Abstract
Background:
Crimean Congo hemorrhagic fever virus (CCHFV) is a member of the Bunyaviridae family and Nairovirus genus. It has a negative-sense, single stranded RNA genome approximately 19.2 kb, containing the Small, Medium, and Large segments. CCHFVs are relatively divergent in their genome sequence and grouped in seven distinct clades based on S-segment sequence analysis and six clades based on M-segment sequences. Our aim was to obtain new insights into the molecular epidemiology of CCHFV in Iran.
Methods:
We analyzed partial and complete nucleotide sequences of the S and M segments derived from 50 Iranian patients. The extracted RNA was amplified using one-step RT-PCR and then sequenced. The sequences were analyzed using Mega5 software.
Results:
Phylogenetic analysis of partial S segment sequences demonstrated that clade IV-(Asia 1), clade IV-(Asia 2) and clade V-(Europe) accounted for 80 %, 4 % and 14 % of the circulating genomic variants of CCHFV in Iran respectively. However, one of the Iranian strains (Iran-Kerman/22) was associated with none of other sequences and formed a new clade (VII). The phylogenetic analysis of complete S-segment nucleotide sequences from selected Iranian CCHFV strains complemented with representative strains from GenBank revealed similar topology as partial sequences with eight major clusters. A partial M segment phylogeny positioned the Iranian strains in either association with clade III (Asia-Africa) or clade V (Europe).
Conclusion:
The phylogenetic analysis revealed subtle links between distant geographic locations, which we propose might originate either from international livestock trade or from long-distance carriage of CCHFV by infected ticks via bird migration.
Keywords: CCHFV, Molecular epidemiology, RT-PCR, Phylogeny, Reassortant virus, Iran
Introduction
Endemic regions of Crimean Congo Hemorrhagic Fever (CCHF) have been reported in Africa, the Middle East, Eastern Europe and Western Asia (Elevli et al. 2010). In the past 10 years, Turkey, Bosnia, and Iran have been reported the most frequent outbreaks of CCHF worldwide (Mohammadi and Razmi 2014).
The virus is transmitted to humans through the bite of infected Ixodid ticks (mostly of the Hyalomma genus) (Mehravaran et al. 2013, Champour et al. 2014), or direct contact with infected blood, by meat from infected animals or by nosocomial transmission (Chinikar et al. 2012c).
CCHFV is a single stranded RNA virus with segmented negative sense genome consisting of a small (S), a medium (M) and a large (L) segment. The S RNA segment encodes the nucleocapsid (N) protein, and the M RNA segment encodes the glycoprotein precursor, resulting in the two envelope glycoproteins Gn and Gc, while the L segment encodes the putative RNA-dependent “RNA” polymerase (Papa et al. 2002). It has been proved that segmented RNA viruses have the potential of segment reassortment and form new distinct genomic variants if the host cells are subject to dual infection by more than one genetically distinct virus (Hewson et al. 2004b, Lukashev 2005).
CCHFVs are relatively divergent in their genome sequence and grouped geographically into seven distinct clades based on the S-segment sequence analysis (Han and Rayner, 2011). West-Africa in clade I, Central Africa in clade II, South-Africa and West-Africa in clade III, Middle-East and Asia in clade IV, Europe in clade V and Greece in clade VI (Deyde et al. 2006, Hewson et al. 2004a). The clade IV may be devided into two distinct clades, Asia-1 and Asia-2 (Hewson et al. 2004a).
Beside the genome analysis on S segment, CCHFVs are divided into six clades based on M-RNA sequences; clade IV (Asia or Middle East) comprising strains in China, Pakistan, Oman, and South Africa, clade III (S Africa or W Africa 2) comprising those in Uzbekistan, Tajikistan, China, Pakistan, Iran, Iraq, South Africa and Nigeria, clade I (W Africa1) comprising those in Congo, Senegal, China, clade V (Europe) comprising those in Russia, Kosovo and Turkey, clade VI (Greece) including isolated strain from Greece, and clade VII (Mauritania) including isolated strain from Mauritania (Papa et al. 2005, Morikawa et al. 2002, Morikawa et al. 2007).
In Iran, the first phylogenetic study on S-segment of CCHFV in 2004 showed that the CCHFV strains were clustered within clade Asia 1 with the highest similarity to Matin strain from Pakistan (Chinikar et al. 2004), previously a pioneering genetic characterization study had showed that the Iranian strain (ArTeh 193-3) obtained from ticks in the North-Eastern region of Khorasan Province in 1978 clustered within clade Africa I and had similarity with CCHFV strains from West Africa, mainly from Senegal and Mauritania (Sureau and Klein 1980).
In 2008, further phylogenetic analysis in tick populations in Isfahan province in Central Iran, demonstrated that a variant strain clustered within clade IV (Asia 1) with the highest similarity to an Iraqi strain (Chinikar et al. 2012a).
Genetic analyses in 2012 of one strain from Northern Iran illustrated that the Russian CCHFV genome is circulating in this area, and accordingly the existence of European clade V in Iran was confirmed (Chinikar et al. 2013).
According to the previous phylogenetic studies, there are seven clades of CCHFV and Iranian strains of CCHFV distributed within three clade I (Africa I), clade IV (Asia 1) and clade V (Europe) (Chinikar et al. 2013).
The main objective of this study was to obtain new insights into the molecular epidemiology of CCHFV in Iran by analyzing the partial and complete nucleotide sequences of the S and M segments of CCHFV genome obtained from Iranian patients and several sequences available from GenBank.
Materials and Methods
Investigation areas and sampling
According to the distribution of CCHFV in Iran, CCHFV have been reported from 27 of 31 provinces until now. Nineteen provinces encompassed all geographical areas and were randomly selected and investigated in a span of 11 years between 2002 and 2013. A total of 50 human sera samples were selected among previously RT-PCR positive samples which were stored in the Arboviruses and Viral Hemorrhagic Fevers Laboratory (National Ref. Lab) sample bank; Pasteur Institute of Iran and also transferred samples to the this laboratory during the study. Geographical distribution and the year of sampling related to each strain are shown in Table 1.
Table 1.
Details of the Iranian CCHFV strains isolated from human serum used in this study. The sequences used for analysis of partial S, complete S and partial M segments are indicated in table
| Strain | Location | Date | Partial S | Complete S | Partial M |
|---|---|---|---|---|---|
| Iran-KhRazavi/1 | Northeast | 2013 | * | ||
| Iran-KhRazavi/2 | Northeast | 2012 | * | ||
| Iran-KhRazavi/3 | Northeast | 2012 | * | ||
| Iran-KhRazavi/4 | Northeast | 2012 | * | ||
| Iran-KhRazavi/5 | Northeast | 2012 | * | ||
| Iran-KhRazavi/16 | Northeast | 2012 | * | ||
| Iran-KhRazavi/17 | Northeast | 2012 | * | ||
| Iran-KhRazavi/80 | Northeast | 2012 | * | ||
| Iran-KhRazavi/72 | Northeast | 2012 | * | * | * |
| Iran-Kerman/22 | Southeast | 2012 | * | * | * |
| Iran-Kerman/27 | Southeast | 2012 | * | ||
| Iran-Kerman/24 | Southeast | 2006 | * | ||
| Iran-Zahedan/25 | Southeast | 2012 | * | ||
| Iran-Zahedan29 | Southeast | 2012 | * | ||
| Iran-Kerman/43 | Southeast | 2013 | * | * | * |
| Iran-Zahedan/74 | Southeast | 2012 | * | ||
| Iran-Kerman/77 | Southeast | 2013 | * | ||
| Iran-SistanBalochestan/85 | Southeast | 2011 | * | ||
| Iran-Zahedan/19 | Southeast | 2012 | * | * | |
| Iran-Zahedan/20 | Southeast | 2012 | * | ||
| Iran-Shiraz/39 | South | 2012 | * | ||
| Iran-Minab/49 | South | 2007 | * | ||
| Iran-BandarAbas/50 | South | 2011 | * | ||
| Iran-Hormozgan/84 | South | 2011 | * | ||
| Iran-Booshehr/52 | South | 2010 | * | ||
| Iran-Hormozgan/87 | South | 2011 | * | ||
| Iran-Fars/89 | South | 2011 | * | ||
| Iran-Booshehr/97 | South | 2002 | * | ||
| Iran-KhJonobi/73 | East | 2012 | * | ||
| Iran-Zanjan/23 | West | 2012 | * | ||
| Iran-Zanjan/41 | West | 2006 | * | ||
| Iran-Kermanshah/55 | West | 2008 | * | ||
| Iran-KhorramAbad/56 | West | 2010 | * | ||
| Iran-Ahvaz/54 | Southwest | 2010 | * | ||
| Iran-Yazd/57 | Central | 2009 | * | ||
| Iran-Qom/58 | Central | 2011 | * | ||
| Iran-Tehran/65 | Central | 2011 | * | * | * |
| Iran-Isfahan/78 | Central | 2013 | * | * | * |
| Iran-Isfahan/81 | Central | 2011 | * | ||
| Iran-Kashan/15 | Central | 2005 | * | ||
| Iran-Qom/71 | Central | 2008 | * | ||
| Iran-Yazd/86 | Central | 2010 | * | ||
| Iran-Tehran/90 | Central | 2012 | * | ||
| Iran-Tehran/91 | Central | 2010 | * | ||
| Iran-Tabriz/99 | Northwest | 2003 | * | ||
| Iran-Oroomieh/100 | Northwest | 2003 | * | ||
| Iran-Tabriz/102 | Northwest | 2004 | * | ||
| Iran-Babol/14 | North | 2012 | * | ||
| Iran-Amlash/21 | North | 2012 | * | ||
| Iran-Gilan/69 | North | 2012 | * | * | * |
Serological and molecular identification
To investigate each human serum sample for the presence of CCHFV-specific antibodies, an ELISA was initially used to detect IgM (Garcia et al. 2006). Molecular analysis was subsequently applied. For this purpose, viral RNA was extracted from 140 μl of serum using a QIAamp RNA Mini kit, according to of the manufacturer’s instructions (QI Agen GmbH, Hilden, Germany) (Yashina et al. 2003). Specific primers for amplification of partial S, whole S and M segments were designed by CLC main workbench software version 5.0, based on available CCHFV sequences in GenBank.
To amplify the partial of the S-segment, a primer pair (PSF5′-GAATGTGCATGGGTT AGCTC-3′) and (PSR 5′-GACATCACAA TTTCACCAGG-3′) was designed and used to amplify a 536 bp section. For reverse transcription, 50 °C (30 min) used. An initial enzyme activation step at 95 °C for 5 min was succeeded by 40 reaction cycles carried out with 30 sec at 94 °C, 30 sec at 50 °C, and 45 sec at 72 °C followed by a final incubation at 72 °C for 10 min.
For amplification of the full-length of the S-segment, a touch down RT-PCR used with the following primers: WSF: 5′-TCTCAAAGAAACACGTGCCGC-3′ and WSR: 5′-TCTCAAAGATATCGTTGCCGC-3′ to amplify a 1680 bp section of the S-segment. Thermal cycle condition designed as 45 °C (30 min) for reverse transcription and 95 °C (15 min) as an initial enzyme activation, and then followed by 40 reaction cycles at 94 °C (10 sec), 66-52 °C (30 sec), 68 °C (100 sec), eventually, 68 °C (10 min) as final extension.
For amplification of the partial of the M-segment, the primers of PMF 5′-TGCAC TTGAGCATCTGCAA-3′ and PMR 5′-AG CTGATTCCTGTCCTTTC-3′ was designed and used to amplify a 557 bp section of the M-segment. For reverse transcription, 50 °C (30 min) used An initial enzyme activation step at 95 °C for 15 min was succeeded by 40 reaction cycles carried out with 30 sec at 94 °C, 30 sec at 45 °C, and 1 min at 72 °C followed by a final incubation at 72 °C for 10 min (Rodriguez et al. 1997). PCR products were amplified using one-step RT-PCR, according to Rodriguez et al. The amplified products were visualized by ethidium bromide agarose gel staining (Yadav et al. 2012).
Sequencing
The PCR products were then sequenced using Big Dye Terminator V3.1 Cycle sequencing Kit with Modified Sanger Sequencing Method by ABI Genetic Analyzer 3130. For full-length S-segment sequencing, internal primers were designed as follow: Forward: 3′AATGCAAACACGGCAGCTTT5′ and Reverse: 3′GGAACTGTGAGACAG TCGGG5′. The obtained sequences were optimized (gaps and noises were deleted) (Chinikar et al. 2010).
Nucleotide sequence accession numbers
Whole S segment isolates Iran-Gilan 69, Iran-Isfahan 78, Iran-Kerman 43, Iran-Kh Razavi 72, Iran-Tehran 65 and Iran-Zahedan 19 submitted to GenBank and assigned accession No. KJ027521, KJ027522, KJ196326, KJ485700, KJ566219 and KJ676542 respectively.
Sequence alignments and phylogeny analysis
In addition to the 50 CCHFV sequences obtained, several sequences available from GenBank at www.ncbi.nih.gov were incorporated into the alignments for phylogenetic analyses (Table 2).
Table 2.
Details of CCHF virus strains originated from Iran and other countries retrieved from GenBank and used for sequence analysis
| Virus strains | Location | Date | Clade/Name (S segment) | GenBank accession no S segment | Clade/Name (M segment) | GenBank accession no M segment |
|---|---|---|---|---|---|---|
| NIV 112143 | India | 2011 | IV/ASI-1 | JN572089 | - | - |
| SCT ex Afghanistan | Afghanistan | 2012 | IV/ASI-1 | JX908640 | - | - |
| Afg09-2990 | Afghanistan | 2009 | IV/ASI-1 | HM452305 | - | - |
| ArTec193-3 | Iran | 1978 | I/WAFR-1 | U15022 | - | - |
| 766/02 | Iran | 2004 | IV/ASI-1 | AY366373 | - | - |
| 787/02 | Iran | 2004 | IV/ASI-1 | AY366379 | - | - |
| Iran 52 | Iran | 2002 | - | - | III/S AFR,WAFR-2 | DQ446215 |
| Iran53 | Iran | 2002 | - | - | III/S AFR,WAFR-2 | DQ446216 |
| Baghadad12 | Iraq | 1979 | IV/ASI-1 | AJ538196 | III/S AFR,WAFR-2 | AJ538197 |
| China79121 | China | 1979 | IV/ASI-2 | AF358784 | I/W AFR-1 | AB069673 |
| ChinaC68031 | China | 1968 | - | - | IV/ASI | DQ211629 |
| Kosovo 1917 | Kosovo | 2009 | V/EUR | JN173797 | - | - |
| Kososvo9553 | Kosovo | 2001 | - | - | V/EUR | AY675511 |
| KosovoHoti | Kosovo | 2001 | - | - | V/EUR | EU037902 |
| ArMg951 | Madagaskar | 1994 | IV/ASI-1 | U15024 | - | - |
| IbAr10200 | Nigeria | 1966 | III/S AFR,W AFR-2 | U88410 | III/S AFR,W AFR-2 | AF467768 |
| Matin | Pakistan | 1976 | IV/ASI-1 | AF527810 | 4/ASI | AF467769 |
| PakistanSR3 | Pakistan | 2000 | IV/ASI-1 | AJ538198 | III/S AFR,W AFR-2 | AJ538199 |
| Drosdov | Russia | 1967 | V/EUR | DQ211643 | V/EUR | DQ211630 |
| Kashmanov | Russia | 1967 | V/EUR | DQ211644 | V/EUR | DQ211631 |
| SPU 128817 | South Africa | 1981 | III/S AFR,W AFR-2 | DQ076415 | III/S AFR,W AFR-2 | DQ157174 |
| SPU9785 | South Africa | 1985 | III/S AFR,W AFR-2 | DQ211646 | IV/ASI | DQ211633 |
| ArD39554 | Mauritania | 1984 | III/S AFR,W AFR-2 | DQ211641 | VII/MAURITANIA | DQ211628 |
| AB1-2009 | Sudan | 2009 | - | - | III/S AFR,W AFR-2 | HQ378187 |
| ArD8194 | Senegal | 1969 | I/W AFR-1 | DQ211639 | I/W AFR-1 | DQ211626 |
| ArD15786 | Senegal | 1972 | I/W AFR-1 | DQ211640 | - | - |
| Semunya | Uganda | 1958 | II/CENTRAL AFR | DQ076413 | ||
| UG3010 | Congo | 1956 | II/CENTRAL AFR | DQ211650 | I/W AFR-1 | DQ211637 |
| Hodzha | Uzbekistan | 1967 | IV/ASI-2 | AY223475 | III/S AFR,W AFR-2 | AY223476 |
| HU8966 | Tajikistan | 1990 | - | - | III/S AFR,WAFR-2 | AY179962 |
| AP92 | Greece | 1975 | VI/GREECE | DQ211638 | VI/GREECE | DQ211625 |
| Oman | Oman | 1997 | IV/ASI-1 | DQ211645 | IV/ASI | DQ211632 |
| Dubai 616 | Dubai | 1979 | IV/ASI-2 | JN108025 | - | - |
| Kelkit06 | Turkey | 2006 | V/EUR | GQ337053 | - | - |
| 200310849 | Turkey | 2003 | V/EUR | DQ211649 | V/EUR | DQ211636 |
| KT281/75 | - | - | Dugbe virus | AF434165 | - | - |
| JC280 | Pakistan | 1992 | Hazara virus | M86624 | - | - |
The sequence alignment was performed by ClustalW and a scaled phylogenetic tree generated by the Maximum Likelihood (ML) with Kimura 2-parameter distance using Mega5 software. Bootstrap confidence limits were based on 1000 replicates. This method evaluates the topologies of different trees and chooses the best tree based on a specified model. This model is based on the evolutionary process that can account for the conversion of one sequence into another (Tonbak et al. 2006, Aradaib et al. 2011).
Results
Viral RNA was extracted from 50 selected sera of infected patients originated from 19 provinces of Iran including Khorasane-Razavi, Khorasane-Jonobi, SistanvaBalochestan, Kerman, Tehran, Gilan, Mazandaran, Isfahan, Yazd, Qom, Azarbaijane-Sharghi, Azarbaijane-Gharbi, Zanjan, Kermanshah, Hormozgan, Fars, Boshehr, Khuzestan and Lorestan collected between 2002 and 2013.
Samples included in this study were correlated to the prevalence of the CCHFV from each region. Accordingly, 11 sera were selected from the Southeast of Iran, 10 sera from Central Iran, 9 sera from Northeast Iran, 8 sera from Southern Iran, 4 sera from West, 3 sera from Northwest, 3 serum samples from North, 1 serum sample from East and 1 serum sample from Southwest.
Partial sequence phylogenetic analysis of S segment
The phylogenetic analysis conducted by the Neighbor Joining (NJ) and Maximum-Likelihood (ML). Only ML are presented here, however phylogenetic tree that were created using the NJ algorithm had the same topology and strongly supported the same phylogenetic groups as the ML tree.
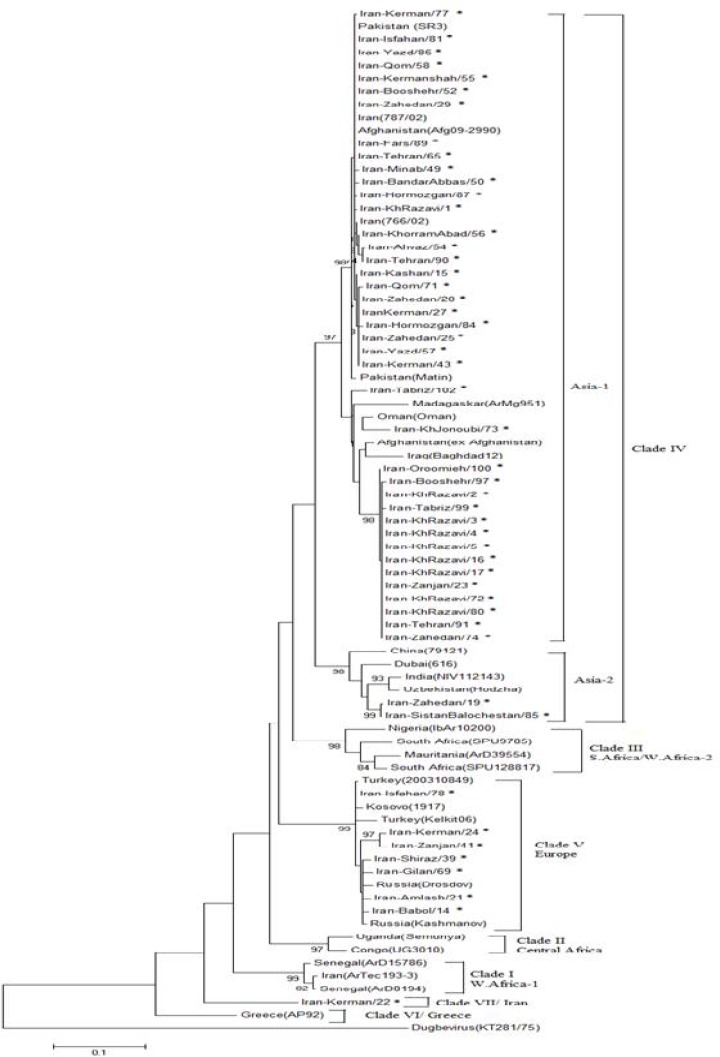
Phylogenetic analysis using the partial S-segment (∼500 bp) demonstrated, of 50 sequenced CCHFV obtained from Iranian patients, 40 sequences (80%), mainly in the Southeast and East of Iran, were located within clade IV (Asia 1). Two Iranian sequences (4%), both in the Southeast of Iran, formed a distinct cluster in the clade IV (Asia 2). These two Iranian sequences fell in the subgroup consisting of the India. Notably, two sequences had strong similarity to each other with 100 % bootstrap support. Seven sequences (14%) of Iranian strains, mainly in the Northwest of Iran, were located within clade V (Europe). One sequence (Iran-Kerman/22) showed itself as out-group and had the highest differences with other clades.
No Iranian sequences were positioned within clade III (S Africa and/or W Africa 2), clade II (Central Africa), clade I (W Africa 1), and clade VI (Greece AP92).
As expected, all sequences from an outbreak in Mashhad city, Khorasane-Razavi province, in 2012 grouped in a cluster with the highest similarity to each other (Iran-KhRazavi 2, 3, 4, 5, 16 and 17) (Fig. 1).
Fig. 1.
Phylogenetic tree of CCHFVs based on the 520-nucleotide S RNA sequences. The tree was constructed by using the maximum likelihood method with Mega 5. The sequences obtained from this study are shown by asterisk. The numbers above the branches indicate the bootstrap values in percentages (of 1000 replicates)
Complete genome sequence phylogenetic analysis of S segment
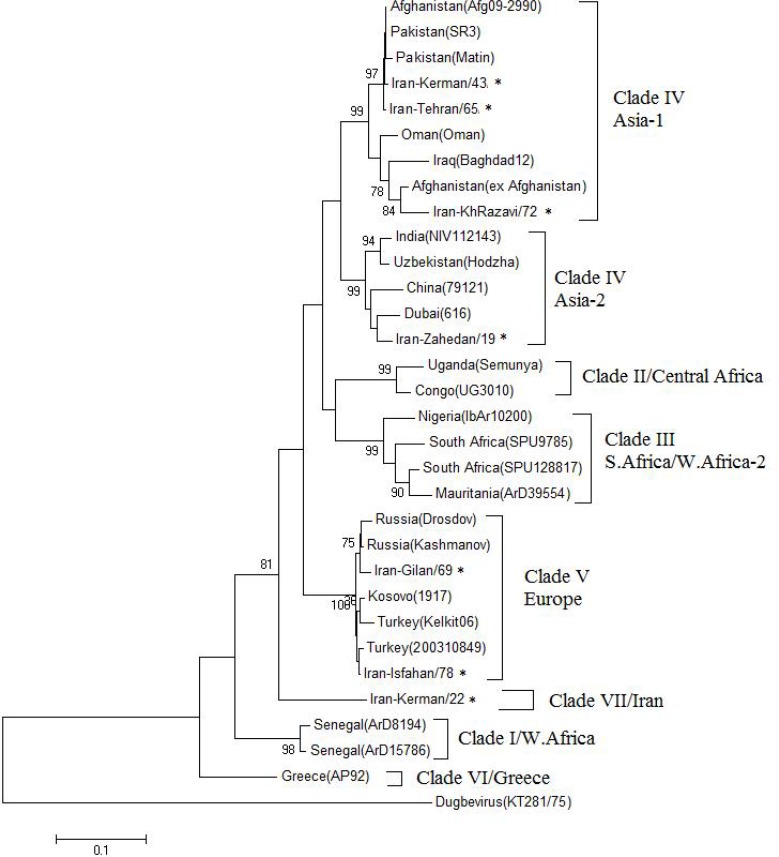
The topology of complete genome sequence analysis of S-segment (∼1500 bp) was in accordance with partial genome sequence analysis of the S-segment. The phylogeny of the complete S segment nucleotide sequences from strains obtained in Gene-Bank indicated that eight major groups could be recognized. The Iranian S segment from strain Iran-Kerman/22 was least similar to all other S segments and stands as an out-group and considered as clade VII (Iran) (Fig. 2).
Fig. 2.
Phylogenetic tree of CCHFVs full-length S RNA sequences (1500 bp). The tree was constructed by using the maximum likelihood method with Mega 5. The sequences obtained from this study are shown by asterisk. The numbers above the branches indicate the bootstrap values in percentages (of 1000 replicates)
Partial sequence phylogenetic analysis of M segment
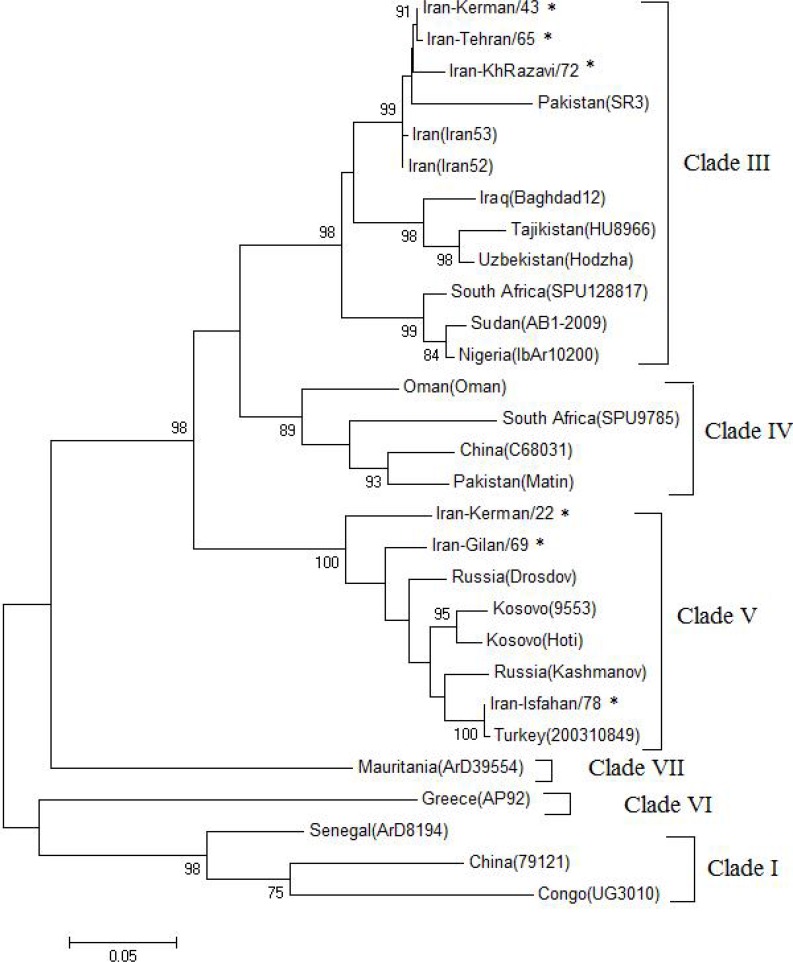
Based on phylogenetic analysis on the partial sequence of M segment (∼520 bp), six clades were distinguishable. The tree showed that seven Iranian strains clustered in the clade III (S Africa and/or W Africa) and V (Europe) with other strains of Middle East and Europe.
The Iranian CCHFV strains Iran-Kerman/43, Iran-Tehran/65 and Iran-KhRazavi/72 in accordance with the Iranian strains (DQ 446216 and DQ446215) formed a separate cluster within the clades III and showed maximum closeness with the Pakistan (AJ 538199). In addition, Iran-Gilan/69 and Iran-Isfahan/78 strains fell in clade V (Europe) and showed maximum association with the isolates from Russia, Kosovo and Turkey. Interestingly, Iran-Kerman/22 showed the highest similarity to clade V in the M segment analysis (Fig. 3).
Fig. 3.
Phylogenetic tree of CCHFVs partial M RNA sequences (500 nucleotides). The tree was constructed by using the maximum likelihood method with Mega 5. The sequences obtained from this study are shown by asterisk symbol. The numbers above the branches indicate the bootstrap values in percentages (of 1000 replicates)
Discussion
In 1992, the first complete nucleotide sequence of S-RNA segment of Chinese strain C68031 of CCHFV was determined (Marriott and Nuttall 1992). Afterwards, during an outbreak of CCHF in the United Arab Emirates between 1994 and 1995, nested reverse transcriptase polymerase chain reaction (RTPCR) amplifying of the partial S-RNA segment of CCHF virus was developed and used for analysis of CCHFVs (Schwarz et al. 1996).
Phylogenetic studies relied on the sequence data of the S-RNA segment have demonstrated genetic diversity for many CCHFV strains from different regions of the world (Iashina et al. 2002). Despite the fact that recombination is relatively scarce in CCHFV genome (Chare et al. 2003) and a partial sequence of S-RNA segment can be used for phylogenetic analyses, but it is recommended to use the full sequence data of the SRNA segment for obtaining more precise phylogenetic analysis, as the possibility of recombination is still expected (Lukashev 2005).
There have been several genetic analyses of CCHFVs obtained from Iran; however, all the reported Iranian strains were obtained mostly from the South-East of Iran and just four obtained from Central and recently one strain from the Northern regions of Iran (Chinikar et al. 2012a, Chinikar et al. 2004, Chinikar et al. 2010, Chinikar et al. 2013). What makes this study unique is in that a large number of CCHFV strains have been characterized genetically by partial and complete nucleotide sequencing of virus small (S) and medium (M) segments with a wide geographical distribution. Fifty partial sequences of S segment and six partial sequences of M segment and seven complete sequences of S segments from various locations have been studied.
In this study, we have used a bioinformatics approach to analyze an alignment by estimating the phylogenetic relationship between the obtained sequences from Iranian patients and GenBank available data. Phylo-genetic analysis based on large collections of partial and complete sequences of the S segment has indicated the existence of eight distinct clades for CCHFV (Drosten et al. 2002). Accordingly, the most surprising finding in this study is discovery of a novel genomic variant of CCHFV. In other words, this study suggests the emergence of VII-IRAN clade for CCHFV in Iran as a novel clade based on S segment analysis.
In general, this phylogenetic analysis based on sequences of S-RNA of CCHFVs reveals that the majority of CCHFV sequences from Iran belongs to clade IV (Asia 1), and clade V (Europe), which is in consistent with previous reports from Iran (Chinikar et al. 2004, Chinikar et al. 2012b). In addition, for the first time, two sequences (Iran-Zahedan/19 and Iran-SistanBalochestan/85, which were obtained from the South-East Iran, were seen within clade IV (Asia 2) with the highest proximity to strains from India (JN572089).
The data of this study are in conformity with previous studies which showed similarity between Iranian S segment of CCHFV strains with Pakistan, Afghanistan (Chinikar et al. 2004) and Iraq (Chinikar et al. 2012a), Interestingly findings regarding all phylogeny studies during these years is that CCHFV strain Iran/ArTec 193-3 is likely fade away from Iran and it has been never reported since 35 years ago when it was reported as the first CCHFV strain from Iran by Sureau (Sureau and Klein 1980).
Our data based on the S segment shows that CCHFVs are grouped in eight different clades and have correlation with their geographical location (Morikawa et al. 2007). Previous phylogenetic studies based on LRNA segment sequences have showed that the L tree topology is similar to the S tree topology (Hewson et al. 2004b). However, the phylogenetic topology based on M-RNA segment sequences of CCHFVs is different from that of S-RNA segments (Seregin et al. 2004, Ahmed et al. 2005). These analyses show that CCHFVs are likely to be grouped in six different phylogenetic clades based on M-RNA sequences (Deyde et al. 2006, Carroll et al. 2010).
To obtain a phylogenetic tree based on M segment, a partial sequence data can be used (Morikawa et al. 2007), indicating that recombination within the M segments is not common during evolution of the CCHFVs. However, genetic re-assortment occurs frequently in CCHFVs, when host ticks co-infected with different types of CCHFVs (Deyde et al. 2006). The reason why M-RNA segment re-assortment is more frequently observed is not clear, however, strong interrelation between N protein encoded in the SRNA segment and RNA polymerase encoded in the L-RNA segment may be required to produce viable virus (Chamberlain et al. 2005).
Different examples of RNA segment re-assortment can be seen in the M segment phylogenetic tree. Tree based on M segment nucleotide and deduced amino acid sequence differences were very similar to each other. Like Mauritanian strain ArD39554 that belongs to group III in S and stands as an out-group VII in M segment trees, surprisingly in our obtained strains, the Iranian strain Iran-Kerman/22 forms a unique group and considered as clade VII in S tree (Fig. 2), whereas it grouped within group V (Europe) with 100 % bootstrap support for this topology in M tree (Fig. 3), suggesting that it representing M segment re-assortment.
Other potential M segment re-assortment events include Iranian strains Iran-Kerman/43 and Iran-KhRazavi/72 that were in group IV (Asia 1) in S tree, while cluster within group III in the M segment tree.
Conclusion
The CCHFVs in Iran have multiple origins with vivid geographical relationships between virus strains. Moreover, phylogenetic results reveal subtle links between distant geographical locations, which may originate either from livestock trade or from long-distance carriage of virus by infected ticks during bird migration.
According to all previous phylogeny studies, four genetic lineages of CCHF viruses, clade IV (Asian 1 and II), clade V (Europe) and new clade VII (Iran) exist in Iran. Based on mentioned above data, having a high genomic variation of CCHFV, have been made Iran as a miniature model of the world for genetic analysis of CCHFV with various strains for CCHFV.
Acknowledgements
This manuscript is based on the results of corresponding author’s PhD thesis. We thank all members of the Arboviruses and Viral Hemorrhagic Fevers Laboratory (National Reference Lab), Pasteur Institute of Iran, and Keyhan Azadmanesh, and Ms Niknam and Mohsen Chiani for their technical assistance. Corresponding author would like to express his sincere gratitude to Nariman Shah-Hosseini, as this research could not be done possible without his assistance and support. This research has been financially supported by Pasteur Institute of Iran. The authors declare that there is no conflict of interests.
References
- Ahmed AA, Mcfalls JM, Hoffmann C, Filone CM, Stewart SM, Paragas J, Khodjaev S, Shermukhamedova D, Schmaljohn CS, Doms RW. (2005) Presence of broadly reactive and group-specific neutralizing epitopes on newly described isolates of Crimean-Congo hemorrhagic fever virus. J Gen Virol. 86: 3327– 3336. [DOI] [PubMed] [Google Scholar]
- Aradaib IE, Erickson BR, Karsany MS, Khristova ML, Elageb RM, Mohamed ME, Nichol ST. (2011) Multiple Crimean-Congo hemorrhagic fever virus strains are associated with disease outbreaks in Sudan, 2008–2009. PLoS Negl Trop Dis. 5(5): e1159– 1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SA, Bird BH, Rollin PE, Nichol ST. (2010) Ancient common ancestry of Crimean-Congo hemorrhagic fever virus. Mol Phylogenet Evol. 55: 1103– 1110. [DOI] [PubMed] [Google Scholar]
- Chamberlain J, Cook N, Lloyd G, Mioulet V, Tolley H, Hewson R. (2005) Co-evolutionary patterns of variation in small and large RNA segments of Crimean-Congo hemorrhagic fever virus. J Gen Virol. 86: 3337– 3341. [DOI] [PubMed] [Google Scholar]
- Champour M, Chinikar S, Mohammadi G, Razmi G, Shah-Hosseini N, Khakifirouz S, Mostafavi E, Jalali T. (2016) Molecular epidemiology of Crimean–Congo hemorrhagic fever virus detected from ticks of one humped camels (Camelus dromedarius) population in northeastern Iran. J Parasit Dis. 40: 110– 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chare ER, Gould EA, Holmes EC. (2003) Phylogenetic analysis reveals a low rate of homologous recombination in negative-sense RNA viruses. J Gen Virol. 84: 2691– 2703. [DOI] [PubMed] [Google Scholar]
- Chinikar S, Shah-Hosseini N, Bouzari S, Jalali T, Shokrgozar MA, Mostafavi E. (2013) New circulating genomic variant of Crimean-Congo hemorrhagic fever virus in Iran. Arch Virol. 158: 1085– 1088. [DOI] [PubMed] [Google Scholar]
- Chinikar S, Ghiasi SM, Naddaf S, Piazak N, Moradi M, Razavi MR, Afzali N, Haeri A, Mostafavizadeh K, Ataei B. (2012a) Serological evaluation of Crimean-Congo hemorrhagic fever in humans with high-risk professions living in enzootic regions of Isfahan Province of Iran and genetic analysis of circulating strains. Vector Borne Zoonotic Dis. 12( 9): 733– 738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinikar S, Shayesteh M, Khakifirouz S, Jalali T, Rasi Varaie FS, Rafigh M, Mostafavi E, Shah-Hosseini N. (2013) Nosocomial infection of Crimean-Congo haemorrhagic fever in eastern Iran: Case report. Travel Med Infect Dis. 11( 4): 252– 255. [DOI] [PubMed] [Google Scholar]
- Chinikar S, Mojtaba Ghiasi S, Moradi M, Goya M, Reza Shirzadi M, Zeinali M, Mostafavi E, Pourahmad M, Haeri A. (2010) Phylogenetic analysis in a recent controlled outbreak of Crimean-Congo haemorrhagic fever in the south of Iran, December 2008. Euro Surveill. 15 ( 47): 19720. [DOI] [PubMed] [Google Scholar]
- Chinikar S, Persson SM, Johansson M, Bladh L, Goya M, Houshmand B, Mirazimi A, Plyusnin A, Lundkvist, Nilsson M. (2004) Genetic analysis of Crimean-Congo hemorrhagic fever virus in Iran. J Med Virol. 73: 404– 411. [DOI] [PubMed] [Google Scholar]
- Deyde VM, Khristova ML, Rollin PE, Ksiazek TG, Nichol ST. (2006) Crimean-Congo hemorrhagic fever virus genomics and global diversity. J Virol. 80: 8834– 8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C, Minnak D, Emmerich P, Schmitz H, Reinicke T. (2002) Crimean-Congo hemorrhagic fever in Kosovo. J Clin Microbiol. 40: 1122– 1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elevli M, Ozkul AA, Civilibal M, Midilli K, Gargili A, Duru NS. (2010) A newly identified Crimean-Congo hemorrhagic fever virus strain in Turkey. Int J Infect Dis. 14: e213– 216. [DOI] [PubMed] [Google Scholar]
- Garcia S, Chinikar S, Coudrier D, Billecocq A, Hooshmand B, Crance J, Garin D, Bouloy M. (2006) Evaluation of a Crimean-Congo hemorrhagic fever virus recombinant antigen expressed by Semliki Forest suicide virus for IgM and IgG antibody detection in human and animal sera collected in Iran. J Clin Virol. 35: 154– 159. [DOI] [PubMed] [Google Scholar]
- Han N, Rayner S. (2011) Epidemiology and mutational analysis of global strains of Crimean-Congo haemorrhagic fever virus. Virol Sin. 26: 229– 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewson R, Chamberlain J, Mioulet V, Lloyd G, Jamil B, Hasan R, Gmyl A, Gmyl L, Smirnova S, Lukashev A. (2004a) Crimean-Congo haemorrhagic fever virus: sequence analysis of the small RNA segments from a collection of viruses world wide. Virus Res. 102: 185– 189. [DOI] [PubMed] [Google Scholar]
- Hewson R, Gmyl A, Gmyl L, Smirnova S E, Karganova G, Jamil B, Hasan R, Chamberlain J, Clegg C. (2004b) Evidence of segment reassortment in Crimean-Congo haemorrhagic fever virus. J Gen Virol. 85: 3059– 3070. [DOI] [PubMed] [Google Scholar]
- Iashina L, Petrov V, Petrova I, Gutorov V, Kazakov S, Ospanov K, Karimov S, Tiunnikov G, Seregin S, Kuzina I. (2002) Genetic identification of the Crimean-Congo hemorrhagic fever virus during epidemic outbreak in Kazakhstan in 2000. Mol Gen Mikrobiol Virusol. 4: 31– 35. [PubMed] [Google Scholar]
- Lukashev AN. (2005) Evidence for recombination in Crimean-Congo hemorrhagic fever virus. J Gen Virol. 86: 2333– 2338. [DOI] [PubMed] [Google Scholar]
- Marriott AC, Nuttall PA. (1992) Comparison of the S RNA segments and nucleoprotein sequences of Crimean-Congo hemorrhagic fever, Hazara, and Dugbe viruses. Virology. 189: 795– 799. [DOI] [PubMed] [Google Scholar]
- Mehravaran A, Moradi M, Telmadarraiy Z, Mostafavi E, Moradi AR, Khakifirouz S, Shah-Hosseini N, Varaie FSR, Jalali T, Hekmat S. (2013) Molecular detection of Crimean-Congo haemorrhagic fever (CCHF) virus in ticks from southeastern Iran. Ticks Tick Borne Dis. 4: 35– 38. [DOI] [PubMed] [Google Scholar]
- Mohammadi GR, Razmi GR. (2014) Seroepidemiology of Crimean-Congo hemorrhagic fever virus in one-humped camels (Camelus dromedarius) population in northeast of Iran. J Vector Borne Dis. 51: 62– 65. [PubMed] [Google Scholar]
- Morikawa S, Saijo M, Kurane I. (2007) Recent progress in molecular biology of Crimean-Congo hemorrhagic fever. Comp Immunol Microbiol Infect Dis. 30: 375– 389. [DOI] [PubMed] [Google Scholar]
- Morikawa S, Qing T, Xinqin Z, Saijo M, Kurane I. (2002) Genetic diversity of the M RNA segment among Crimean-Congo hemorrhagic fever virus isolates in China. Virology. 296: 159– 164. [DOI] [PubMed] [Google Scholar]
- Papa A, Papadimitriou E, Bozovic B, Antoniadis A. (2005) Genetic characterization of the M RNA segment of a Balkan Crimean-Congo hemorrhagic fever virus strain. J Med Virol. 75: 466– 469. [DOI] [PubMed] [Google Scholar]
- Papa A, Ma B, Kouidou S, Tang Q, Hang C, Antoniadis A. (2002) Genetic characterization of the M RNA segment of Crimean Congo hemorrhagic fever virus strains, China. Emerg Infect Dis. 8( 1): 50– 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez LL, Maupin GO, Ksiazek TG, Rollin PE, Khan AS, Schwarz TF, Lofts RS, Smith JF, Noor AM, Peters CJ. (1997) Molecular investigation of a multisource outbreak of Crimean-Congo hemorrhagic fever in the United Arab Emirates. Am J Trop Med Hyg. 57: 512– 518. [DOI] [PubMed] [Google Scholar]
- Schwarz TF, Nsanze H, Longson M, Nitschko H, Gilch S, Shurie H, Ameen A, Zahir ARM, Acharya UG, Jager G. (1996) Polymerase chain reaction for diagnosis and identification of distinct variants of Crimean-Congo hemorrhagic fever virus in the United Arab Emirates. Am J Trop Med Hyg. 55: 190– 196. [DOI] [PubMed] [Google Scholar]
- Seregin SV, Samokhvalov EI, Petrova ID, Vyshemirskii OI, Samokhvalova EG, Lvov DK, Gutorov VV, Tyunnikov GI, Shchelkunov SN, Netesov SV. (2004) Genetic characterization of the M RNA segment of Crimean-Congo hemorrhagic fever virus strains isolated in Russia and Tajikistan. Virus Genes. 28: 187– 193. [DOI] [PubMed] [Google Scholar]
- Sureau P, Klein J. (1980) Arboviruses in Iran. Med Trop. 40: 549– 554. [PubMed] [Google Scholar]
- Tonbak S, Aktas M, Altay K, Azkur A K, Kalkan A, Bolat Y, Dumanli N, Ozdarendeli A. (2006) Crimean-Congo hemorrhagic fever virus: genetic analysis and tick survey in Turkey. J Clin Microbiol. 44: 4120– 4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav PD, Cherian SS, Zawar D, Kokate P, Gunjikar R, Jadhav S, Mishra AC, Mourya DT. (2012) Genetic characterization and molecular clock analyses of the Crimean-Congo Hemorrhagic Fever virus from human and ticks in India, 2010–2011. Infect Genet Evol. 14: 223– 231 [DOI] [PubMed] [Google Scholar]
- Yashina L, Vyshemirskii O, Seregin S, Petrova I, Samokhvalov E, Lvov D, Gutorov V, Kuzina I, Tyunnikov G, Tang Y W. (2003). Genetic analysis of Crimean-Congo hemorrhagic fever virus in Russia. J Clin Microbiol. 41: 860– 862. [DOI] [PMC free article] [PubMed] [Google Scholar]