Abstract
Background:
Wolbachia are common intracellular bacteria that infect different groups of arthropods including mosquitoes. These bacteria modify host biology and may induce feminization, parthenogenesis, male killing and cytoplasmic incompatibility (CI). Recently Wolbachia is being nominated as a bio-agent and paratransgenic candidate to control mosquito borne diseases.
Methods:
Here we report the results of a survey for presence, frequency, and phylogenetic congruence of these endosymbiont bacteria in Culex pipiens populations in Northern, Central, and Southern parts of Iran using nested-PCR amplification of wsp gene.
Results:
Wolbachia DNA were found in 227 (87.3%) out of 260 wild-caught mosquitoes. The rate of infection in adult females ranged from 61.5% to 100%, while in males were from 80% to 100%. The Blast search and phylogenetic analysis of the wsp gene sequence revealed that the Wolbachia strain from Iranian Cx. pipiens was identical to the Wolbachia strains of supergroup B previously reported in members of the Cx. pipiens complex. They had also identical sequence homology with the Wolbachia strains from a group of distinct arthropods including lepidopteran, wasps, flies, damselfly, thrips, and mites from remote geographical areas of the world.
Conclusion:
It is suggested that Wolbachia strains horizontally transfer between unrelated host organisms over evolutionary time. Also results of this study indicates that Wolbachia infections were highly prevalent infecting all Cx. pipiens populations throughout the country, however further study needs to define Wolbachia inter-population reproductive incompatibility pattern and its usefulness as a bio-agent control measure.
Keywords: Culex pipiens, Wolbachia, Cytoplasmic incompatibility, Nested-PCR, Iran
Introduction
Mosquitoes including Culex pipiens complex with global distribution are vectors of arboviral pathogens and parasites such as West Nile, St Louis, Sindbis, Wuchereria bancrofti, Dirofilaria immitis, D. repens, Plasmodium relictum, and P. gallinaceum (Vinogradova 2000, Pawelek et al. 2014). Among the ‘neglected’ mosquito-borne diseases, lymphatic filariasis continues to be a hazard to over a billion people in 83 countries (O'Connor et al. 2012). Culex pipiens is a species complex and comprise Cx. quinquefasciatus and Cx. pipiens in South and North America, Asia and Africa, as well as Cx. globocoxitus and Cx. australicus in Australia (Farajollahi et al. 2011). Culex pipiens and Cx. quinquefasciatus are distributed in most parts of Iran ranging from north to south (Zaim 1986, Azari-Hamidian 2007, Nikookar et al. 2010, Khoshdel-Nezamiha et al. 2013, Banafshi et al. 2013, Dehghan et al. 2013, 2014).
The raising of resistance to current insecticides by insect vectors (Hemingway and Ranson 2000), the progress of drug resistance in parasites (Talisuna et al. 2004) and lack of clinical cures or vaccines for many vector borne diseases have led researchers to develop urgently new and advanced approaches to control of the diseases. Paratransgenesis, as a new approach, direct towards reducing vector competence through genetically manipulated symbionts (Coutinho-Abreu et al. 2010). Transformed symbionts are distributed across the insect population via transovarial or transstadial transmision routs (Durvasula et al. 1997, Chavshin et al. 2012, 2014, 2015, Maleki-Ravasan et al. 2015). Symbionts currently aimed at in paratransgenesis include fungi (Rasgon 2011), symbiont bacteria of triatomine bugs (Durvasula et al. 1997, Durvasula et al. 1999, Durvasula et al. 2008), tsetse flies (Cheng and Aksoy 1999), sandflies (Maleki-Ravasan et al. 2015) and mosquitoes (Favia et al. 2007, Chavshin et al. 2014), and densoviruses infecting An. gambiae and Ae. aegypti mosquitoes (Ward et al. 2001, Ren et al. 2008). Recently, paratransgenesis have been successfully employed to reduce vector competence of the triatomine bug, Rhodnius prolixus, vector of Trypanosoma cruzi, the causative agent of Chagas disease (Durvasula et al. 1997), and Anopheles gambiae and An. stephensi, two main malaria vectors (Rasgon 2011, Wang and Jacobs-Lorena 2013). These data showed that the genetically manipulated symbionts could interfere with the development of the parasites in the vectors and provide the groundwork for the use of genetically modified symbionts as a potent tool to battle vector borne diseases.
The bacterium of Wolbachia pipientis is an intracellular organism and inherited maternally. It is established in more than 20% of all insects and a vast majority of other arthropods as well as filarial nematodes (Werren 1997a, Dobson 2004, Lo and Evans 2007). Recent studies imply that 20–76% of investigated insects give shelter to Wolbachia (Hilgenboecker et al. 2008), as well as many arachnids, terrestrial crustaceans, and mites (Cordaux et al. 2001, Gotoh et al. 2003, Rowley et al. 2004). This unique endosymbiont species was originally found in Cx pipiens but later molecular studies have discovered a number of phylogenetically diverse strains within the species (Lo et al. 2007). This endosymbiont bacterium has significant effects on its arthropod hosts and nominated as a bioagent to control important arthropod pests.
Wolbachia is the cause of various modifications in insect reproductive arrangement, comprising male-killing, feminization, cytoplasmic incompatibility (CI), and parthenogenesis (Werren et al. 2008). When CI occurs, sperm and eggs are not able to produce feasible progeny (Werren 1997b, Clark et al. 2003, Beckmann and Fallon 2013). Infected females relative to uninfected ones, participate more in offspring production, which permit Wolbachia to take up by all of host individuals even if it cases fitness costs (Field et al. 1999). The bacterium also can be used as a vector for delivering desirable genetic modifications in insect populations (Werren 1997b). As reviewed by Werren (1997a), Wolbachia have potential roles in the rapid speciation of their hosts. Also as a pandemic endosymbiont, Wolbachia can be recruited to control of a large number of human infectious diseases (Slatko et al. 2014). In filarial nematodes comprising Wuchereria bancrofti, Brugia malayi, Brugia timori and Onchocerca volvulus that infect humans, Wolbachia are obligated for proper development, fertility and survival, whereas in arthropods, although they can affect development and reproduction, but are not required for host survival. So Wolbachia have been a target for drug discovery against filariasis. In vivo/vitro experiments indicate that antibiotics such as doxycycline and tetracycline can kill both adults and immature nematodes through depletion of Wolbachia (Foster et al. 2013, Taylor et al. 2014). It is also shown that, Wolbachia spp where naturally infected or artificially introduced into vector population can affect and decrease the mosquitoes competence carrying of viruses, such as Yellow Fever, Chikungunya, Dengue, West Nile, as well as ones transmitting of the Plasmodium protozoans and filarial nematodes (Bourtzis et al. 2014).
Due to the fact that Wolbachia is an obligate endosymbiont that cannot be cultured exterior their hosts, recognition of infection has been based vastly on amplification of Wolbachia DNA using PCR. Until now a number of loci including wsp, 16S rDNA, coxA, ftsZ, hcpA, gatB, groEL, fbpA, gltA and dnaA genes have been studied and evaluated in the phylogenetic studies (Zhou et al. 1998, Ravikumar et al. 2011). The sequences from Wolbachia surface protein (wsp) gene were extremely mutable and could be used to recognition and to re solve the phylogenetic relationships of different Wolbachia strains (Zhou et al. 1998).
In the present study we used a nested PCR assay to detect and investigate the prevalence of Wolbachia endobacteria using the partial genomic nucleotide sequence of wsp gene in twelve field populations of Culex pipiens in various geographical regions across Iran ranging from north to south. Results of this study will provide fundamental background for understanding ecology, distribution, and potential utility of Wolbachia as bio-control agent of Cx. pipiens.
Materials and Methods
Study areas
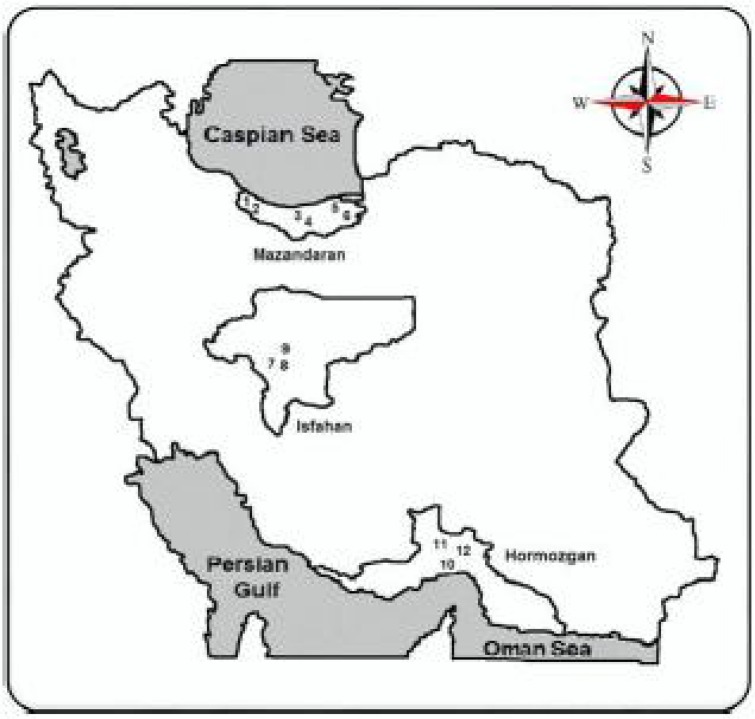
The study was conducted in twelve locations belong to three provinces of Iran, Mazandaran in the North (six locations), Isfahan in the center (3 locations) and Hormozgan in the South (3 locations) of the country (Fig. 1). Live larvae, pupae, and adult mosquitoes were collected from different biotypes including plane, jungle, riverside, rice field and human dwellings.
Fig. 1.
Map of study areas for collection of Culex pipiens specimens in Iran. Nos, 1–2: Ramsar, 3–4: Amol, 5–6: Behshahr in Mazandaran Province, 7: Vinicheh, 8: Dizicheh, 9: Dorcheh in Isfahan Province, 10: Hormodar, 11: Siahoo, and 12: Shamil in Hormozgan Province
Mosquito collection
Adult mosquitoes were collected in human dwellings monthly for a period of five months (June to late October, 2014) by hand-catch collection method using mouth aspirator. Also live larvae and pupae were collected from mosquito breeding sites locating in plane, jungle, riverside and rice field using dipping method, transferred to insectary, and allowed them to grow till adult emergence. Adult specimens were keyed to species level using standard morphological keys (Zaim 1986, Azari-Hamidian and Harbach 2009). The male and female mosquito specimens belong to Cx. pipiens were selected and stored individually at −20 °C for further molecular investigations. Double distilled water and mix of 10 adult male and female specimens of Anopheles maculipennis were collected from Mazanderan Province and used as negative controls.
DNA extraction and PCR
Totally 260 (120 males and 140 females) Cx. pipiens specimens originated from different biotopes from north to south of Iran were randomly subjected to genomic DNA extraction. Genomic DNA of An. maculipennis ss was extracted and used in all PCR assays as negative control. Total DNA of individual mosquitoes was extracted using Collins DNA extraction method (Collins et al. 1987). Previously a PCR based method for the classification of Wolbachia has been described (Zhou et al. 1998). In that method, group-specific wsp PCR primers have been used to identify Wolbachia strains without the need to clone and sequence individual Wolbachia genes. Here in detection of Wolbachia infection in the mosquitoes was performed by a nested-PCR assay on the basis of Zhou introduced primers. Initially, a set of primers including 81F: 5′–TGGTCCA ATAAGTGATGAAGAAAC–3′ and 691R: 5′–AAAAATTAAACGCTACTCCA–3′ were recruited to amplify 632 bp of partial sequence of the wsp gene. The PCR product of the first step was applied as a template for second step. In the second step, another pairs of the primers, 183F: 5′–AAGGAACCG AAGTTCATG–3′ and 691R: 5′–AAAAA TTAAACGCTACTCCA–3′, were used to amplify a 501 bp fragment.
The PCR amplification was performed using Maxime PCR PreMix Kit (i-Taq) Cat. No. 25026 in 20 μl reaction mixtures containing 2.5 μl of 10 μM both forward and reverse primers and 5 μl (∼0.5 μg) of genomic DNA and 2.5 μl PCR product for the first and second step of nested-PCR reactions respectively. An individual specimen of Anopheles maculipennis s.s. was used as DNA extraction and PCR negative controls. The PCR conditions were set as an initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min, and extension at 72 °C for 1 min, followed by a final extension at 72 °C for 7 min. PCR products were visualized on a 1% agarose gel containing ethidium bromide and using an UV transilluminator.
Wsp gene sequencing and analyzing
Representative specimens with clear and sharp wsp gene amplicons of the twelve Cx. pipiens populations were sequenced via the same amplification primers by Bioneer Company (S. Korea). The consensus of confident sequences was analyzed using NCBI (Nucleotide collection) database.
The wsp gene sequences determined in this study were subjected to molecular phylogenetic analysis together with 44 wsp gene sequences of Wolbachia from various arthropod host species retrieved from the Gen-bank database (Table 1). A multiple alignment of the wsp sequences was generated by the program package Clustal W (Thompson et al. 1994). Phylogenetic trees were constructed using the neighbor-joining method embedded in MEGA5 software. Bootstrap tests were performed with 1,000 replications.
Table 1.
Description of Wolbachia strains used for phylogenetic analysis in this study
| No | Wolbachia Strain | Host | Common name | Accession number | References |
|---|---|---|---|---|---|
| 1 | wPip/B | Culex pipiens | Mosquito | KM401552 | This study |
| 2 | wPip/B | Cx. pipiens | Mosquito | KM401553 | This study |
| 3 | wPip/B | Cx. pipiens | Mosquito | KM401556 | This study |
| 4 | wPip/B | Cx. pipiens | Mosquito | JX474753 | Direct Submission |
| 5 | wPip/B | Cx. pipiens (syn. pallens) | Mosquito | AF216860 | Direct Submission |
| 6 | wPip/B | Cx. pipiens form molestus | Mosquito | HG428761 | (Pinto et al. 2013) |
| 7 | wPip/B | Cx. quinquefasciatus | Mosquito | AF020060 | (Zhou et al. 1998) |
| 8 | wPip/B | Cx. quinquefasciatus | Mosquito | KJ140126 | Direct Submission |
| 9 | wAlbB/B | Aedes albopictus | Mosquito | AF020059 | (Zhou et al. 1998) |
| 10 | wPip/B | Ae. punctor | Mosquito | AJ311040 | (Ricci et al. 2002) |
| 11 | w AlbA/A | Ae. albopictus | Mosquito | AF020059 | (Zhou et al. 1998) |
| 12 | wNo/B | Drosophila simulans | Fruit Fly | AF020074 | (Zhou et al. 1998) |
| 13 | wMel/A | D. melanogaster | Fruit Fly | AF020072 | (Zhou et al. 1998) |
| 14 | wAus/A | Glossina austeni | Tsetse fly | AF020077 | (Zhou et al. 1998) |
| 15 | wMors/A | G. morsitans morsitans | Tsetse fly | AF020079 | (Zhou et al. 1998) |
| 16 | N.S | Protocalliphora sialia | Blow fly | DQ842482 | (Baldo et al. 2006) |
| 17 | wPak-B1 | Hydrellia pakistanae | Leaf mining fly | AF217718 | (Jeyaprakash and Hoy, 2000) |
| 18 | papa01/A | Phlebotomus papatasi | Sand fly | EU780683 | (Parvizi et al. 2013) |
| 19 | Turk 07 | Ph. mongolensis | Sand Fly | KC576916 | (Parvizi et al. 2013) |
| 20 | wCon/B | Tribolium confusum | Flour Beetle | AF020083 | (Zhou et al. 1998) |
| 21 | N.S | Chelymorpha alternans | Leaf Beetle | DQ842458 | (Baldo et al., 2006) |
| 22 | wOri/B | Tagosodes orizicolus | Plant hopper | AF020085 | (Zhou et al. 1998) |
| 23 | wStri/B | Laodelphax striatellus | Plant hopper | AF020080 | (Zhou et al. 1998) |
| 24 | F | Cimex lectularius | Bed Bug | DQ842459 | (Baldo et al. 2006) |
| 25 | wDei/B | Trichogramma deion | Wasp | AF020084 | (Zhou et al. 1998) |
| 26 | wTde-HEB | T. dendrolimi | Wasp | JX027991 | Direct Submission |
| 27 | wkue/A | Spalangia cameroni | Wasp | AF289668 | Direct Submission |
| 28 | N.S | Encarsia formosa | Wasp | DQ842471 | (Baldo et al. 2006) |
| 29 | wNPan/A | Nomada panzeri | Red Wasp | KC798315 | (Gerth et al. 2013) |
| 30 | A | Solenopsis invicta | Fire Ant | DQ842483 | (Baldo et al. 2006) |
| 31 | A | Formica truncorum | Ant | AF326978 | (Wenseleers et al. 2002) |
| 32 | wCauB/B | Ephestia cautella | Moth | AF020076 | (Zhou et al. 1998) |
| 33 | wCauA/A | Ephestia cautella | Moth | AF020075 | (Baldo et al. 2006) |
| 34 | B | Ostrinia scapulalis | Moth | DQ842481 | (Baldo et al. 2006) |
| 35 | NS | Eurema hecabe | Butterfly | AB285478 | (Narita et al. 2007) |
| 36 | NS | Udaspes folus | Butterfly | JN236179 | (Salunke et al. 2012) |
| 37 | NS | Agriocnemis femina | Damselfly | AY173939 | (Thipaksorn et al. 2003) |
| 38 | NS | Gryllus firmus | Cricket | DQ842474 | (Baldo et al. 2006) |
| 39 | A | Incisitermes snyderii | Termite | DQ842475 | (Baldo et al. 2006) |
| 40 | F | Coptotermes acinaciformis | Termite | AJ833931 | (Baldo et al. 2006) |
| 41 | NS | Hercinothrips femoralis | Thrips | AB245521 | Direct Submission |
| 42 | NS | Nephila clavata | Spider | EF612772 | Direct Submission |
| 43 | NS | Oxyopes sertatus | Spider | EF612771 | Direct Submission |
| 44 | NS | Eriovixia cavaleriei | Spider | DQ778738 | Direct Submission |
| 45 | NS | Tetranychus urticae | Two-spotted spider mite | AJ437290 | Direct Submission |
| 46 | NS | Bryobia berlesei | Mite | JN572865 | (Ros et al. 2012) |
| 47 | NS | Armadillidium vulgare | Pill woodlouse | DQ842457 | (Baldo et al. 2006) |
| 48 | Outgroup | Dirofilaria immitis | Nematode | AJ252062 | (Bazzocchi et al. 2000) |
NS: Not stated.
Statistics analyzing
Wolbachia infection data in Culex pipiens specimens were analyzed using SPSS 22.0 and Chi square (χ2) test to make comparisons and evaluate variation in infection rates between the males and females and among the twelve populations. The P-value more than 5% was considered as significant.
Results
Wolbachia detection in Cx. pipiens
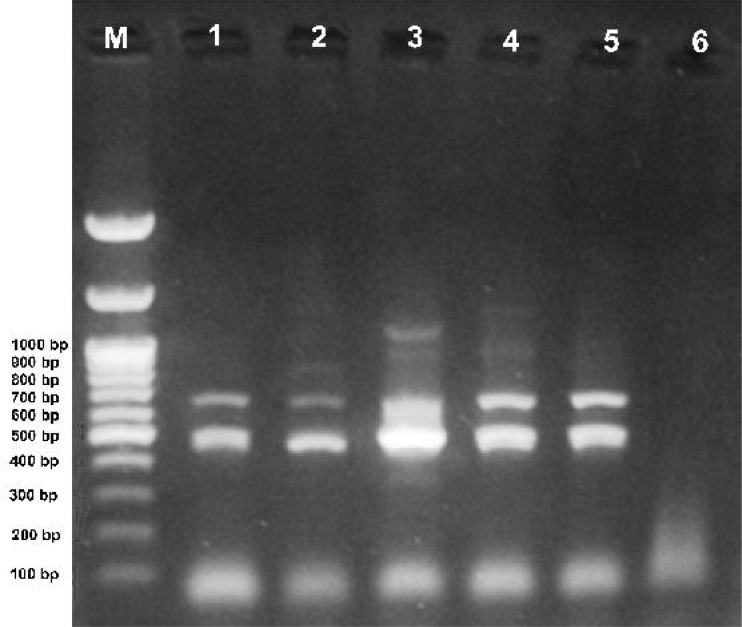
The infection of Wolbachia in different Cx. pipiens populations was detected by the nested-PCR assay using wsp gene. The amplicons of first and second runs of nested-PCR assay were ∼ 650 and 500 bp respectively (Fig. 2).
Fig. 2.
Species-specific nested-PCR products (∼ 500 bp) of Wolbachia wsp gene of Culex pipiens specimens. Lanes: M, 1 Kbp molecular weight marker (Fermentas), 1–2: Mazandaran Provine, 3: Isfahan Provine, 4–5: Hormozgan Provine, 6: Anopheles maculipennis as negative control
Wolbachia infection rate
Results of the study demonstrated that in total, 227 (87.3%) out of 260 individual adult mosquitoes belonged to 12 distinct populations were positive against wsp gene (Table 2). All the infected mosquitoes were found to harbor a single wPip strain. Infection rate in adult females and males were 61.5–100% and 80–100% respectively. There were no significant differences between total infection rates of either sexes (Female= 89.2%, Male = 85.7%, df= 1, P> 0.05) or zones (df= 3, P> 0.05).
Table 2.
Prevalence of Wolbachia pipientis infection in the Culex pipiens collected from North, Center and South of Iran, 2014
| Province | Location | Biotope | Males tested (% P +) | Females tested (% P+) | Total (% P+) |
|---|---|---|---|---|---|
| Mazandaran (North) | Amol 1 | Plane | 10(90) | 13(61.5) | 74 |
| Amol 2 | Jungle | 10(80) | 10(100) | 90 | |
| Behshar 1 | Plane | 10(100) | 10(100) | 100 | |
| Behshar 2 | Jungle | 10(90) | 10(90) | 90 | |
| Ramsar 1 | Plane | 10(90) | 10(80) | 85 | |
| Ramsar 2 | Jungle | 10(100) | 14(100) | 100 | |
| Isfahan (Center) | Dizicheh | Rice fields | 10(90) | 10(90) | 90 |
| Vinicheh | Rice fields | 10(80) | 10(70) | 75 | |
| Dorcheh | Rice fields | 10(100) | 15(100) | 100 | |
| Hormozgan (South) | Shamil | Date Groves | 10(80) | 13(61.5) | 70 |
| Siahoo | Riverside | 10(80) | 10(90) | 85 | |
| Hormoodar | Date Groves | 10(90) | 15(86.7) | 88 | |
| Total | 120(89.2) | 140(85.7) | 87.3 (260) | ||
Wolbachia wsp sequences
Seven nested–PCR products the wsp gene of Wolbachia found in different Iranian populations of Cx. pipiens were successfully sequenced and submitted to Genbank (Accession Numbers (ANs): KM401551–7). The nested primers we used were only able to amplify fragments from infected specimens and not from uninfected An. maculipennis ss hosts. The sequences were A-T rich (61%) with only 39% GC content. The BLAST results indicated that all the wsp sequences of Wolbachia detected from the Iranian Cx. pipiens were 100% identical to each other and to the Wolbachia strains found in other members of the Cx. pipiens complex including Cx. pipiens, Cx. pipiens form molestus, Cx. pipiens (syn. pallens), and Cx. quinquefasciatus from remote geographical areas of the world (Table 3). Since the Wolbachia strain that infects Cx. pipiens complex belongs to Pip group of B supergroup (wPipB) (Zhou et al. 1998, Pidiyar et al. 2003), we can conclude that the Wolbachia strains from Iranian Cx. pipiens specimens belongs to wPipB strain. In addition, the sequences of Wolbachia wsp gene of Iranian Cx. pipiens were 100% identical to the wsp gene of Wolbachia strains found in divers insect or arthropod groups particularly to the order of Lepidoptera comprising 18 different butterfly and moth species, as well as to wasps, thrips, damselflies, Aedes mosquito, Three-striped fruit fly, leaf-mining fly, and mite. These Wolbachia host species belong to geographically remote regions of Asian, European, and African countries (Table 3). A comparison of the wsp sequences from the arthropod hosts showed up to 30.67% genetic diversity between taxa, in which the wsp sequence from bedbug was the most diverged one.
Table 3.
Details of arthropods have identical Wolbachia wsp sequences with the Iranian Culex pipiens
| Arthropod group | Species | Accession Number | Country | Reference |
|---|---|---|---|---|
| Mosquito | Culex pipiens | JX474753 | Turkey | Direct Submission |
| Cx. pipiens form molestus | HG428761 | NS | (Pinto et al. 2013) | |
| Cx. pipiens (Syn. pallens) | AF216860 | China | Direct Submission | |
| Cx. quinquefasciatus | KJ140126 | China | Direct Submission | |
| Cx. quinquefasciatus | EU194487 | India | Direct Submission | |
| Cx. quinquefasciatus | AF397413, | India | Direct Submission | |
| Cx. quinquefasciatus | AF397412 | India | Direct Submission | |
| Cx. quinquefasciatus | AY462861 | Taiwan | (Tsai et al. 2004) | |
| Cx. quinquefasciatus | AM999887 | NS | (Klasson et al. 2008) | |
| Aedes punctor | AJ311040 | Italy | (Ricci et al. 2002) | |
| Butterfly | Udaspes folus | JN236179 | India | (Salunke et al. 2012) |
| Hypolimnas bolina | JN236180 | India | (Salunke et al. 2012) | |
| Castalius rosimon | JN236182 | India | (Salunke et al. 2012) | |
| Eurema hecabe | JN236189 | India | (Salunke et al. 2012) | |
| Ypthima asterope | JN236192 | India | (Salunke et al. 2012) | |
| Papilio demoleus | JN236193 | India | (Salunke et al. 2012) | |
| Zizeeria knysna | JN236194 | India | (Salunke et al. 2012) | |
| Colotis amata | JN236195 | India | (Salunke et al. 2012) | |
| Pseudozizeeria maha | JN236205 | India | (Salunke et al. 2012) | |
| Leptidea sinapis | KC137222 | NS | (Russell et al. 2012) | |
| Pararge aegeria | KC137224 | NS | (Russell et al. 2012) | |
| Polygonia calbum | JN093149 | NS | (Kodandaramaiah et al. 2011) | |
| Hypolimnas bolina | AJ307076 | Fiji | (Dyson et al. 2002) | |
| Moth | Corcyra cephalonica | KC844060 | China | Direct Submission |
| Epirrita autumnata | JX310335 | NS | (Kvie et al. 2012) | |
| Spodoptera exempta | JN656943 | Tanzania | Direct Submission | |
| Corcyra cephalonica | AY634679 | China | Direct Submission | |
| Acraea encedon | AJ271198 | Tanzania | Direct Submission | |
| Wasp | Trichogramma chilonis | AY311486 | China | Direct Submission |
| T. dendrolimi | JX027991 | China | Direct Submission | |
| T. brassicae | AF452646 | China | Direct Submission | |
| T. dendrolimi | DQ017751 | China | Direct Submission | |
| T. japonicum | KC161917 | China | Direct Submission | |
| Tropobracon schoenobii | AF481194 | NS | (Kittayapong et al. 2003) | |
| Thrips | Hercinothrips femoralis | AB245521 | Japan | Direct Submission |
| Damselfly | Agriocnemis femina | AY173939 | NS | (Thipaksorn et al. 2003) |
| Coenagrionidae sp | KC161926 | China | Direct Submission | |
| Fruit fly | Bactocera diversa | AF295353 | NS | (Jamnongluk et al. 2002) |
| Leaf-mining fly | Hydrellia pakistanae | AF217718) | NS | (Jeyaprakash and Hoy 2000) |
| Mite | Bryobia berlesei | JN572865 | France | (Ros et al. 2012) |
NS: Not stated.
Phylogenetic analysis
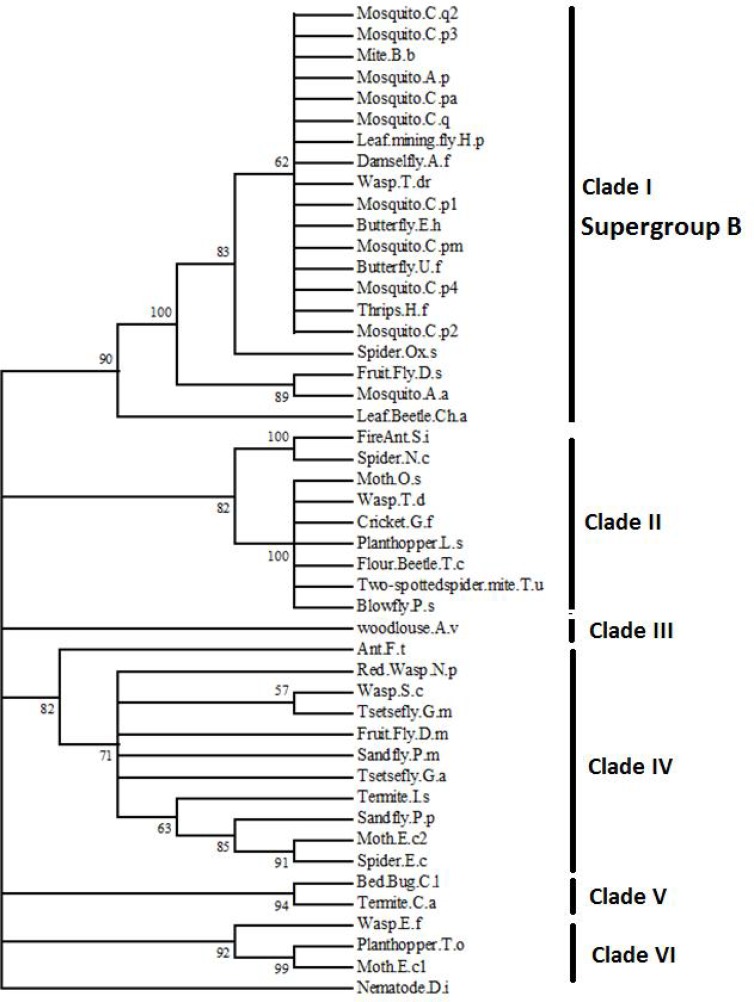
For phylogenetic analysis a subset of the Wolbachia strains identified in this study were combined with a 44 available sequence data of other Wolbachia strains from Genbank. These sequences belonged to twenty different arthropod hosts of Wolbachia including mosquitoes (Culex and Aedes), fruit flies, blow flies, sand flies, tsetse flies, leaf mining flies, bed bugs, thrips, damselflies, plant hoppers, crickets, termites, butterflies, moths, wasps, ants, beetles, pill woodlouse, spiders, and mites (table 1). Phylogenetic tree was constructed using neighbor-joining method, based on the 445–511 bp of wsp sequences (Fig. 3). The length variation between sequence data was due to insertion or deletion (indels) events. We also used Dirofilaria immitis wsp sequence as an out-group in the analysis. Phylogenetic analysis showed that Wolbachia strains from Iranian Cx. pipiens specimens were clustered with Wolbachia strains of other members of the Cx. pipiens complex such as Cx. pipiens, Cx. pipiens (syn. pallens), Cx. pipiens form molestus and Cx. quinquefasciatus (Fig. 3). They also associated with Wolbachia strains found in distinct groups of arthropods not obtained from the same insect genus, family, or even order. In other word, Wolbachia strains obtained from the same insect genus or families were not clustered into distinct groups but were scattered throughout the phylogenetic tree. Except for the congenic clusters of mosquitoes, sand flies, and tsetse flies, there were no other congenic clusters indicating little congruence between Wolbachia phylogeny and host systematics. The phylogenetic analysis revealed six main clades for the wsp sequences of Wolbachia strains analysed (Fig. 3). The first clade was composed of all mosquitoes (eight Culex spp and two Aedes spp) and ten wsp sequences from lepidopteran, wasp, Thrips, damselfly, Three-striped fruit fly, leaf-mining fly, leaf beetle, and mite, all belonged to the known supergroup B of Wolbachia. The second lineage was composed of nine wsp sequences from blowfly, plant hopper, cricket, moth, wasp, fire ant, flour beetle, and mite. Eleven wsp sequences from fruit flies, sand flies (2 species), tsetse flies (2 species), termite, moth, wasps (2 species), ant, and spider, constituted an isolated lineage. The wsp sequences from one of each wasp, plant hopper, and moth formed a distinct clade. Most of strains of second and third clades belong to the known supergroup A of Wolabachia. Notably the bedbug and one termite wsp sequences associated together and formed a well-defined clade, and finally pill wood louse constituted a diverse clade well separated from other five clades. Except for four nodes with 57–71% support, all of the nodes had very high (82–100) bootstrap support values (Fig. 3).
Fig. 3.
The phylogenetic tree inferred from 445–511 bp of wsp sequences of Wolbachia pipientis hosts using the neighbor-Joining method embedded in MEGA 5.0. C.p1–3 (Culex pipiens from this study), C.p4 (Culex pipiens), C.pm (Culex pipiens form molestus), C.q and C.q2 (Culex quinquefasciatus), C.pa (Culex pipiens, syn.: pallens), A.a (Aedes albopictus), D.m (Drosophila melanogaster), D.s (Drosophila simulans), G.m (Glossina morsitans morsitans), G.a (Glossina austeni), P.s (Protocalliphora sialia), P.p (Phlebotomus papatasi), P.m (Phlebotomus mongolensis), T.c (Tribolium confusum), Ch.a (Chelymorpha alternans), L.s (Laodelphax striatellus), T.o (Tagosodes orizicolus), C.l (Cimex lectularius), T.d (Trichogramma deion), T.dr (T.dendrolimi), S.c (Spalangia cameroni), E.f (Encarsia formosa), N.p (Nomada panzeri), S.i (Solenopsis invicta), , F.t (Formica truncorum), E.c1–2 (Ephestia cautella), O.s (Ostrinia scapulalis), E.h (Eurema hecabe), G.f (Gryllus firmus), I.s (Incisitermes snyderii), C.a (Coptotermes acinaciformis), N.c (Nephila clavata), Ox.s (Oxyopes sertatus), E.c (Eriovixia cavaleriei), T.u (Tetranychus urticae), A.v (Armadillidium vulgare), A.f (Agriocnemis femina), H.f (Hercinothrips femoralis), B.b (Bryobia berlesei), A.p (Aedes punctor), U.f (Udaspes folus), H.p (Hydrellia pakistanae), and D.i (Dirofilaria immitis). The bootstrap values are shown as numbers on the nodes
Discussion
This is the first report on Wolbachia infection from Cx. pipiens populations of Iran. In our study, 260 specimens of Cx. pipiens collected from the 12 villages were individually assayed for Wolbachia, and the overall rate of infection was determined to be 87.3%. This result is in agreement with previous study conducted in South West Iran revealed 100 percent Wolbachia infection in Cx. quinquefasciatus specimens (Behbahani 2012). In California, Wolbachia infection frequency in Cx. pipiens complex during 1999 and 2000 was 99.4% (Rasgon and Scott, 2003). Also Sunish et al. (2011) found an overall prevalence of 91.2% Wolbachia infections in Cx. quinquefasciatus mosquitoes from south India. Study of Chen et al (2013) revealed that three Cx. pipiens (Syn. pallens) populations of China were all infected with Wolbachia. This rate was reported between 10–100% in members of Cx. pipiens complex mosquitoes from the Upper Rhine Valley in Germany and Cebu City in Philippines (Mahilum et al. 2003).
In this study we found no Wolbachia infection in An. maculippenis ss specimens which is in concurrence of study of Rasgon and Scott (2004) who tested five genera of mosquito (Aedes, Anopheles, Culiseta, Culex, and Ochlerotatus) for Wolbachia, and infections was only detected in members of the Cx. pipiens complex. Also study of Kittayapong et al. (2000) detected Wolbachia infection in all main disease vector genera excluding Anopheles. In our study, the percentage prevalence in adult males was 80–100%, while in females were 61.5–100%. However the difference was not significant between males and females. In contrast, in the study of Sunish et al. (2011) the rate of Wolbachia infection in females of Cx. quinquefasciatus was found slightly higher than in males but like our study it was not statistically significant.
This study showed no sequence variation in wsp gene of Wolbachia from Cx. pipiens populations across geographical regions of Iran, which is similar to the results of Morais et al. (2012) which showed that both Cx. quinquefasciatus and Cx. pipiens × Cx. quinquefasciatus hybrids collected Brazil and Argentina were infected with a single Wolbachia strain. The genetic similarity detected among Wolbachia samples in the Culex mosquitoes from geographically scattered regions may be explained by either Wolbachia host-endosymbiont specificity (Werren et al. 2008) or recently Wolbachia infection in Culex populations (Morais et al. 2012).
High sequence homology and close phylogenetic relationships of Wolbachia strains from mosquitoes, spider, wasp, mite, damselfly, butterfly, thrips, fruit fly, and leaf mining fly indicate that Wolbachia endosymbionts not only are maternally transmitted through host generations by vertical transmission but also horizontally transfer between unrelated host organisms (i.e. shift host species or “jumping”) (Van Meer et al. 1999, Baldo et al. 2005). Although the mechanisms of jumping are still unclear, it is believed that parasitoids may involve (Heath et al. 1999, Huigens et al. 2000, Noda et al. 2001, Kikuchi and Fukatsu 2003). Recombination in wsp gene of Wolbachia strains has been evidenced by other researchers (Werren and Bartos 2001, Jiggins 2002, Reuter and Keller 2003). For example, Werren and Bartos (2001) reported recombination within supergroup B, occurring between the two Wolbachia strains of a parasitoid wasp and the fly it parasitizes. More recently it is shown that hypervariable regions of wsp gene of Wolbachia strains have got a complex mosaic structure, suggesting a clear intragenic recombination of segments among several divergent strains, both within and between the arthropod supergroups (Baldo et al. 2005).
The phylogenetic analysis of wsp sequences of Wolbachia from 20 different arthropod hosts scattered the sequences into five main clades that in some parts, topographically matched well with the tree of Zhou et al. (1998). Based on Wolbachia ftsZ gene sequences, two major supergroups A and B were reported within the Wolbachia strains (Werren and Jaenike 1995) where the type strain from Cx. pipiens was placed within supergroup B. In the tree we obtained in this study, two main clades represent supergroups A and B (Fig. 3). In addition to the Wolbachia strains from mosquitoes, the strains from spider, wasp, mite, damselfly, butterfly, thrips, fruit fly, and leaf mining fly also placed in supergroup B. Interestingly the Wolbachia strain from bedbug was associated with the one from termite of supergroup F or H. As reviewed by Lo et al. (2007), currently the genus Wolbachia was divided into eight taxonomic supergroups (A to H) where A and B are the two major groups established in arthropods, C and D are found in filarial nematodes, E infecting springtails and F contains Wolbachia bacteria that infect termites and filarial species. Supergroup G and H were reported in spiders and termites respectively. In addition other divergent lineages, such as those from various flea species and the filarial nematode Dirofilaria repens, might be added to the list of supergroups. Therefore, as more sequence information becomes available the number of clades, groups, or supergroups might be increased. For example, in our analysis the Wolbachia from woodlouse construct a single clade and might be considered as a separate clade.
Conclusion
In this study we found a single Wolbachia strain from Cx. pipiens populations across the country. Although it is suggested that a large set of compatible Wolbachia strains are always locally dominate within mosquito populations (Duron et al. 2011), however, several studies have showed that some wPip strains are reciprocally incompatible but also that some others, although genetically distinct, are fully compatible (Duron et al. 2006, Duron et al. 2007, Atyame et al. 2011). Therefore, it is worth to test cytoplasmic incompatibility (CI) between the Iranian populations. In case of having CI, it can be used as a form of sterile-insect technique (SIT), to suppress, to replace, or to reduce the survival of mosquito populations and thereby control them or reduce their ability to transmit the infection (Townson 2002).
Acknowledgements
Tehran University of Medical Sciences (TUMS) financially supported this work (Grant No. 22738). Also it is noteworthy that this research has been done by support of the Babol University of Medical Sciences (BUMS). Special thanks to Roghayeh Pourbagher, Seyedeh Narges Mousavi Kani, Zeinab Abedian, and Seyed Mohsen Aghajanpour Mir for helping in Cellular and Molecular Laboratory. The authors appreciate Mr Bagheri and Mr Hosseintabar for their cooperation in the laboratory. We would like to thank Mr Pakari, Mr Salari and Mr Arandian for helping in field collections.
References
- Atyame CM, Delsuc F, Pasteur N, Weill M, Duron O. ( 2011) Diversification of Wolbachia endosymbiont in the Culex pipiens mosquito. Mol Biol Evol. 28 ( 10): 2761– 2772. [DOI] [PubMed] [Google Scholar]
- Azari-Hamidian S. ( 2007) Larval habitat characteristics of mosquitoes of the genus Culex (Diptera: Culicidae) in Guilan Province, Iran. J Arthropod Borne Dis. 1( 1): 9– 20. [PMC free article] [PubMed] [Google Scholar]
- Azari-Hamidian S, Harbach RE. ( 2009) Keys to the adult females and fourth-instar larvae of the mosquitoes of Iran (Diptera: Culicidae). Zootaxa. 2078: 1– 33. [Google Scholar]
- Baldo L, Dunning Hotopp JC, Jolley KA, Bordenstein SR, Biber SA, Choudhury RR, Hayashi C, Maiden MC, Tettelin H, Werren J. H. ( 2006) Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol. 72( 11): 7098– 7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo L, Lo N, Werren JH. ( 2005) Mosaic nature of the wolbachia surface protein. J Bacteriol. 187( 15): 5406– 5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banafshi O, Abai MR, Ladonni H, Bakhshi H, Karami H, Azari-Hamidian S. ( 2013) The fauna and ecology of mosquito larvae (Diptera: Culicidae) in western Iran. Turk J Zool. 37( 3): 298– 307. [Google Scholar]
- Bazzocchi C, Jamnongluk W, O'Neill SL, Anderson TJ, Genchi C, Bandi C. ( 2000) wsp gene sequences from the Wolbachia of filarial nematodes. Curr Microbiol. 41( 2): 96– 100. [DOI] [PubMed] [Google Scholar]
- Beckmann JF, Fallon AM. ( 2013) Detection of the Wolbachia protein WPIP0282 in mosquito spermathecae: implications for cytoplasmic incompatibility. Insect Biochem Mol Biol. 43( 9): 867– 878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behbahani A. ( 2012) Wolbachia infection and mitochondrial DNA comparisons among Culex mosquitoes in South West Iran. Pak J Biol Sci. 15( 1): 54– 57. [DOI] [PubMed] [Google Scholar]
- Bourtzis K, Dobson SL, Xi Z, Rasgon JL, Calvitti M, Moreira LA, Bossin HC, Moretti R, Baton LA, Hughes GL, Mavingui P, Gilles JR. ( 2014) Harnessing mosquito-Wolbachia symbiosis for vector and disease control. Acta Trop. 132: S150– 163. [DOI] [PubMed] [Google Scholar]
- Chavshin A, Oshaghi M, Vatandoost H, Yakhchali B, Zarenejad F, Terenius O. ( 2015) Malpighian tubules are important determinants of Pseudomonas transstadial transmission and longtime persistence in Anopheles stephensi. Parasit Vectors. 8( 1): 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavshin AR, Oshaghi MA, Vatandoost H, Pourmand MR, Raeisi A, Enayati AA, Mardani N, Ghoorchian S. ( 2012) Identification of bacterial microflora in the midgut of the larvae and adult of wild caught Anopheles stephensi: a step toward finding suitable paratransgenesis candidates. Acta Trop. 121( 2): 129– 134. [DOI] [PubMed] [Google Scholar]
- Chavshin AR, Oshaghi MA, Vatandoost H, Pourmand MR, Raeisi A, Terenius O. ( 2014) Isolation and identification of culturable bacteria from wild Anopheles culicifacies, a first step in a paratransgenesis approach. Parasit Vectors. 7: 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhu C, Zhang D. ( 2013) Naturally occurring incompatibilities between different Culex pipiens pallens populations as the basis of potential mosquito control measures. PLoS Negl Trop Dis. 7( 1): e2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Aksoy S. ( 1999) Tissue tropism, transmission and expression of foreign genes in vivo in midgut symbionts of tsetse flies. Insect Mol Biol. 8( 1): 125– 132. [DOI] [PubMed] [Google Scholar]
- Clark ME, Veneti Z, Bourtzis K, Karr TL. ( 2003) Wolbachia distribution and cytoplasmic incompatibility during sperm development: the cyst as the basic cellular unit of CI expression. Mech Dev. 120( 2): 185– 198. [DOI] [PubMed] [Google Scholar]
- Collins FH, Mendez MA, Rasmussen MO, Mehaffey PC, Besansky NJ, Finnerty V. ( 1987) A ribosomal RNA gene probe differentiates member species of the Anopheles gambiae complex. Am J Trop Med Hyg. 37( 1): 37– 41. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Michel-Salzat A, Bouchon D. ( 2001) Wolbachia infection in crustaceans: novel hosts and potential routes for horizontal transmission. J Evol Biol. 14( 2): 237– 243. [Google Scholar]
- Coutinho-Abreu IV, Zhu KY, Ramalho-Ortigao M. ( 2010) Transgenesis and paratransgenesis to control insect-borne diseases: current status and future challenges. Parasitol Int. 59( 1): 1– 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehghan H, Moosa-Kazemi SH, Sadraei J, Soleimani H. ( 2014) The Ecological Aspects of Culex pipiens (Diptera: Culicidae) in Central Iran. J Arthropod Borne Dis. 8( 1): 35– 42. [PMC free article] [PubMed] [Google Scholar]
- Dehghan H, Sadraei J, Moosa-Kazemi SH, Baniani NA, Nowruzi F. ( 2013) The molecular and morphological variations of Culex pipiens complex (Diptera: Culicidae) in Iran. J Vector Borne Dis. 50( 2): 111– 120. [PubMed] [Google Scholar]
- Dobson SL. ( 2004) Evolution of Wolbachia cytoplasmic incompatibility types. Evolution. 58( 10): 2156– 2166. [DOI] [PubMed] [Google Scholar]
- Duron O, Bernard C, Unal S, Berthomieu A, Berticat C, Weill M. ( 2006) Tracking factors modulating cytoplasmic incompatibilities in the mosquito Culex pipiens. Mol Ecol. 15( 10): 3061– 3071. [DOI] [PubMed] [Google Scholar]
- Duron O, Boureux A, Echaubard P, Berthomieu A, Berticat C, Fort P, Weill M. ( 2007) Variability and expression of ankyrin domain genes in Wolbachia variants infecting the mosquito Culex pipiens. J Bacteriol. 189 ( 12): 4442– 4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duron O, Raymond M, Weill M. ( 2011) Many compatible Wolbachia strains coexist within natural populations of Culex pipiens mosquito. Heredity. 106 ( 6): 986– 993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durvasula RV, Gumbs A, Panackal A, Kruglov O, Aksoy S, Merrifield RB, Richards FF, Beard CB. ( 1997) Prevention of insect-borne disease: an approach using transgenic symbiotic bacteria. Proc Natl Acad Sci U S A. 94( 7): 3274– 3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durvasula RV, Gumbs A, Panackal A, Kruglov O, Taneja J, Kang AS, Cordon-Rosales C, Richards FF, Whitham RG, Beard CB. ( 1999) Expression of a functional antibody fragment in the gut of Rhodnius prolixus via transgenic bacterial symbiont Rhodococcus rhodnii. Med Vet Entomol. 13( 2): 115– 119. [DOI] [PubMed] [Google Scholar]
- Durvasula RV, Sundaram RK, Kirsch P, Hurwitz I, Crawford CV, Dotson E, Beard CB. ( 2008) Genetic transformation of a Corynebacterial symbiont from the Chagas disease vector Triatoma infestans. Exp Parasitol. 119( 1): 94– 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson EA, Kamath MK, Hurst GD. ( 2002) Wolbachia infection associated with all-female broods in Hypolimnas bolina (Lepidoptera: Nymphalidae): evidence for horizontal transmission of a butterfly male killer. Heredity (Edinb). 88( 3): 166– 171. [DOI] [PubMed] [Google Scholar]
- Farajollahi A, Fonseca DM, Kramer LD, Marm Kilpatrick A. ( 2011) “Bird biting” mosquitoes and human disease: a review of the role of Culex pipiens complex mosquitoes in epidemiology. Infect Genet Evol. 11( 7): 1577– 1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favia G, Ricci I, Damiani C, Raddadi N, Crotti E, Marzorati M, Rizzi A, Urso R, Brusetti L, Borin S, Mora D, Scuppa P, Pasqualini L, Clementi L, Genchi A, Corona S, Negri I, Grandi G, Alma A, Kramer L, Esposito F, Bandi C, Sacchi L, Daffonchio D. ( 2007) Bacteria of the genus Asaia stably associate with Anopheles stephensi an Asian malarial mosquito vector. Proc Natl Acad Sci U S A. 104( 21): 9047– 9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field L, James A, Turelli M, Hoffmann A. ( 1999) Microbe-induced cytoplasmic incompatibility as a mechanism for introducing transgenes into arthropod populations. Insect Mol Biol. 8( 2): 243– 255. [DOI] [PubMed] [Google Scholar]
- Foster JM, Hoerauf A, Slatko BE, Taylor MJ. ( 2011) The molecular biology, immunologyand chemotherapy of Wolbachia bacterial endosymbionts of filarial nematodes. In: Kennedy M, Harnett W, (Eds): Parasitic nematodes: molecular biology, biochemistry and immunology. Wallingford, UK, CABI, pp. 308– 336. [Google Scholar]
- Gerth M, Rothe J, Bleidorn C. ( 2013) Tracing horizontal Wolbachia movements among bees (Anthophila): a combined approach using multilocus sequence typing data and host phylogeny. Mol Ecol. 22( 24): 6149– 6162. [DOI] [PubMed] [Google Scholar]
- Gotoh T, Noda H, Hong XY. ( 2003) Wolbachia distribution and cytoplasmic incompatibility based on a survey of 42 spider mite species (Acari: Tetranychidae) in Japan. Heredity. 91( 3): 208– 216. [DOI] [PubMed] [Google Scholar]
- Heath BD, Butcher RD, Whitfield WG, Hubbard SF. ( 1999) Horizontal transfer of Wolbachia between phylogenetically distant insect species by a naturally occurring mechanism. Curr Biol. 9( 6): 313– 316. [DOI] [PubMed] [Google Scholar]
- Hemingway J, Ranson H. ( 2000) Insecticide resistance in insect vectors of human disease. Annu Rev Entomol. 45: 371– 391. [DOI] [PubMed] [Google Scholar]
- Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. ( 2008) How many species are infected with Wolbachia? A statistical analysis of current data. FEMS Microbiol Lett. 281( 2): 215– 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huigens ME, Luck RF, Klaassen RH, Maas MF, Timmermans MJ, Stouthamer R. ( 2000) Infectious parthenogenesis. Nature. 405( 6783): 178– 179. [DOI] [PubMed] [Google Scholar]
- Jamnongluk W, Kittayapong P, Baimai V, O'Neill SL. ( 2002) Wolbachia infections of tephritid fruit flies: molecular evidence for five distinct strains in a single host species. Curr Microbiol. 45( 4): 255– 260. [DOI] [PubMed] [Google Scholar]
- Jeyaprakash A, Hoy MA. ( 2000) Long PCR improves Wolbachia DNA amplification: wsp sequences found in 76% of sixty-three arthropod species. Insect Mol Biol. 9( 4): 393– 405. [DOI] [PubMed] [Google Scholar]
- Jiggins FM. ( 2002) The rate of recombination in Wolbachia bacteria. Mol Biol Evol. 19( 9): 1640– 1643. [DOI] [PubMed] [Google Scholar]
- Khoshdel-Nezamiha F, Vatandoost H, Azari-Hamidian S, Bavani MM, Dabiri F, Entezar-Mahdi R, Chavshin AR. ( 2013) Fauna and Larval Habitats of Mosquitoes (Diptera: Culicidae) of West Azerbaijan Province, Northwestern Iran. J Arthropod Borne Dis. 8( 2): 163– 173. [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Fukatsu T. ( 2003) Diversity of Wolbachia endosymbionts in heteropteran bugs. Appl Environ Microbiol. 69( 10): 6082– 6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittayapong P, Baisley KJ, Baimai V, O'Neill SL. ( 2000) Distribution and diversity of Wolbachia infections in Southeast Asian mosquitoes (Diptera: Culicidae). J Med Entomol. 37( 3): 340– 345. [DOI] [PubMed] [Google Scholar]
- Kittayapong P, Jamnongluk W, Thipaksorn A, Milne JR, Sindhusake C. ( 2003) Wolbachia infection complexity among insects in the tropical rice-field community. Mol Ecol. 12( 4): 1049– 1060. [DOI] [PubMed] [Google Scholar]
- Klasson L, Walker T, Sebaihia M, Sanders MJ, Quail MA, Lord A, Sanders S, Earl J, O'Neill SL, Thomson N, Sinkins SP, Parkhill J. ( 2008) Genome evolution of Wolbachia strain wPip from the Culex pipiens group. Mol Biol Evol. 25( 9): 1877– 1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodandaramaiah U, Weingartner E, Janz N, Dalen L, Nylin S. ( 2011) Population structure in relation to host-plant ecology and Wolbachia infestation in the comma butterfly. J Evol Biol. 24( 10): 2173– 2185. [DOI] [PubMed] [Google Scholar]
- Kvie KS, Hogner S, Aarvik L, Lifjeld JT, Johnsen A. ( 2012) Deep sympatric mtDNA divergence in the autumnal moth (Epirrita autumnata). Ecol Evol. 3( 1): 126– 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo N, Evans TA. ( 2007) Phylogenetic diversity of the intracellular symbiont Wolbachia in termites. Mol Phylogenet Evol. 44( 1): 461– 466. [DOI] [PubMed] [Google Scholar]
- Lo N, Paraskevopoulos C, Bourtzis K, O'Neill SL, Werren JH, Bordenstein SR, Bandi C. ( 2007) Taxonomic status of the intracellular bacterium Wolbachia pipientis. Int J Syst Evol Microbiol. 57( 3): 654– 657. [DOI] [PubMed] [Google Scholar]
- Mahilum MM, Storch V, Becker N. ( 2003) Molecular and electron microscopic identification of Wolbachia in Culex pipiens complex populations from the Upper Rhine Valley, Germany, and Cebu City, Philippines. J Am Mosq Control Assoc. 19( 3): 206– 210. [PubMed] [Google Scholar]
- Maleki-Ravasan N, Oshaghi MA, Afshar D, Arandian MH, Hajikhani S, Akhavan AA, Yakhchali B, Shirazi MH, Rassi Y, Jafari R, Aminian K, Fazeli-Varzaneh RA, Durvasula R. ( 2015) Aerobic bacterial flora of biotic and abiotic compartments of a hyperendemic Zoonotic Cutaneous Leishmaniasis (ZCL) focus. Parasit Vectors 8( 1): 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais SA, Almeida F, Suesdek L, Marrelli MT. ( 2012) Low genetic diversity in Wolbachia-Infected Culex quinquefasciatus (Diptera: Culicidae) from Brazil and Argentina. Rev Inst Med Trop Sao Paulo. 54( 6): 325– 329. [DOI] [PubMed] [Google Scholar]
- Narita S, Nomura M, Kageyama D. ( 2007) A natural population of the butterfly Eurema hecabe with Wolbachia-induced female-biased sex ratio not by feminization. Genome. 50( 4): 365– 372. [DOI] [PubMed] [Google Scholar]
- Nikookar S, Moosa-Kazemi S, Oshaghi M, Yaghoobi-Ershadi M, Vatandoost H, Kianinasab A. ( 2010) Species composition and diversity of mosquitoes in Neka county, Mazandaran Province, northern Iran. Iran J Arthropod Borne Dis. 4( 2): 26– 34. [PMC free article] [PubMed] [Google Scholar]
- Noda H, Miyoshi T, Zhang Q, Watanabe K, Deng Km, Hoshizaki S. ( 2001) Wolbachia infection shared among planthoppers (Homoptera: Delphacidae) and their endoparasite (Strepsiptera: Elenchidae): a probable case of inter-species transmission. Mol Ecol. 10( 8): 2101– 2106. [DOI] [PubMed] [Google Scholar]
- O'Connor L, Plichart C, Sang AC, Brelsfoard CL, Bossin HC, Dobson SL. ( 2012) Open release of male mosquitoes infected with a wolbachia biopesticide: field performance and infection containment. PLoS Negl Trop Dis. 6( 11): e1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvizi P, Fardid F, Soleimani S. ( 2013) Detection of a New Strain of Wolbachia pipientis in Phlebotomus perfiliewi transcaucasicus, a Potential Vector of Visceral Leishmaniasis in North West of Iran, by Targeting the Major Surface Protein Gene. J Arthropod Borne Dis. 7( 1): 46– 55. [PMC free article] [PubMed] [Google Scholar]
- Pawelek KA, Niehaus P, Salmeron C, Hager EJ, Hunt GJ. ( 2014) Modeling dynamics of culex pipiens complex populations and assessing abatement strategies for West Nile Virus. PLoS One. 9( 9): e108452. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pidiyar VJ, Jangid K, Dayananda KM, Kaznowski A, Gonzalez JM, Patole MS, Shouche YS. ( 2003) Phylogenetic affiliation of Aeromonas culicicola MTCC 3249(T) based on gyrB gene sequence and PCR-amplicon sequence analysis of cytolytic enterotoxin gene. Syst Appl Microbiol. 26( 2): 197– 202. [DOI] [PubMed] [Google Scholar]
- Pinto SB, Stainton K, Harris S, Kambris Z, Sutton ER, Bonsall MB, Parkhill J, Sinkins SP. ( 2013) Transcriptional regulation of Culex pipiens mosquitoes by Wolbachia influences cytoplasmic incompatibility. PLoS Pathog. 9( 10): e1003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasgon JL. ( 2011) Using infections to fight infections: paratransgenic fungi can block malaria transmission in mosquitoes. Future Microbiol. 6( 8): 851– 853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasgon JL, Scott TW. ( 2003) Wolbachia and cytoplasmic incompatibility in the California Culex pipiens mosquito species complex: parameter estimates and infection dynamics in natural populations. Genetics. 165( 4): 2029– 2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasgon JL, Scott TW. ( 2004) An initial survey for Wolbachia (Rickettsiales: Rick-ettsiaceae) infections in selected California mosquitoes (Diptera: Culicidae). J Med Entomol. 41( 2): 255– 257. [DOI] [PubMed] [Google Scholar]
- Ravikumar H, Prakash BM, Sampathkumar S, Puttaraju HP. ( 2011) Molecular subgrouping of Wolbachia and bacteriophage WO infection among some Indian Drosophila species. J Genet. 90 ( 3): 507– 510. [DOI] [PubMed] [Google Scholar]
- Ren X, Hoiczyk E, Rasgon JL. ( 2008) Viral paratransgenesis in the malaria vector Anopheles gambiae. PLoS Pathog. 4( 8): e1000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Keller L. ( 2003) High levels of multiple Wolbachia infection and recombination in the ant Formica exsecta. Mol Biol Evol. 20( 5): 748– 753. [DOI] [PubMed] [Google Scholar]
- Ricci I, Cancrini G, Gabrielli S, D'Amelio S, Favi G. ( 2002) Searching for Wolbachia (Rickettsiales: Rickettsiaceae) in mosquitoes (Diptera: Culicidae): large polymerase chain reaction survey and new identifications. J Med Entomol. 39( 4): 562– 567. [DOI] [PubMed] [Google Scholar]
- Ros VI, Fleming VM, Feil EJ, Breeuwer JA. ( 2012) Diversity and recombination in Wolbachia and Cardinium from Bryobia spider mites. BMC Microbiol. 12( 1): S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley SM, Raven RJ, McGraw EA. ( 2004) Wolbachia pipientis in Australian spiders. Curr Microbiol. 49( 3): 208– 214. [DOI] [PubMed] [Google Scholar]
- Russell JA, Funaro CF, Giraldo YM, Goldman-Huertas B, Suh D, Kronauer DJ, Moreau CS, Pierce NE. ( 2012) A veritable menagerie of heritable bacteria from ants, butterflies, and beyond: broad molecular surveys and a systematic review. PLoS One. 7( 12): e51027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salunke BK, Salunkhe RC, Dhotre DP, Walujkar SA, Khandagale AB, Chaudhari R, Chandode RK, Ghate HV, Patole MS, Werren JH, Shouche YS. ( 2012) Determination of Wolbachia diversity in butterflies from Western Ghats, India, by a multigene approach. Appl Environ Microbiol. 78( 12): 4458– 4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatko BE, Luck AN, Dobson SL, Foster JM. ( 2014) Wolbachia endosymbionts and human disease control. Mol Biochem Parasitol. 195( 2): 88– 95. [DOI] [PubMed] [Google Scholar]
- Sunish IP, Rajendran R, Paramasivan R, Dhananjeyan KJ, Tyagi BK. ( 2011) Wolbachia endobacteria in a natural population of Culex quinquefasciatus from filariasis endemic villages of south India and its phylogenetic implication. Trop Biomed. 28( 3): 569– 576. [PubMed] [Google Scholar]
- Talisuna AO, Bloland P, D'Alessandro U. ( 2004) History, dynamics, and public health importance of malaria parasite resistance. Clin Microbiol Rev. 17( 1): 235– 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Hoerauf A, Townson S, Slatko BE, Ward SA. ( 2014) Anti-Wolbachia drug discovery and development: safe macrofilaricides for onchocerciasis and lymphatic filariasis. Parasitology. 141 ( 1): 119– 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thipaksorn A, Jamnongluk W, Kittayapong P. ( 2003) Molecular evidence of Wolbachia infection in natural populations of tropical odonates. Curr Microbiol. 47( 4): 314– 318. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. ( 1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22( 22): 4673– 4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townson H. ( 2002) Wolbachia as a potential tool for suppressing filarial transmission. Ann Trop Med Parasitol. 96( 2): S117– 127. [DOI] [PubMed] [Google Scholar]
- Tsai KH, Lien JC, Huang CG, Wu WJ, Chen WJ. ( 2004) Molecular (sub) grouping of endosymbiont Wolbachia infection among mosquitoes of Taiwan. J Med Entomol. 41( 4): 677– 683. [DOI] [PubMed] [Google Scholar]
- Van Meer MM, Witteveldt J, Stouthamer R. ( 1999) Phylogeny of the arthropod endosymbiont Wolbachia based on the wsp gene. Insect Mol Biol. 8( 3): 399– 408. [DOI] [PubMed] [Google Scholar]
- Vinogradova EB. ( 2000) Culex pipiens pipiens mosquitoes: taxonomy, distribution, ecology, physiology, genetics, applied importance and control. Pensoft Series Parasitologica No 2. Pensoft Publishers, Sofia-Moscow. [Google Scholar]
- Wang S, Jacobs-Lorena M. ( 2013) Genetic approaches to interfere with malaria transmission by vector mosquitoes. Trends Biotechnol. 31( 3): 185– 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward TW, Jenkins MS, Afanasiev BN, Edwards M, Duda BA, Suchman E, Jacobs-Lorena M, Beaty BJ, Carlson JO. ( 2001) Aedes aegypti transducing densovirus pathogenesis and expression in Aedes aegypti and Anopheles gambiae larvae. Insect Mol Biol. 10( 5): 397– 405. [DOI] [PubMed] [Google Scholar]
- Wenseleers T, Sundstrom L, Billen J. ( 2002) Deleterious Wolbachia in the ant Formica truncorum. Proc Biol Sci. 269 ( 1491): 623– 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH. ( 1997a) Biology of Wolbachia. Annu Rev Entomol. 42: 587– 609. [DOI] [PubMed] [Google Scholar]
- Werren JH. ( 1997b) Wolbachia run amok. Proc Natl Acad Sci U S A. 94( 21): 11154– 11155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Baldo L, Clark ME. ( 2008) Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 6( 10): 741– 751. [DOI] [PubMed] [Google Scholar]
- Werren JH, Bartos JD. ( 2001) Recombination in Wolbachia. Current Biology. 11( 6): 431– 435. [DOI] [PubMed] [Google Scholar]
- Werren JH, Jaenike J. ( 1995) Wolbachia and cytoplasmic incompatibility in mycophagous Drosophila and their relatives. Heredity. 75( 3): 320– 326. [DOI] [PubMed] [Google Scholar]
- Zaim M, Cranston PS, ( 1986) Checklist and keys to the Culicinae of Iran (Diptera: Culicidae). Mosq Syst. 18: 233– 245. [Google Scholar]
- Zhou W, Rousset F, O'Neil S. ( 1998) Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc Biol Sci. 265( 1395): 509– 515. [DOI] [PMC free article] [PubMed] [Google Scholar]