: role of the low affinity heparin-like bFGF receptors. J Cell Physiol. 1993;154:152–61. doi: 10.1002/jcp.1041540119. [DOI] [PubMed] [Google Scholar]
- [90].Quarto N, Amalric F. Heparan sulfate proteoglycans as transducers of FGF-2 signalling. J Cell Sci. 1994;107(Pt 11):3201–12. doi: 10.1242/jcs.107.11.3201. [DOI] [PubMed] [Google Scholar]
- [91].Christianson HC, Belting M. Heparan sulfate proteoglycan as a cell-surface endocytosis receptor. Matrix Biol. 2014;35:51–5. doi: 10.1016/j.matbio.2013.10.004. [DOI] [PubMed] [Google Scholar]
- [92].Gleizes PE, Noaillac-Depeyre J, Amalric F, Gas N. Basic fibroblast growth factor (FGF-2) internalization through the heparan sulfate proteoglycans-mediated pathway: an ultrastructural approach. Eur J Cell Biol. 1995;66:47–59. [PubMed] [Google Scholar]
- [93].Citores L, Khnykin D, Sorensen V, Wesche J, Klingenberg O, Wiedlocha A, et al. Modulation of intracellular transport of acidic fibroblast growth factor by mutations in the cytoplasmic receptor domain. J Cell Sci. 2001;114:1677–89. doi: 10.1242/jcs.114.9.1677. [DOI] [PubMed] [Google Scholar]
- [94].Hsia E, Richardson TP, Nugent MA. Nuclear localization of basic fibroblast growth factor is mediated by heparan sulfate proteoglycans through protein kinase C signaling. J Cell Biochem. 2003;88:1214–25. doi: 10.1002/jcb.10470. [DOI] [PubMed] [Google Scholar]
- [95].Elfenbein A, Lanahan A, Zhou TX, Yamasaki A, Tkachenko E, Matsuda M, et al. Syndecan 4 regulates FGFR1 signaling in endothelial cells by directing macropinocytosis. Sci Signal. 2012;5:ra36. doi: 10.1126/scisignal.2002495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Auciello G, Cunningham DL, Tatar T, Heath JK, Rappoport JZ. Regulation of fibroblast growth factor receptor signalling and trafficking by Src and Eps8. J Cell Sci. 2013;126:613–24. doi: 10.1242/jcs.116228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Sandilands E, Akbarzadeh S, Vecchione A, McEwan DG, Frame MC, Heath JK. Src kinase modulates the activation, transport and signalling dynamics of fibroblast growth factor receptors. EMBO Rep. 2007;8:1162–9. doi: 10.1038/sj.embor.7401097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Haugsten EM, Malecki J, Bjorklund SM, Olsnes S, Wesche J. Ubiquitination of fibroblast growth factor receptor 1 is required for its intracellular sorting but not for its endocytosis. Mol Biol Cell. 2008;19:3390–403. doi: 10.1091/mbc.E07-12-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Persaud A, Alberts P, Hayes M, Guettler S, Clarke I, Sicheri F, et al. Nedd4-1 binds and ubiquitylates activated FGFR1 to control its endocytosis and function. EMBO J. 2011;30:3259–73. doi: 10.1038/emboj.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Nowak M, Machate A, Yu SR, Gupta M, Brand M. Interpretation of the FGF8 morphogen gradient is regulated by endocytic trafficking. Nat Cell Biol. 2011;13:153–8. doi: 10.1038/ncb2155. [DOI] [PubMed] [Google Scholar]
- [101].Persaud A, Alberts P, Mari S, Tong J, Murchie R, Maspero E, et al. Tyrosine phosphorylation of NEDD4 activates its ubiquitin ligase activity. Sci Signal. 2014;7:ra95. doi: 10.1126/scisignal.2005290. [DOI] [PubMed] [Google Scholar]
- [102].Nadratowska-Wesolowska B, Haugsten EM, Zakrzewska M, Jakimowicz P, Zhen Y, Pajdzik D, et al. RSK2 regulates endocytosis of FGF receptor 1 by phosphorylation on serine 789. Oncogene. 2014;33:4823–36. doi: 10.1038/onc.2013.425. [DOI] [PubMed] [Google Scholar]
- [103].Jean S, Mikryukov A, Tremblay MG, Baril J, Guillou F, Bellenfant S, et al. Extended-synaptotagmin-2 mediates FGF receptor endocytosis and ERK activation in vivo. Dev Cell. 2010;19:426–39. doi: 10.1016/j.devcel.2010.08.007. [DOI] [PubMed] [Google Scholar]
- [104].Tremblay MG, Herdman C, Guillou F, Mishra PK, Baril J, Bellenfant S, et al. Extended synaptotagmin interaction with the FGF receptor depends on receptor conformation, not catalytic activity. J Biol Chem. 2015 doi: 10.1074/jbc.M115.656918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Jean S, Tremblay MG, Herdman C, Guillou F, Moss T. The endocytic adapter E-Syt2 recruits the p21 GTPase activated kinase PAK1 to mediate actin dynamics and FGF signalling. Biol Open. 2012;1:731–8. doi: 10.1242/bio.2012968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Haugsten EM, Zakrzewska M, Brech A, Pust S, Olsnes S, Sandvig K, et al. Clathrin- and dynamin-independent endocytosis of FGFR3–implications for signalling. PLoS ONE. 2011;6:e21708. doi: 10.1371/journal.pone.0021708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Francavilla C, Cattaneo P, Berezin V, Bock E, Ami D, de Marco A, et al. The binding of NCAM to FGFR1 induces a specific cellular response mediated by receptor trafficking. J Cell Biol. 2009;187:1101–16. doi: 10.1083/jcb.200903030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Ueno H, Huang X, Tanaka Y, Hirokawa N. KIF16B/Rab14 molecular motor complex is critical for early embryonic development by transporting FGF receptor. Dev Cell. 2011;20:60–71. doi: 10.1016/j.devcel.2010.11.008. [DOI] [PubMed] [Google Scholar]
- [109].Dubrulle J, Pourquie O. fgf8 mRNA decay establishes a gradient that couples axial elongation to patterning in the vertebrate embryo. Nature. 2004;427:419–22. doi: 10.1038/nature02216. [DOI] [PubMed] [Google Scholar]
- [110].Casci T, Vinos J, Freeman M. Sprouty, an intracellular inhibitor of Ras signaling. Cell. 1999;96:655–65. doi: 10.1016/s0092-8674(00)80576-0. [DOI] [PubMed] [Google Scholar]
- [111].Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA. Sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell. 1998;92:253–63. doi: 10.1016/s0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- [112].Wakioka T, Sasaki A, Kato R, Shouda T, Matsumoto A, Miyoshi K, et al. Spred is a Sprouty-related suppressor of Ras signalling. Nature. 2001;412:647–51. doi: 10.1038/35088082. [DOI] [PubMed] [Google Scholar]
- [113].Sivak JM, Petersen LF, Amaya E. FGF signal interpretation is directed by Sprouty and Spred proteins during mesoderm formation. Dev Cell. 2005;8:689–701. doi: 10.1016/j.devcel.2005.02.011. [DOI] [PubMed] [Google Scholar]
- [114].Tsang M, Friesel R, Kudoh T, Dawid IB. Identification of Sef, a novel modulator of FGF signalling. Nat Cell Biol. 2002;4:165–9. doi: 10.1038/ncb749. [DOI] [PubMed] [Google Scholar]
- [115].Furthauer M, Lin W, Ang SL, Thisse B, Thisse C. Sef is a feedback-induced antagonist of Ras/MAPK-mediated FGF signalling. Nat Cell Biol. 2002;4:170–4. doi: 10.1038/ncb750. [DOI] [PubMed] [Google Scholar]
- [116].Newitt P, Boros J, Madakashira BP, Robinson ML, Reneker LW, McAvoy JW, et al. Sef is a negative regulator of fiber cell differentiation in the ocular lens. Differentiation. 2010;80:53–67. doi: 10.1016/j.diff.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Abraira VE, Hyun N, Tucker AF, Coling DE, Brown MC, Lu C, et al. Changes in Sef levels influence auditory brainstem development and function. J Neurosci. 2007;27:4273–82. doi: 10.1523/JNEUROSCI.3477-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Harduf H, Halperin E, Reshef R, Ron D. Sef is synexpressed with FGFs during chick embryogenesis and its expression is differentially regulated by FGFs in the developing limb. Dev Dyn. 2005;233:301–12. doi: 10.1002/dvdy.20364. [DOI] [PubMed] [Google Scholar]
- [119].Acosta H, Iliev D, Grahn TH, Gouignard N, Maccarana M, Griesbach J, et al. The serpin PN1 is a feedback regulator of FGF signaling in germ layer and primary axis formation. Development. 2015;142:1146–58. doi: 10.1242/dev.113886. [DOI] [PubMed] [Google Scholar]