Abstract
It has been reported previously that the expression of glucose transporter member 3 (GLUT3) is increased in malignant glioma cells compared with normal glial cells. However, the regulating mechanism that causes this phenomenon remains unknown. The present study investigated the regulating role of transcription factor specific protein 1 (Sp1) in GLUT3 expression in a human glioma cell line. In the present study, Sp1 was identified to directly bind to the GLUT3 5′-untranslated region in human glioma U251 cells. Small interfering RNA- and the Sp1-inhibitor mithramycin A-mediated Sp1 knockdown experiments revealed that Sp1 depletion decreased glucose uptake and inhibited cell growth and invasion of U251 cells by downregulating GLUT3 expression. Therefore Sp1 is an important positive regulator for the expression of GLUT3 in human glioma cells, and may explain the overexpression of GLUT3 in U251 cells. These results suggest that Sp1 may have a role in glioma treatment.
Keywords: malignant gliomas, glucose uptake, SP1, GLUT3
Introduction
Malignant glioma is one of the most common types of malignant primary brain tumor, and has high morbidity and mortality. Even with optimal treatment, which consists of surgery, chemotherapy and radiotherapy, the median survival time of patients with malignant glioma is extremely short; only 12–15 months for patients with glioblastomas and 2–5 years for patients with anaplastic gliomas (1). It is important to understand the genetic and epigenetic factors that cause the activation of oncogenes and inactivation of tumor suppressor genes in cancer cells. An increasing number of studies indicate that accelerated glycolysis is one of the most important and vital biochemical characteristics of cancer cells (2,3). Glucose is the major energy substrate of most eukaryotic cells, and tumors, particularly solid cancer cells, often possess a growth advantage compared with non-tumorous cells due to increased anaerobic glycolysis (4,5).
Glucose transporter (GLUT) proteins allow the energy independent transport of glucose across the hydrophobic cell membrane and against a concentration gradient, and this is the first rate-limiting step for sugar metabolism in cells (6). GLUTs are regarded as important regulators of glucose metabolism in transformed cells. Among the 14 members of GLUTs, GLUT member 3 (GLUT3), which has a high affinity for glucose, is reported to be upregulated in samples obtained from patients with brain tumors (7,8). Furthermore, the level of GLUT3 expression in gliomas has a significant association with pathological grading of tumors (7,9). It has been reported previously that GLUT3 is highly expressed in glioblastoma, and promotes the growth of brain tumor initiating cells (3). Therefore, it is probable that GLUT3 may become a therapeutic target in cancer treatment. However, the mechanism by which GLUT3 is upregulated remains unknown.
Transcription factor Sp1 (Sp1) was the first transcription factor to be isolated, and it regulates the expression of numerous genes involved in cell proliferation, apoptosis and differentiation (10). Previous studies have demonstrated that an increase in Sp1 transcriptional activity is associated with tumorigenesis (11–13). In addition, Sp1, has been suggested as a therapeutic target for cancer (14), and the Sp1 inhibitor mithramycin A and analogues have been used as a novel strategy for the treatment of various types of cancer (15–17). In muscle cells, insulin-like growth factor-1 controls GLUT3 expression via Sp1 (17). In human glioma, it has been reported that Sp1 is upregulated, which promotes matrix metalloproteinase (MMP)-2-mediated cell invasion and predicts poor clinical outcomes of patients (18). However, the clinical significance and biological role of the association between Sp1 and GLUT3 expression in glioma requires further investigation. The aim of the current study was to identify whether the expression of GLUT3 is regulated by Sp1, and to investigate the role of Sp1 in regulating glucose transport and the biological function of glioma cells.
Materials and methods
Cell culture and materials
Human embryonic kidney (HEK)-293T and human glioma U251 cells were obtained from the American Type Culture Collection (Manassas, VA, USA). HEK-293T and U251 cells were maintained in Dulbecco's Modified Eagle Medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum and 100 U/ml penicillin (Gibco®; Thermo Fisher Scientific, Inc.). The cells were free from mycoplasma contamination. All cell lines were incubated in a humidified atmosphere of 5% CO2 at 37°C. Mithramycin A, a selective inhibitor of Sp1-mediated transcriptional activation, was assayed according to previous report (15). The cells were treated with mithramycin A (300 nM; Sigma-Aldrich, St. Louis, MO, USA) and were then harvested and lysed for either RNA or protein extraction.
Plasmids constructs and luciferase assay
To determine the possible effect of the Sp1 binding site on the solute carrier family 2 (facilitated glucose transporter), member 3 (SLC2A3) gene, which encodes GLUT3 and GLUT3 5′-untranslated region (5′-UTR) containing putative binding sites for Sp1 (−306 to-297 nt and −312 to-303 nt) was amplified from normal human genomic DNA using the PCR and the following primers: Forward, 5′-TACGGTACCAAACCCAGGGTGGAGAGAG-3′ and reverse, 5′-TAGCTCGAGAGCCTGAAAGGGCGAC-3′. The binding sites were cloned into the KpnI and XhoI sites of the firefly luciferase reporter gene vector pGL3-Basic (Promega Corporation, Madison, WI, USA), which was named as pGL3-GLUT3-WT. GLUT3 mutant 5′-UTR was generated using site-directed mutagenesis (Forward, 5′-GGGGCGGGG-3′ and reverse, 5′-GGTTTTTGG) and was recombined into pGL3-Basic, which was named as pGL3-GLUT3-MUT. Silencing of Sp1 expression was achieved using small interfering (si) RNA duplex sequences according to a previous study by Garcia-Huerta et al (19): Short hairpin (sh) Sp1.1, 5′-CCTTGCTACCTGTCAACAGCGTTTCTGCA-3′; and shSp1.2, 5′-AGGACAGACTCAGTATGTGACCAATGTAC-3′. The siRNA sequence was synthesized by Genetimes Technology, Inc. (Shanghai, China) and named as siRNA-S1. All constructs were confirmed by DNA sequencing.
All GLUT3-associated promoter-luciferase constructs were co-transfected into U251 cells using Lipofectamine® 2000 Transfection Reagent (Thermo Fisher Scientific, Inc.). For the reporter assay, 100 ng pGL3-GLUT3-WT or pGL3-GLUT3-MUT were co-transfected together with 50 ng p-renilla luciferase reporter vector-TK with or without siRNA-S1. The U251 cells were cultured in 24-well plates under serum-free conditions. The cells were harvested 48 h following transfection, and luciferase activity was measured using the Dual-Luciferase® Reporter Assay System (Promega Corporation) according to the manufacturer's protocol. Firefly luciferase activity was normalized to renilla luciferase activity.
Chromatin immunoprecipitation (ChIP) assay
To detect any in vivo binding of Sp1 to the SLC2A3 gene, a ChIP assay was performed using a ChIP Assay kit (EMD Millipore, Billerica, MA, USA). Briefly, U251 cells (1×106 cells) were cross-linked with 1% formaldehyde (Sigma-Aldrich) at 37°C for 10 min. The cross-linked chromatin was subsequently extracted, diluted with lysis buffer and sheared by sonication. Following centrifugation (Eppendorf 5415R; 7,500 × g, 10 min), supernatants were diluted 10-fold with ChIP dilution buffer and an aliquot of the diluted supernatant (1%) was saved as a positive control for polymerase chain reaction (PCR). Subsequent to pre-clearing the lysate with protein A and protein G-agarose beads (1:2 ratio; EMD Millipore), the chromatin was divided into equal samples for immunoprecipitation with either anti-Sp1 (mouse monoclonal IgG2a; #sc-17824) or anti-immunoglobulin (Ig)G (negative control; mouse monoclonal IgG2a; #sc-53740) antibodies (purchased from Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Immunoprecipitated DNA was recovered using the QIAquick PCR Purification kit (Qiagen, Inc., Valencia, CA, USA) and 1:100 of the precipitated DNA was used for PCR. Prior to adding the antibody, 1:100 of the solution was used as an internal control for the quantitative accuracy of the DNA. Purified DNA was subjected to PCR with primers specific for a region in the SLC2A3 promoter (−381 to −200 nt), which spanned two putative Sp1-binding sites. The primer sequences used for the PCR reactions were as follows: Forward, 5′-AAACCCAGGGTGGAGAGAG-3′ and reverse, 5′-AGCCTGAAAGGGCGAC-3′. Quantitative (q) PCR was performed using SYBR green. The following cycling conditions were used: 95°C for 1 min; and amplification for 40 cycles of 95°C for 30 sec and 58°C for 45 sec. The comparative threshold cycle method was used to calculate the relative alterations in the SLC2A3 gene. The data were normalized using the negative control, an untranscribed region upstream of glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and was represented as fold-change of the negative control.
Virus production and infection
The role of GLUT3 in glioma cell proliferation was examined using RNA interference-mediated gene silencing. The siRNA sequence that targeted the GLUT3 gene plasmid (pLV-SLC2A3-shRNA) were synthesized by Genetimes Technology, Inc. as follows: 5′-CGGTGCAGATAGATCTGGAAACTCGAGTTTCCAGATCTATCTGCACCGTTTTT-3′. HEK-293T cells were transfected using the calcium-phosphate precipitation method, co-transfecting the lentiviral plasmid of interest in conjunction with proteolipid protein (pLP)-1, vesicular stomatitis virus (VSV-G) and pLP2. Viral supernatants were harvested 48 h post-transfection, filtered and used directly for infection of U251 cells. Puromycin selection was performed to select cells with stable pLV genomic integration (with control or GLUT3 shRNA). Fluorescence-activated cell sorting was used to select cells with stable constructs, based on green fluorescent protein expression.
[3H]-2-deoxyglucose ([3H]-2-DG) uptake experiment
The glucose uptake experiment was measured by modifying the method described previously (20). Briefly, U251 cells were plated in 24-well plates at a density of 2×105 cells/well 48 h prior to the uptake experiment. The cells were washed with phosphate-buffered saline (PBS) and uptake was initiated by the addition of 0.5 ml incubation buffer containing [3H]-2-DG (DuPont USA, Wilmington, DE, USA) at a final concentration of 600 nM (0.1 µCi/500 µl media in well) for 20 min at 37°C. Uptake was halted by washing the cells with ice-cold 0.1 M PBS. The cells were dissolved in 200 µl of lysis buffer [10 mM Tris-HCl (pH 8.0) and 0.2% sodium dodecyl sulfate (SDS)] and the incorporated radioactivity was measured by liquid scintillation spectrometry. Non-specific uptake was determined in the presence of cytochalasin B (10 µmol/l; control), which was subtracted from all values.
RNA purification and qPCR amplification
Total RNA was extracted from the U251 cells using TRIzol® Reagent (Invitrogen™; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Nucleic acid concentrations were determined using Quant-iT™ RiboGreen® RNA Assay kit (Invitrogen™; Molecular Probes®; Thermo Fisher Scientific, Inc.). The RNA was stored at −80°C until required.
SLC2A3 expression level was detected by one-step qPCR using TaqMan probe specific for SLC2A3 mRNA. SLC2A3 primers and probe were as follows: Forward 5′-CTCTTCGTCAACCGCTTTGG-3′, reverse 5′-TCAACCGACTTAGCTACTTTACAC-3′ and TaqMan probe, 5′-FAM-AGCAGCCACCAGTGACAGCCAACA-BHQ1-3′. One-step qPCR was performed on ABI 7500 Read-Time PCR system (Applied Biosystems®; Thermo Fisher Scientific, Inc.) using 25 µl QuantiTect Probe RT-PCR kit (Qiagen, Inc.) on 1–2 µg of total RNA. PCR conditions were as follows: 50°C for 15 min; 95°C for 15 min; 45 cycles at 94°C for 30 sec; annealing at 55°C for 45 sec. A no-template control was included in each assay. β-actin was used as an endogenous control and vehicle control was used as a calibrator. Each sample was run three times. The comparative threshold cycle method was used to calculate the relative changes in SLC2A3 gene expression. The relative changes of gene expression were calculated using the 2−ΔΔCq method, where ΔCq = Cq (detected gene) - Cq (β-actin), and Cq represents threshold cycle number.
Western blot analysis
U251 cells were cultured in 6-well plates to 70–80% confluence. The cells were scraped and homogenized with lysis buffer [150 mM NaCl, 100 mM Tris-HCl (pH 8.0), 1 mM ethylenediaminetetraacetic acid, 0.5% Triton X-100] and a protease inhibitor cocktail (Roche Diagnostics GmbH, Mannheim, Germany). The cells were lysed in the same way 2–3 days following transfection or Sp1 inhibitor treatment. The amount of soluble protein was determined using a modified Lowry assay (DC™ Protein Assay kit; Bio-Rad Laboratories, Inc., Hercules, CA, USA). Each lane was loaded with 20 µg total protein. Proteins were electrophoretically resolved by 9% SDS-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride (PVDF) membranes (EMD Millipore). Following blocking with 5% dry milk for 1 h at room temperature, the PVDF membranes were incubated overnight with anti-GLUT3 (mouse monoclonal IgG1; #sc-74399; dilution, 1:200) and for 4 h with anti-GAPDH (mouse monoclonal IgG1; #sc-365062; dilution, 1:200) antibodies, which were purchased from Santa Cruz Biotechnology, Inc. Following 3 washes with Tris-buffered saline and Tween 20, the membranes were incubated with a sheep anti-mouse IgG conjugated to horseradish peroxidase secondary antibody (#NA931; GE Healthcare Life Sciences, Chalfont, UK; dilution, 1:1,000) for 1 h at room temperature. The protein blots were visualized using Pierce™ ECL Western Blotting Substrate (Thermo Fisher Scientific, Inc.) on a Kodak Image Station (Kodak, Rochester, NY, USA).
Cell viability assay
In vitro cell viability was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Briefly, 2×105 cells, including shRNA cotransfection or mithramycin A treated cells, were seeded into 96-well culture plates. In total, 20 µl MTT (5 mg/ml; Sigma-Aldrich) was added to the media at 37°C in 5% CO2 and 95% air for 72 h. Following removal of the culture media, the remaining crystals were dissolved in 200 µl dimethyl sulfoxide (DMSO; Sigma-Aldrich) and absorbance at 490 nm was measured. Cell viability was calculated relative to a DMSO control.
Transwell assay
U251 cells were cultured for 72 h with 10 pmol shRNA-GLUT3, 20 pmol siRNA-Sp1 or Sp1 inhibitor, and subsequently cell invasion was assayed three times using a 24-well Transwell plate (Neuro Probe, Inc., Gaithersburg, MD, USA) using polycarbonate nucleopore filters (pore size, 8-µm). The cells were centrifuged, resuspended in serum-free medium and 1.5×104 cells were seeded in the upper chamber of each well of the Transwell plate. The lower chambers contained serum-free medium. The invasive cells that had moved through the membrane were rinsed with PBS, fixed, and stained with an ethanol-based crystal violet solution following 24 h of incubation. The cells were examined under a microscope and all cells in a specified region in the middle of the membrane were counted. Experiments were repeated at least three times.
Scratch assay
The scratch motility assay was used to measure two-dimensional movement of the cells. Aliquots of 1×106 cells were plated in individual wells of 6-well tissue culture plates. Following 48 h, a line of adherent cells was scraped from the bottom of each well using a sterile 200 µl pipette tip to generate a wound, and medium containing serum and blasticidin (Thermo Fisher Scientific, Inc.) was added and cells were incubated at 37°C. The cells were allowed to proliferate and migrate into the wound for 36 h. The extent of migration of cells into the region from which cells had been scraped was determined using photographs. The experiment was repeated three times with multiple scratches each time.
Statistical analysis
Data are presented as the mean ± standard error. Statistical significance was determined by Student's t test using SPSS version 17.0 was used (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Sp1 induces GLUT3 through transcriptional activation in glioma cells
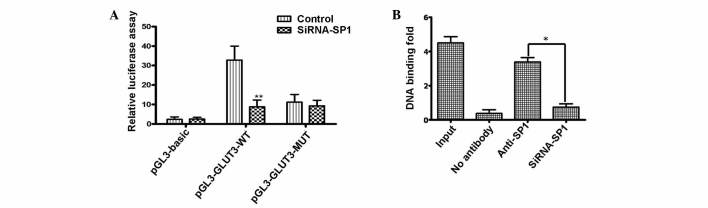
To test the hypothesis that Sp1 is critical for the production of GLUT3, which is encoded by the SLC2A3 gene, putative Sp1 binding site sequences of the GLUT3 promoter were transfected into a luciferase reporter plasmid. Compared with the empty luciferase vector, pGL3-GLUT3-WT reporters, which comprised nucleotides (nt) −381 to −200, generated a high level of luciferase activity [32.8-fold compared with pGL3-Basic (empty vector); P<0.001] in U251 cells. However, this increase was greatly attenuated in Sp1-depleted cells (11.2-fold compared with pGL3-GLUT3-WT; P<0.001) using Sp1 siRNA (Fig. 1A). Using reporters containing mutated binding sites, the present study identified that the Sp1-binding site −306 to −297 nt was required for activation of the GLUT3 promoter. The pGL3-GLUT3-MUT reporters containing a mutated Sp1 binding site was unable to respond to Sp1 deletion, indicating that the putative Sp1 binding site is critical in Sp1 activation of GLUT3 transcription (Fig 1A).
Figure 1.
(A) Assay for promoter responsiveness to SP1. Human glioma U251 cells were co-transfected with the indicated luciferase reporter plasmids, and control or siRNA-SP1. Luciferase activity was analyzed 24 h later. (B) Effect of SP1 binding to GLUT3 chromatin. Extracts from U251 cells were used for chromatin immunoprecipitation followed by quantitative polymerase chain reaction. Data represent the means of three experiments. Error bars indicate the standard deviation from the mean; *P<0.05;**P<0.01 vs. control. GLUT3, glucose transporter 3; pGL3-GLUT3-WT, wild-type reporter; pGL3-GLUT3-MUT, mutated reporter; pGL3-Basic, control; SP1, specific protein 1; siRNA, small interfering RNA; anti-SP1, antibody to SP1
Subsequently, the Sp1 putative binding site within the promoter of SLC2A3, composed of a GC-box (GGGGGCGGGGG), was analyzed using ChIP. The assay revealed that a 182 bp DNA fragment corresponding to the putative Sp1 binding site of the SLC2A3 gene (−381 to −200 nt) was amplified from U251 cells when chromatin was immunoprecipitated with a Sp1 antibody (Fig. 1B).
Sp1 activate GLUT3 expression in glioma cells
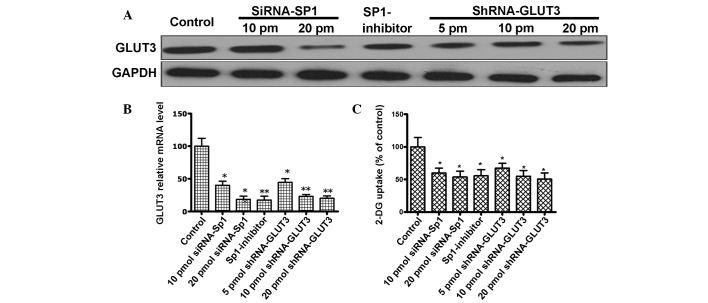
The present study transduced U251 cells with lentiviruses, which delivered shRNAs targeting GLUT3, to generate cells with stable GLUT3 knockdown. The expression level of GLUT3 in U251 cells were clearly reduced following an increase in the GLUT3 shRNA treatment concentration (Fig. 2A). By contrast, there was no reduction in GLUT3 expression observed compared with untransfected cells when the cells were treated with the negative control shRNA. Furthermore, Sp1 depletion decreased GLUT3 mRNA expression with an increasing concentration of siRNA-Sp1. Mithramycin A may also lead to a decrease in GLUT3 mRNA. Therefore, the present study analyzed if GLUT3 expression was affected by self-targeted shRNA or an Sp1-regulated mechanism, either transfection with siRNA-Sp1 or treatment with mithramycin A (Fig. 2B).
Figure 2.
(A) Western blot analysis for the expression of GLUT3 in control and human glioma U251 cells treated with siRNA-SP1 (10 and 20 pmol), shRNA-GLUT3 (10 and 20 pmol) and SP1-inhibitor (300 nM). Notably, GLUT3 expression is decreased following treatment with siRNA-SP1, shRNA-GLUT3 and SP1-inhibitor. (B) GLUT3 relative mRNA level. GLUT3 mRNA expression was detected using quantitative polymerase chain reaction in control and U251 cells treated with GLUT3 shRNA (5, 10 and 20 pmol), SP1 siRNA (10 and 20 pmol) and SP1-inhibitor (300 nM). (C) Glucose uptake in U251 cells. Uptake of-2-DG in control and U251 cells treated with GLUT3 shRNA (5, 10 and 20 pmol), SP1 siRNA (10 and 20 pmol) and SP1-inhibitor (300 nM). Data are the means of three independent experiments performed in triplicate. Error bars indicate the standard deviation from the mean; *P<0.05;**P<0.01 vs. control. GLUT3, glucose transporter 3; control, untreated U251 cells; siRNA, small interfering RNA; SP1, specific protein 1; shRNA, short hairpin RNA; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; SP1-inhibitor, mithramycin A; mRNA, messenger RNA; 2-DG, [3H]-2-deoxyglucose; pm, picomole.
GLUT3 promotes glucose uptake
The present study used stable lentivirus-transfected U251 cells to assay the contribution of GLUT3 to the import of glucose. Silencing of GLUT3 expression led to a decrease of ~50% in the efficiency of radioactive 2-DG incorporation, revealing that GLUT3 is important in glucose uptake in glioma cells (Fig. 2C). Furthermore, the present study demonstrated that Sp1 expression silencing and mithramycin A also led to a reduction in the efficiency of radioactive 2-DG incorporation in the cells (Fig. 2C). The present data indicates that Sp1 participates in the GLUT3-dependent glucose uptake by regulating GLUT3 expression.
Reduced GLUT3 expression attenuated the invasive potential of glioma cells in vitro by Sp1 regulation
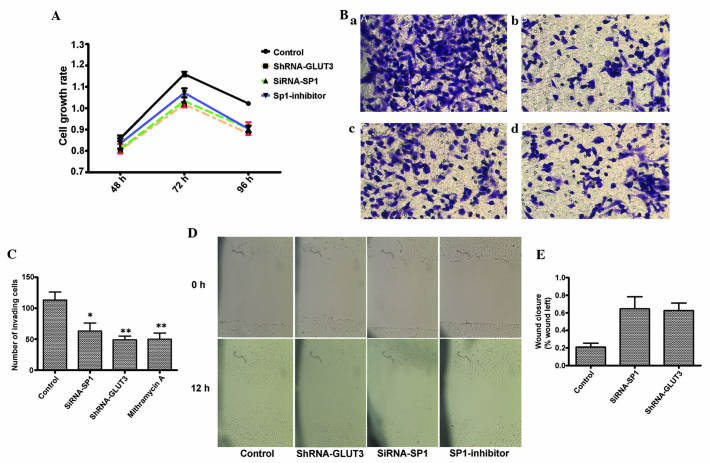
Subsequently, the effect of Sp1 depletion and silencing of GLUT3 expression on the biological characteristics of glioma cells was evaluated. The proliferation rates of U251 cells treated with shRNA-GLUT3 (10 pmol), siRNA-Sp1 (20 pmol) or mithramycin A (300 nM) were detected using an MTT assay. The treated cells demonstrated significant growth inhibition compared with the untreated cells (P<0.05; Fig. 3A).
Figure 3.
(A) 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay was used as an in vitro cell proliferation assay following GLUT3 knockdown or SP1 depletion in human glioma U251 cells. The control cells were transfected with non-targeting siRNA or were untreated. (B) Transwell assay. U251 cells transfected with (a) control, (b) shRNA-GLUT3 and (c) siRNA-SP1 and (d) treated with mithramycin A (300 nM) were plated into Transwell plates. Tumor cells invaded through matrigel were photographed. (C) Quantification of the Transwell assay. (D) Scratch assay. U251 cells treated with shRNA-GLUT3, siRNA-SP1 mithramycin A migrated more slowly compared with control cells. (E) Quantification of the scratch assay. Data represent the mean of three experiments. Error bars indicate the standard deviation from the mean; *P<0.05;**P<0.01 vs. control. GLUT3, glucose transporter 3; SP1, specific protein 1; shRNA, short hairpin RNA; siRNA, small interfering RNA; control, untreated cells.
The present study next examined the role of Sp1 in GLUT3-mediated glioma invasion using Transwell assay. Sp1 depletion using Sp1 RNAi or mithramycin A clearly inhibited glioma invasion and the role of shRNA-GLUT3. Representative photomicrographs and quantification of tumor cell invasion through matrigel are shown in Fig. 3B and C.
In the scratch assay, the present study observed that shRNA-GLUT3 transfected and Sp1 depleted cell migrated more slowly compared with control cells (Fig. 3D). The values at 0 and 12 h demonstrated a significant difference (Fig. 3E). The present data confirmed that siRNA-Sp1 inhibits glioma cell migration in the in vitro assay.
Discussion
The present study identified that in glioma cells Sp1 is a transcription factor for GLUT3, and GLUT3 expression is regulated by the Sp1 interaction with the GLUT3 promoter. The present data also suggest that Sp1 knockdown or the use of an Sp1 inhibitor decreases glucose uptake and inhibits the biological function of glioma cells and the GLUT3 blockade. Therefore, demonstrating the contribution of Sp1 to tumor proliferation through the regulation of GLUT3-mediated glucose metabolism.
It is well known that a variety of cancer cells exhibit increased levels of glucose uptake and glycolysis as a fundamental metabolic alteration, which is often associated with elevated levels of GLUT family members, including GLUT1 and GLUT3 (9,21). In normal brain cells, GLUT3 gene expression is confined to neurons; however, in highly malignant glial cells of the human brain, GLUT3 may be the predominant glucose transporter (7). Furthermore, GLUT3 may contribute to the maintenance of human brain tumors, and the expression of GLUT3 may be closely associated with various pathological types of tumor (9). However, the regulation of GLUT3 expression and its biological significance in glioma tumorigenesis requires clarification. The present study demonstrated that knockdown of GLUT3 expression decreased the proliferation, invasion and migration of human glioma cells. This indicates that GLUT3 is important in glioma cell survival, growth and invasion. Furthermore, knockdown of GLUT3 expression decreased glucose uptake of the cells, suggesting that GLUT3 may be critical in sugar metabolism.
It has been reported that Sp1 is an important transcription factor, which enhances or represses the activity of gene promoters involved in cell differentiation, cell cycle progression and oncogenesis (10). Sp1 is upregulated in human glioma and predicts a poor clinical outcome (18). Sp1 mediates transcriptional regulation of numerous genes, including MMP-2 and purinergic receptor P2X7 in brain cells (18,19). In addition, the Sp1 inhibitor mithramycin A has therapeutic effects in numerous diseases, including brain cancer and neurodegeneration (14,15,22,23). Mithramycin A competitively binds to GC-rich promoter regions; thereby replacing the transcription factor Sp1 (24,25). Through this mechanism, mithramycin A inhibits the transcription of Sp1-regulated genes, including v-myc avian myelocytomatosis viral oncogene homolog and SRC proto-oncogene, non-receptor tyrosine kinase (24,26). However, few studies demonstrate the role of Sp1 in the regulation of glioma cell metabolism through GULT3 expression.
The present data identify that Sp1 regulates the expression of GLUT3 directly, and mithramycin A inhibits GLUT3 expression by binding to GGGGGCGGGGG regions of the GLUT3 promoter in glioma cells. The present results were different from a previous study in mice, in which Sp1 bound to the mouse Glut3 gene that mediated the suppression of Glut3 gene expression in murine neuroblasts and trophoblasts. Furthermore, in the present study glucose uptake was decreased in Sp1 depleted cells or Sp1 inhibitor treated cells. This suggests that Sp1 may also have a role in glioma cell metabolism through GULT3 regulation. Additionally, Sp1 depletion or inhibition decreases glioma cells proliferation, invasion and immigration. This indicates that Sp1 is important in glioma cells survival, growth and invasion, which has been revealed by certain studies in other types of tumors (14,16,17,26). Indeed, mithramycin A is approved by the Food and Drug Administration for the treatment of leukemia and testicular cancer. In the future, it is possible that Sp1 may become a crucial therapeutic target.
In summary, the present study suggests that GLUT3 is extremely important in human glioma, and that its knockdown may contribute to decreased biological activity and glucose uptake in glioma cells. In addition, the present study identified that the role of GLUT3 in glioma may be regulated by the Sp1 transcription factor directly through binding to Sp1-sites in the GLUT3 promoter. These results may provide a novel insight into glioma drug treatment.
Acknowledgements
The present study was supported by National Natural Science Foundation of China (grant no., 81260371).
References
- 1.Ahmed R, Oborski MJ, Hwang M, Lieberman FS, Mountz JM. Malignant gliomas: Current perspectives in diagnosis, treatment and early response assessment using advanced quantitative imaging methods. Cancer Manag Res. 2014;6:149–170. doi: 10.2147/CMAR.S54726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung WJ, Lyons SA, Nelson GM, Hamza H, Gladson CL, Gillespie GY, Sontheimer H. Inhibition of cystine uptake disrupts the growth of primary brain tumors. J Neurosci. 2005;25:7101–7110. doi: 10.1523/JNEUROSCI.5258-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flavahan WA, Wu Q, Hitomi M, Rahim N, Kim Y, Sloan AE, Weil RJ, Nakano I, Sarkaria JN, Stringer BW, et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat Neurosci. 2013;16:1373–1382. doi: 10.1038/nn.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng J. Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation (Review) Oncol Lett. 2012;4:1151–1157. doi: 10.3892/ol.2012.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker-Samuel S, Ramasawmy R, Torrealdea F, Rega M, Rajkumar V, Johnson SP, Richardson S, Gonçalves M, Parkes HG, Arstad E, et al. In vivo imaging of glucose uptake and metabolism in tumors. Nat Med. 2013;19:1067–1072. doi: 10.1038/nm.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodríguez-Enríquez S, Marin-Hernández A, Gallardo-Pérez JC, Moreno-Sánchez R. Kinetics of transport and phosphorylation of glucose in cancer cells. J Cell Physiol. 2009;221:552–559. doi: 10.1002/jcp.21885. [DOI] [PubMed] [Google Scholar]
- 7.Boado RJ, Black KL, Pardridge WM. Gene expression of GLUT3 and GLUT1 glucose transporters in human brain tumors. Brain Res Mol Brain Res. 1994;27:51–57. doi: 10.1016/0169-328X(94)90183-X. [DOI] [PubMed] [Google Scholar]
- 8.Nagamatsu S, Sawa H, Wakizaka A, Hoshino T. Expression of facilitative glucose transporter isoforms in human brain tumors. J Neurochem. 1993;61:2048–2053. doi: 10.1111/j.1471-4159.1993.tb07441.x. [DOI] [PubMed] [Google Scholar]
- 9.Nishioka T, Oda Y, Seino Y, Yamamoto T, Inagaki N, Yano H, Imura H, et al. Distribution of the glucose transporters in human brain tumors. Cancer Res. 1992;52:3972–3979. [PubMed] [Google Scholar]
- 10.Li L, Davie JR. The role of Sp1 and Sp3 in normal and cancer cell biology. Ann Anat. 2010;192:275–283. doi: 10.1016/j.aanat.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 11.Copland JA, Pardini AW, Wood TG, Yin D, Green A, Bodenburg YH, Urban RJ, Stuart CA. IGF-1 controls GLUT3 expression in muscle via the transcriptional factor Sp1. Biochim Biophys Acta. 2007;1769:631–640. doi: 10.1016/j.bbaexp.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Hsu TI, Wang MC, Chen SY, Yeh YM, Su WC, Chang WC, Hung JJ. Sp1 expression regulates lung tumor progression. Oncogene. 2012;31:3973–3988. doi: 10.1038/onc.2011.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kou XX, Hao T, Meng Z, Zhou YH, Gan YH. Acetylated Sp1 inhibits PTEN expression through binding to PTEN core promoter and recruitment of HDAC1 and promotes cancer cell migration and invasion. Carcinogenesis. 2013;34:58–67. doi: 10.1093/carcin/bgs336. [DOI] [PubMed] [Google Scholar]
- 14.Safe S, Imanirad P, Sreevalsan S, Nair V, Jutooru I. Transcription factor Sp1, also known as specificity protein 1 as a therapeutic target. Expert Opin Ther Targets. 2014;18:759–769. doi: 10.1517/14728222.2014.914173. [DOI] [PubMed] [Google Scholar]
- 15.Sleiman SF, Langley BC, Basso M, Berlin J, Xia L, Payappilly JB, Kharel MK, et al. Mithramycin is a gene-selective Sp1 inhibitor that identifies a biological intersection between cancer and neurodegeneration. J Neurosci. 2011;31:6858–6870. doi: 10.1523/JNEUROSCI.0710-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malek A, Núñez LE, Magistri M, Brambilla L, Jovic S, Carbone GM, Morís F, Catapano CV. Modulation of the activity of Sp transcription factors by mithramycin analogues as a new strategy for treatment of metastatic prostate cancer. PLoS One. 2012;7:e35130. doi: 10.1371/journal.pone.0035130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Copland JA, Pardini AW, Wood TG, Yin D, Green A, Bodenburg YH, Urban RJ, Stuart CA. IGF-1 controls GLUT3 expression in muscle via the transcriptional factor Sp1. Biochim Biophys Acta. 2007;1769:631–640. doi: 10.1016/j.bbaexp.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Guan H, Cai J, Zhang N, Wu J, Yuan J, Li J, Li M. Sp1 is upregulated in human glioma, promotes MMP-2-mediated cell invasion and predicts poor clinical outcome. Int J Cancer. 2012;130:593–601. doi: 10.1002/ijc.26049. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Huerta P, Díaz-Hernandez M, Delicado EG, Pimentel-Santillana M, Miras-Portugal MT, Gómez-Villafuertes R. The specificity protein factor Sp1 mediates transcriptional regulation of P2X7 receptors in the nervous system. J Biol Chem. 2012;287:44628–44644. doi: 10.1074/jbc.M112.390971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maher F, Davies-Hill TM, Lysko PG, Henneberry RC, Simpson IA. Expression of two glucose transporters, GLUT1 and GLUT3, in cultured cerebellar neurons: Evidence for neuron-specific expression of GLUT3. Mol Cell Neurosci. 1991;2:351–360. doi: 10.1016/1044-7431(91)90066-W. [DOI] [PubMed] [Google Scholar]
- 21.Macheda ML, Rogers S, Best JD. Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. J Cell Physiol. 2005;202:654–662. doi: 10.1002/jcp.20166. [DOI] [PubMed] [Google Scholar]
- 22.Seznec J, Silkenstedt B, Naumann U. Therapeutic effects of the Sp1 inhibitor mithramycin A in glioblastoma. J Neurooncol. 2011;101:365–377. doi: 10.1007/s11060-010-0266-x. [DOI] [PubMed] [Google Scholar]
- 23.Amann T, Hellerbrand C. GLUT1 as a therapeutic target in hepatocellular carcinoma. Expert Opin Ther Targets. 2009;13:1411–1427l. doi: 10.1517/14728220903307509. [DOI] [PubMed] [Google Scholar]
- 24.Blume SW, Snyder RC, Ray R, Thomas S, Koller CA, Miller DM. Mithramycin inhibits Sp1 binding and selectively inhibits transcriptional activity of the dihydrofolate reductase gene in vitro and in vivo. J Clin Invest. 1991;88:1613–1621. doi: 10.1172/JCI115474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Dyke MW, Dervan PB. Chromomycin, mithramycin and olivomycin binding sites on heterogeneous deoxyribonucleic acid. Footprinting with (methidiumpropyl-EDTA) iron (II) Biochemistry. 1983;22:2373–2377. doi: 10.1021/bi00279a011. [DOI] [PubMed] [Google Scholar]
- 26.Remsing LL, Bahadori HR, Carbone GM, McGuffie EM, Catapano CV, Rohr J. Inhibition of c-src transcription by mithramycin: Structure-activity relationships of biosynthetically produced mithramycin analogues using the c-src promoter as target. Biochemistry. 2003;42:8313–8324. doi: 10.1021/bi034091z. [DOI] [PubMed] [Google Scholar]