Abstract
The purpose of the present study was to investigate the serum levels of microRNA (miRNA/miR)-382-3p, −598-3p, −1246 and −184 in breast cancer patients and to assess their feasibility as biomarkers for breast cancer screening. Serum samples were obtained from 100 breast cancer patients and 40 age-matched healthy control subjects in Taizhou Central Hospital (Taizhou, Zhejiang, China) between January 2013 and September 2014. The serum expression levels of miR-382-3p, −598-3p, −1246 and −184 were determined by stem-loop reverse transcription-quantitative polymerase chain reaction. Receiver operating characteristic curves were drawn to evaluate the sensitivity and specificity of the serum miRNA expression levels for the screening of breast cancer. miR-382-3p and −1246 were significantly upregulated in the serum of the breast cancer patients, while miR-598-3p and −184 were significantly downregulated. The sensitivity and specificity to detect breast cancer were as follows: miR-382-3p, 52.0 and 92.5%; miR-598-3p, 95.0 and 85.0%; miR-1246, 93.0 and 75.0%; and miR-184, 87.5 and 71.0%, respectively. The expression levels of the four serum miRNAs were not correlated with the patients' clinical stage. In summary, miR-382-3p, −598-3p, −1246 and −184 are all involved in the development of breast cancer, and are promising biomarkers for breast cancer detection.
Keywords: breast cancer, microRNAs, quantitative polymerase chain reaction, biomarker
Introduction
Breast cancer has become the most common cancer in women in China, accounting for 12.2% of all newly diagnosed breast cancers and 9.6% of all mortalities from breast cancer worldwide (1). Early detection of breast cancer is the key to successful treatment and patient survival. Mammographies, magnetic resonance imaging and ultrasound are being used for the screening of breast cancer, but these techniques have limitations such as low sensitivity and specificity (2). Finding effective biomarkers is vital for the screening of breast cancer.
MicroRNAs (miRNAs/miRs) are a group of small non-coding RNAs (3), which can bind to the 3′ untranslated region (UTR), the 5′ UTR (4) or the coding region (5) of target mRNA. miRNAs play important roles in the negative regulation of gene expression in a post-transcriptional manner, and are involved in a number of signal transduction pathways, including cell proliferation, differentiation, apoptosis, immune response and angiogenesis (6). Previous studies have revealed the abnormal expression of miRNAs in the development and progression of breast cancer (7), and miRNAs are closely linked with tumor-associated signal transduction pathways (8). In our previous study, 10 miRNAs that were significantly differentially expressed were found by the second generation of high-throughput sequencing technology, Illumina Hiseq2500 (9). In the present study, 4 miRNAs, miR-382-3p, −598-3p, −1246 and −184, were further investigated. The purpose of this study was to investigate the serum levels of these miRNAs in breast cancer patients and to assess their feasibility as biomarkers for breast cancer screening.
Materials and methods
Subjects
A total of 140 serum samples were obtained in Taizhou Central Hospital (Taizhou, Zhejiang, China) between January 2013 and September 2014, including 100 serum samples from breast cancer patients and 40 samples from 40 age-matched healthy women. The clinical characteristics of the breast cancer patients are listed in Table I. The present study was approved by the Ethics Committee of Taizhou Central Hospital, Taizhou, China.
Table I.
Clinical features of the breast cancer patients.
| Patient characteristics | Value |
|---|---|
| Mean age (range), years | 50.17 (25–80) |
| Stages, n | |
| I | 15 |
| II | 46 |
| III | 24 |
| IV | 6 |
| Unknown | 9 |
| Lymph nodes, n | |
| Positive | 37 |
| Negative | 57 |
| Unknown | 6 |
| ER, n | |
| Positive | 54 |
| Negative | 30 |
| Unknown | 16 |
| PR, n | |
| Positive | 46 |
| Negative | 38 |
| Unknown | 16 |
| HER2, n | |
| Positive | 56 |
| Negative | 26 |
| Unknown | 18 |
ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
Reagents and instruments
The QIAzol Lysis Reagent, miRNeasy Serum/Plasma kit, miScript II Reverse Transcription (RT) kit, miScript Primer assays and miScript SYBR Green Polymerase Chain Reaction (PCR) kit were purchased from Qiagen GmbH (Hilden, Germany). Ethanol and chloroform were purchased from Adamas-beta (Shanghai, China). Primers were purchased from Sangon Biotech (Shanghai, China) and the StepOnePlus PCR thermal cycler was purchased form Applied Biosystems (Foster City, CA, USA).
RNA extraction and purification
Total RNA was isolated from 200 µl of plasma using the miRNeasy Serum/Plasma kit according to the manufacturer's protocols. RNA was dissolved in RNase-free water. Following the use of QIAzol Lysis Reagent, exogenous cel-miR-39 (Sangon Biotech) was added as an internal standard.
RT-quantitative (q)PCR
RT-qPCR was performed using the ABI StepOne system (Qiagen GmbH). All experiments were performed as specified in the manufacturer's protocols. RT reactions were performed with a reaction mixture that included 4 µl 5X miScript HiSpec Buffer, 2 µl 10X miScript Nucleics Mix, 10 µl RNase-free water, 2 µl miScript Reverse Transcriptase Mix and 2 µl RNA. The reaction occurred at 37°C for 60 min and then 95°C for 5 min. The obtained cDNA was stored at −80°C.
Amplifications were performed using the miScript SYBR Green PCR kit (Qiagen GmbH) with a reaction mixture that included 12.5 µl 2X QuantiTect SYBR Green PCR Master Mix, 2.5 µl 10X miScript Universal Primer, 2.5 µl primers, 5 µl RNase-free water and 2.5 µl cDNA. The PCR conditions were as follows: Initial denaturation at 95°C for 15 min; then 94°C for 15 sec, 55°C for 30 sec and 70°C for 30 sec for 40 cycles. The melting curve was obtained from 65°C to 95°C. A quantitation cycle (Cq) value >35 was regarded as impossible for amplification. qPCR primers are shown in Table II. Each sample was analyzed in triplicate. The relative expression of the miRNA (10) was calculated using the 2−ΔΔCq method as follows: ΔΔCq = (CqmiRNA - CqmiR-39)cancer group - (CqmiRNA - CqmiR-39)control group.
Table II.
Polymerase chain reaction primers.
| Products | Primers (5′-3′) |
|---|---|
| miR-382–3p-F | CTGCAATCATTCACGGACAAC |
| miR-598–3p-F | AGCTACGTCATCGTTGTCATC |
| miR-1246-F | GCCGAATGGATTTTTGGAGC |
| miR-184-F | ATGGACGGAGAACTGATAAGG |
| Reverse primer | GTGTCGTCGAGTCGGCAATTC |
F, forward primer. The miScript Universal Primer in the miScript SYBR Green PCR kit was used as the reverse primer.
Statistical analysis
Statistical analyses were performed using SPSS 17.0. software (SPSS Inc., Chicago, IL, USA). An independent sample t-test, Mann-Whitney U test and Kruskal-Wallis t-test were performed accordingly. P<0.05 was considered to indicate a statistically significant difference.
Results
Serum levels of cel-miR-39
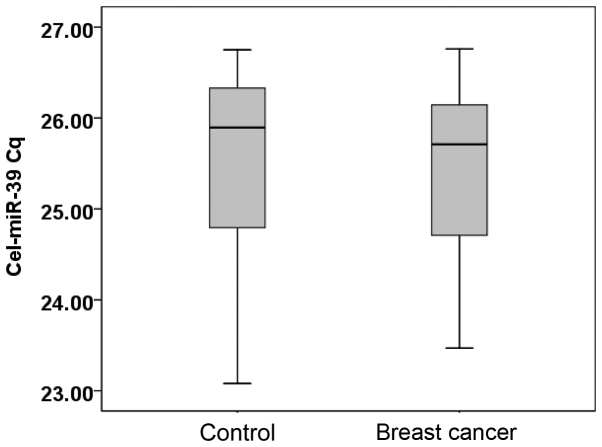
The serum levels of cel-miR-39 in 20 randomly selected breast cancer patients and 20 healthy individuals were examined. The median Cq value for cel-miR-39 in the healthy individuals was 25.90 (range, 23.08–26.61), while in patients with breast cancer, the value was 25.71 (range, 23.47–26.76) (Fig. 1).
Figure 1.
Cel-miR-39 Cq value in healthy controls and breast cancer patients. Cq, quantitation cycle; miR, microRNA.
Cel-miR-39 expression was calculated using the 2−Cq equation (11). The Mann-Whitney U test showed that there was no statistically significant difference between the healthy controls and breast cancer patients (P=0.543). Cel-miR-39 expression is stable, and can be used as an exogenous reference in qPCR.
Serum levels of miR-382-3p, −598-3p, −1246 and −184
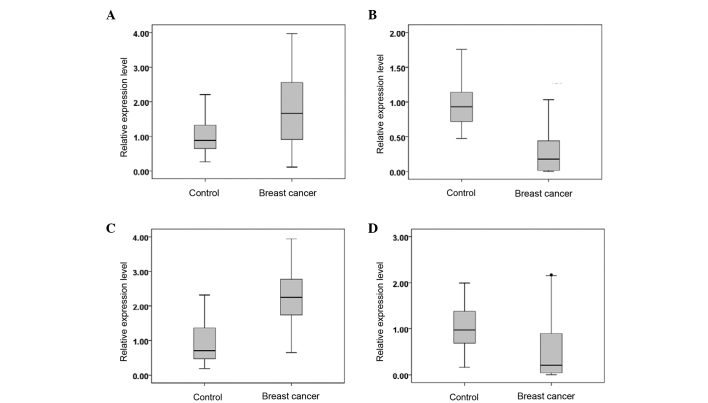
The serum miR-382-3p, −598-3p, −1246 and −184 expression levels in 100 cases of breast cancer and 40 healthy individuals are shown in Fig. 2. Compared with the serum of the healthy controls, the serum of the breast cancer patients exhibited upregulated levels of miR-382-3p and −1246, while the levels of miR-598-3p and −184 were downregulated (P<0.05).
Figure 2.
Relative expression levels of (A) miR-382-3p, (B) miR-598-3p, (C) miR-1246 and (D) miR-184 in the serum of breast cancer patients and control subjects. miR, microRNA.
Serum miRNAs in breast cancer patients with different tumor-node-metastasis (TNM) stages
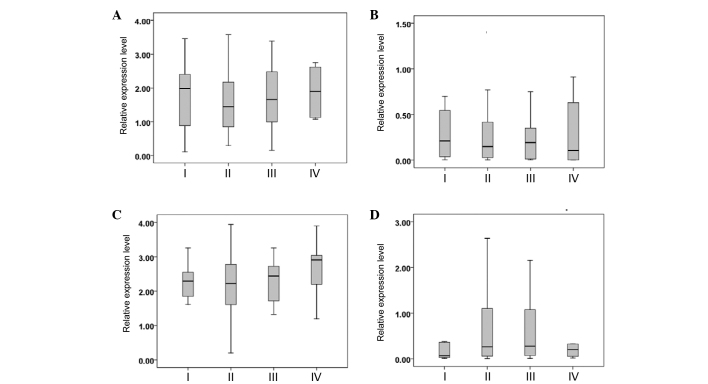
The breast cancer patients were divided into TNM stages I, II, III and IV according to the National Comprehensive Cancer Network (NCCN) Clinical Practice Guidelines in Oncology: Breast Cancer (version 3.2014) (12). The serum miR-382-3p, −598-3p, −1246 and −184 levels were not significantly different between the patients with different TNM stages (P>0.05) (Fig. 3).
Figure 3.
Relative expression of serum (A) miR-382-3p, (B) miR-598-3p, (C) miR-1246 and (D) miR-184 in breast cancer patients with different tumor-node-metastasis stages. miR, microRNA.
Receiver operating characteristic (ROC) curve analysis
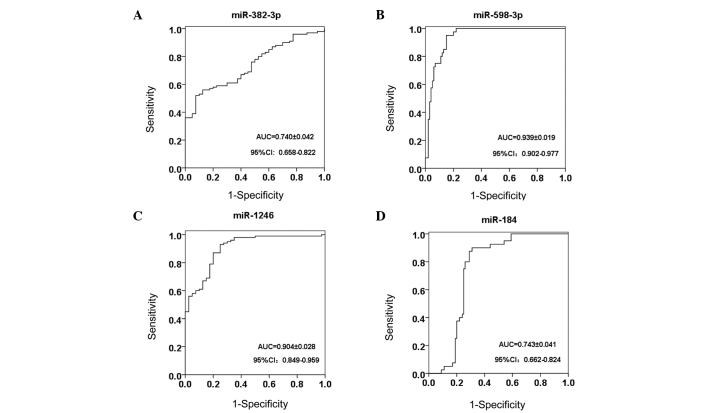
The area under the ROC curve for miR-382-3p, −598-3p, −1246 and −184 was 0.740, 0.939, 0.904 and 0.743, respectively (Fig. 4). Notably, serum miR-598-3p showed the highest sensitivity and specificity of 95.0 and 85.0%, respectively, while miR-1246 exhibited a lower specificity of 75.0%, and miR-382-3p showed the lowest sensitivity at 52.0% (Table III).
Figure 4.
ROCs for (A) miR-382-3p, (B) miR-598-3p, (C) miR-1246 and (D) miR-184. miR, microRNA; ROC, receiver operating curve; AUC, area under the ROC; CI, confidence interval.
Table III.
Diagnostic performance of serum miR-382–3p, −598–3p, −1246 and −184.
| miRNA | AUC | 95% CI | Sensitivity, % | Specificity, % | Cut-off value |
|---|---|---|---|---|---|
| miR-382–3p | 0.740 | 0.658–0.822 | 52.0 | 92.5 | 1.611 |
| miR-598–3p | 0.939 | 0.902–0.977 | 95.0 | 85.0 | 0.549 |
| miR-1246 | 0.904 | 0.849–0.959 | 93.0 | 75.0 | 1.318 |
| miR-184 | 0.743 | 0.663–0.824 | 87.5 | 71.0 | 0.484 |
miR/miRNA, microRNA; AUC, area under receiver operating characteristic curve; CI, confidence interval.
Discussion
The early detection of breast cancer remains a challenge. Mammography X-ray photography is widely used for breast cancer screening and detection, but this technology has limitations, such as irradiation and a high false-positive rate. miRNAs are expressed in the serum and a variety of other bodily fluids (13). The molecules are stable enough for long-term preservation (14) and can tolerate repeated freezing and thawing, which are promising characteristics for tumor markers. A number of studies have shown that serum miRNAs are promising biomarkers for cancer detection (15). Sun et al showed that serum miRNA-155 can be used as a diagnostic marker for breast cancer (16). It has also been demonstrated that serum miRNA-10b expression is significantly higher in breast cancer patients with bone metastases, and that it can therefore be used as a tumor marker for breast cancer bone metastasis (17). However, previous studies on serum miRNAs have not provided a standardized internal standard. A number of studies have used miR-16 as a standardized internal control for serum miRNA experiments (18,19), however, the expression of serum miR-16 is not stable in a number of tumors (20). Furthermore, in miR-16 derived from red blood cells, the occurrence of hemolysis would result in 20–30-fold increase in blood miR-16, which would affect the results (21). In the present study, exogenous RNA cel-miR-39 was added during serum RNA extraction. The results showed that cel-miR-39 expression was stable in the serum of the breast cancer patients and the healthy controls. Therefore, cel-miR-39 can be used as a standard for serum miRNA detection.
The upregulation of miR-598 was previously found in patients with esophageal cancer (22). miR-184 was observed to be highly expressed in prostate cancer, while it was significantly downregulated in renal cell carcinoma patients, and was not correlated with patient age and gender or clinical stage of renal cell carcinoma (23). The study by Mar-Aguilar et al confirmed the upregulation of miR-382 in the serum of patients with breast cancer; a sensitivity of 94.40% and a specificity of 90.00% was found for the diagnosis of breast cancer (24). A study by Li et al showed that miR-382-5p expression was significantly downregulated in patients with ductal carcinoma in situ (25), while miR-1246 was demonstrated to exhibit high expression in colorectal cancer (26), esophageal squamous cell carcinoma (27), cervical carcinoma (28), hepatocellular carcinoma (29) and other cancers. The expression profiles of miRNAs in the serum and tissues are different. miR-1246 is produced by mammary gland epithelial cells and then stored; when the mammary epithelial cells become malignant, miR-1246 is selectively released into the circulating blood, resulted in elevated levels of miR-1246 in the circulation (30).
The results of the present study also showed that miR-382-3p and miR-1246 are highly expressed in the serum of breast cancer patients. To the best of our knowledge, there have been no studies on the serum miR-598 and miR-184 levels in breast cancer. The present study results showed that breast cancer patients had downregulated serum miR-598-3p and miR-184 expression. When using miR-598-3p as a biomarker with a cutoff value of 0.549, the sensitivity and specificity for the detection of breast cancer was 95.0 and 87.5%, respectively, indicating that miR-598-3p is a promising biomarker for breast cancer. Furthermore, the expression levels were not correlated with breast cancer stage.
In summary, in the present study, it was found that the expression of miR-382-3p and −1246 in the serum of breast cancer patients was upregulated, while the expression of miR-598-3p and −184 was downregulated. There were no significant differences in the expression levels among different TNM stages. miR-598-3p was shown to be a promising marker with a high sensitivity and specificity for the diagnosis of breast cancer. This study provides novel insights into serum miRNA expression profiles in breast cancer.
Acknowledgements
This study was supported by grants from the Zhejiang Science and Technology Program (no. 2012C37037), and the Taizhou Science and Technology Program (no. 2011A33212).
References
- 1.Fan L, Strasser-Weippl K, Li JJ, St Louis J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM, Goss PE. Breast cancer in China. Lancet Oncol. 2014;15:e279–e289. doi: 10.1016/S1470-2045(13)70567-9. [DOI] [PubMed] [Google Scholar]
- 2.Mishra S, Srivastava AK, Suman S, Kumar V, Shukla Y. Circulating miRNAs revealed as surrogate molecular signatures for the early detection of breast cancer. Cancer Lett. 2015;369:67–75. doi: 10.1016/j.canlet.2015.07.045. [DOI] [PubMed] [Google Scholar]
- 3.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 4.Chen H, Xu GZ, Zhao Y, Tian B, Lu H, Yu X, Xu Z, Ying N, Hu S, Hua Y. A novel OxyR sensor and regulator of hydrogen peroxide stress with one cysteine residue in Deinococcus radiodurans. Plos One. 2008;3:e1602. doi: 10.1371/journal.pone.0001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H, Huang LF, Hua XT, Yin L, Hu Y, Wang C, Chen W, Yu X, Xu Z, Tian B, et al. Pleiotropic effects of RecQ in Deinococcus radiodurans. Genomics. 2009;94:333–340. doi: 10.1016/j.ygeno.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Bethke A, Fielenbach N, Wang Z, Mangelsdorf DJ, Antebi A. Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science. 2009;324:95–98. doi: 10.1126/science.1164899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 8.Ng EK, Wong CL, Ma ES, Kwong A. MicroRNAs as new players for diagnosis, prognosis and therapeutic targets in breast cancer. J Oncol. 2009;2009:305420. doi: 10.1155/2009/305420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu J, Zheng Z, Wang J, Sun J, Wang P, Cheng X, Fu L, Zhang L, Wang Z, Li Z. Different miRNA expression profiles between human breast cancer tumors and serum. Front Genet. 2014;5:149. doi: 10.3389/fgene.2014.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 11.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 12.Gradishar WJ, Anderson BO, Blair SL, Burstein HJ, Cyr A, Elias AD, Farrar WB, Forero A, Giordano SH, Goldstein LJ, et al. Breast cancer version 3.2014. J Natl Compr Canc Netw. 2014;12:542–590. doi: 10.6004/jnccn.2014.0058. [DOI] [PubMed] [Google Scholar]
- 13.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N, et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 15.Brase JC, Wuttig D, Kuner R, Sültmann H. Serum microRNAs as non-invasive biomarkers for cancer. Mol Cancer. 2010;9:306. doi: 10.1186/1476-4598-9-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Y, Wang M, Lin G, Sun S, Li X, Qi J, Li J. Serum MicroRNA-155 as a potential biomarker to track disease in breast cancer. PLoS One. 2012;7:e47003. doi: 10.1371/journal.pone.0047003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao FL, Hua GD, Wang XF, Zhang XH, Zhang YK, Yu ZS. Serum overexpression of microRNA-10b in patients with bone metastatic primary breast cancer. J Int Med Res. 2012;40:859–866. doi: 10.1177/147323001204000304. [DOI] [PubMed] [Google Scholar]
- 18.Zearo S, Kim E, Zhu Y, Zhao JT, Sidhu SB, Robinson BG, Soon PSh. MicroRNA-484 is more highly expressed in serum of early breast cancer patients compared to healthy volunteers. BMC Cancer. 2014;14:200. doi: 10.1186/1471-2407-14-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Si H, Sun X, Chen Y, Cao Y, Chen S, Wang H, Hu C. Circulating microRNA-92a and microRNA-21 as novel minimally invasive biomarkers for primary breast cancer. J Cancer Res Clin Oncol. 2013;139:223–229. doi: 10.1007/s00432-012-1315-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan M, Liaw CS, Ji SM, Tan HH, Wong CY, Thike AA, Tan PH, Ho GH, Lee AS. Identification of circulating microRNA signatures for breast cancer detection. Clin Cancer Res. 2013;19:4477–4487. doi: 10.1158/1078-0432.CCR-12-3401. [DOI] [PubMed] [Google Scholar]
- 21.Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, Tait JF, Tewari M. Blood cell origin of circulating microRNAs: A cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 2012;5:492–497. doi: 10.1158/1940-6207.CAPR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao BS, Liu SG, Wang TY, Ji YH, Qi B, Tao YP, Li HC, Wu XN. Screening of microRNA in patients with esophageal cancer at same tumor node metastasis stage with different prognoses. Asian Pac J Cancer Prev. 2013;14:139–143. doi: 10.7314/APJCP.2013.14.1.139. [DOI] [PubMed] [Google Scholar]
- 23.Walter BA, Valera VA, Pinto PA, Merino MJ. Comprehensive microRNA profiling of prostate cancer. J Cancer. 2013;4:350–357. doi: 10.7150/jca.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mar-Aguilar F, Mendoza-Ramírez JA, Malagón-Santiago I, Espino-Silva PK, Santuario-Facio SK, Ruiz-Flores P, Rodríguez-Padilla C, Reséndez-Pérez D. Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis Markers. 2013;34:163–169. doi: 10.1155/2013/259454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S, Meng H, Zhou F, Zhai L, Zhang L, Gu F, Fan Y, Lang R, Fu L, Gu L, Qi L. MicroRNA-132 is frequently down-regulated in ductal carcinoma in situ (DCIS) of breast and acts as a tumor suppressor by inhibiting cell proliferation. Pathol Res Pract. 2013;209:179–183. doi: 10.1016/j.prp.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Piepoli A, Tavano F, Copetti M, Mazza T, Palumbo O, Panza A, di Mola FF, Pazienza V, Mazzoccoli G, Biscaglia G, et al. Mirna expression profiles identify drivers in colorectal and pancreatic cancers. PLoS One. 2012;7:e33663. doi: 10.1371/journal.pone.0033663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeshita N, Hoshino I, Mori M, Akutsu Y, Hanari N, Yoneyama Y, Ikeda N, Isozaki Y, Maruyama T, Akanuma N, et al. Serum microRNA expression profile: MiR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Brit J Cancer. 2013;108:644–652. doi: 10.1038/bjc.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, Yao D, Li Y, Chen H, He C, Ding N, Lu Y, Ou T, Zhao S, Li L, Long F. Serum microRNA expression levels can predict lymph node metastasis in patients with early-stage cervical squamous cell carcinoma. Int J Mol Med. 2013;32:557–567. doi: 10.3892/ijmm.2013.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Z, Meng C, Wang S, Zhou N, Guan M, Bai C, Lu S, Han Q, Zhao RC. MicroRNA-1246 enhances migration and invasion through CADM1 in hepatocellular carcinoma. BMC Cancer. 2014;14:616. doi: 10.1186/1471-2407-14-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pigati L, Yaddanapudi SC, Iyengar R, Kim DJ, Hearn SA, Danforth D, Hastings ML, Duelli DM. Selective release of MicroRNA species from normal and malignant mammary epithelial cells. PLoS One. 2010;5:e13515. doi: 10.1371/journal.pone.0013515. [DOI] [PMC free article] [PubMed] [Google Scholar]