Abstract
MicroRNAs (miRNAs or miRs) are a family of small non-coding RNAs that function as oncogenes or tumor suppressor genes. Recent evidence suggests that the pri-miR-34b/c rs4938723 variant is associated with the development of cancer. At present, there is an inconsistent association between the single-nucleotide polymorphism in pri-miR-34b/c and cancer in the limited studies. The present study is a case-control investigation, with 263 breast cancer (BC) patients and 221 control women, which examined the potential association of the pri-miR-34b/c rs4938723 polymorphisms with BC susceptibility. The polymorphisms were genotyped by the polymerase chain reaction restriction fragment length polymorphism method. No significant association between the pri-miR-34b/c rs4938723 variant and BC was identified [TC vs. TT: Odds ratio (OR), 0.87; 95% confidence interval (CI), 0.60–1.26; P=0.506; CC vs. TT: OR, 1.22; 95% CI, 0.61–2.47; P=0.600; TC+CC vs. TT: OR, 0.91; 95% CI, 0.64–1.31; P=0.648; CC vs. TT+TC: OR, 1.32; 95% CI, 0.67–2.59; P=0.498; C vs. T: OR, 0.99; 95% CI, 0.75–1.31; P=0.986]. However, a significant association was observed between the pri-miR-34b/c rs4938723 genotypes and clinicopathological characteristics, such a grade, progesterone receptor and human epidermal growth factor receptor 2 status were observed (P<0.05). These findings suggest that the pri-miR-34b/c rs4938723 variant may not be a risk factor for the development of BC.
Keywords: pri-miR-34b/c, breast cancer, polymorphism
Introduction
Breast cancer (BC), the most prevalent type of cancer in women, is a major public and global health problem and accounts for 14% of total annual cancer fatalities worldwide (1). Similarly, BC is the most common malignancy affecting Iranian women (2). Although the etiology of BC remains to be identified, genetic factors are shown to have important roles in the pathogenesis and progress of this malignancy (3–8).
MicroRNAs (miRNAs or miRs) are a class of single-stranded non-coding RNA typically 17–25 nucleotides in length that have key roles in the regulation of cellular processes by targeting mRNAs for cleavage or translational repression (9,10). They regulate the expression of genes at the post-transcriptional level by targeting the 3′ untranslated regions of mRNAs. Cumulative evidence suggests that the dysregulation of miRNA expression is involved in the tumorigenesis by acting as tumor suppressors or oncogenes (11–15). Single-nucleotide polymorphisms (SNPs) or mutations in miRNA genes can affect the miRNA biosynthesis and expression of target genes, therefore resulting in diverse functional consequences and thereby possibly representing potentially important biomarkers for the prognosis of cancer (7,16–18).
The pri-miR-34b/c gene resides in a CpG island within the intron of the B-cell translocation gene 4. Therefore, it is expected that pri-miR-34b/c is co-transcribed by either the promoter of the protein-coding gene or by its own transcription initiation region, as is the case with the majority of miRNAs, whether they are intergenic or located within the introns of protein coding genes (9). A putatively functional rs4938723 C>T is located within the CpG island of the promoter of pri-miR-34b/c and is a 423-base pair (bp) upstream from the transcription start site (19). The variation of rs4938723 C to T may affect a predicted GATA-X transcription factor binding and subsequently affect the expression and carcinogenesis (19–21).
It is well known that the expression of miRNAs, particularly the miR-34 family members (such as miR-34a, miR-34b and miR-34c) can be regulated by p53. These three mature miRNAs are encoded by two different primary miRNAs. miR-34a is encoded by its own transcript, while miR-34b and miR-34c share a common primary transcript (pri-miR-34b/c) (9). The promoter region of miR-34b/c transcripts contains p53-binding sites (22).
Numerous studies investigated the impact of miR-34b/c rs4938723 on the risk of various cancers, but the results are inconsistent (19,23–31). To the best of our knowledge, there is only one previous study regarding the impact of the miR-34b/c variant on BC risk (32). Therefore, the present case-control study aimed to evaluate the possible association between the pri-miR-34b/c rs4938723 polymorphism and susceptibility to BC in a sample of Iranian population.
Materials and methods
Patients
This case-control study consisted of 263 BC patients and 221 age-matched healthy women with no history of cancer of any type (as the control group) in Zahedan (southeast Iran). The enrolment procedure and study design were as described previously (7,33). Ethical approvals for recruitment were provided from the local Ethics Committee of Zahedan University of Medical Sciences, and informed consent was obtained from all patients and healthy individuals. Blood samples were collected in EDTA-containing tubes from the cases and controls, and genomic DNA was extracted using the salting out method, as described previously (34).
Genotyping
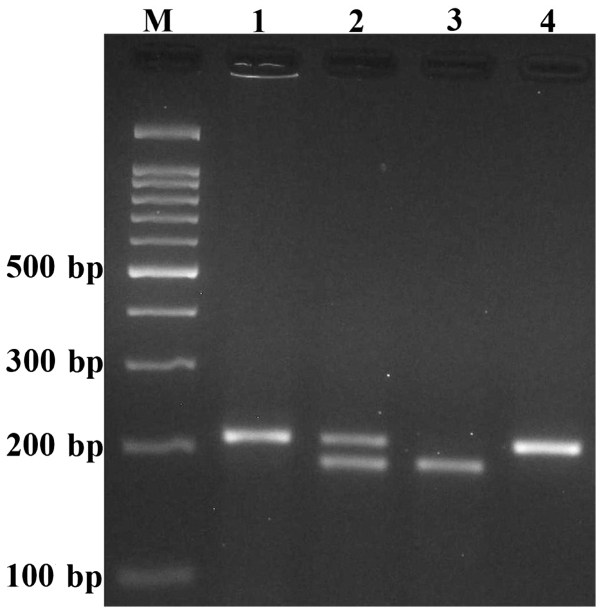
Genotyping of pri-miR-34b/c rs4938723 was analyzed by polymerase chain reaction (PCR) restriction fragment length polymorphism methods. Briefly, forward and reverse primers were 5′-CCTCTGGGAACCTTCTTTGACCTGT-3′ and 5′-CCTGGGCCTTCTAGTCAAATAGTGA-3′, respectively. A 0.20-ml reaction solution included 1 µl genomic DNA (~100 ng/ml), 1 µl forward and reverse primers and 10 µl 2X Prime Taq Premix (Genet Bio, Chungnam, Korea) and 7 µl double-distilled H2O. The PCR conditions were justified as follows: 5 min preheating at 95°C, 30 cycles of 95°C for 30 sec, 57°C for 30 sec and 72°C for 30 sec, followed by a final extension step for 10 min at 72°C. Following this, 10 µl of the PCR product was digested by the NmuCI restriction enzyme (Fermentas, Thermo Fisher Scientific, Inc., Waltham, MA, USA). The C allele produced a 26- and 186-bp pattern, while the T allele was undigested and produced a 212-bp fragment (Fig. 1).
Figure 1.
Image of the pri-miR-34b/c rs4938723 T>C polymorphism using the polymerase chain reaction restriction fragment length polymorphism method. The C allele was digested by the NmuCI restriction enzyme and produced a 26- and 186-base pair (bp) pattern, while the T allele was undigested (212-bp fragment). M, DNA marker; lanes 1 and 4, TT; lane 2, TC; lane 3, CC.
Statistical analysis
Statistical analysis was performed using statistical package SPSS 20 software (IBM, Corp., Armonk, NY, USA). The categorical and continuous data were analyzed using χ2 and t-test, respectively. The association between genotypes and BC were assessed by computing the odds ratio (OR) and 95% confidence intervals (CIs) from logistic regression analyses. P<0.05 was considered to indicate a statistically significant difference.
Results
Patient characteristics
The study group consisted of 256 BC patients with an average age of 49.0±11.1 years and 221 healthy women with a mean age of 49.7±12.6 years. No significant difference was identified between the groups regarding age (P=0.549).
The genotype and allele frequencies of the pri-miR-34b/c rs4938723 T>C polymorphism in the BC patients and healthy women are shown in Table I. The results indicated that the rs4938723 polymorphism was not significantly associated with the BC risk in the codominant (TC vs. TT: OR, 0.87; 95% CI, 0.60–1.26; P=0.506; CC vs. TT: OR, 1.22; 95% CI, 0.61–2.47; P=0.600), dominant (TC+CC vs. TT: OR, 0.91; 95% CI, 0.64–1.31; P=0.648), recessive (CC vs. TT+TC: OR, 1.32; 95% CI, 0.67–2.59; P=0.498) and overdominant (TC vs. TT+CC: OR, 0.84; 95% CI, 0.59–1.21; P=0.361) inheritance models tested. Furthermore, the rs4938723 C allele was not a risk factor for BC (OR, 0.99; 95% CI, 0.75–1.31; P=0.986).
Table I.
Association of the pri-miR-34b/c rs4938723 T>C polymorphism and risk of breast cancer.
| pri-miR-34b/c rs4938723 | Case, n (%) | Control, n (%) | OR (95% CI) | P-value |
|---|---|---|---|---|
| Codominant | ||||
| TT | 125 (47.5) | 100 (45.2) | 1.00 | – |
| TC | 115 (43.7) | 106 (48.0) | 0.87 (0.60–1.26) | 0.506 |
| CC | 23 (8.7) | 15 (6.8) | 1.22 (0.61–2.47) | 0.600 |
| Dominant | ||||
| TT | 125 (47.5) | 100 (45.2) | 1.00 | – |
| TC+CC | 138 (52.5) | 121 (54.8) | 0.91 (0.64–1.31) | 0.648 |
| Recessive | ||||
| TT+TC | 240 (91.3) | 206 (93.2) | 1.00 | – |
| CC | 23 (8.7) | 15 (6.8) | 1.32 (0.67–2.59) | 0.498 |
| Overdominant | ||||
| TT+CC | 148 (56.3) | 115 (52.0) | 1.00 | – |
| TC | 115 (43.7) | 106 (48.0) | 0.84 (0.59–1.21) | 0.361 |
| Allele | ||||
| T | 365 (69.4) | 306 (69.2) | 1.00 | – |
| C | 161 (30.6) | 136 (30.8) | 0.99 (0.75–1.31) | 0.986 |
OR, odds ratio; CI, confidence interval.
The genotype of the polymorphism in the controls and cases were in Hardy-Weinberg equilibrium (χ2=3.49, P=0.061 and χ2=0.227, P=0.624, respectively).
Associations between the variant and clinicopathological characteristics
The association between the pri-miR-34b/c rs4938723 variant and clinicopathological characteristics, including age, body mass index (BMI), tumor size, tumor stage, tumor grade, lymph node metastasis, estrogen and progesterone receptors (ER and PgR), and human epidermal growth factor receptor 2 (HER2) are shown in Table II. The results showed a significant association between the pri-miR-34b/c rs4938723 variant and grade, PgR and HER2 status of BC patients (P<0.05). No significant association was identified between the rs4938723 polymorphism and age, BMI, tumor size, lymph node metastasis, stage and histological type of BC patients (P>0.05).
Table II.
Correlation between the miR-34b/c rs4938723 T>C genotypes and clinical characteristics of breast cancer patients.
| miR-34b/c rs4938723 T>C genotypes | ||||
|---|---|---|---|---|
| Variables | TT | CT | CC | P-value |
| Age, years | 0.535 | |||
| ≤50 | 63 | 68 | 14 | |
| >50 | 56 | 46 | 9 | |
| Mean BMI ± SD, kg/m2 | 25.8±3.2 | 25.6±3.3 | 26.4±2.5 | 0.489 |
| Tumor size, cm | 0.453 | |||
| ≤2 | 44 | 34 | 9 | |
| >2 | 72 | 76 | 13 | |
| Lymph node metastasis status, n | 0.848 | |||
| Yes | 34 | 30 | 5 | |
| No | 44 | 43 | 9 | |
| Grade, n | 0.028 | |||
| I | 17 | 23 | 5 | |
| II | 74 | 56 | 7 | |
| III+IV | 16 | 21 | 8 | |
| Stage, n | 0.941 | |||
| I | 21 | 20 | 2 | |
| II | 44 | 44 | 10 | |
| III | 34 | 28 | 7 | |
| IV | 19 | 18 | 3 | |
| Histology, n | 0.890 | |||
| Ductal carcinoma | 85 | 72 | 16 | |
| Other | 29 | 36 | 7 | |
| Estrogen receptor, n | 0.004 | |||
| Positive | 65 | 67 | 21 | |
| Negative | 46 | 35 | 1 | |
| Progesterone receptor, n | 0.007 | |||
| Positive | 58 | 72 | 16 | |
| Negative | 53 | 28 | 6 | |
| HER2, n | 0.031 | |||
| Positive | 56 | 64 | 7 | |
| Negative | 61 | 45 | 16 | |
BMI, body mass index; SD, standard deviation; HER2, human epidermal growth factor receptor 2.
Discussion
The present study investigated the impact of the pri-miR-34b/c rs4938723 polymorphism on the BC risk in a sample of the Iranian population. The data showed that the rs4938723 variant of pri-miR-34b/c was not associated with the risk of BC population. By contrast, a significant association was identified between the pri-miR-34b/c rs4938723 genotypes and clinicopathological characteristics, such as grade, PgR and HER2 status (P<0.05). In agreement to this finding, Bensen et al (32) identified no significant association between the pri-miR-34b/c rs4938723 variant and risk of BC, and reported that this polymorphism was associated with BC survival (CC vs. TT+TC: hazard ratio, 0.57; 95% CI, 0.37–0.89; P=0.01).
Recently, Chen et al (26) performed a case-control study and revealed that pri-miR-34b/c rs4938723 significantly increased the risk of papillary thyroid carcinoma (PTC). In addition, a significant upregulation of pri-miR-34b was observed in PTC patients. Zhu et al (23) found no significant association between the pri-miR-34b/c rs4938723 variant and esophageal squamous cell carcinoma (ESCC) in Kazakh patients in northwest China. However, Zhang et al (24) reported that pri-miR-34b/c rs4938723 significantly decreased the risk of ESCC in Chinese populations.
It has been reported that the potentially functional SNP rs4938723 in the promoter region of pri-miR-34b/c increased the risk of hepatocellular carcinoma (HCC) in Chinese and Korean populations (19,35). However, the results of a meta-analysis performed by Liang et al (31) did not support an association between the rs4938723 variant and HCC risk. A meta-analysis performed by Liu et al (36) reported a significant association between the rs4938723 variant and cancer risk. Subsequent to stratifying by ethnicity and cancer type, the CT genotype of rs4938723 was significantly associated with an increased cancer risk in the Asian population, and the C allele and CT genotype significantly increased the risk of HCC, but the rs4938723 CT genotype significantly decreased the risk of colorectal cancer. Yi et al (37) conducted a meta-analysis and identified that the pri-miR-34b/c rs4938723 TC heterozygote increased the overall cancer risk. However, the CC genotypes of rs4938723 were associated with an increased HCC risk but also with a decreased colorectal cancer risk (37). Tao et al (29) found no significant association between the hsa-miR-34b/c rs4938723 polymorphism and overall cancer risk. Furthermore, subgroup analysis suggested that the variant CT genotypes were associated with an increased risk of HCC compared with the wild-type TT genotype. However, the CC/TT genotype decreased the risk of colorectal cancer compared with TT genotype.
A meta-analysis performed by Ji et al (25) showed that the pri-miR-34b/c rs4938723 variant significantly decreased the risk of digestive tract cancer. Pan et al (27) reported that the CT and CT/CC genotypes of the miR-34b/c rs4938723 were associated with a significantly decreased risk of gastric cancer compared with the TT genotype. It has been shown that the pri-miR-34b/c rs4938723 C allele may increase the susceptibility to renal cell carcinoma by decreasing the activity of pri-miR-34b/c promoter (30).
The results of a previous meta-analysis indicated that the pri-miR-34b/c rs4938723 polymorphism may decrease the susceptibility to colorectal cancer (28). A meta-analysis performed by Wang et al (38) suggests that the pri-miR-34b/c rs4938723 polymorphism may be associated with the risk of cancer, including nasopharyngeal cancer, osteosarcoma and renal cell cancer. Tong et al (39) reported that the rs4938723 variant significantly decreased the risk of Chinese childhood acute lymphoblastic leukemia.
There is no clear explanation for the controversy of the results regarding the impact of the pri-miR-34b/c rs4938723 variant on different types of cancer. Ethnicity, genetics and/or environmental factors, as well as gene-diet interaction, may interact in diverse modes to either increase or decrease the risk of various types of cancers in different areas.
In conclusion, no significant association was identified between pri-miR-34b/c rs4938723 polymorphism and the risk of BC. However the present findings indicate that this variant may be associated with clinicopathological characteristics. Population-based studies with larger sample sizes of different ethnicities and long-term follow-up are required to confirm this finding.
Acknowledgements
The present study was funded as a dissertation grant (MSc Thesis of SS) from the Deputy for Research, Zahedan University of Medical Sciences.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Harirchi I, Kolahdoozan S, Karbakhsh M, Chegini N, Mohseni SM, Montazeri A, Momtahen AJ, Kashefi A, Ebrahimi M. Twenty years of breast cancer in Iran: Downstaging without a formal screening program. Ann Oncol. 2011;22:93–97. doi: 10.1093/annonc/mdq303. [DOI] [PubMed] [Google Scholar]
- 3.Hashemi M, Fazaeli A, Ghavami S, Eskandari-Nasab E, Arbabi F, Mashhadi MA, Taheri M, Chaabane W, Jain MV, Łos MJ. Functional polymorphisms of FAS and FASL gene and risk of breast cancer - pilot study of 134 cases. PLoS One. 2013;8:e53075. doi: 10.1371/journal.pone.0053075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hashemi M, Eskandari-Nasab E, Fazaeli A, Taheri M, Rezaei H, Mashhadi M, Arbabi F, Kaykhaei MA, Jahantigh M, Bahari G. Association between polymorphisms of glutathione S-transferase genes (GSTM1, GSTP1 and GSTT1) and breast cancer risk in a sample Iranian population. Biomarkers Med. 2012;6:797–803. doi: 10.2217/bmm.12.61. [DOI] [PubMed] [Google Scholar]
- 5.Hashemi M, Eskandari-Nasab E, Fazaeli A, Rezaei H, Mashhadi MA, Arbabi F, Taheri M. Bi-directional PCR allele-specific amplification (bi-PASA) for detection of caspase-8 −652 6N ins/del promoter polymorphism (rs3834129) in breast cancer. Gene. 2012;505:176–179. doi: 10.1016/j.gene.2012.05.043. [DOI] [PubMed] [Google Scholar]
- 6.Eskandari-Nasab E, Hashemi M, Rezaei H, Fazaeli A, Mashhadi MA, Moghaddam SS, Arbabi F, Jahantigh M, Taheri M. Evaluation of UDP-glucuronosyltransferase 2B17 (UGT2B17) and dihydrofolate reductase (DHFR) genes deletion and the expression level of NGX6 mRNA in breast cancer. Mol Biol Rep. 2012;39:10531–10539. doi: 10.1007/s11033-012-1938-8. [DOI] [PubMed] [Google Scholar]
- 7.Omrani M, Hashemi M, Eskandari-Nasab E, Hasani SS, Mashhadi MA, Arbabi F, Taheri M. hsa-mir-499 rs3746444 gene polymorphism is associated with susceptibility to breast cancer in an Iranian population. Biomarkers Med. 2014;8:259–267. doi: 10.2217/bmm.13.118. [DOI] [PubMed] [Google Scholar]
- 8.Amininia S, Hashemi M, Ebrahimi M, Mashhadi MA, Hashemi SM, Taheri M, Ghavami S. Association between CCNE1 polymorphisms and the risk of breast cancer in a sample of southeast Iranian population. Med Oncol. 2014;31:189. doi: 10.1007/s12032-014-0189-z. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 11.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 13.Li YQ, Lu JH, Bao XM, Wang XF, Wu JH, Hong WQ. miR-24 functions as a tumor suppressor in nasopharyngeal carcinoma through targeting FSCN1. J Exp Clin Cancer Res. 2015;34:130. doi: 10.1186/s13046-015-0242-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruoming W, Zhen Y, Tengteng Z, Jisheng H. Tumor suppressor microRNA-31 inhibits gastric carcinogenesis by targeting Smad4 and SGPP2. Cancer Gene Ther. 2015;22:564–572. doi: 10.1038/cgt.2015.41. [DOI] [PubMed] [Google Scholar]
- 15.Glover AR, Zhao JT, Gill AJ, Weiss J, Mugridge N, Kim E, Feeney AL, Ip JC, Reid G, Clarke S, et al. MicroRNA-7 as a tumor suppressor and novel therapeutic for adrenocortical carcinoma. Oncotarget. 2015;6:36675–36688. doi: 10.18632/oncotarget.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 17.Buscaglia LE, Li Y. Apoptosis and the target genes of microRNA-21. Chin J Cancer. 2011;30:371–380. doi: 10.5732/cjc.30.0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hashemi M, Sheybani-Nasab M, Naderi M, Roodbari F, Taheri M. Association of functional polymorphism at the miR-502-binding site in the 3′ untranslated region of the SETD8 gene with risk of childhood acute lymphoblastic leukemia, a preliminary report. Tumour Biol. 2014;35:10375–10379. doi: 10.1007/s13277-014-2359-1. [DOI] [PubMed] [Google Scholar]
- 19.Xu Y, Liu L, Liu J, Zhang Y, Zhu J, Chen J, Liu S, Liu Z, Shi H, Shen H, et al. A potentially functional polymorphism in the promoter region of miR-34b/c is associated with an increased risk for primary hepatocellular carcinoma. Int J Cancer. 2011;128:412–417. doi: 10.1002/ijc.25342. [DOI] [PubMed] [Google Scholar]
- 20.Bossard P, Zaret KS. GATA transcription factors as potentiators of gut endoderm differentiation. Development. 1998;125:4909–4917. doi: 10.1242/dev.125.24.4909. [DOI] [PubMed] [Google Scholar]
- 21.Chou J, Provot S, Werb Z. GATA3 in development and cancer differentiation: Cells GATA have it! J Cell Physiol. 2010;222:42–49. doi: 10.1002/jcp.21943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu J, Yang L, You W, Cui X, Chen Y, Hu J, Liu W, Li S, Song X, Wei Y, et al. Genetic variation in miR-100 rs1834306 is associated with decreased risk for esophageal squamous cell carcinoma in Kazakh patients in northwest China. Int J Clin Exp Pathol. 2015;8:7332–7340. [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Huang X, Xiao J, Yang Y, Zhou Y, Wang X, Liu Q, Yang J, Wang M, Qiu L, et al. Pri-miR-124 rs531564 and pri-miR-34b/c rs4938723 polymorphisms are associated with decreased risk of esophageal squamous cell carcinoma in Chinese populations. PLoS One. 2014;9:e100055. doi: 10.1371/journal.pone.0100055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji TX, Zhi C, Guo XG, Zhou Q, Wang GQ, Chen B, Ma FF. miR-34b/c rs4938723 polymorphism significantly decreases the risk of digestive tract cancer: Meta-analysis. Asian Pac J Cancer Prev. 2015;16:6099–6104. doi: 10.7314/APJCP.2015.16.14.6099. [DOI] [PubMed] [Google Scholar]
- 26.Chen P, Sun R, Pu Y, et al. Pri-Mir-34b/C and Tp-53 polymorphisms are associated with the susceptibility of papillary thyroid carcinoma: A case-control study. Medicine (Baltimore) 2015;94:e1536. doi: 10.1097/MD.0000000000001536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan XM, Sun RF, Li ZH, Guo XM, Qin HJ, Gao LB. Pri-miR-34b/c rs4938723 polymorphism is associated with a decreased risk of gastric cancer. Genet Test Mol Biomarkers. 2015;19:198–202. doi: 10.1089/gtmb.2014.0287. [DOI] [PubMed] [Google Scholar]
- 28.Lin Z, Chen L, Song M, Shi KQ, Tang KF. Association between a polymorphism in miR-34b/c and susceptibility to cancer - a meta-analysis. Asian Pac J Cancer Prev. 2014;15:7251–7255. doi: 10.7314/APJCP.2014.15.17.7251. [DOI] [PubMed] [Google Scholar]
- 29.Tao T, Chen S, Xu B, Liu C, Wang Y, Huang Y, Chen M. Association between hsa-miR-34b/c rs4938723 T > C promoter polymorphism and cancer risk: A meta-analysis based on 6,036 cases and 6,204 controls. Chin J Cancer Res. 2014;26:315–322. doi: 10.3978/j.issn.1000-9604.2014.06.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang S, Qian J, Cao Q, Li P, Wang M, Wang J, Ju X, Meng X, Lu Q, Shao P, et al. A potentially functional polymorphism in the promoter region of miR-34b/c is associated with renal cell cancer risk in a Chinese population. Mutagenesis. 2014;29:149–154. doi: 10.1093/mutage/geu001. [DOI] [PubMed] [Google Scholar]
- 31.Liang TJ, Liu HJ, Zhao XQ, Yu CH, Li CS. Lack of association of miR-34b/c polymorphism (rs4938723) with hepatocellular carcinoma: A meta-analysis. PLoS One. 2013;8:e68588. doi: 10.1371/journal.pone.0068588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bensen JT, Tse CK, Nyante SJ, Barnholtz-Sloan JS, Cole SR, Millikan RC. Association of germline microRNA SNPs in pre-miRNA flanking region and breast cancer risk and survival: The Carolina Breast Cancer Study. Cancer Causes Control. 2013;24:1099–1109. doi: 10.1007/s10552-013-0187-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rezaei M, Hashemi M, Hashemi SM, Mashhadi MA, Taheri M. APOBEC3 deletion is associated with breast cancer risk in a sample of southeast Iranian population. Int J Mol Cell Med. 2015;4:103–108. [PMC free article] [PubMed] [Google Scholar]
- 34.Hashemi M, Hanafi Bojd H, Eskandari Nasab E, Bahari A, Hashemzehi NA, Shafieipour S, Narouie B, Taheri M, Ghavami S. Association of adiponectin rs1501299 and rs266729 gene polymorphisms with nonalcoholic fatty liver disease. Hepat Mon. 2013;13:e9527. doi: 10.5812/hepatmon.9527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Son MS, Jang MJ, Jeon YJ, Kim WH, Kwon CI, Ko KH, Park PW, Hong SP, Rim KS, Kwon SW, et al. Promoter polymorphisms of pri-miR-34b/c are associated with hepatocellular carcinoma. Gene. 2013;524:156–160. doi: 10.1016/j.gene.2013.04.042. [DOI] [PubMed] [Google Scholar]
- 36.Liu Q, Yang G, Song XL, Wang Z, Shi G. Association between rs4938723 functional polymorphism in the promoter region of miR-34b/c gene and cancer risk. Clin Res Hepatol Gastroenterol. 2015;39:526–533. doi: 10.1016/j.clinre.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 37.Yi DH, Wang BG, Zhong XP, Liu H, Liu YF. Pri-miR-34b/c rs4938723 TC heterozygote is associated with increased cancer risks: Evidence from published data. Tumour Biol. 2014;35:11967–11975. doi: 10.1007/s13277-014-2493-9. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Lu X, Fang Y, Chen H, Deng X, Peng C, Li H, Shen B. Association between miR34b/c polymorphism rs4938723 and cancer risk: A meta-analysis of 11 studies including 6169 cases and 6337 controls. Med Sci Monit. 2014;20:1977–1982. doi: 10.12659/MSM.892350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tong N, Chu H, Wang M, Xue Y, Du M, Lu L, Zhang H, Wang F, Fang Y, Li J, et al. Pri-miR-34b/c rs4938723 polymorphism contributes to acute lymphoblastic leukemia susceptibility in Chinese children. Leuk Lymphoma. 2016;57:1436–1441. doi: 10.3109/10428194.2015.1092528. [DOI] [PubMed] [Google Scholar]