Abstract
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors in China and the third leading cause of cancer-associated morality. The aim of the present study was to investigate and analyze differentially-expressed genes (DEGs) between cirrhosis and HCC, in order to screen the key genes involved in the transformation from cirrhosis to HCC and provide novel targets for the diagnosis and treatment of HCC in patients with cirrhosis. The gene expression profile, GSE17548, was obtained from Gene Expression Omnibus database and the DEGs were identified by LIMMA package in R language. Kyoto Encyclopedia of Genes and Genomes and gene ontology biology process analysis were performed for the DEGs. Differential co-expression network (DEN) analysis was conducted and the network was visualized using Cytoscape. Small molecule drugs were also screened from the Comparative Toxicogenomics Database for higher degree DEGs. A total of 95 DEGs were obtained, including 46 upregulated and 49 downregulated genes. The upregulated DEGs were primarily involved in biological processes and pathways associated with the cell cycle, while the downregulated DEGs were primarily involved in immune-associated biological processes. A total of 22 key DEGs were identified by DEN analysis, which distinguished HCC from cirrhosis samples. Furthermore, estradiol, benzo(a)pyrene, acetaminophen, copper sulfate and bisphenol A were identified as the five most associated chemicals to these 22 DEGs. In conclusion, the hub genes and chemicals identified by the present study may provide a theoretical basis for additional research on diagnosis and treatment of HCC transformed from cirrhosis.
Keywords: hepatocellular carcinoma, cirrhosis, differentially-expressed genes, differential co-expression network, cancer-associated chemicals
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors in China and the third leading cause of cancer-associated mortality (1,2). Notably, China accounts for >60% of the global incidence of HCC (3,4). In the past 15 years, the incidence of HCC has increased, and is a serious threat to human health (5).
Risk factors of HCC include hepatitis B and C infections, cirrhosis and alcohol intake (6). As a common chronic and progressive disease with extensive liver parenchymal cells damage (7), cirrhosis is the largest risk factor, which accounts for between 80 and 90% of the total number of HCC cases (8). The pathogenesis from cirrhosis to HCC appears to arise from the development of regenerative nodules with dysplasia of hepatic cells (7). In total, ~1–5% of cirrhosis cases transform to HCC every year (9). The overall survival time of patients with cirrhosis that receive early detection and treatment may be extended to ~5 years (7); however, symptomatic HCC patients may only survive for ~3 months post-diagnosis, and the 1-year survival rate is 44% for these patients (10,11). Therefore, early detection of HCC in patients with cirrhosis is crucial for improving the survival time of patients and preventing the progression of HCC (12,13).
Currently, molecular biomarkers have been developed for diagnostic use in numerous diseases (14–16). For HCC, α-fetoprotein (17), des-carboxyprothrombin (18), insulin-like growth factor (19), osteopontin (20) and glypican-3 (21) have been identified as potential biomarkers. However, none of these markers is capable of distinguishing HCC from cirrhosis. Therefore, the present study aimed to investigate potential biomarkers for HCC in patients with cirrhosis. The present study screened differentially-expressed genes (DEGs) by comparing the expression data of cirrhosis and HCC, and identified key DEGs using co-expression network analysis. In addition, five chemicals most associated with these key DEGs were identified, based on the Comparative Toxicogenomics Database (CTD), to investigate effective treatments for HCC transformed from cirrhosis.
Materials and methods
Affymetrix microarray data
To identify the DEGs between cirrhosis and HCC, the present study obtained the publicly available microarray data GSE17548 (22) from the Gene Expression Omnibus database, which is based on the Affymetrix GPL 571 platform data (Affymetrix Human Genome U133A 2.0 Array; Affymetrix, Thermo Fisher Scientific, Inc., Waltham, MA, USA). A total of 37 chips were available for analysis, including 20 cirrhotic tissue samples and 17 HCC tissue samples.
Identification of DEGs
The original microarray expression data was preprocessed by Affymetrix Bioconductor package in R (www.bioconductor.org/help/workflows/arrays/; v1.30.0) (23), and probe annotation was performed using an annotation file supplied by Affymetrix. Subsequently, LIMMA Bioconductor package in R (bioconductor.org/packages/release/bioc/html/limma.html; v2.8) (24) was used to identify the DEGs. A false discovery rate (FDR) of <0.05 and |log2 fold change|>1 were chosen as the cut-off criteria to select genes that were differentially-expressed in HCC compared with cirrhotic samples.
Functional enrichment analysis of DEGs
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of DEGs were performed using the Database for Annotation, Visualization and Integrated Discovery software (david.ncifcrf.gov/; v6.7) (25). P<0.05 was considered significantly enriched, and the results were visualized with Enrichment Map (26) by Cytoscape software (www.cytoscape.org/; v2.8.3) (27).
Co-expression network construction of DEGs and identification of hub nodes
A differential co-expression network (DEN) was constructed using the following method. The targets that interacted with the encoding proteins of DEGs were selected in the Search Tool for the Retrieval of Interacting Genes/Proteins database (string-db.org/) (28). The protein-protein interaction (PPI) network was constructed with a cut-off criterion of a combined score of >0.6. Subsequently, Spearman's correlation analysis was used to analyze the correlation between the pairs in the PPI network for cirrhotic and HCC tissues, using the cor.test function in R software (www.r-project.org/; v2.15.3). Pairs with a FDR of <0.05 were considered as co-expressed correlations, and the DEN was constructed. Three sub-networks of DEN were constructed, namely C.DEN for unique co-expressed pairs in cirrhosis, H.DEN for unique co-expressed pairs in HCC and S.DEN for co-expressed pairs shared by cirrhosis and HCC. The three DENs were visualized by Cytoscape software. Nodes in the sub-networks of DENs with degrees larger than the average degree of the DEN were considered as hub nodes. Two-way hierarchical clustering dendrograms were generated using the heatmap.2 function of the plots package in R software.
Chemical-disease-inference gene symbol analysis
The CTD (ctdbase.org/downloads/) provided the association between Chemical-Disease-Inference Gene Symbol and the inference scores of chemical-gene interactions (29,30). Chemical-DEG interactions, chemical-cancer interactions and DEG-cancer interactions were searched from this database. Chains with inference scores of ≥21 were extracted to build chemical-cancer-gene interaction pairs.
Results
Identification of DEGs in HCC and cirrhosis
A total of 95 DEGs were identified from the analysis of the gene expression profile of GSE17548, including 46 upregulated and 49 downregulated DEGs in HCC compared with cirrhosis.
Enrichment analysis of DEGs
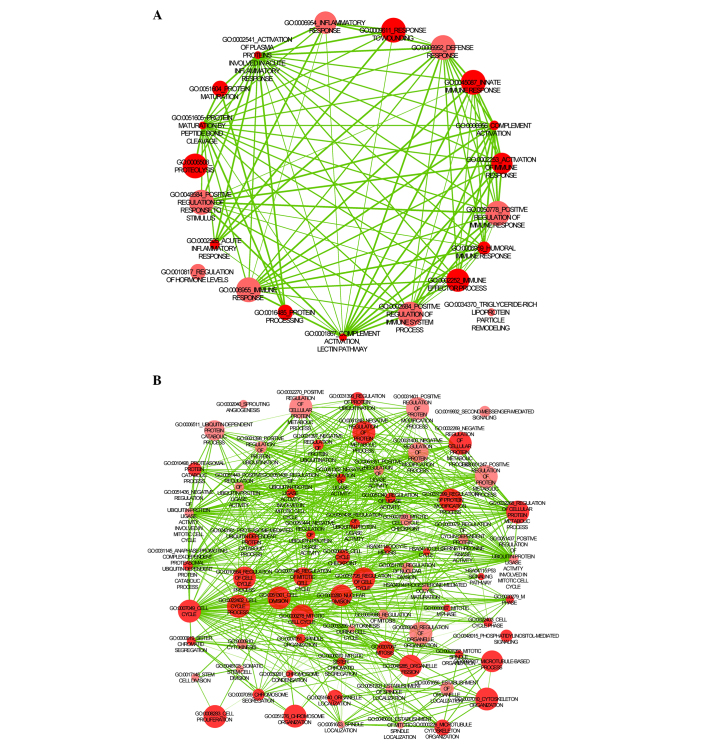
The results of GO BP and KEGG enrichment analysis for the unregulated and downregulated genes are presented in Fig. 1. The upregulated DEGs were primarily enriched in biological processes and pathways associated with the cell cycle and DNA and protein synthesis (Fig. 1A), while the downregulated DEGs were primarily enriched in immune-associated biological processes and pathways, including complement activation and the immune response (Fig. 1B). The top 10 GO BP terms and all the KEGG terms are presented in Table I.
Figure 1.
Functional enrichment of (A) upregulated and (B) downregulated differentially-expressed genes.
Table I.
Enrichment analysis of differentially-expressed genes in hepatocellular carcinoma compared with cirrhosis.
| Term | Associated pathway | Count | P-value |
|---|---|---|---|
| Upregulated genes | |||
| GO:0022403 | Cell cycle | 20 | 1.44×10−18 |
| GO:0007049 | Cell cycle | 24 | 1.48×10−18 |
| GO:0051301 | Cell division | 18 | 3.31×10−18 |
| GO:0000278 | Mitotic cell cycle | 19 | 5.36×10−18 |
| GO:0000279 | M phase of cell cycle | 18 | 2.09×10−17 |
| GO:0022402 | Cell cycle process | 21 | 2.13×10−17 |
| GO:0007067 | Mitosis | 15 | 1.39×10−15 |
| GO:0000280 | Nuclear division | 15 | 1.39×10−15 |
| GO:0000087 | M phase of mitotic cell cycle | 15 | 1.79×10−15 |
| GO:0048285 | Organelle fission | 15 | 2.54×10−15 |
| hsa04110 | Cell cycle | 8 | 6.96×10−9 |
| hsa04115 | p53 signaling pathway | 5 | 1.91×10−5 |
| hsa04114 | Oocyte meiosis | 4 | 2.40×10−3 |
| hsa04914 | Progesterone-mediated oocyte maturation | 3 | 1.96×10−2 |
| Downregulated genes | |||
| GO:0051605 | Protein maturation by peptide bond cleavage | 5 | 6.86×10−5 |
| GO:0001867 | Complement activation, lectin pathway | 3 | 9.69×10−5 |
| GO:0006956 | Complement activation | 4 | 1.70×10−4 |
| GO:0002541 | Activation of plasma proteins in acute inflammatory response | 4 | 1.82×10−4 |
| GO:0016485 | Protein processing | 5 | 1.91×10−4 |
| GO:0051604 | Protein maturation | 5 | 2.66×10−4 |
| GO:0009611 | Response to skin wounds | 8 | 3.52×10−4 |
| GO:0045087 | Innate immune response | 5 | 4.25×10−4 |
| GO:0006959 | Humoral immune response | 4 | 1.10×10−4 |
| GO:0006508 | Proteolysis | 10 | 1.13×10−4 |
Top 10 GO and all Kyoto Encyclopedia of Genes and Genomes terms are presented. GO, gene ontology.
Identification of hub nodes according to DENs
There were 1.469 edges and 127 nodes in the DEN, including 35 upregulated genes, 9 downregulated genes and 83 non-differentially-expressed genes in HCC (Fig. 2A). The three sub-networks of DENs, C.DEN, S.DEN and H.DEN, are presented in Fig. 2B. The edges, nodes and average degrees of the three sub-networks are presented in Table II.
Figure 2.
(A) DEN. (B) Three sub-networks of DEN, C.DEN, S.DEN and H.DEN. Green, red and blue nodes represent high-expressed DEGs, low-expressed DEGs and non-DEGs in HCC, respectively. C.DEN, unique co-expressed pairs in cirrhosis; H.DEN, unique co-expressed pairs in HCC; S.DEN, co-expressed pairs shared by cirrhosis and HCC. DEN, differential expression network; HCC, hepatocellular carcinoma.
Table II.
Parameters of C.DEN, S.DEN and H.DEN.
| Objects | C.DEN | S.DEN | H.DEN |
|---|---|---|---|
| Edges | 868 | 389 | 212 |
| Nodes | 143 | 70 | 90 |
| Average degree | 12.1 | 11.1 | 4.7 |
C.DEN, unique co-expressed pairs in cirrhosis; H.DEN, unique coexpressed pairs in HCC; S.DEN, co-expressed pairs shared by cirrhosis and heptocellular carcinoma. DEN, differential expression network.
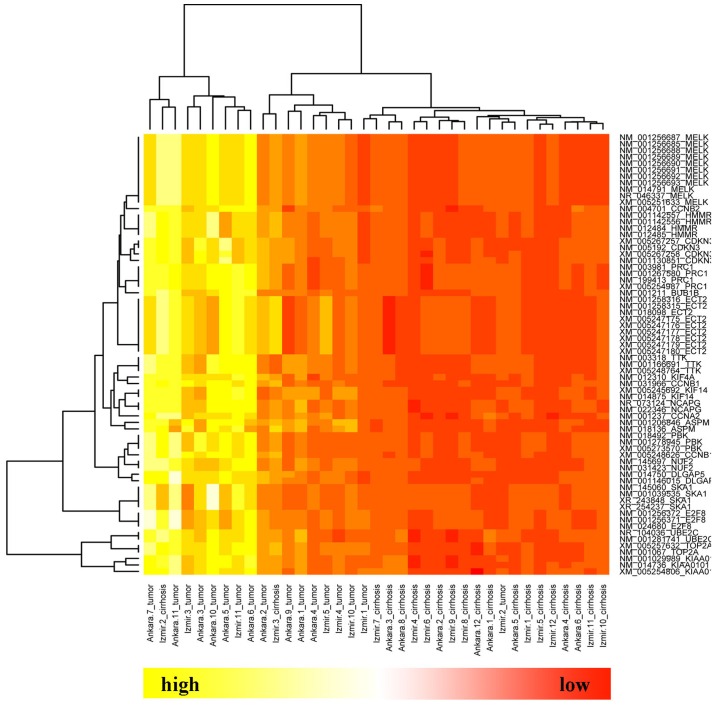
Since the average degree of the nodes in the DEN was 23.1, the DEGs with degrees of >23.1 were extracted from the three sub-networks. As a result, a total of 22 genes were obtained, which were all highly-expressed in HCC (Table III). Two-way hierarchical clustering analysis demonstrated that the expression of these 22 key DEGs distinguished between HCC and cirrhosis samples (Fig. 3).
Table III.
Differentially-expressed genes with high degrees from the three sub-networks of DEN.
| Degree | ||||
|---|---|---|---|---|
| NM | Gene | C.DEN | S.DEN | H.DEN |
| NM_005192 | CDKN3 | 72 | 10 | 1 |
| NM_014875 | KIF14 | 63 | 4 | 2 |
| NM_018136 | ASPM | 56 | 0 | 0 |
| NM_024680 | E2F8 | 51 | 13 | 2 |
| NM_003981 | PRC1 | 44 | 17 | 7 |
| NM_012484 | HMMR | 35 | 9 | 8 |
| NM_145060 | SKA1 | 31 | 0 | 5 |
| NM_018098 | ECT2 | 26 | 2 | 3 |
| NM_003318 | TTK | 51 | 21 | 7 |
| NM_001237 | CCNA2 | 51 | 28 | 5 |
| NM_001211 | BUB1B | 50 | 31 | 3 |
| NM_022346 | NCAPG | 49 | 28 | 8 |
| NM_014750 | DLGAP5 | 48 | 25 | 9 |
| NM_014791 | MELK | 43 | 29 | 7 |
| NM_001281741 | UBE2C | 36 | 32 | 15 |
| NM_145697 | NUF2 | 35 | 33 | 6 |
| NM_012310 | KIF4A | 34 | 27 | 5 |
| NM_031966 | CCNB1 | 31 | 32 | 13 |
| NM_004701 | CCNB2 | 31 | 38 | 6 |
| NM_018492 | PBK | 27 | 29 | 7 |
| NM_001067 | TOP2A | 19 | 32 | 6 |
| NM_014736 | KIAA0101 | 32 | 38 | 32 |
C.DEN, unique co-expressed pairs in cirrhosis; H.DEN, unique co-expressed pairs in HCC; S.DEN, co-expressed pairs shared by cirrhosis and HCC. DEN, differential expression network; NM, number; HCC, hepatocellular carcinoma.
Figure 3.
Hierarchial clustering of 22 key nodes.
Network of the key gene-chemical-cancer interactions
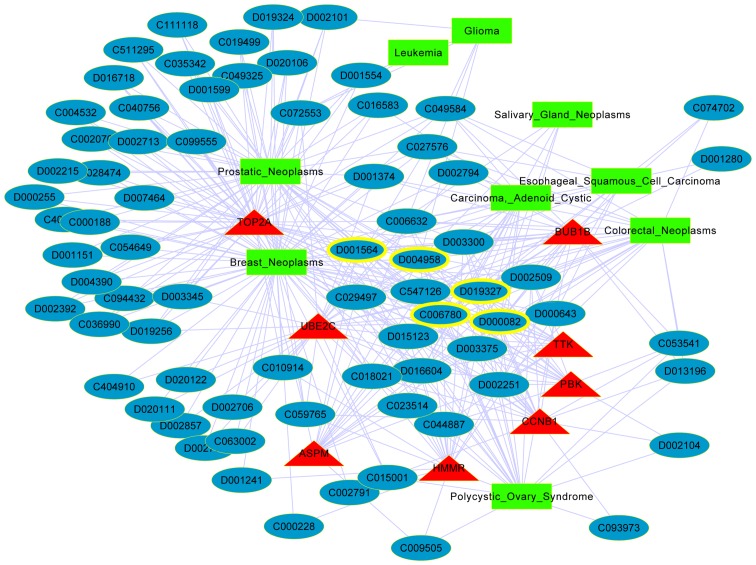
The network of the key gene-chemical-cancer interactions was constructed based on the CTD (Fig. 4). The network included 8 key DEGs, 9 diseases and 74 chemicals. The 8 key DEGs were DNA topoisomerase 2-alpha (TOP2A), budding uninhibited by benzimidazoles 1 homolog beta (BUB1B), ubiquitin-conjugating enzyme E2 C (UBE2C), TTK protein kinase (TTK), PDZ binding kinase (PBK), cyclin B1 (CCNB1), hyaluronan-mediated motility receptor (HMMR) and abnormal spindle-like microcephaly-associated protein (ASPM). The top 5 chemicals with high degrees in the interaction network are presented in Table IV, and are estradiol, benzo(a)pyrene, acetaminophen, copper sulfate and bisphenol A.
Figure 4.
Network of the key gene-chemical-cancer interactions. Red triangle nodes, green square nodes and blue oval nodes represent key genes, cancers and chemical IDs, respectively. Yellow ringed nodes are the chemicals with the highest degrees.
Table IV.
Top 5 chemicals with high degrees associated with differentially-expressed genes.
| Chemical ID | Chemical name | Degree |
|---|---|---|
| D004958 | Estradiol | 17 |
| D001564 | Benzo(a)pyrene | 16 |
| D000082 | Acetaminophen | 16 |
| D019327 | Copper sulfate | 14 |
| C006780 | Bisphenol A | 14 |
Discussion
Currently, numerous studies focus on the difference between molecular biomarkers in patients with liver cancer compared with healthy individuals, while there are limited studies comparing the molecular differences between HCC and cirrhosis (31–33). Therefore, the present study aimed to investigate biomarkers that may distinguish HCC from cirrhosis, using bioinformatics methods.
The present results demonstrated that there were 46 upregulated and 49 downregulated DEGs in HCC compared with cirrhosis. In cancer, the cell cycle, nucleotide replication and protein synthesis are increased, which is consistent with the results of the present functional enrichment analysis. Further analysis of the genes revealed that there were 22 DEGs with high degrees, and 8 of these were reported to be associated with cancer, including TOP2A, BUB1B, UBE2C, TTK, PBK, CCNB1, HMMR and ASPM. Hierarchical clustering analysis indicated that the expression of these 22 key DEGs were capable of distinguishing HCC from cirrhosis samples.
HMMR is a key member of the hyaluronan-mediated motility receptor family, which has been associated with various malignant processes, including cell invasiveness and metastasis in certain tumors (34). The present results suggest that HMMR was upregulated in HCC compared with cirrhosis. The DEN in the present study revealed that HMMR directly interacted with a number of DEGs, including CCNB1, ASPM and UBE2C. In a previous study, the mRNA level of ASPM was significantly increased in HCC compared with healthy samples, and ASPM has been reported as a novel marker for vascular invasion (35). Therefore, the present study additionally confirmed that the upregulated expression of HMMR may promote cell invasiveness and metastasis in HCC transformed from cirrhosis, and an interaction between HMMR and ASPM may be involved. In addition, higher transcript levels of HMMR and CCNB1 have been associated with more advanced systemic progression of prostate cancer (36), and increased levels of CCNB1 promote colorectal carcinogenesis and metastasis (37). Therefore, the increase of these genes may promote cell metastasis in HCC. Additionally, it was reported that a potential tumor suppressor, C2ORF40, inhibited cell invasion and migration by blocking cell cycle progression at the G2/M phase by suppressing the expression of UBE2C (38). Considering the upregulated expression of UBE2C from cirrhosis to HCC identified in the present study, it may be hypothesized that there may be a switch mechanism of HMMR-C2ORF40 in cell invasion and migration. In summary, HMMR and UBE2C may be key genes in the invasion/migration pathogenesis of HCC from cirrhosis. This suggests the theoretical basis for HMMR and UBE2C to be studied as molecular biomarkers for HCC in cirrhosis patients. Furthermore, the present study showed the high expression of TOP2A, BUB1B, TTK and PBK in HCC compared with cirrhosis. TOP2A, which regulates the topological states of DNA, has been shown to be associated with tumor advancement and recurrence in HCC (39). BUB1B, as a key gene in the mitotic spindle checkpoint, is overexpressed and closely linked to cell cycle proliferation in HCC (40). TTK has been reported to participate in the regulation of the DNA damage checkpoint and is also overexpressed in HCC (40). PBK has been demonstrated to play a critical role in an early step of mitosis and the inhibition of tumor growth (41). This data suggests that TOP2A, BUB1B, TTK and PBK may play an important role in promoting the proliferation of malignant tumors by affecting the cell cycle process.
Although there are several medications for the treatment of HCC, including chemotherapy drugs, such as cisplatin (42), and oral drugs, such as fluorouracil (43), these medicines are do not provide enough benefit to patients with HCC and more effective drugs are required. In the present study, 5 medication candidates were selected with high degrees, including estradiol, benzo(a)pyrene, acetaminophen, copper sulfate and bisphenol A. It has been previously confirmed that estradiol treatment inhibits cancer cell migration and invasion to a ceratin degree (44,45). In addition, short-interfering RNA-mediated metastasis associated lung adenocarcinoma transcript 1 (MALAT-1) silencing may impair lung cancer cell metastasis and affect the expression of numerous genes, including HMMR (46). A previous study has identified that 17β-estradiol treatment inhibits breast cell invasion and migration by decreasing the MALAT-1 RNA level (47). Therefore, it is reasonable to hypothesize that estradiol may be used as a treatment for HCC targeted to HMMR, by decreasing the MALAT-1 RNA level. Bisphenol A promotes cell invasion and migration and triggers the transformation of colorectal cancer cells from epithelial to mesenchymal transitions via protein kinase B (AKT)/glycogen synthase kinase-3β-mediated stabilization of Snail (48). Phosphoinositide 3-kinase/AKT phosphorylates mediator complex subunit 1, resulting in UBE2C locus looping (49). Therefore, the present study hypothesizes that the potential mechanism of bisphenol A in the treatment for HCC transformed from cirrhosis may be used as a regulator of cell invasion and migration by AKT-mediated disruption of the expression of UBE2C.
In addition, benzo(a)pyrene is a common environmental and foodborne pollutant, which could promote HCC cell migration and invasion by the nuclear factor-κB pathway (50). Acetaminophen is the most widely used analgesic; however, it could cause severe hepatic necrosis, thereby leading to acute liver failure (51). Excess copper sulfate has been considered as a potent oxidant and causes the generation of reactive oxygen species, which could accelerate the cellular damage induced by oxidative stress in cancer (52). All these studies suggested that novel drugs could be designed against benzo(a)pyrene exposure, acetaminophen and copper sulfate for the treatment of HCC patients.
In conclusion, the present study identified 22 key genes in the transformation from cirrhosis to HCC using bioinformatics analysis of microarray data, and these results have the potential to aid in the diagnosis and development of biomarkers for HCC in cirrhosis patients. The identified chemicals, estradiol, benzo(a)pyrene, acetaminophen, copper sulfate and bisphenol A, should be additionally studied for HCC treatment. Overall, the present results provides novel targets for HCC; however, the molecular mechanisms underlying the progression from cirrhosis to HCC require additional investigation, since liver cancerization is a complex process. Additional investigation is particularly required, since there is a lack of effective therapies for this disease.
Acknowledgements
The present study was supported by the Liaoning Province Technology Project (Lianoning, China; grant no., 2013225021).
References
- 1.Gandhi S, Khubchandani S, Iyer R. Quality of life and hepatocellular carcinoma. J Gastrointest Oncol. 2014;5:296–317. doi: 10.3978/j.issn.2078-6891.2014.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mise Y, Satou S, Ishizawa T, Kaneko J, Aoki T, Hasegawa K, Sugawara Y, Makuuchi M, Kokudo N. Impact of surgery on quality of life in patients with hepatocellular carcinoma. World J Surg. 2014;38:958–967. doi: 10.1007/s00268-013-2342-9. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 4.Bertino G, Demma S, Ardiri A, Proiti M, Gruttadauria S, Toro A, Malaguarnera G, Bertino N, Malaguarnera M, Malaguarnera M, Di Carlo I. Hepatocellular carcinoma: Novel molecular targets in carcinogenesis for future therapies. Biomed Res Int. 2014;2014:203693. doi: 10.1155/2014/203693. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Tanaka M, Katayama F, Kato H, Tanaka H, Wang J, Qiao YL, Inoue M. Hepatitis B and C virus infection and hepatocellular carcinoma in China: A review of epidemiology and control measures. J Epidemiol. 2011;21:401–416. doi: 10.2188/jea.JE20100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sherman M. Hepatocellular carcinoma: Epidemiology, risk factors, and screening. Semin Liver Dis. 2005;25:143–154. doi: 10.1055/s-2005-871194. [DOI] [PubMed] [Google Scholar]
- 7.Schuppan D, Afdhal NH. Liver cirrhosis. Lancet. 2008;371:838–851. doi: 10.1016/S0140-6736(08)60383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nordenstedt H, White DL, El-Serag HB. The changing pattern of epidemiology in hepatocellular carcinoma. Dig Liver Dis. 2010;42(Suppl 3):S206–S214. doi: 10.1016/S1590-8658(10)60507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu CJ, Kao JH. Hepatitis B virus-related hepatocellular carcinoma: Epidemiology and pathogenic role of viral factors. J Chin Med Assoc. 2007;70:141–145. doi: 10.1016/S1726-4901(09)70346-6. [DOI] [PubMed] [Google Scholar]
- 10.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 11.Wörns MA, Weinmann A, Schuchmann M, Galle PR. Systemic therapies in hepatocellular carcinoma. Dig Dis. 2009;27:175–188. doi: 10.1159/000218351. [DOI] [PubMed] [Google Scholar]
- 12.Walther Z, Jain D. Molecular pathology of hepatic neoplasms: Classification and clinical significance. Patholog Res Int. 2011;2011:403929. doi: 10.4061/2011/403929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stravitz RT, Heuman DM, Chand N, et al. Surveillance for hepatocellular carcinoma in patients with cirrhosis improves outcome. Am J Med. 2008;121:119–126. doi: 10.1016/j.amjmed.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 14.Pinheiro Ddo R, Ferreira WA, Barros MB, et al. Perspectives on new biomarkers in gastric cancer: Diagnostic and prognostic applications. World J Gastroenterol. 2014;20:11574–11585. doi: 10.3748/wjg.v20.i33.11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rouzier R, Pronzato P, Chéreau E, Carlson J, Hunt B, Valentine WJ. Multigene assays and molecular markers in breast cancer: Systematic review of health economic analyses. Breast Cancer Res Treat. 2013;139:621–637. doi: 10.1007/s10549-013-2559-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang AY, Wang M. Molecular mechanisms of action and potential biomarkers of growth inhibition of dasatinib (BMS-354825) on hepatocellular carcinoma cells. BMC Cancer. 2013;13:267. doi: 10.1186/1471-2407-13-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim BK, Ahn SH, Seong JS, Park JY, Kim Do Y, Kim JK, Lee Do Y, Lee KH, Han KH. Early α-fetoprotein response as a predictor for clinical outcome after localized concurrent chemoradiotherapy for advanced hepatocellular carcinoma. Liver Int. 2011;31:369–376. doi: 10.1111/j.1478-3231.2010.02368.x. [DOI] [PubMed] [Google Scholar]
- 18.Song P, Feng X, Zhang K, et al. Perspectives on using des-γ-carboxyprothrombin (DCP) as a serum biomarker: Facilitating early detection of hepatocellular carcinoma in China. Hepatobiliary Surg Nutr. 2013;2:227–231. doi: 10.3978/j.issn.2304-3881.2013.08.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Q, Mao YQ, Jiang WD, Chen YR, Huang RY, Zhou XB, Wang YF, Shi Z, Wang ZS, Huang RP. Development of IGF signaling antibody arrays for the identification of hepatocellular carcinoma biomarkers. PLoS One. 2012;7:e46851. doi: 10.1371/journal.pone.0046851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Li HY, Yin XL, Xu YP, Chen Y, Xie XY, Shen YF, Ye QH, Ren ZG. Relative analysis of OPN and its related signal molecules in hepatocellular carcinoma. Zhonghua Gan Zang Bing Za Zhi. 2011;19:48–51. doi: 10.3760/cma.j.issn.1007-3418.2011.01.013. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 21.Zou ZQ, Ding YP, Long B, Yuh JG, Xu AL, Lang ZW, Zou SY, Liu YD, Ding K, Li YY. Gpc-3 is a notable diagnostic, prognostic and a latent targeted therapy marker in hepatocellular carcinoma. Hepatogastroenterology. 2010;57:1285–1290. [PubMed] [Google Scholar]
- 22.Yildiz G, Arslan-Ergul A, Bagislar S, Konu O, Yuzugullu H, Gursoy-Yuzugullu O, Ozturk N, Ozen C, Ozdag H, Erdal E, et al. Genome-wide transcriptional reorganization associated with senescence-to-immortality switch during human hepatocellular carcinogenesis. PLoS One. 2013;8:e64016. doi: 10.1371/journal.pone.0064016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 24.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 25.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 26.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: A network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database Issue):D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis AP, Murphy CG, Johnson R, Lay JM, Lennon-Hopkins K, Saraceni-Richards C, Sciaky D, King BL, Rosenstein MC, Wiegers TC, Mattingly CJ. The Comparative Toxicogenomics Database: Update 2013. Nucleic Acids Res. 2013;41(Database Issue):D1104–D1114. doi: 10.1093/nar/gks994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis AP, Murphy CG, Saraceni-Richards CA, Rosenstein MC, Wiegers TC, Mattingly CJ. Comparative Toxicogenomics Database: A knowledgebase and discovery tool for chemical-gene-disease networks. Nucleic Acids Res. 2009;37(Database Issue):D786–D792. doi: 10.1093/nar/gkn580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gehrmann M, Cervello M, Montalto G, Cappello F, Gulino A, Knape C, Specht HM, Multhoff G. Heat shock protein 70 serum levels differ significantly in patients with chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Front Immunol. 2014;5:307. doi: 10.3389/fimmu.2014.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sterling RK, Wright EC, Morgan TR, et al. Frequency of elevated hepatocellular carcinoma (HCC) biomarkers in patients with advanced hepatitis C. Am J Gastroenterol. 2012;107:64–74. doi: 10.1038/ajg.2011.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nahon P, Amathieu R, Triba MN, Bouchemal N, Nault JC, Ziol M, Seror O, Dhonneur G, Trinchet JC, Beaugrand M, Le Moyec L. Identification of serum proton NMR metabolomic fingerprints associated with hepatocellular carcinoma in patients with alcoholic cirrhosis. Clin Cancer Res. 2012;18:6714–6722. doi: 10.1158/1078-0432.CCR-12-1099. [DOI] [PubMed] [Google Scholar]
- 34.Villegas-Ruíz V, Salcedo M, Zentella-Dehesa A, de Oca EV, Román-Basaure E, Mantilla-Morales A, Dávila-Borja VM, Juárez-Méndez S. A case of cervical cancer expressed three mRNA variant of Hyaluronan-mediated motility receptor. Int J Clin Exp Pathol. 2014;7:2256–2264. [PMC free article] [PubMed] [Google Scholar]
- 35.Lin SY, Pan HW, Liu SH, Jeng YM, Hu FC, Peng SY, Lai PL, Hsu HC. ASPM is a novel marker for vascular invasion, early recurrence, and poor prognosis of hepatocellular carcinoma. Clin Cancer Res. 2008;14:4814–4820. doi: 10.1158/1078-0432.CCR-07-5262. [DOI] [PubMed] [Google Scholar]
- 36.Gomez CR, Kosari F, Munz JM, Schreiber CA, Knutson GJ, Ida CM, El Khattouti A, Karnes RJ, Cheville JC, Vasmatzis G, Vuk-Pavlović S. Prognostic value of discs large homolog 7 transcript levels in prostate cancer. PLoS One. 2013;8:e82833. doi: 10.1371/journal.pone.0082833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li JQ, Kubo A, Wu F, Usuki H, Fujita J, Bandoh S, Masaki T, Saoo K, Takeuchi H, Kobayashi S, et al. Cyclin B1, unlike cyclin G1, increases significantly during colorectal carcinogenesis and during later metastasis to lymph nodes. Int J Oncol. 2003;22:1101–1110. [PubMed] [Google Scholar]
- 38.Lu J, Wen M, Huang Y, He X, Wang Y, Wu Q, Li Z, Castellanos-Martin A, Abad M, Cruz-Hernandez JJ, et al. C2ORF40 suppresses breast cancer cell proliferation and invasion through modulating expression of M phase cell cycle genes. Epigenetics. 2013;8:571–583. doi: 10.4161/epi.24626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong N, Yeo W, Wong WL, Wong NL, Chan KY, Mo FK, Koh J, Chan SL, Chan AT, Lai PB, et al. TOP2A overexpression in hepatocellular carcinoma correlates with early age onset, shorter patients survival and chemoresistance. Int J Cancer. 2009;124:644–652. doi: 10.1002/ijc.23968. [DOI] [PubMed] [Google Scholar]
- 40.Wang L, Sun L, Huang J, Jiang M. Cyclin-dependent kinase inhibitor 3 (CDKN3) novel cell cycle computational network between human non-malignancy associated hepatitis/cirrhosis and hepatocellular carcinoma (HCC) transformation. Cell Prolif. 2011;44:291–299. doi: 10.1111/j.1365-2184.2011.00752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nandi AK, Rapoport AP. Expression of PDZ-binding kinase (PBK) is regulated by cell cycle-specific transcription factors E2F and CREB/ATF. Leuk Res. 2006;30:437–447. doi: 10.1016/j.leukres.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 42.Chen X, Hou Y, Duan L, Tang M, Kang Q, Shu J, Peng Z, Li S. Inhibitory effect of 131I-CD133mAb combined with cisplatin on liver cancer cells in vitro and in a tumor-bearing mouse model. Nan Fang Yi Ke Da Xue Xue Bao. 2014;34:934–938. [PubMed] [Google Scholar]
- 43.Gu W, Fang FF, Li B, Cheng BB, Ling CQ. Characterization and resistance mechanisms of a 5-fluorouracil-resistant hepatocellular carcinoma cell line. Asian Pac J Cancer Prev. 2012;13:4807–4814. doi: 10.7314/APJCP.2012.13.9.4807. [DOI] [PubMed] [Google Scholar]
- 44.Piccolella M, Crippa V, Messi E, Tetel MJ, Poletti A. Modulators of estrogen receptor inhibit proliferation and migration of prostate cancer cells. Pharmacol Res. 2014;79:13–20. doi: 10.1016/j.phrs.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 45.Giretti MS, Montt Guevara MM, Cecchi E, Mannella P, Palla G, Spina S, Bernacchi G, Di Bello S, Genazzani AR, Genazzani AD, Simoncini T. Effects of estetrol on migration and invasion in T47-D breast cancer cells through the actin cytoskeleton. Front Endocrinol (Lausanne) 2014;5:80. doi: 10.3389/fendo.2014.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, Ijiri K, Akimitsu N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584:4575–4580. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 47.Zhao Z, Chen C, Liu Y, Wu C. 17β-Estradiol treatment inhibits breast cell proliferation, migration and invasion by decreasing MALAT-1 RNA level. Biochem Biophys Res Commun. 2014;445:388–393. doi: 10.1016/j.bbrc.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 48.Chen ZJ, Yang XL, Liu H, Wei W, Zhang KS, Huang HB, Giesy JP, Liu HL, Du J, Wang HS. Bisphenol A modulates colorectal cancer protein profile and promotes the metastasis via induction of epithelial to mesenchymal transitions. Arch Toxicol. 2015;89:1371–1381. doi: 10.1007/s00204-014-1301-z. [DOI] [PubMed] [Google Scholar]
- 49.Chen Z, Zhang C, Wu D, Chen H, Rorick A, Zhang X, Wang Q. Phospho-MED1-enhanced UBE2C locus looping drives castration-resistant prostate cancer growth. EMBO J. 2011;30:2405–2419. doi: 10.1038/emboj.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ba Q, Li J, Huang C, Qiu H, Li J, Chu R, Zhang W, Xie D, Wu Y, Wang H. Effects of benzo[a]pyrene exposure on human hepatocellular carcinoma cell angiogenesis, metastasis, and NF-κB signaling. Environ Health Perspect. 2015;123:246–254. doi: 10.1289/ehp.1408524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Larson AM, Polson J, Fontana RJ, Davern TJ, Lalani E, Hynan LS, Reisch JS, Schiødt FV, Ostapowicz G, Shakil AO, et al. Acetaminophen-induced acute liver failure: Results of a United States multicenter, prospective study. Hepatology. 2005;42:1364–1372. doi: 10.1002/hep.20948. [DOI] [PubMed] [Google Scholar]
- 52.Gupte A, Mumper RJ. Elevated copper and oxidative stress in cancer cells as a target for cancer treatment. Cancer Treat Rev. 2009;35:32–46. doi: 10.1016/j.ctrv.2008.07.004. [DOI] [PubMed] [Google Scholar]