Abstract
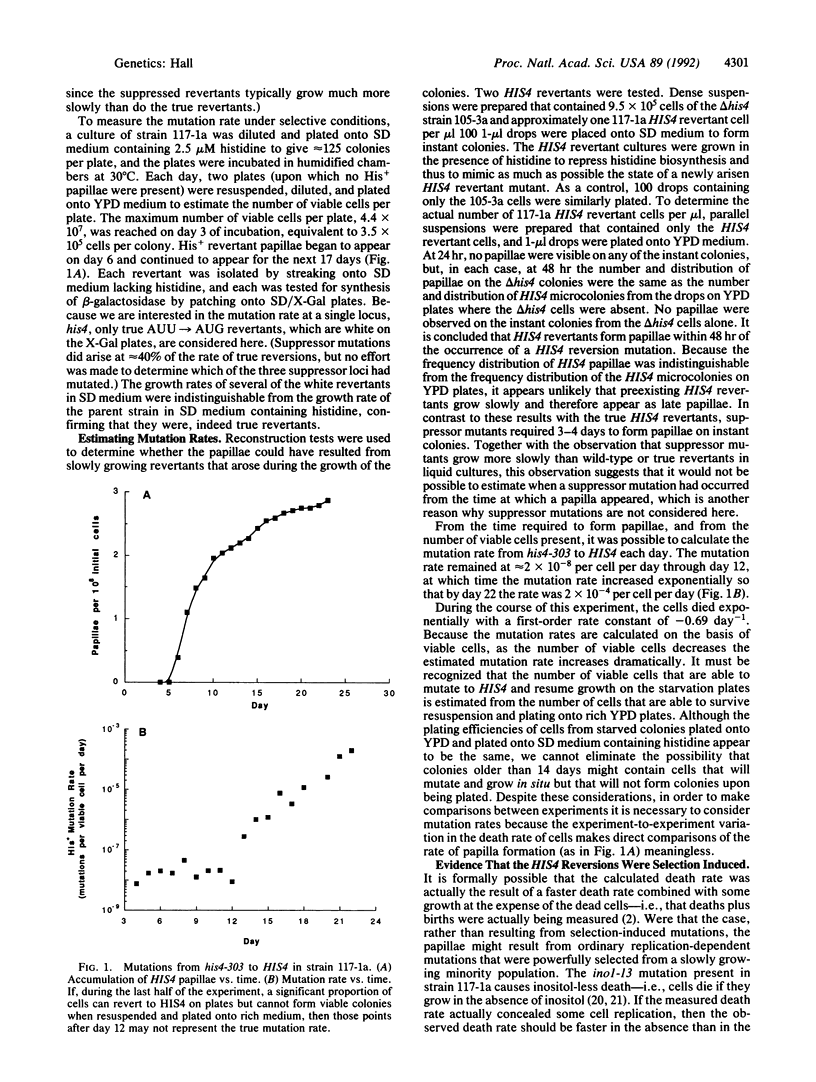
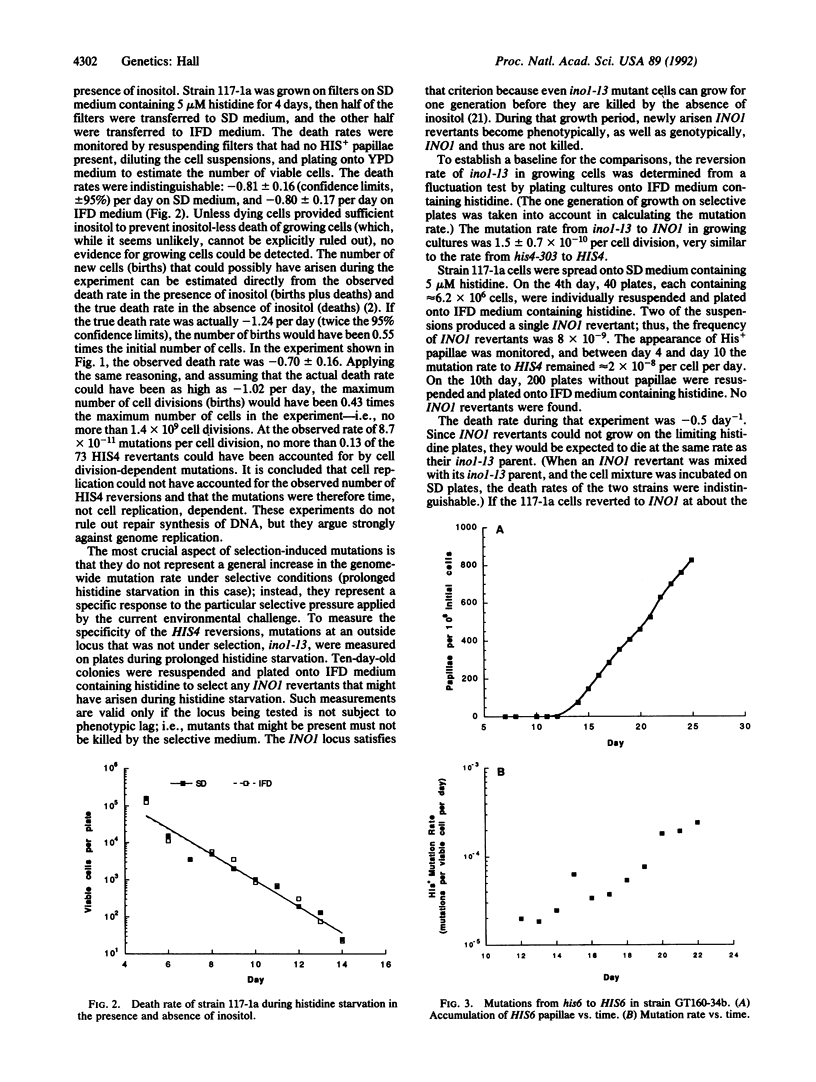
Selection-induced mutations are nonrandom mutations that occur as specific and direct responses to environmental challenges and primarily in nondividing cells under conditions of intense prolonged selection. Selection-induced mutations have been shown to occur at six loci in Escherichia coli, but their existence has not previously been demonstrated in any eukaryotic organism. Here it is shown that selection-induced mutations occur at the HIS4 locus in the eukaryotic microorganism Saccharomyces cerevisiae.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cairns J., Foster P. L. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics. 1991 Aug;128(4):695–701. doi: 10.1093/genetics/128.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J., Overbaugh J., Miller S. The origin of mutants. Nature. 1988 Sep 8;335(6186):142–145. doi: 10.1038/335142a0. [DOI] [PubMed] [Google Scholar]
- Campbell D., Doctor J. S., Feuersanger J. H., Doolittle M. M. Differential mitotic stability of yeast disomes derived from triploid meiosis. Genetics. 1981 Jun;98(2):239–255. doi: 10.1093/genetics/98.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho-Valavicius B., Yoon H., Donahue T. F. Genetic characterization of the Saccharomyces cerevisiae translational initiation suppressors sui1, sui2 and SUI3 and their effects on HIS4 expression. Genetics. 1990 Mar;124(3):483–495. doi: 10.1093/genetics/124.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson M. R., Henry S. A. Inositol-requiring mutants of Saccharomyces cerevisiae. Genetics. 1975 May;80(1):23–40. doi: 10.1093/genetics/80.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B. D. Transcriptional bias: a non-Lamarckian mechanism for substrate-induced mutations. Proc Natl Acad Sci U S A. 1989 Jul;86(13):5005–5009. doi: 10.1073/pnas.86.13.5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Cigan A. M. Genetic selection for mutations that reduce or abolish ribosomal recognition of the HIS4 translational initiator region. Mol Cell Biol. 1988 Jul;8(7):2955–2963. doi: 10.1128/mcb.8.7.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. G. Adaptive evolution that requires multiple spontaneous mutations. I. Mutations involving an insertion sequence. Genetics. 1988 Dec;120(4):887–897. doi: 10.1093/genetics/120.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. G. Adaptive evolution that requires multiple spontaneous mutations: mutations involving base substitutions. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5882–5886. doi: 10.1073/pnas.88.13.5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. G. Directed evolution of a bacterial operon. Bioessays. 1990 Nov;12(11):551–558. doi: 10.1002/bies.950121109. [DOI] [PubMed] [Google Scholar]
- Hall B. G. Spectrum of mutations that occur under selective and non-selective conditions in E. coli. Genetica. 1991;84(2):73–76. doi: 10.1007/BF00116545. [DOI] [PubMed] [Google Scholar]
- Hall B. G. Spontaneous point mutations that occur more often when advantageous than when neutral. Genetics. 1990 Sep;126(1):5–16. doi: 10.1093/genetics/126.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry S. A., Atkinson K. D., Kolat A. I., Culbertson M. R. Growth and metabolism of inositol-starved Saccharomyces cerevisiae. J Bacteriol. 1977 Apr;130(1):472–484. doi: 10.1128/jb.130.1.472-484.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. W. Classical mutagenesis techniques. Methods Enzymol. 1991;194:273–281. doi: 10.1016/0076-6879(91)94021-4. [DOI] [PubMed] [Google Scholar]
- Lenski R. E., Slatkin M., Ayala F. J. Another alternative to directed mutation. Nature. 1989 Jan 12;337(6203):123–124. doi: 10.1038/337123b0. [DOI] [PubMed] [Google Scholar]
- Lenski R. E., Slatkin M., Ayala F. J. Mutation and selection in bacterial populations: alternatives to the hypothesis of directed mutation. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2775–2778. doi: 10.1073/pnas.86.8.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria S. E., Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943 Nov;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Origin of mutants disputed. Nature. 1988 Dec 8;336(6199):525–528. doi: 10.1038/336525a0. [DOI] [PubMed] [Google Scholar]
- Ryan F. J. Spontaneous Mutation in Non-Dividing Bacteria. Genetics. 1955 Sep;40(5):726–738. doi: 10.1093/genetics/40.5.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Stahl F. W. Bacterial genetics. A unicorn in the garden. Nature. 1988 Sep 8;335(6186):112–113. doi: 10.1038/335112a0. [DOI] [PubMed] [Google Scholar]
- Symonds N. Evolution. Anticipatory mutagenesis. Nature. 1989 Jan 12;337(6203):119–120. doi: 10.1038/337119a0. [DOI] [PubMed] [Google Scholar]



