Abstract
Breast cancer cells are heterogeneous in their ability to invade and fully metastasize, and thus also in their capacity to survive the numerous stresses encountered throughout the multiple steps of the metastatic cascade. Considering the role of autophagy as a survival response to stress, the present study hypothesized that distinct populations of breast cancer cells may possess an altered autophagic capacity that influences their metastatic potential. It was observed that a metastatic breast cancer cell line, MDA-MB-231, that was sensitive to autophagic induction additionally possessed the ability to proliferate following nutrient deprivation. Furthermore, a selected subpopulation of these cells that survived multiple exposures to starvation conditions demonstrated a heightened response to autophagic induction compared to their parent cells. Although this subpopulation maintained a more grape-like pattern in three-dimensional culture compared to the extended spikes of the parent population, autophagic induction in this subpopulation elicited an invasive phenotype with extended spikes. Taken together, these results suggest that autophagic induction may contribute to the ability of distinct breast cancer cell populations to survive and invade.
Keywords: autophagy, heterogeneity, breast cancer, invasion, metastasis
Introduction
It is widely appreciated that cancer cells in a growing tumor are heterogeneous (1,2). Only a subset of these cells possesses invasive qualities that drive their detachment from the primary tumor and movement through the surrounding microenvironment. Among these invasive cells, an even smaller subset has the capacity to fully metastasize (3,4). However, it remains to be elucidated how phenotypic differences between individual cells or cell populations translate to variations in metastatic ability.
Among the myriad cellular functions that contribute to cell phenotype is autophagy. Autophagy, here connoting macroautophagy, is a catabolic process during which a cell encloses cytoplasmic components within double-membrane vesicles (autophagosomes) that subsequently fuse with the lysosomal compartment, where the autophagosomal cargo is degraded enzymatically (5). Under normal conditions, a low level of basal autophagy serves to eliminate invading microorganisms and unnecessary, old or damaged proteins and organelles (6,7). During times of stress and starvation, autophagy is upregulated and the degraded autophagic cargo is recycled, generating substrates that enable cells to preserve intra- and extracellular homeostasis. The function of autophagy as a stress response makes it particularly notable in the framework of metastasis, an inherently stressful process that requires enhanced survival capabilities in the very select few cells that complete it (3,8). A necessary characteristic of invasive cancer cells that metastasize successfully is an exceptional ability to weather stress; throughout the multi-stage metastatic cascade, these cells must survive a wide range of factors, including nutrient deprivation, hypoxia, acidosis, extracellular matrix (ECM) detachment and shear force in the vasculature (4).
The present study investigated heterogeneity among breast cancer cell populations in terms of their autophagic capacities, proposing that perhaps the cancer cells with the most robust abilities to respond to autophagic induction are those that have the ability to withstand the pressures associated with the metastatic process. Thus, the present study hypothesized that distinct populations of breast cancer cells have increased autophagic potential that, in turn, leads to increased metastatic potential. The focus of the present study was the ability of breast cancer cells to endure nutrient deprivation, which is one of the pressures encountered throughout the metastatic process. The present study revealed that the metastatic breast cancer cell line that was sensitive to autophagic induction was also the cell line that maintained its ability to proliferate following nutrient deprivation. Furthermore, a subpopulation of this cell line comprised of cells that survived multiple exposures to nutrient deprivation was more responsive to autophagic induction compared with its parent population. The results of the present study suggest that, within a growing tumor, the autophagic response may contribute to the capacity of breast cancer cell subpopulations to endure starvation stress and better weather the metastatic process.
Materials and methods
Cells and cell culture
The human breast metastatic carcinoma cells lines, MDA-MB-231 (231) and MDA-MB-435 (435) (9); non-metastatic carcinoma cell line, MDA-MB-436 (436) (9); and epithelial cell line, MCF10A (10A) (10) were cultured as previously described. All cell lines were maintained at 37°C with 5% CO2 in a humidified atmosphere, and were tested regularly and confirmed as negative for Mycoplasma spp. contamination (PlasmoTest kit; InvivoGen, San Diego, CA).
Cell subpopulation selection
To select a distinct subpopulation of 231 cells that exhibited the capacity to survive nutrient deprivation, parental 231 cells were cultured to 80–90% confluency in complete culture medium consisting of a 1:1 (v/v) mixture of Dulbecco's modified Eagle's medium and Ham's F12 nutrient mixture (Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 5% fetal bovine serum (Thermo Fisher Scientific, Inc.), 2 mM L-glutamine (Thermo Fisher Scientific, Inc.), and 0.02 mM non-essential amino acids (Thermo Fisher Scientific, Inc.). Cells were subsequently washed twice with phosphate-buffered saline (PBS; Thermo Fisher Scientific, Inc.), incubated in Earle's balanced salt solution (EBSS; Thermo Fisher Scientific, Inc.) for 24 h, rewashed in PBS and cultured again to 80–90% confluency in complete culture medium. This procedure was repeated 4 additional times. The final subpopulation (231.EB5) was comprised of adherent cells that had survived all 5 starvation rounds.
Proliferation assay
Proliferation assays were performed as described previously (11). Cells were seeded (5×102 cells/well) in complete culture medium in a 96-well tissue culture plate. To assess innate proliferation capacity, fluorescence was measured after 1, 2, 3, 4 and 6 days with the addition of AlamarBlue reagent (Thermo Fisher Scientific, Inc.; DAL 1025). To assess proliferation capacity upon nutrient depletion, the complete medium in each well was replaced with EBSS 24 h after seeding and cells were incubated for 24 h, after which complete medium was returned to the wells. Proliferation was measured 1, 2, 3, 4 and 6 days following EBSS treatment. Fluorescence intensity at 570/580 nm excitation/emission was determined using a Hitachi F-7000 fluorescence spectrophotometer.
Immunoblot analysis
Cells were seeded in complete medium and treated for 0.5, 3, 6 and/or 12 h with 100 nM rapamycin (Sigma-Aldrich, St. Louis, MO, USA) and 50 µm chloroquine (Sigma-Aldrich), individually or in combination. Whole-cell lysates were collected with 1X radioimmunoprecipitation assay lysis buffer (EMD Millipore, Billerica, MA, USA) containing 1X Halt™ protease and phosphatase single-use inhibitor cocktail (Thermo Fisher Scientific, Inc.). Proteins (30 µg) were separated by SDS-PAGE (4–12% criterion XT precast gel; Bio-Rad Laboratories, Inc., Hercules, CA, USA) and transferred to nitrocellulose membranes. Membranes were blocked for 1 h at room temperature with 5% non-fat milk in Tris-buffered saline supplemented with 0.05% Tween 20 (TBST; Bio-Rad Laboratories, Inc., Hercules, CA, USA) and incubated overnight at 4°C with primary antibody. Membranes were washed in TBST, and incubated with the corresponding monoclonal donkey anti-rabbit (cat no. NA934V) or sheep anti-mouse (cat no. NA931V) IgG secondary antibody (GE Healthcare Life Sciences, Chalfont, UK; dilution, 1:10,000) for 1 h at room temperature, and rewashed before blots were developed with ECL Western Blotting Substrate (ThermoFisher Scientific, Inc.) and Supersignal West Dura Extended Duration Substrate (Thermo Fisher Scientific, Inc.). Results were quantified with Image Studio version 4.0 software (LI-COR Biosciences, Lincoln, NE, USA). The following primary antibodies were used: Anti-LC3B (Abcam, Cambridge, MA, USA; dilution, 1:3,000; cat no. ab51520), anti-β-actin (Abcam; dilution, 1:10,000), and anti-GAPDH (Cell Signaling Technology, Inc., Danvers, MA, USA; dilution, 1:2,000; cat no. 2118S).
Immunofluorescence studies
Cells were seeded and grown to 90% confluence on glass coverslips that were pretreated with 0.01% poly-L-lysine and placed in 6-well culture plates. Cells were incubated for 3 and 12 h in complete culture medium containing 100 nM rapamycin individually or in combination with 50 µM chloroquine. Each condition was performed in triplicate. Cells were subsequently washed 4 times for 3 min each in PBS, fixed in 3% formaldehyde in PBS for 45 min at room temperature, permeabilized with 0.5% Triton X-100 (Sigma-Aldrich) for 3 min at room temperature, blocked with 1% bovine serum albumin in PBS for 1 h and incubated with rabbit polyclonal anti-human LC3B antibody (Abcam; dilution, 1:2,000; cat no. ab51520) in 1% BSA in PBS at 4°C overnight. Secondary incubation was performed for 1 h at room temperature using a goat anti-rabbit immunoglobulin G fluorescein isothiocyanate-conjugated secondary antibody (Abcam; dilution, 1:2,000; cat no. ab6717). Vectashield with DAPI H-1200 (Vector Laboratories, Inc., Burlingame, CA, USA) was used to mount the coverslips on glass slides. Immunofluorescent images were captured with a Nikon Eclipse TE2000-U fluorescent microscope and archived using QCapture Pro software, version 5.1.1.14 (QImaging, Surrey, BC, Canada).
Three-dimensional (3D) cell morphology studies
For 3D cell culture assays, 400 µl/well of reduced growth factor Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) was placed in 24-well plates and allowed to solidify at 37°C for 40 min. Cells were plated (2×103 cells/well) in complete culture medium supplemented with 2% Matrigel and incubated at 37°C. After 4 d, the media was replaced with a fresh layer of Matrigel-supplemented medium only, or medium containing 100 nM rapamycin or 50 µM chloroquine. Media was replaced every 3 d. Each condition was performed in triplicate. Images were captured at 10 d with a Nikon Eclipse TE2000-U fluorescent microscope and archived using TSView software (Tucsen photonics Co., Ltd., Fuzhou, China).
Results
MDA-MB-231 cells are sensitive to autophagic induction
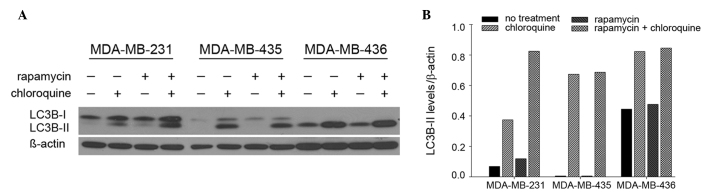
The presence of LC3B in autophagosomes and the conversion of LC3B-I to its lipidated form, LC3B-II, are used routinely as indicators of autophagic induction (12). The present study evaluated the levels of LC3B-II in 231, 435 and 436 cells by immunoblot (Fig. 1A). Treatment with rapamycin, a well-accepted autophagy inducer via inhibition of mammalian target of rapamycin (mTOR), altered LC3B-II levels in 231 cells, but not 435 and 436 cells. Specifically, in 231 cells, LC3B-II levels were 74% higher following rapamycin treatment compared with no treatment, and 120% higher following co-treatment with rapamycin and chloroquine, an autophagy-blocking agent (lysosomal inhibitor), compared with chloroquine treatment alone (Fig. 1B). By contrast, rapamycin did not appreciably modulate LC3B-II levels in either 435 or 436 cells. Notably, 231 cells exhibited the lowest levels of basal autophagosome formation, indicated by LC3B-II levels apparent only upon the addition of chloroquine. By contrast, basal autophagosome levels were relatively high in 435 and 436 cells. Overall, these results indicate that 231 cells are most sensitive to autophagic induction compared with 435 or 436 cells, which suggests that 435 and 436 cells have intrinsically high levels of autophagy that an additional stimulus may not robustly alter.
Figure 1.
MDA-MB-231 cells are sensitive to autophagic induction. MDA-MB-231, MDA-MB-435 and MDA-MB-436 cells were seeded in complete medium and treated with 100 nM rapamycin and/or 50 µm chloroquine. Levels of LC3B-II were (A) probed by immunoblotting and (B) quantified by densitometry and comparing LC3B-II levels with the loading control beta-actin.
MDA-MB-231 cells proliferate following nutrient depletion
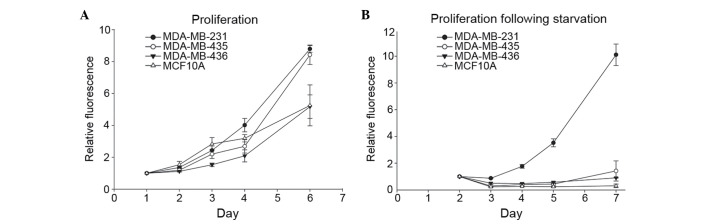
For cells to metastasize successfully, the capacity to survive in suboptimal environments is essential. To begin investigating the association between variations in autophagic capacity and abilities to withstand challenging conditions, the present study used the same cell lines to test how nutrient deprivation altered cell proliferation. Cells were grown in complete medium to determine their innate proliferation rate. As expected, the metastatic cell lines, 231 and 435, exhibited a distinctly increased proliferation rate compared with the non-metastatic 436 breast cancer cell line and the immortalized, but otherwise normal, MCF10A breast epithelial cell line (Fig. 2A). Proliferation rates following nutrient deprivation were subsequently assessed by culturing cells in EBSS for 24 h and then returning them to complete medium. The 231 cells maintained their proliferative ability, whereas 435, 436 and MCF10A cells did not recover from starvation conditions (Fig. 2B). Together with the results presented in Fig. 1, these results suggest that the cells that are most responsive to autophagic induction are also able to best withstand the stress of nutrient deprivation.
Figure 2.
MDA-MB-231 cells proliferate following nutrient depletion. (A) Relative cell number in complete culture medium of MDA-MB-231, MDA-MB-435, MDA-MB-436 and MCF10A cell lines was assessed with AlamarBlue and revealed that the metastatic cell lines (MDA-MB-231 and MDA-MB-435) had higher innate proliferation rates. (B) Relative cell number of MDA-MB-231, MDA-MB-435, MDA-MB-436 and MCF10A cells following 24 h of culture in Earle's balanced salt solution indicated that MDA-MB-231 cells were uniquely able to maintain their innate proliferation rate following starvation treatment.
Autophagic induction is more efficient in a select MDA-MB-231 subpopulation
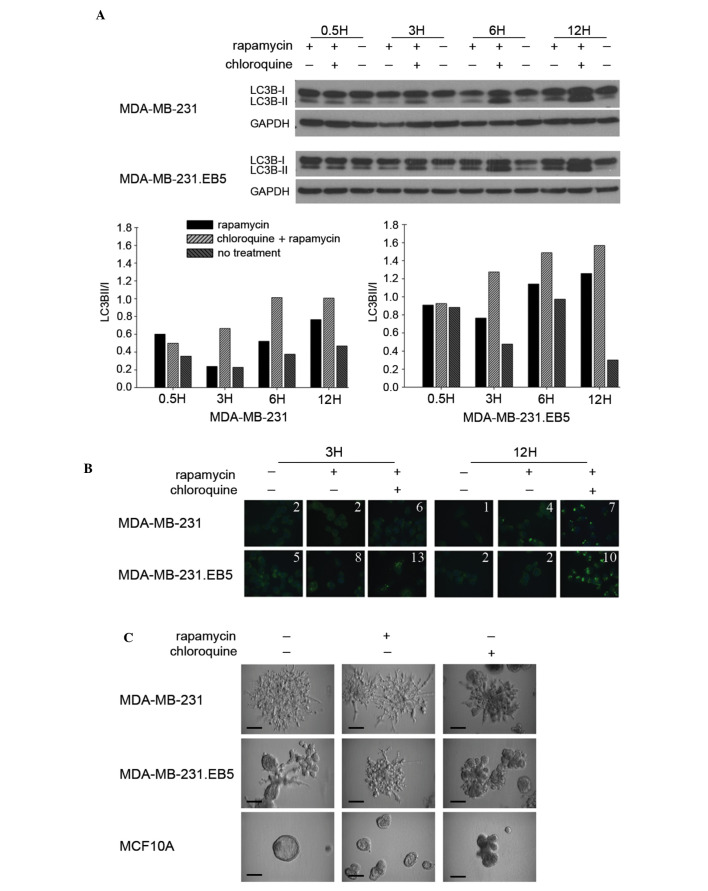
Noting that 231 cells were unique in both their sensitivity to autophagic induction and their sustained proliferation following stress, the present study assessed whether a subpopulation of these cells possessed a capacity for autophagic induction distinct from parental cells. It was reasoned that it may be this type of subset of invasive cells that has the ability to fully metastasize. Parental 231 cells were subjected to 5 rounds of culture in EBSS for 24 h followed by 24 h of culture in complete medium. The surviving cells were selected (231.EB5). The five rounds of selection were performed to ensure the generation of a distinct cell subpopulation. LC3B-II levels by immunoblot (Fig. 3A) and LC3B puncta formation by immunofluorescence staining (Fig. 3B) were subsequently compared between the parental and selected cell lines. Notably, 231.EB5 cells displayed the most marked changes in LC3B-II levels in response to rapamycin treatment compared with untreated conditions, particularly at 3 and 12 h (Fig. 3A). Supporting the immunoblot results, the number of LC3B puncta was increased with no treatment and in response to rapamycin in 231.EB5 cells compared with parent 231 cells (Fig. 3B).
Figure 3.
Autophagic induction is more efficient in a select subpopulation of MDA-MB-231 cells. (A) MDA-MB-231 and MDA-MB-231.EB5 cells were seeded in complete medium and treated for 0.5, 3, 6 and 12 h with 100 nM rapamycin individually and in combination with 50 µm chloroquine. LC3B-I and -II expression was assayed by immunoblotting and quantified by densitometry. (B) MDA-MB-231 and MDA-MB-231.EB5 cells were incubated in complete culture medium containing 100 nM rapamycin individually and in combination with 50 µm chloroquine for 3 and 12 h. Immunofluorescence images of LC3B puncta were captured in triplicate and representative images are shown. The average number of puncta per cell is indicated for each condition. (C) MDA-MB-231, MDA-MB-231.EB5 and MCF10A cells were seeded in complete culture medium on Matrigel-coated cell culture plates and treated with 100 nM rapamycin with or without 50 µM chloroquine. All conditions were performed in triplicate. Images were captured 10 d after seeding and representative images are shown. Scale bar=30 µm. GAPDH, glyceraldehyde-3-phosphate.
Parent 231 cells grown in 3D Matrigel with complete medium exhibit spindled branches characteristic of highly invasive cells (Fig. 3C). Notably, 231.EB5 cells were rounded with few branched spikes, which is typical of less invasive cells. However, in response to rapamycin treatment, 231.EB5 cells became substantially more invasive, developing a phenotype reminiscent of the parent population. Minimal changes were observed in parent 231 cells and MCF10A control cells following rapamycin treatment compared with no treatment. Chloroquine reduced the invasive nature of both 231 and 231.EB5 cells, eliciting smaller and more rounded colonies. Overall, the results of the present study demonstrate distinct phenotypic differences between a select population of 231 cells compared with the parent population following a stimulus of autophagic induction, and reiterate prior observations that responsiveness to autophagic induction may be correlated with the ability to endure nutrient deprivation.
Discussion
It is well-acknowledged that cancer cells within a tumor vary widely in their functions and attributes, yet how traits of individual cells or cell populations drive tumor progression, including invasion and metastasis, remains to be fully elucidated (1). In the present study, the association between the response to autophagic induction and the capacity to endure a type of stress encountered during the metastatic process was investigated in various breast cancer cell populations.
The role of autophagy is particularly multifarious in the framework of cancer, in which the matter of whether autophagy is anti- or protumorigenic is associated with that of whether it is cytoprotective or cytotoxic (13). It is increasingly appreciated that autophagic functioning is dependent on numerous factors, notably cancer stage (14). While it has been demonstrated that autophagy may act to suppress tumor growth during tumorigenesis (6,15,16), autophagy is able to promote tumor growth in transformed cells, which may exploit autophagy as a protective mechanism against stresses in the tumor microenvironment, including glucose deprivation and hypoxia, as well as against therapeutic insult (5,6,17–20). Activation of autophagy additionally has been observed in response to stresses characteristic of the metastatic cascade (21). For instance, autophagy is upregulated following loss of ECM contact (22–24), which typically induces programmed cell death (anoikis) in normal cells, yet is an essential event for metastasizing tumor cells (3).
The present study observed that 231 cells were sensitive to autophagic induction (that is, increases in markers of autophagosome formation in response to an agent, rapamycin, known to induce autophagy were observed), and that autophagic induction was associated with increased metastatic potential as assessed by the ability to weather nutrient deprivation stress. Rapamycin treatment increased LC3B-II levels in 231 cells, but LC3B-II levels did not change appreciably in 435 or 436 cells. Furthermore, 231 cells, but not 435 or 436 cells, maintained their innate ability to grow following a period of nutrient deprivation. The use of nutrient deprivation to investigate whether cells with sensitivity to autophagic induction are able to overcome difficult conditions is consistent with a previous study by Wojtkowiak et al (19), who demonstrated that an acidic environment triggered chronic upregulation of autophagy as a survival mechanism. While direct links cannot be made between the autophagic response to rapamycin and the response to nutrient deprivation, there is logic in juxtaposing rapamycin treatment with starvation. These conditions may promote autophagic induction via similar signaling pathways: Rapamycin by inhibiting mTORC1 activity, and starvation by upregulating 5′ AMP-activated protein kinase (AMPK), which in turn may inhibit mTORC1 activity downstream. Inhibition of mTORC1 and upregulation of AMPK may promote unc-51 like autophagy activating kinase 1 complexing and thus autophagy initiation (25–28).
As it cannot be concluded that the presently investigated cell lines had an identical response to autophagic induction by nutrient deprivation as they did to that by rapamycin, there exists an alternative possibility to consider. The 436 cells, followed by 435 cells, had the highest levels of basal autophagosome formation compared with the lowest observed in 231 cells; 436 and 435 cells additionally had the lowest numbers of cells that proliferated following starvation. Autophagy has been linked not only to cell survival, but also to cell death (13,29,30). Thus, it cannot be ruled out that the additional stimulation of nutrient deprivation in the cells with high basal levels of autophagosome formation resulted in excessive levels that provoked a cytotoxic response. The importance of considering basal autophagy was underscored in an investigation by Maycotte et al (31), although their findings associated autophagy with cell survival, not death. These investigators reported that breast cell lines differed in their dependency on autophagy for survival in complete medium under conditions with no added stress; upon autophagy inhibition with chloroquine and small hairpin RNA knockdown of autophagy protein 5 (ATG5), ATG7 and Beclin 1, triple negative breast cancer cell lines (including 231) displayed the greatest decreases in proliferation and cell viability. Whether basal dependency on autophagy under normal conditions is associated with dependency on autophagy for survival under stress was not assessed.
The results of the present study in 231.EB5 cells, a subpopulation of 231 cells that survived multiple rounds of starvation, provide additional evidence in support of the association between sensitivity to autophagic induction and metastatic potential. While previous studies have reported the link between autophagy and nutrient deprivation (32–34), to the best of our knowledge, the present study is the first to generate a subpopulation through nutrient deprivation and to investigate differences in autophagic capacity based on this factor. These 231.EB5 cells with apparently superior aptitude for survival exhibited the highest overall ability to respond to rapamycin, as seen by increases in LC3B-II conversion at 3 h and particularly at 12 h. The fact that sensitivity to rapamycin varied by time point may be associated with the observation that autophagy is regulated through a negative feedback loop based on the sensitivity of mTORC1 to nutrient levels (7). Yu et al (35) reported that the intracellular nutrients produced during the autophagic process may reactivate mTORC1 signaling and inhibit autophagy over time even in the event of ongoing starvation, a condition that initially inhibits mTORC1 activity and upregulates autophagy. Similar to starvation, rapamycin treatment upregulates autophagy through mTORC1 inhibition. Therefore, perhaps the fluctuations in LC3B-II conversion that were observed following prolonged rapamycin treatment represent cycles of rapamycin-induced autophagic upregulation counteracted by the subsequent generation of nutrients. This requires additional investigation, particularly considering the fact that consistent changes in LC3B-II conversion in response to rapamycin were noted in parental 231 cells. However, this observation in itself is in line with the fact that 231.EB5 cells were most sensitive to autophagic induction, which would in turn incite more robust negative feedback and variation by time point.
The fact that 231.EB5 cells exhibited a less invasive phenotype in 3D when cultured in complete medium with no treatment was unexpected, as it was predicted that these cells would have heightened invasive qualities based on their high survival capacity. It was notable that these cells displayed a markedly more invasive phenotype following autophagic induction. At this point, the factors responsible for these observations remain to be elucidated. One key avenue to be investigated is that cancer stem-like cells (CSCs) may have been enriched during cell subpopulation selection. That greater changes were observed in LC3B-II and increased numbers of LC3B puncta were observed in 231.EB5 cells is in line with an observation reported by Gong et al (36), who identified significantly increased autophagic flux in breast CSC-enriched MCF-7 breast cancer cells compared with non-enriched adherent cells in basal and starvation (EBSS culture) conditions. If the present study did enrich CSCs in the cell subpopulation, it is possible that this is the reason for the higher baseline levels of LC3B-II and that the increased invasive 3D morphology that was observed upon autophagic induction is associated with generation of migrating CSCs (37). Additional potential explanations include stimulation of quiescence and triggering of an epithelial-to-mesenchymal switch (5). All of these have been associated with autophagy but none have been completely characterized. Additional experiments will be required to fully elucidate the reasons for the difference in phenotype between 231.EB5 and parent 231 cells.
In conclusion, the present study demonstrated that the populations of metastatic cells that displayed the most robust reactions to autophagic induction additionally best withstood starvation stress. It must be acknowledged that nutrient deprivation and autophagy are not the only factors governing the progression of a growing tumor. It must also be acknowledged that autophagic induction does not denote completion of the entire autophagic process, and thus that future studies should establish the contribution of autophagic flux. However, the present study is significant, noting that autophagy inducers and inhibitors are among the developing therapeutic approaches in breast cancer (38,39). The results of the present study suggest that these agents may cause differential responses depending on the properties of the unique cancer cell populations and their surrounding environments.
Acknowledgements
Funding for the present study was provided in part by the American Cancer Society (grant no., RSG-11-259-01-CSM) and METAvivor Research and Support, Inc., as well as the University of Alabama at Birmingham Cancer Prevention and Control Training Program (grant no., R25 CA047888).
Glossary
Abbreviations
- 231
MDA-MB-231
- 231.EB5
MDA-MB-231.EB5
- 435
MDA-MB-435
- 436
MDA-MB-436
- 3D
three-dimensional
References
- 1.Almendro V, Marusyk A, Polyak K. Cellular heterogeneity and molecular evolution in cancer. Annu Rev Pathol. 2013;8:277–302. doi: 10.1146/annurev-pathol-020712-163923. [DOI] [PubMed] [Google Scholar]
- 2.Heppner GH, Loveless SE, Miller FR, Mahoney KH, Fulton AM. Mammary tumor heterogeneity. Symp Fundam Cancer Res. 1983;36:209–221. [PubMed] [Google Scholar]
- 3.Talmadge JE, Fidler IJ. AACR centennial series: The biology of cancer metastasis: Historical perspective. Cancer Res. 2010;70:5649–5669. doi: 10.1158/0008-5472.CAN-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Welch DR, Steeg PS, Rinker-Schaeffer CW. Molecular biology of breast cancer metastasis. Genetic regulation of human breast carcinoma metastasis. Breast Cancer Res. 2000;2:408–416. doi: 10.1186/bcr87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: Therapeutic implications. Mol Cancer Ther. 2011;10:1533–1541. doi: 10.1158/1535-7163.MCT-11-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Z, Klionsky DJ. Eaten alive: A history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss L. Metastatic inefficiency: Intravascular and intraperitoneal implantation of cancer cells. Cancer Treat Res. 1996;82:1–11. doi: 10.1007/978-1-4613-1247-5_1. [DOI] [PubMed] [Google Scholar]
- 9.Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 2009;69:1279–1283. doi: 10.1158/0008-5472.CAN-08-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hurst DR, Xie Y, Edmonds MD, Welch DR. Multiple forms of BRMS1 are differentially expressed in the MCF10 isogenic breast cancer progression model. Clin Exp Metastasis. 2009;26:89–96. doi: 10.1007/s10585-008-9216-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cody JJ, Markert JM, Hurst DR. Histone deacetylase inhibitors improve the replication of oncolytic herpes simplex virus in breast cancer cells. PLoS One. 2014;9:e92919. doi: 10.1371/journal.pone.0092919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:1845–1846. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 14.Roy S, Debnath J. Autophagy and tumorigenesis. Semin Immunopathol. 2010;32:383–396. doi: 10.1007/s00281-010-0213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gélinas C, Fan Y, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu YL, DeLay M, Jahangiri A, Molinaro AM, Rose SD, Carbonell WS, Aghi MK. Hypoxia-induced autophagy promotes tumor cell survival and adaptation to antiangiogenic treatment in glioblastoma. Cancer Res. 2012;72:1773–1783. doi: 10.1158/0008-5472.CAN-11-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wojtkowiak JW, Rothberg JM, Kumar V, Schramm KJ, Haller E, Proemsey JB, Lloyd MC, Sloane BF, Gillies RJ. Chronic autophagy is a cellular adaptation to tumor acidic pH microenvironments. Cancer Res. 2012;72:3938–3947. doi: 10.1158/0008-5472.CAN-11-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455:396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- 21.Kenific CM, Thorburn A, Debnath J. Autophagy and metastasis: Another double-edged sword. Curr Opin Cell Biol. 2010;22:241–245. doi: 10.1016/j.ceb.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertrand K. Survival of exfoliated epithelial cells: A delicate balance between anoikis and apoptosis. J Biomed Biotechnol. 2011;2011:534139. doi: 10.1155/2011/534139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lock R, Debnath J. Extracellular matrix regulation of autophagy. Curr Opin Cell Biol. 2008;20:583–588. doi: 10.1016/j.ceb.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Debnath J. Detachment-induced autophagy during anoikis and lumen formation in epithelial acini. Autophagy. 2008;4:351–353. doi: 10.4161/auto.5523. [DOI] [PubMed] [Google Scholar]
- 25.Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, Cecconi F. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol. 2013;15:406–416. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- 26.Alers S, Löffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: Cross talk, shortcuts, and feedbacks. Mol Cell Biol. 2012;32:2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee EJ, Tournier C. The requirement of uncoordinated 51-like kinase 1 (ULK1) and ULK2 in the regulation of autophagy. Autophagy. 2011;7:689–695. doi: 10.4161/auto.7.7.15450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohli L, Kaza N, Carroll SL, Roth KA. Protector turns predator: Autophagic death via selective degradation of KRAS. Autophagy. 2013;9:1438–1439. doi: 10.4161/auto.25837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eisenberg-Lerner A, Bialik S, Simon HU, Kimchi A. Life and death partners: Apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009;16:966–975. doi: 10.1038/cdd.2009.33. [DOI] [PubMed] [Google Scholar]
- 31.Maycotte P, Gearheart CM, Barnard R, Aryal S, Mulcahy Levy JM, Fosmire SP, Hansen RJ, Morgan MJ, Porter CC, Gustafson DL, Thorburn A. STAT3-mediated autophagy dependence identifies subtypes of breast cancer where autophagy inhibition can be efficacious. Cancer Res. 2014;74:2579–2590. doi: 10.1158/0008-5472.CAN-13-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong PM, Puente C, Ganley IG, Jiang X. The ULK1 complex: Sensing nutrient signals for autophagy activation. Autophagy. 2013;9:124–137. doi: 10.4161/auto.23323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L, Chen Y, Gibson SB. Starvation-induced autophagy is regulated by mitochondrial reactive oxygen species leading to AMPK activation. Cell Signal. 2013;25:50–65. doi: 10.1186/1478-811X-11-50. [DOI] [PubMed] [Google Scholar]
- 34.Li Y, Zhang Q, Tian R, Wang Q, Zhao JJ, Iglehart JD, Wang ZC, Richardson AL. Lysosomal transmembrane protein LAPTM4B promotes autophagy and tolerance to metabolic stress in cancer cells. Cancer Res. 2011;71:7481–7489. doi: 10.1158/0008-5472.CAN-11-0940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong C, Bauvy C, Tonelli G, Yue W, Deloménie C, Nicolas V, Zhu Y, Domergue V, Marin-Esteban V, Tharinger H, et al. Beclin 1 and autophagy are required for the tumorigenicity of breast cancer stem-like/progenitor cells. Oncogene. 2013;32:2261–2272. doi: 10.1038/onc.2012.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: Migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744–749. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 38.Gewirtz DA. The four faces of autophagy: Implications for cancer therapy. Cancer Res. 2014;74:647–651. doi: 10.1158/0008-5472.CAN-13-2966. [DOI] [PubMed] [Google Scholar]
- 39.Vinayak S, Carlson RW. mTOR inhibitors in the treatment of breast cancer. Oncology (Williston Park) 2013;27:38–44. [PubMed] [Google Scholar]