Abstract
AML1-ETO is the translational product of a chimeric gene created by the stable chromosome translocation t (8;21)(q22;q22). It causes acute myeloid leukemia (AML) by dysregulating the expression of genes critical for myeloid cell development and differentiation and recently has been reported to bind multiple subunits of the mammalian cytosolic chaperonin TRiC (or CCT), primarily through its DNA binding domain (AML1-175). Through these interactions, TRiC plays an important role in the synthesis, folding, and activity of AML1-ETO. Using single-particle cryo-electron microscopy, we demonstrate here that a folding intermediate of AML1-ETO’s DNA-binding domain (AML1-175) forms a stable complex with apo-TRiC. Our structure reveals that AML1-175 associates directly with a specific subset of TRiC subunits in the open conformation.
Introduction
AML1-ETO is a rogue transcription factor that results from chromosomal translocation, t (8;21)(q22;q22), and causes acute myeloid leukemia (AML), a disease characterized by the uncontrolled proliferation and incomplete differentiation of a malignant clone of myeloid stem or progenitor cells (1). The chimeric protein comprises the conserved DNA binding domain from the hematopoietic transcription factor AML1 (also known as RUNX1) joined to most of the sequence of the ETO (2). The AML1 gene encodes a critical transcription factor that regulates a variety of genes involved in proliferation and differentiation of many cell types, including those within the hematopoietic system (3, 4). On the other hand, ETO possesses dimerization and zinc finger motifs (5) and appears to function as a scaffold for the assembly of corepressors including mSin3A, SMRT, and histone deacetylases (6, 7, 8). AML1-ETO retains the ability to bind AML1 DNA binding sites through its DNA binding domain while gaining the capacity to recruit additional cofactors (mostly corepressors) through the ETO moiety. Thus, AML1-ETO has been predicted to act as a transcriptional repressor for AML1 target genes (2).
Although there are currently no specific treatments for AML1-ETO-positive leukemia, it has been demonstrated that AML1-ETO suppression by small interfering RNAs (siRNAs) supports normal myeloid differentiation of t (8;21)-positive leukemic cells (9), which highlights AML1-ETO as a major clinical target to treat AML. Interestingly, inhibition of HSP70/90, two major proteostasis regulators, has shown antileukemic effects in AML1-ETO-positive cells (10, 11). Moreover, it has recently been reported that the mammalian cytosolic chaperonin TRiC (or CCT) modulates the synthesis, folding and activity of AML1-ETO by direct association, primarily through its DNA-binding domain (AML1-175), and that HSP70 promotes this interaction (12). These studies suggest that AML1-ETO relies on the molecular chaperone network to fold and function properly. However, few details are known about the interplay between AML1-ETO and the proteostasis system, which may be targeted for therapeutic benefit.
The proper folding of proteins into their native conformation in the crowded milieu of the cell is facilitated by molecular chaperones, a network of cellular proteostasis regulators (13). Among these, the essential mammalian cytosolic chaperonin TRiC (T-complex polypeptide-1 ring complex, also known as CCT or chaperonin containing TCP1) is an ATPase with an intricate architecture, which allows it to fold many essential proteins. TRiC substrates include cell cycle regulators, signaling proteins, and cytoskeletal components (14, 15). Furthermore, TRiC has been suggested to play a critical role in cancer cell development by modulating the folding and activity of client proteins involved in oncogenesis (16), such as the tumor suppressor proteins Von Hippel-Lindau (VHL) (17, 18) and p53 (19), as well as the oncogenic protein STAT3 and AML1-ETO (12, 20). Structurally, TRiC/CCT is a 1 MDa hetero-oligomeric complex composed of two rings with eight different, yet paralogous, subunits each (CCT1–CCT8) arranged in a specific order (21, 22). The rings are stacked back-to-back, forming a double-ring structure with an empty central cavity in each ring (23, 24). During a folding reaction, the rings of the chaperonin can close through large conformational changes driven by ATP hydrolysis, thereby providing an isolated environment for client proteins to fold (25, 26). TRiC has the unique ability to fold certain proteins that cannot be folded by simpler chaperone systems; Indeed, TRiC plays a central role in the cytosol by assisting the folding of ∼10% of all newly synthesized polypeptides (27).
All of TRiC’s subunits are structural homologs that consist of an ATP-binding equatorial domain and a substrate-binding apical domain linked by an intermediate domain (14). However, the apical domains of the eight subunits are highly divergent, which allows them to recognize different motifs in substrate proteins (24, 28, 29). The diversification of TRiC subunits thus provides a modular array of binding specificities that allows for a combinatorial recognition of substrate polypeptides (i.e., different substrates are hypothesized to engage TRiC through different combinations of subunits). In this report, we demonstrate that TRiC contributes to the activity of AML1-ETO’s DNA-binding domain (AML1-175) through direct association, consistent with the association between TRiC and full-length AML1-ETO (12). In addition, single-particle cryo-electron microscopy (cryo-EM) reveals that AML1-175 associates specifically with the apical domains of adjacent subunits CCT6 and CCT8, which may be potential therapeutic targets to treat AML1-ETO-positive AML.
Materials and Methods
Protein purifications
Human TRiC
Human TRiC was purified from HELA cells as previously described (30, 31). To achieve extra purity, TRiC fractions were incubated with 1 mM ATP for 15 min at 37°C and then reprocessed by Mono-Q HR 16/10 (GE Healthcare, Chicago, IL) and Superose 6 10/300 GL columns (GE Healthcare) in sequence. TRiC’s folding activity was assessed by luciferase refolding as described (32).
DNA-binding domain (AML1-175)
The coding sequence of full-length human AML1-ETO (pCDNA3-flag-AML1-ETO: Addgene) was subcloned with a C-terminal 6His-tag in pET28 (Novagen, Billerica, MA) expression vector and the domain sequence corresponding to 1-175 AML1-ETO was inserted with an N-terminal flag tag into a pET-His-2HR-T (Addgene) expression vector. Each plasmid was delivered to Rosetta DE3 (EMD Biosciences, Billerica, MA) cells cultured at 37°C. The cells were induced with 1 mM isopropyl-β-d-thiogalactopyranoside at an optical density of 0.7 at 600 nm. After growing for 4 h at 37°C, the cells were harvested, resuspended in buffer at pH 8.0 containing 20 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 1% Triton X-100 and 100 μg/ml lysozyme, and lysed by sonication. The insoluble fraction containing AML1-175 as inclusion bodies was collected by centrifugation. The harvested pellet was washed five times through cycles of sonication and centrifugation in resuspending buffer. The washed pellet was solubilized in buffer A at pH 8.0 (1X phosphate buffered saline, 100 mM NaCl, 5 mM imidazole, 10% glycerol, and 6 M guanidinium hydrochloride (Gn-HCl)). After centrifugation, the supernatant was applied onto a Ni-NTA column (GE his-trap HP) equilibrated with the same buffer. The column was washed successively with buffer A and proteins were eluted from the column with buffer B (buffer A with 300 mM imidazole). Subsequently, concentrated proteins using a centrifugal concentrator (Millipore, Billerica, MA) were applied to Superose 6(GE), preequilibrated with buffer 1X phosphate buffered saline, at pH 8.0, 100 mM NaCl, 10% glycerol, and 6 M Gn-HCl. Corresponding AML1-175 fractions were concentrated and stored at −80°C until further use.
In vitro protein refolding (Gel-shift DNA-binding assays)
Chemically denatured AML1-175 (25 μM) in 6 M Gn-HCl was rapidly diluted 100-fold into refolding buffer (25 mM HEPES at pH 7.5, 100 mM KOAc, 10 mM Mg(OAc)2, 2 mM DTT, 10 mM creatine phosphate, 200 mM NaCl, 10% glycerol), containing 25 μM TRiC oligomer, HSP70 and/or 1 mM ATP resulting in a 5:1 molar ratio of monomeric substrate to TRiC oligomer. The mixture was incubated at 30°C for 1 h and examined by gel shift DNA-binding assay as described previously (12).
Size exclusion chromatography
Analytical gel filtrations of protein complex were performed using Superose 6 10/300 GL (GE Healthcare) with a MQ buffer (20 mM HEPES, 0.1 mM EDTA, 5 mM MgCl2, and pH 7.4), with 50 mM NaCl, 1 mM DTT, and 10% glycerol. Experiments were conducted with a flow rate of 0.2 ml/min and 1.5× column volume. The molecular masses of separated fractions were deduced from the calibration curve of Superose 6 10/300 GL column. Fractions were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS PAGE) and analyzed by Western blot with antibodies for HSP70, Flag and CCT2.
Cryo-specimen preparation and cryo-EM imaging
AML1-175 denatured in 6 M Gn-HCl was diluted 100-fold into MQ buffer containing 0.5 μM purified human TRiC and 0.5 μM HSP70, resulting in a 10:1 molar ratio of substrate to TRiC. The complex mixture was applied to a Superose 6 column. The eluted fractions containing TRiC were concentrated with Q Sepharose resin and eluted with MQ with 300 mM NaCl, 2% glycerol, and 1% PEG8000. Octyl glucoside was added to a concentration of 0.05% before grid placement to increase the yield of side views in electron micrographs. For the nickel nanogold labeling experiment, ∼20 μM nanogold prepared in MQ buffer with 0.2% bovine serum albumin, 20% MeOH, and 600 mM imidazole were diluted 10 times into 1 μM TRiC-AML1-175 complex resulting in 2:1 molar ratio to TRiC at 60 mM final imidazole concentration. The resulting samples were applied to 200-mesh R1.2/1.3 holey-carbon grids (Quantifoil, Großlöbichau, Germany) and vitrified using an EM GP (Leica, Wetzlar, Germany). Images were collected on a JEM2010F electron microscope (JEOL (Freising, Germany), 200 kV) equipped with a field emission gun and a US4000 (Gatan, Pleasanton, CA). The charge-coupled device detector magnification was 55 K× (2.17Å/pixel) with a dose of 20∼25 e−/Å2. Images were captured across an approximate defocus range of 1.5–4.0 μm.
Three-dimensional cryo-EM map reconstruction
EMAN2.1 [62] and Relion1.2 (33) software packages were used in tandem for image processing and three-dimensional (3D) map reconstruction without imposing any symmetry. A total of 32,143 particles of TRiC-AML1-175 (apo-TRiC: 15,340) were boxed out and CTF-corrected in EMAN2.1. Two-dimensional (2D) class averages were computed and an initial 3D model was generated from scratch (34). We computed a single-model reconstruction using EMAN2.1, following the gold standard procedure (35, 36). This single model was filtered to 50 Å and used as an initial model for 3D classification of the raw particles into two groups using Relion1.2. Each of the two classified subgroups were then independently refined using Relion to generate a final map from each group (33). The resolution of the final map for each group was determined by Fourier shell correlation between two independent reconstructions using the 0.143 cutoff criterion.
Particle-based mean intensity analysis
To compute mean intensity histogram plots, the classified individual particles were edge-mean normalized and masked; the mean of nonzero values was then computed for each particle in each of the classified groups and plotted as one histogram per group on the same plot. A t-test was conducted to measure the z-score between the populations being compared.
Map deposition
The final maps for substrate-free TRiC, AML1-175-bound TRiC, group I/II of control apo-hTRiC, and the complex with Ni2+nanogold particle have been deposited into the Electron Microscopy Data Bank (EMData Bank) (EMD6226 ∼6230).
Results
Active AML1-175 forms a stable complex with TRiC
AML1-ETO consists of five distinct functional domains: DNA-binding (DBD), transactivation, linker, oligomerization, and zinc-finger (6). The DBD (or AML1-175) has been identified as the primary site of association with TRiC (12, 37, 38, 39, 40). Of note, the DBD of AML1-ETO is composed exclusively of β-strands, a common structural motif among TRiC substrates known to require assistance from chaperones to fold properly (27). As previously demonstrated for full-length AML1-ETO (12), we examined the DNA binding ability of AML1-175 in the presence of TRiC. Denatured AML1-175 was diluted into refolding buffer containing purified human TRiC. The reaction mixture was incubated with an AML1-binding DNA duplex oligonucleotide labeled with 32P and analyzed by electrophoresis mobility shift assay with native PAGE (41). Rapid dilution of denatured AML1-175 in physiological buffer led to immediate precipitation and no DNA binding activity in the absence of TRiC. However, when incubated with TRiC, AML1-175 displayed DNA binding activity (Fig. 1 A). These findings are in agreement with recent results showing that TRiC is required for the proper folding and activity of full-length AML1-ETO (12). Here, we demonstrated that TRiC can refold denatured AML1-175 (DBD) and restore its DNA binding activity in vitro. The results show that the activity of denatured AML1-175 is directly dependent on its ability to bind TRiC, implying that the chaperonin promotes folding of the DBD of AML1-ETO.
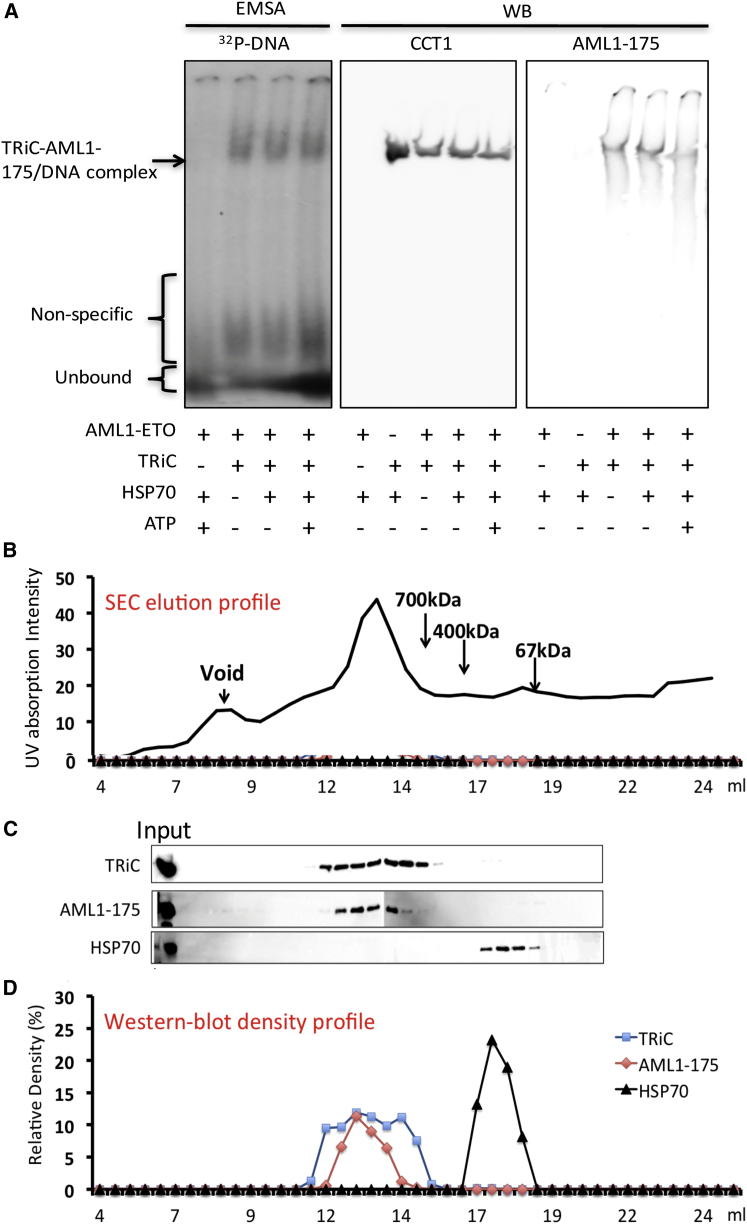
Figure 1.
TRiC forms a stable complex with denatured AML1-175 reconstituting its ability to bind DNA. AML1-binding duplex DNA oligonucleotide (32P-labeled) was incubated with or without denatured AML1-175, TRiC, HSP70, and ATP, as indicated, and the reaction mixtures separated using a 4% native PAGE for electrophoresis mobility shift assay analysis (left panel). As an independent experiment, we analyzed the same reaction mixture without oligonucleotide using native gel and Western blot (middle and right panel) with specific antibodies for CCT1 and FLAG, as indicated (A). Denatured AML1-175 was incubated with TRiC, centrifuged, and the supernatant separated by Superpose 6 gel chromatography (B) and the fractions from 7 to 20 ml were analyzed by Western blot (C). Relative density of the bands for TRiC (anti-CCT1), AML1-175, and HSP70 were measured and plotted (D).
In addition, Western blot analysis of the native gel using specific antibodies to TRiC and AML1-175 shows that, DNA, TRIC, and AML1-175 comigrate, suggesting that they form a complex. Consistent with previous results, renatured AML1-175 by itself was not soluble and could not enter the gel in the absence of TRiC. However, in the presence of TRiC, AML1-175 entered the gel and migrated down, as expected. Interestingly, the positions on the native gel for DNA, AML1-175, and TRiC overlapped and remained unchanged in the presence of ATP, indicating that the AML1-DNA complex comigrated with TRiC independently of TRiC’s ATP usage (Fig. 1 A). This comigration has also been observed with full-length AML1-ETO (12). Of note, ATP hydrolysis by TRiC is typically required for substrate folding and release (42, 43). Yet, our results suggest that while AML1-175 is folded by TRiC to achieve its native function, ATP hydrolysis does not suffice to trigger its release from the chaperonin.
To further characterize the association of AML1-175 with the entire TRiC complex, we used size exclusion chromatography. First, the TRiC-AML1-175 complex was prepared as previously described with the assistance of HSP70 (12), the insoluble fraction was eliminated by centrifugation and filtration. The protein mixture was then applied to a Superose 6 column, which allows for a broad protein fractionation range from 5 kDa to 5 MDa. AML1-175 (∼20 kDa) largely coeluted with TRiC (∼1 MDa), suggesting an interaction between AML1-175 and the entire TRiC complex (Fig. 1 B). Understandably, AML1-175 alone was not detected because of its strict insolubility when not bound to TRiC (Fig. 1 C). Comparing the area under the elution curve corresponding to TRiC and AML1-175, the approximate occupancy of AML1-175 on TRiC was calculated at ∼50% (Fig. 1 D). Taken together, these results indicate that folded AML1-175 forms a stable complex with the entire TRiC complex in vitro.
Cryo-EM reveals that AML1-175 localizes to specific TRiC subunits
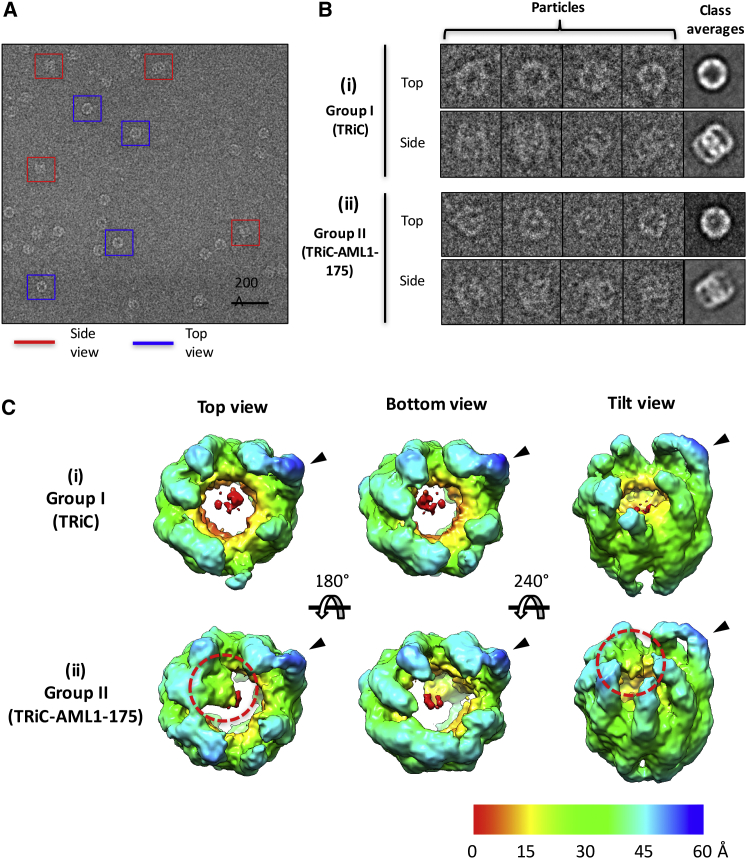
To determine the structural basis for TRiC’s interaction with AML1-ETO, we mixed human apo-TRiC with denatured AML1-175 in the absence of nucleotide and imaged this specimen using single-particle cryo-EM. Judging from our biochemical experiments, this data set is likely to be heterogeneous, with different compositional populations including free unbound TRiC and AML1-175 bound TRiC. Therefore, we processed our data set using a multiple-model-refinement strategy based on maximum-likelihood (33, 34, 44). A total of 32,143 particle images were sorted into two major subpopulations (group I and group II) through iterative refinement until particle assignment to either one of the groups converged. The resulting maps generated from each of the two groups (Fig. 2) had a resolution of ∼25 Å (Fig. S1 in the Supporting Material), as measured by the gold standard criterion (35, 36, 45). Both structures display TRiC-like architecture remarkably similar to that previously reported for bovine apo-TRiC (46). The TRiC structure from group I appears to be indistinguishable from our apo-TRiC control structures, reconstructed independently using a substrate-free human TRiC (hTRiC) specimen (Fig. S2) and the same data processing protocol. In contrast, there is a prominent density protruding centrally from the apical domains of the TRiC structure from group II. This density spans two subunits on one ring and cannot be accounted for by TRiC alone (Fig. 2 C ii). Segmentation analysis (47) suggests a mass of ∼20 kDa for the bound density, corresponding well with the molecular weight of AML1-175. Of importance, this density is also visible in 2D class averages of the raw particle images (Fig. 2 B ii). On the other hand, the 2D class averages (Fig. 2 B i) and 3D map from group I (Fig. 2 C i) do not display an extra density anywhere. Similarly, the apo-hTRiC maps do not display a significant extra density anywhere either.
Figure 2.
TRiC interacts with AML1-175 in a subunit-specific manner. (A) Representative area of a micrograph of a vitrified sample of TRiC-AML1-175 complex. (B) Gallery of selected images and class averages of group I (i) and II (ii) in the respective data set. (C) Top, bottom, and tilted views of the 3D reconstructions from two subpopulations, group I (i) and II (ii). The same radial color scheme of distance (Å) from the center is used throughout. Extra density attributed to AML1-175 is shown in red circles and the black arrowheads indicate the protruding subunit.
Affinity gold nanoparticle validates AML1-75 in TRiC
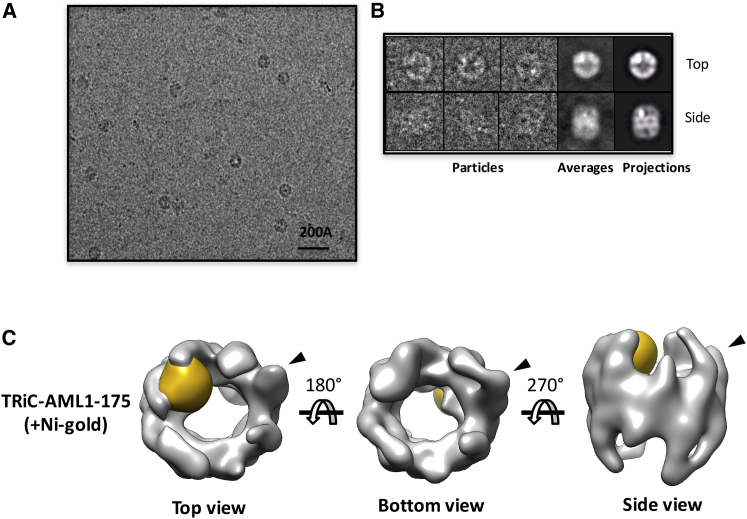
To validate that the observed extra density is attributed by AML1-175, we reconstructed the TRiC-AML1-175 complex formed in the presence of Ni2+-gold nanoparticles capable of binding to the His-tag on AML1-175. The resulting 3D cryo-EM map (Fig. 3) has an overall TRiC-like morphology and, most importantly, clearly exhibits an even larger extra density in the same location as the structure from group II. This unambiguously confirms the specific localization of AML1-175 bound to TRiC. Therefore, the extra density identified in the group II structure from the TRiC+AML1-175 specimen can confidently be attributed to AML1-175. Furthermore, the distribution of particles with the group II population consisted of ∼48% of the total number of particles, which agrees well with our estimates for AML1-175 occupancy on TRiC (∼50%) based on size-exclusion chromatography analyses (Fig. 1 D). Taken together, our results indicate that our computational data processing protocol successfully isolated a subpopulation of TRiC bound to AML1-175 from a heterogeneous data set and reliably reconstructed symmetry-free and model-bias free cryo-EM structures. Therefore, we hereafter refer to group I and group II as substrate-free (Fig. 2 C i) and AML1-175-bound (Fig. 2 C ii) TRiC, respectively.
Figure 3.
Ni-affinity nanogold particle revealed the location of AML1-175 on TRiC. (A) Representative area of a micrograph of a vitrified sample of AML1-175-TRiC-Ni-nanogold complex. (B) Gallery of selected images, class averages, and projection of AML1-175-TRiC-Ni-nanogold complex. (C) Top, bottom, and side views of the 3D reconstruction of TRiC-AML1-175 labeled with Ni-nanogold. The density from nanogold particles is colored in gold and the black arrowheads indicate the protruding subunit.
Particle-based intensity analysis shows higher mean intensity in the subpopulation of AML1-175
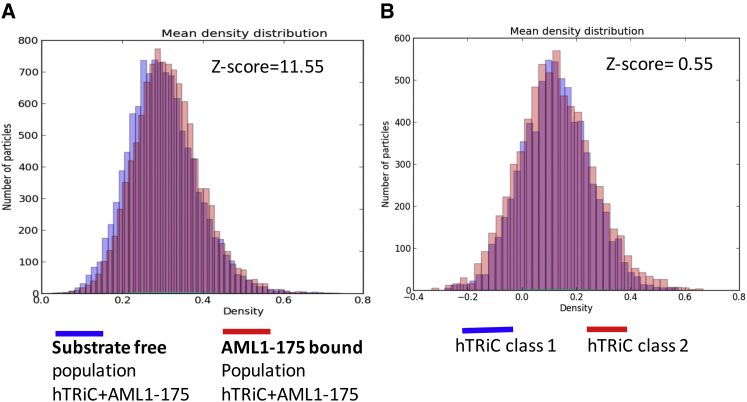
To further evaluate the statistical significance of our classification into substrate-free and AML1-175-bound groups, we performed t-tests to examine the mean intensity distribution of the individual particle images comprising each group (Fig. 4). Notably, the distribution for AML1-175-bound TRiC displays a shift toward higher mass, not seen for substrate-free TRiC. This shift is statistically significant, as indicated by a z-score of 11.55, markedly above the 95% confidence interval (z = 1.96). As a control, we repeated the same analysis using the control apo-hTRiC data set. In this case, there was no significant shift between the mean intensity distributions of the two classified groups, as indicated by the z-score of 0.55, well below the 95% confidence interval (z = 1.96). This indicates that the two classified groups from the apo-hTRiC control data are statistically indistinguishable based on mean intensity distribution and rather comprise one homogeneous class of particles. This further substantiates the success of our 3D classification strategy in separating TRiC + AML1-175 particles into two significantly different groups, thereby resolving the structures of substrate-free and AML1-175-bound TRiC (Fig. 2).
Figure 4.
Mean intensity distribution of individual cryo-EM particles. (A) Mean intensity distribution plot for substrate free (blue) and AML1-175 bound human TRiC (red). (B) Group I (blue) and group II (red) of apo-TRiC; overlapped area shown in purple.
AML1-175 binds to the apical domains of CCT6 and 8
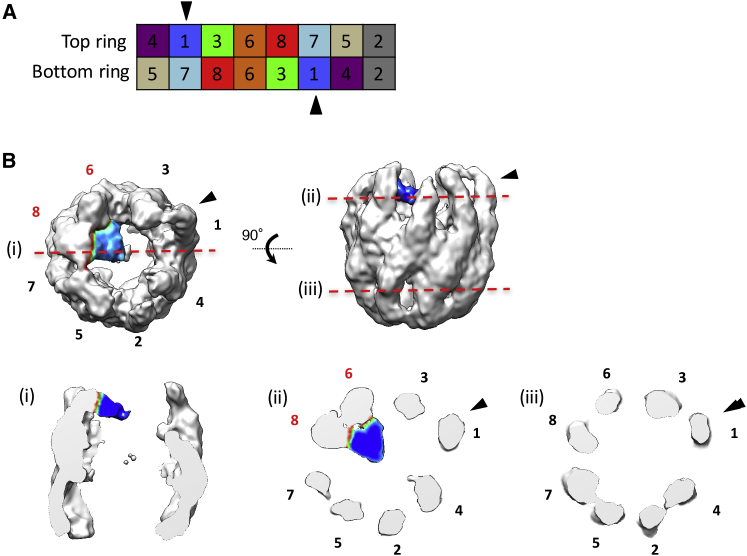
Multiple structures have been reported for TRiC (46, 48, 49), with the x-ray crystal structure of bovine TRiC in the nucleotide-free, open conformation (Protein Data Bank (PDB): 2XSM) (49) being the one closest to the biochemical conditions of our cryo-EM studies. Two striking structural hallmarks are shared by our cryo-EM map and the crystal structure of TRiC. First, the subunits in each ring are arranged as a quasitetramer of dimers. Second, the apical domain of one subunit protrudes much further outward than any other subunit (46). Based on these shared structural features, the x-ray crystal structure of TRiC (PDB: 2XSM) in its open conformation (49) was docked into the TRiC-AML1-175 cryo-EM map using rigid-body fitting (Fig. S3). Combining this result with the recently reported intraring subunit arrangement that is based on cross-linking and refinement of the x-ray structure (21, 22), we were able to determine the identity of each TRiC subunit that engaged AML1-175, namely CCT6 and CCT8 (45) (Fig. 5). It has been suggested that the apical domain of each CCT subunit within TRiC contains a substrate-recognition site that participates in specific interactions with substrates in their intermediate folding states (28). Similar to results reported for tubulin’s interactions with TRiC, the initial contact of AML1-175 with the apical domain of the two CCT subunits would be expected to precede the subsequent delivery of the substrate to the equatorial sensing loops, which is facilitated by large conformational changes in TRiC driven by ATP hydrolysis (49).
Figure 5.
AML1-175 binds to the apical domains of CCT6 and 8. (A) The subunit order of TRiC has been determined using chemical cross-linking and mass spectroscopy. Black arrowheads indicate CCT1 whose apical domain is protruding outward in the 3D structure. (B) AML1-175 bound TRiC. Numbers depict the TRiC subunit order and AML1-175 is colored in blue. Black arrowheads indicate the location of the protruding subunit. Cutaway views on side (i), top (ii), and bottom ring (iii).
Discussion
In living cells, the folding of proteins into their functional conformation largely depends on assistance from molecular chaperones, a network of cellular proteostasis regulators (13). Among these, the mammalian cytosolic chaperonin TRiC/CCT is distinguished by its complex architecture and mechanism of action, which allow it to fold many proteins that cannot be folded by simpler chaperone systems. Of importance, TRiC plays a central role in the cell by assisting the folding of ∼10% of all newly synthesized polypeptides (13, 27). Furthermore, TRiC has been suggested to play a critical role in cancer cell development by modulating the folding and activity of client proteins involved in oncogenesis (16), such as the tumor suppressor proteins VHL (17, 18) and p53 (19), as well as the oncogenic protein STAT3 (20). More recently, AML1-ETO has been identified as the first chimeric oncoprotein that is a TRiC substrate with this chaperonin playing an important role in its synthesis, folding and activity (12). However, little is known concerning the specific mechanisms of interaction between AML1-ETO and TRiC, which may be targeted for therapeutic benefit. In this report, we demonstrate that TRiC’s contribution to the activity of the DBD (AML1-175) of AML1-ETO is consistent with its contribution to the activity of full-length AML1-ETO (12) and is mediated through its direct association with the DBD. In addition, single-particle cryo-EM reveals that AML1-175 associates with the apical domains of adjacent subunits CCT6 and CCT8.
Our structures and gel shift assay show that folded AML1-175 remains bound to TRiC in open conformation of TRiC. This is consistent with our previous refolding study (12) with purified full-length AML1-ETO, which demonstrated that folded AML1-ETO remains associated with TRiC throughout its ATP hydrolysis cycle, in contrast to actin and luciferase, which are released from TRiC in the presence of ATP (32). On the other hand, the interaction of AML1-ETO with TRiC is transient in rabbit reticulocyte lysate (12). These results suggest that rabbit reticulocyte lysate contains cofactors required for AML1-ETO to dissociate from TRiC, as is seen for tumor suppressor VHL, which requires Elongin-B and C to be released from TRiC after protein folding is completed (50). Based on our observations, TRiC not only assists protein folding but may also work as a regulatory hub by holding on to transcription factors like AML1 and VHL after they are folded until other cofactors bind and release them. Therefore, it would be extremely interesting to investigate the question of how AML1 and AML1-ETO are released from TRiC (17).
We identified minor conformational differences between the maps for substrate-free human TRiC and AML1-175-TRiC (Fig. 2). CCT2 previously has been shown to have an extremely flexible apical domain whose density was not resolved in previous studies of bovine apo-TRiC (46, 49). This lack of resolution of the CCT2 apical domain is recapitulated in our maps of human apo-TRiC and substrate free TRiC, but not in the map of AML1-175-TRiC (Fig. 2). In addition, the apical domains of CCT5 and CCT4 make closer contact to CCT2 in the other side of the ring, which does not contain AML1-175. This suggests that substrate binding may stabilize the flexibility of the apical domain of CCT2 and trigger allosterism to enhance the interring negative cooperativity of TRiC (51).
TRiC is a mechanically complex folding machine in that each subunit has a different ATP binding affinity and ATP hydrolysis rate. This results in an asymmetric “ATP-usage gradient,” where half of TRiC’s subunits within each ring that are adjacent to each other have low ATPase activity (CCT1, 3, 6, 8), whereas the other half hydrolyze ATP much more actively (CCT4, 2, 5, 7) (52). Although the subunits with high ATP affinity are more strictly required for cell viability (52), the detailed biological significance of this polarity and its impact on assisted protein folding remain largely unknown. Our cryo-EM map for the TRiC-AML1-175 complex (Figs. 3, 4, and 5) reveals that AML1-175 localizes to the center of TRiC’s low-ATPase activity cluster, which corresponds to approximately the same subunits involved in actin and tubulin binding (CCT6) (45, 49). Taking this coincidence into account, our results support the hypothesis that the ATP-usage gradient of TRiC’s subunits is linked to a functional and evolutional partitioning into a substrate-binding side that is opposite to the high ATP-hydrolyzing side (45). Therefore, our findings provide important biological insight into the mechanistic relationship between TRiC and its oncoprotein client AML1-ETO.
TRiC folds a subset of cellular proteins, suggesting a degree of specificity; however, its substrates are functionally and structurally diverse, indicating the potential to bind a broad array of proteins (27). The apical domains of each TRiC subunit are thought to recognize different motifs in substrates (24, 28). The Frydman group defined the basis of TRiC substrate recognition and suggested that unique subunit-specific patterns of polar and hydrophobic residues on apical domains underlie the distinct substrate binding properties of each subunit in the complex (29). Among the diverse TRiC subunits, subsets of subunits provide specificities for combinatorial recognition of substrate polypeptides. Here, we present the TRiC-AML1-175 complex in the open conformation with AML1-175 binding to the apical domains of CCT6 and CCT8 (Fig. 5), a snapshot of the initial engagement of AML1-175 to TRiC, which could be a basis for development of therapeutic agents that specifically target TRiC-AML1-ETO interactions. This possibility warrants further investigation, including downregulation of CCT6 and/or CCT8 in hematopoietic cell lines, such as KASUMI-1 cells, commonly used as a model system for AML1-ETO-positive AML (53).
Author Contributions
S.R., W.C., and D.J.T designed the project. S.R. and M.K. performed biochemistry experiments. S.R. purified the proteins, performed cryo-EM experiments, and reconstructed the maps. J.G. performed the statistical analysis of the particle images. S.R., J.G., W.C., and D.J.T. wrote the article with input from all other coauthors.
Acknowledgments
This research was supported by National Institutes of Health (NIH) grants (P41GM103832 and PN2EY016525) and a Cancer Prevention Research Institute of Texas (CPRIT) grant RP110291, as well as a training fellowship to J.G.G.M. from the Computational Cancer Biology Training Program of the Gulf Coast Consortia (CPRIT grant RP140113). This work used the Extreme Science and Engineering Discovery Environment (XSEDE) allocation MCB150009, which is supported by National Science Foundation grant No. ACI-1053575.
Editor: Jill Trewhella.
Footnotes
Supporting Materials and Methods and three figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30248-X.
Contributor Information
Wah Chiu, Email: wah@bcm.edu.
David J. Tweardy, Email: djtweardy@mdanderson.org.
Supporting Material
References
- 1.Deschler B., Lübbert M. Acute myeloid leukemia: epidemiology and etiology. Cancer. 2006;107:2099–2107. doi: 10.1002/cncr.22233. [DOI] [PubMed] [Google Scholar]
- 2.Look A.T. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 3.Cameron E.R., Neil J.C. The Runx genes: lineage-specific oncogenes and tumor suppressors. Oncogene. 2004;23:4308–4314. doi: 10.1038/sj.onc.1207130. [DOI] [PubMed] [Google Scholar]
- 4.de Bruijn M.F., Speck N.A. Core-binding factors in hematopoiesis and immune function. Oncogene. 2004;23:4238–4248. doi: 10.1038/sj.onc.1207763. [DOI] [PubMed] [Google Scholar]
- 5.Davis J.N., McGhee L., Meyers S. The ETO (MTG8) gene family. Gene. 2003;303:1–10. doi: 10.1016/s0378-1119(02)01172-1. [DOI] [PubMed] [Google Scholar]
- 6.Downing J.R. The AML1-ETO chimaeric transcription factor in acute myeloid leukaemia: biology and clinical significance. Br. J. Haematol. 1999;106:296–308. doi: 10.1046/j.1365-2141.1999.01377.x. [DOI] [PubMed] [Google Scholar]
- 7.Meyers S., Lenny N., Hiebert S.W. The t(8;21) fusion protein interferes with AML-1B-dependent transcriptional activation. Mol. Cell. Biol. 1995;15:1974–1982. doi: 10.1128/mcb.15.4.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lutterbach B., Hiebert S.W. Role of the transcription factor AML-1 in acute leukemia and hematopoietic differentiation. Gene. 2000;245:223–235. doi: 10.1016/s0378-1119(00)00014-7. [DOI] [PubMed] [Google Scholar]
- 9.Heidenreich O., Krauter J., Nordheim A. AML1/MTG8 oncogene suppression by small interfering RNAs supports myeloid differentiation of t(8;21)-positive leukemic cells. Blood. 2003;101:3157–3163. doi: 10.1182/blood-2002-05-1589. [DOI] [PubMed] [Google Scholar]
- 10.Yu W., Wang J., Wang L. Heat shock protein 90 inhibition results in altered downstream signaling of mutant KIT and exerts synergistic effects on Kasumi-1 cells when combining with histone deacetylase inhibitor. Leuk. Res. 2011;35:1212–1218. doi: 10.1016/j.leukres.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 11.Reikvam H., Hatfield K.J., Bruserud O. Expression profile of heat shock proteins in acute myeloid leukaemia patients reveals a distinct signature strongly associated with FLT3 mutation status--consequences and potentials for pharmacological intervention. Br. J. Haematol. 2012;156:468–480. doi: 10.1111/j.1365-2141.2011.08960.x. [DOI] [PubMed] [Google Scholar]
- 12.Roh S.H., Kasembeli M., Tweardy D.J. Chaperonin TRiC/CCT modulates the folding and activity of leukemogenic fusion oncoprotein AML1-ETO. J. Biol. Chem. 2015;291:4732–4741. doi: 10.1074/jbc.M115.684878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartl F.U., Bracher A., Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 14.Bigotti M.G., Clarke A.R. Chaperonins: the hunt for the Group II mechanism. Arch. Biochem. Biophys. 2008;474:331–339. doi: 10.1016/j.abb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 15.Kim Y.E., Hipp M.S., Hartl F.U. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 16.Roh S.H., Kasembeli M., Tweardy D.J. Contribution of the Type II chaperonin, TRiC/CCT, to oncogenesis. Int. J. Mol. Sci. 2015;16:26706–26720. doi: 10.3390/ijms161125975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melville M.W., McClellan A.J., Frydman J. The Hsp70 and TRiC/CCT chaperone systems cooperate in vivo to assemble the von Hippel-Lindau tumor suppressor complex. Mol. Cell. Biol. 2003;23:3141–3151. doi: 10.1128/MCB.23.9.3141-3151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McClellan A.J., Scott M.D., Frydman J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell. 2005;121:739–748. doi: 10.1016/j.cell.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 19.Trinidad A.G., Muller P.A., Vousden K.H. Interaction of p53 with the CCT complex promotes protein folding and wild-type p53 activity. Mol. Cell. 2013;50:805–817. doi: 10.1016/j.molcel.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasembeli M., Lau W.C., Tweardy D.J. Modulation of STAT3 folding and function by TRiC/CCT chaperonin. PLoS Biol. 2014;12:e1001844. doi: 10.1371/journal.pbio.1001844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalisman N., Adams C.M., Levitt M. Subunit order of eukaryotic TRiC/CCT chaperonin by cross-linking, mass spectrometry, and combinatorial homology modeling. Proc. Natl. Acad. Sci. USA. 2012;109:2884–2889. doi: 10.1073/pnas.1119472109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leitner A., Joachimiak L.A., Frydman J. The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure. 2012;20:814–825. doi: 10.1016/j.str.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- 24.Spiess C., Meyer A.S., Frydman J. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 2004;14:598–604. doi: 10.1016/j.tcb.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saibil H.R., Fenton W.A., Horwich A.L. Structure and allostery of the chaperonin GroEL. J. Mol. Biol. 2013;425:1476–1487. doi: 10.1016/j.jmb.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 26.Yébenes H., Mesa P., Valpuesta J.M. Chaperonins: two rings for folding. Trends Biochem. Sci. 2011;36:424–432. doi: 10.1016/j.tibs.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Yam A.Y., Xia Y., Frydman J. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat. Struct. Mol. Biol. 2008;15:1255–1262. doi: 10.1038/nsmb.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spiess C., Miller E.J., Frydman J. Identification of the TRiC/CCT substrate binding sites uncovers the function of subunit diversity in eukaryotic chaperonins. Mol. Cell. 2006;24:25–37. doi: 10.1016/j.molcel.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joachimiak L.A., Walzthoeni T., Frydman J. The structural basis of substrate recognition by the eukaryotic chaperonin TRiC/CCT. Cell. 2014;159:1042–1055. doi: 10.1016/j.cell.2014.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feldman D.E., Spiess C., Frydman J. Tumorigenic mutations in VHL disrupt folding in vivo by interfering with chaperonin binding. Mol. Cell. 2003;12:1213–1224. doi: 10.1016/s1097-2765(03)00423-4. [DOI] [PubMed] [Google Scholar]
- 31.Knee K.M., Sergeeva O.A., King J.A. Human TRiC complex purified from HeLa cells contains all eight CCT subunits and is active in vitro. Cell Stress Chaperones. 2013;18:137–144. doi: 10.1007/s12192-012-0357-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thulasiraman V., Ferreyra R.G., Frydman J. Folding assays. Assessing the native conformation of proteins. Methods Mol. Biol. 2000;140:169–177. doi: 10.1385/1-59259-061-6:169. [DOI] [PubMed] [Google Scholar]
- 33.Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang G., Peng L., Ludtke S.J. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Scheres S.H., Chen S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henderson R., Sali A., Lawson C.L. Outcome of the first electron microscopy validation task force meeting. Structure. 2012;20:205–214. doi: 10.1016/j.str.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartfeld D., Shimon L., Shakked Z. DNA recognition by the RUNX1 transcription factor is mediated by an allosteric transition in the RUNT domain and by DNA bending. Structure. 2002;10:1395–1407. doi: 10.1016/s0969-2126(02)00853-5. [DOI] [PubMed] [Google Scholar]
- 38.Wei Y., Liu S., Werner M.H. A TAF4-homology domain from the corepressor ETO is a docking platform for positive and negative regulators of transcription. Nat. Struct. Mol. Biol. 2007;14:653–661. doi: 10.1038/nsmb1258. [DOI] [PubMed] [Google Scholar]
- 39.Corpora T., Roudaia L., Bushweller J.H. Structure of the AML1-ETO NHR3-PKA(RIIα) complex and its contribution to AML1-ETO activity. J. Mol. Biol. 2010;402:560–577. doi: 10.1016/j.jmb.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y., Cheney M.D., Bushweller J.H. The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO’s activity. Cancer Cell. 2006;9:249–260. doi: 10.1016/j.ccr.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 41.Qiu J., Wong J., Dong S. Decreased intranuclear mobility of acute myeloid leukemia 1-containing fusion proteins is accompanied by reduced mobility and compartmentalization of core binding factor beta. Oncogene. 2006;25:3982–3993. doi: 10.1038/sj.onc.1209431. [DOI] [PubMed] [Google Scholar]
- 42.Gao Y., Thomas J.O., Cowan N.J. A cytoplasmic chaperonin that catalyzes beta-actin folding. Cell. 1992;69:1043–1050. doi: 10.1016/0092-8674(92)90622-j. [DOI] [PubMed] [Google Scholar]
- 43.Melki R., Batelier G., Williams R.C., Jr. Cytoplasmic chaperonin containing TCP-1: structural and functional characterization. Biochemistry. 1997;36:5817–5826. doi: 10.1021/bi962830o. [DOI] [PubMed] [Google Scholar]
- 44.Scheres S.H. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 2012;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalisman N., Schröder G.F., Levitt M. The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning. Structure. 2013;21:540–549. doi: 10.1016/j.str.2013.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cong Y., Schröder G.F., Chiu W. Symmetry-free cryo-EM structures of the chaperonin TRiC along its ATPase-driven conformational cycle. EMBO J. 2012;31:720–730. doi: 10.1038/emboj.2011.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pintilie G.D., Zhang J., Gossard D.C. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct. Biol. 2010;170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dekker C., Roe S.M., Willison K.R. The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic cytosolic chaperonins. EMBO J. 2011;30:3078–3090. doi: 10.1038/emboj.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muñoz I.G., Yébenes H., Montoya G. Crystal structure of the open conformation of the mammalian chaperonin CCT in complex with tubulin. Nat. Struct. Mol. Biol. 2011;18:14–19. doi: 10.1038/nsmb.1971. [DOI] [PubMed] [Google Scholar]
- 50.Feldman D.E., Thulasiraman V., Frydman J. Formation of the VHL-elongin BC tumor suppressor complex is mediated by the chaperonin TRiC. Mol. Cell. 1999;4:1051–1061. doi: 10.1016/s1097-2765(00)80233-6. [DOI] [PubMed] [Google Scholar]
- 51.Kafri G., Willison K.R., Horovitz A. Nested allosteric interactions in the cytoplasmic chaperonin containing TCP-1. Protein Sci. 2001;10:445–449. doi: 10.1110/ps.44401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reissmann S., Joachimiak L.A., Frydman J. A gradient of ATP affinities generates an asymmetric power stroke driving the chaperonin TRIC/CCT folding cycle. Cell Reports. 2012;2:866–877. doi: 10.1016/j.celrep.2012.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Asou H., Tashiro S., Kamada N. Establishment of a human acute myeloid leukemia cell line (Kasumi-1) with 8;21 chromosome translocation. Blood. 1991;77:2031–2036. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.