Abstract
The anti-malarial agent cladosporin is a nanomolar inhibitor of Plasmodium falciparum lysyl-tRNA synthetase, and exhibits activity against both blood and liver stage infection. Cladosporin can be isolated from the fungus Cladosporium cladosporioides, where it was believed to be biosynthesized by a highly reducing (HR) and non-reducing (NR) iterative type I polyketide synthase (PKS) pair. Genome sequencing of the host organism, and subsequent heterologous expression of these enzymes in Saccharomyces cerevisiae produced cladosporin, confirming the identity of the putative gene cluster. Incorporation of a pentaketide intermediate analog indicated a 5+3 assembly by the HR PKS Cla2 and the NR PKS Cla3 during cladosporin biosynthesis. A putative lysyl-tRNA synthetase resistance gene was also identified in the cladosporin gene cluster. Analysis of the active site emphasizes key structural features thought to be important in resistance to cladosporin.
Graphical Abstract
Cladosporin (1) (also known as asperentin) is a tricyclic octaketide that is produced by several fungal species including Cladosporium,1–4 Chaetomium,5 Penicillium,6 Eurotium,7,8 and Aspergillus.9 Cladosporin exhibits some interesting bioactivity, including antifungal, antibiotic, and plant growth inhibitory properties, as well as anti-inflammatory effects in mouse lung tissue.10 Most recently, cladosporin has been shown to be a potent, nanomolar, inhibitor of Plasmodium falciparum blood- and liver-stage proliferation.11 Although many antimalarial agents currently exist, endoperoxides represent the only class of molecules for which resistance has not significantly developed, and even these do not inhibit the asymptotic liver stage infection. The discovery of the bioactivity of cladosporin represents a promising lead for treatment of malaria, and several studies on the topic have been published since then.12,13
Previously in our group, we investigated the biosynthesis of several related fungal polyketides that belong to the resorcylic acid lactone (RAL)- and dihydroxyphenyl acetic acid lactone (DAL)-containing polyketides, including hypothemycin (RAL type),14 radicicol (RAL type),15 and dehydrocurvularin (DAL type).16–19 Biosynthesis of these polyketides requires cooperative action of two iterative type I polyketide synthases (PKSs): a highly reducing (HR) PKS and a non-reducing (NR) PKS. Based on structural similarities, we hypothesized that cladosporin is also biosynthesized by a HR and NR PKS. Early work in our group assigned the absolute stereochemistry of cladosporin.2,3 More recently, the total synthesis of cladosporin and its diastereomer isocladosporin, has been reported.20,21 To better understand PKS assembly and enable analog production via synthetic biology; we sought to heterologously express, and reconstitute cladosporin expression in Saccharomyces cerevisiae. To this end, we sequenced the genome of the producer organism Cladosporium cladosporioides UAMH 5063. This resulted in 30Mb of genomic information over a total of 764 contigs. The genomic data was annotated using Antibiotics & Secondary Metabolite Analysis Shell (antiSMASH v2.0).22 The software identified 50 putative secondary metabolite gene clusters in the genome of C. cladosporioides, seven of which encode type I iterative PKSs. One gene cluster in particular possessed high sequence homology to those of hypothemycin and zearalenone. Spliced gene sequences contained within this gene cluster were identified using Hidden Markov model (HMM)-based software FGENESH (Softberry),23 and the resulting intron-less sequences analyzed individually using BLAST (NCBI), Figure 1.
Figure 1.
Cladosporin gene cluster in Cladosporium cladosporioides and putative biosynthesis by the HR PKS Cla3 and NR PKS Cla2 contained therein. KS, ketosynthase; MAT, malonyl-CoA:ACP acyltransferase; DH, dehydratase; ψMT, pseudo C-methyltransferase; ψKR, structural ketoreductase; ER, enoylreductase; KRc catalytic ketoreductase; ACP, acyl carrier protein; SAT starter unit:ACP transacylase; PT, product template; TE, thioesterase.
The HR and NR PKS contained within the gene cluster, Cla2 and Cla3 respectively, were cloned and expressed in S. cerevisiae BJ5464-NpgA (Supporting Information S2 and S3). The proteins were successfully expressed from single transformants, and with minimal optimization, cladosporin was isolated from double transformants at a titer of 10 mg/L after RP-HPLC purification (Supporting Information Figure S1). Identity of cladosporin was confirmed by LC-ESI-MS (Supporting Information Figure S2), using combined retention time matching with accurate mass matching, and NMR analysis (Supporting Information Figure S3 and S4). This confirmed our identification of the cladosporin gene cluster in C. cladosporioides. Given that S. cerevisiae is an extremely well-studied organism for heterologous production of other natural products such as artemisinic acid24 and lovastatin,25 this result constituents a significant step toward large scale production of cladosporin.
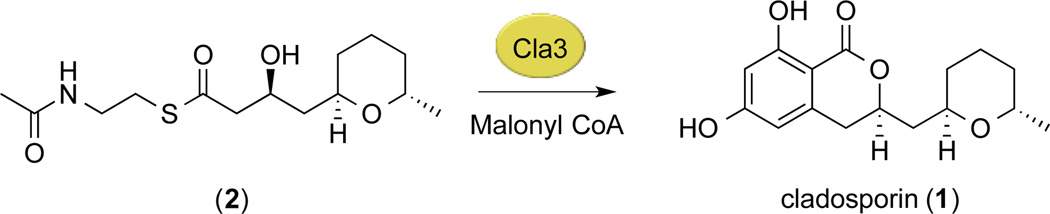
To further probe the biosynthesis of cladosporin in vivo, we conducted advanced precursor feeding studies (see Supporting Information). The necessary reduction of the C3 position of cladosporin led us to hypothesize that the HR PKS, Cla2, is responsible for biosynthesis up to the pentaketide stage, inclusive of the THP ring (Supporting Information Figure S5),3 whereas the three subsequent ketide extensions with no reduction are catalyzed by Cla3. In accord with observations on other NR PKSs that require an HR PKS partner,14 Cla3 is not able to load and produce product on its own from malonyl CoA. Hence, the proposed pentaketide intermediate was synthesized as a N-acetylcysteamine (SNAC) thioester (2), Scheme 1 (see Supporting Information), and fed to purified Cla3, along with malonyl-CoA. After 24-hour incubation at room temperature, metabolites were extracted and analyzed by LC-ESI-MS (Supporting Information Figure S6). Identification of cladosporin (1), using combined retention time matching with accurate mass matching, confirmed that the pentaketide intermediate analog (2) is recognized by Cla3. It is likely the final product of the HR PKS Cla2, which remains covalently bound as a thioester until transfer to the SAT domain of the NR PKS Cla3. This “5+3” ketide assembly of cladosporin represents the first example of its type, with other DAL- and RAL-type polyketides assembled in a “4+4” (dehydrocurvularin)17–19 “6+3” (hypothemycin, zearalenone)14,26 and “5+4” (radicicol)15 fashion. Thus THP ring formation appears to be catalysed by Cla2, where oxa-Michael type cyclization on an unsaturated thioester intermediate at the tetraketide stage would be highly favoured,3 Figure 1.
Scheme 1.
In vitro assay with Cla3. Cladosporin can be synthesized from pentaketide-SNAC thioester (2) and malonyl CoA.
Encouraged by these results, we conducted further feeding studies with Cla3 (Table1, Supporting Information Figures S7 and S8). Surprisingly, Cla3 accepts several unnatural substrate analogs that do not contain the tetrahydropyran ring. The presence of a hydroxyl group is vital but its stereochemical configuration is not important for recognition. However, carbon chains longer than ten carbons are not accepted, suggesting a hydrophobic binding pocket of limited size in the enzyme. Furthermore, the promiscuity of Cla3 could allow for the semi-synthesis of new antimalarial analogs.
Table 1.
Incorporation of unnatural analogs by Cla3.
We then undertook homology modelling of the product template (PT) domain using I-TASSER,27–29 and subsequent docking studies using AutoDock Vina,30 in an effort to better understand how the THP ring of the natural substrate fits in the active site, Figure 2.
Figure 2.
Active site cavity of Cla3 PT with docked thioacid analogs. Residues analogous to Leu1585, Tyr1458 and catalytic His1311, are conserved among all RAL-type PKSs. (A) Pentaketide analog relatively linear in the active site (B) Hexaketide starting to form a bent conformation (C) Constricted heptaketide analog forms a loop structure (D) Uncyclized octaketide thioacid analog. C8a is now only 4Å away from C4a.
The most suitable homology that was used by I-TASSER as the top threading template was the crystal structure of the PT domain from PksA, the NR PKS responsible for biosynthesis of aflatoxin B1 in Aspergillus parasiticus (PDB ID 3HRQ).31 Our homolgy model indicates that the THP ring can be readily accommodated in the PT active site. Furthermore, as the ketide is extended in the PT domain, it appears to curve around on itself, up until the octaketide stage where C8a is now in close enough proximity to C4a to cyclize. The resulting aromatized intermediate could then be transferred to the TE domain, where hydrolysis and/or lactone formation could occur to produce cladosporin (1).
Cladosporin’s proposed antimalarial mode-of-action is inhibition of the P. falciparum lysyl t-RNA synthetase (KRS1).11,12,32 Interestingly, a putative lysyl-tRNA synthetase gene, cla4, is contained within the cladosporin gene cluster. Resistance genes can often be found close to the biosynthetic machinery of natural products. It is possible that Cla4 may be infer cladosporin resistance. Hoephner et al. found that S. cerevisiae lysyl-tRNA synthetase KRS1 is not inhibited by cladosporin, and that this resistance is related to the two key active site residues, Gln324 and Thr340. Replacement of Gln324 with a hydrophobic valine lead to a 5.7-fold increase in cladosporin sensitivity, whereas replacement of Thr340 with a less bulky serine increased sensitivity 10.4-fold. The corresponding double mutant was 38.7-fold more sensitive to cladosporin. Therefore, it appears that a prerequisite of cladosporin resistance is the presence of a polar group at position 324 and a bulky group at position 340, a requirement that is met by the analogous residues in Cla4. This is further supported by the co-crystal structure of P. falciparum KRS1 and cladosporin, wherein the isocoumarin moiety of cladosporin occupies a similar orientation as the adenine of ATP, with its aromatic ring in a hydrophobic interaction with Val328 (Supporting Information Figure S9). Additionally, the THP ring of cladosporin is located directly adjacent to Ser344, where any increase in steric bulk would clash with the methyl substituent on the THP ring. The C. cladosporioides genome contains 3 putative lysyl t-RNA synthetases, Cla4, Lys2 and Lys3. Neither Lys2 nor Lys3 contain the Gln-Thr pair that appear necessary for cladosporin resistance, in contrast to Cla4.
We suggest that Cla4 may not be inhibited by cladosporin, thereby imparting cladosporin resistance to C. cladosporioides. It is likely that cla4 is under the control of the same regulation as cla2 and cla3, and when cladosporin biosynthesis is switched on, transcription of cla4 will then be necessary for continued protein synthesis in C. cladosporioides.
In this work, we present identification, expression and demonstration of functional activity of the HR and NR PKS responsible for cladosporin production in Cladosporium cladosporioides. We have also identified a putative lysyl-tRNA synthetase encoded in the cladosporin gene cluster. This likely indicates a probable resistance mechanism in Cladosporium cladosporioides.
Supplementary Material
Acknowledgments
Funding Sources
These investigations were supported by the Natural Sciences & Engineering Research Council of Canada (NSERC), by the Canada Research Chair in Bioorganic & Medicinal Chemistry. Research in YT lab is supported by the US NIH (1R01GM085128 and 1DP1GM106413).
We would like to thank Mr Jonathan Cartmell for his assistance in using AutoDock Vina.
Footnotes
ASSOCIATED CONTENT
Cloning, protein expression, cladosporin characterization, and chemical synthesis are supplied in the Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org. Sequence data for the cladosporin gene cluster can be accessed using GenBank® Accession numbers KT037691-KT037693 and KT250989-KT250991.
REFERENCES
- 1.Scott PM, Van Walbeek W, MacLean WM. J. Antibiot. 1971;24:747. doi: 10.7164/antibiotics.24.747. [DOI] [PubMed] [Google Scholar]
- 2.Reese PB, Rawlings BJ, Ramer SE, Vederas JC. J. Am. Chem. Soc. 1988;110:316. [Google Scholar]
- 3.Rawlings BJ, Reese PB, Ramer SE, Vederas JC. J. Am. Chem. Soc. 1989;111:3382. [Google Scholar]
- 4.Wang X, Radwan MM, Tarawneh AH, Gao J, Wedge DE, Rosa LH, Cutler HG, Cutler SJ. J. Agric. Food Chem. 2013;61:4551. doi: 10.1021/jf400212y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang S, Li XM, Teuscher F, Li DL, Diesel A, Ebel R, Proksch P, Wang BG. J. Nat. Prod. 2006;69:1622. doi: 10.1021/np060248n. [DOI] [PubMed] [Google Scholar]
- 6.Flewelling AJ, Johnson JA, Gray CA. Nat. Prod. Commun. 2013;8:373. [PubMed] [Google Scholar]
- 7.Anke H, Zahner H. Arch. Microbiol. 1978;116:253. doi: 10.1007/BF00417848. [DOI] [PubMed] [Google Scholar]
- 8.Slack GJ, Puniani E, Frisvad JC, Samson RA, Miller JD. Mycol. Res. 2009;113:480. doi: 10.1016/j.mycres.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 9.Kimura Y, Shimomura N, Tanigawa F, Fujioka S, Shimada A. Z. Naturforsch. C Bio. Sci. 2012;67:587. doi: 10.1515/znc-2012-11-1209. [DOI] [PubMed] [Google Scholar]
- 10.Miller JD, Sun M, Gilyan A, Roy J, Rand TG. Chem. Biol. Interact. 2010;183:113. doi: 10.1016/j.cbi.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 11.Hoepfner D, McNamara CW, Lim CS, Studer C, Riedl R, Aust T, McCormack SL, Plouffe DM, Meister S, Schuierer S, Plikat U, Hartmann N, Staedtler F, Cotesta S, Schmitt EK, Petersen F, Supek F, Glynne RJ, Tallarico JA, Porter JA, Fishman MC, Bodenreider C, Diagana TT, Movva NR, Winzeler EA. Cell Host Microbe. 2012;11:654. doi: 10.1016/j.chom.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan S, Sharma A, Belrhali H, Yogavel M, Sharma A. J. Struct. Funct. Genomics. 2014;15:63. doi: 10.1007/s10969-014-9182-1. [DOI] [PubMed] [Google Scholar]
- 13.Guiguemde WA, Guy RK. Cell Host Microbe. 2012;11:555. doi: 10.1016/j.chom.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Zhou H, Qiao K, Gao Z, Meehan MJ, Li JW, Zhao X, Dorrestein PC, Vederas JC, Tang Y. J. Am. Chem. Soc. 2010;132:4530. doi: 10.1021/ja100060k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou H, Qiao K, Gao Z, Vederas JC, Tang Y. J. Biol. Chem. 2010;285:41412. doi: 10.1074/jbc.M110.183574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arai K, Rawlings BJ, Yoshizawa Y, Vederas JC. J. Am. Chem. Soc. 1989;111:3391. [Google Scholar]
- 17.Yoshizawa Y, Li Z, Reese PB, Vederas JC. J. Am. Chem. Soc. 1990;112:3212. [Google Scholar]
- 18.Li Z, Martin FM, Vederas JC. J. Am. Chem. Soc. 1992;114:1531. [Google Scholar]
- 19.Liu Y, Li Z, Vederas JC. Tetrahedron. 1998;54:15937. [Google Scholar]
- 20.Zheng H, Zhao C, Fang B, Jing P, Yang J, Xie X, She X. J. Org. Chem. 2012;77:5656. doi: 10.1021/jo300805n. [DOI] [PubMed] [Google Scholar]
- 21.Mohapatra DK, Maity S, Rao TS, Yadav JS, Sridhar B. European journal of organic chemistry. 2013;2013:2859–2863. [Google Scholar]
- 22.Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R. Nucleic Acids Res. 2011;39:W339. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salamov AA, Solovyev VV. Genome. Res. 2000;10:516. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MC, Withers ST, Shiba Y, Sarpong R, Keasling JD. Nature. 2006;440:940. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 25.Xu W, Chooi YH, Choi JW, Li S, Vederas JC, Da Silva NA, Tang Y. Angew. Chem. Int. Ed. Engl. 2013;52:6472. doi: 10.1002/anie.201302406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou H, Zhan J, Watanabe K, Xie X, Tang Y. Proc. Natl. Acad. Sci. U. S. A. 2008;105:6249. doi: 10.1073/pnas.0800657105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y. Proteins. 2009;77:100. [Google Scholar]
- 29.Roy A, Kucukural A, Zhang Y. Nat. Protoc. 2010;5:725. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trott O, Olson AJ. J. Comput. Chem. 2010;31:455. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crawford JM, Korman TP, Labonte JW, Vagstad AL, Hill EA, Kamari-Bidkorpeh O, Tsai SC, Townsend CA. Nature. 2009;461:1139. doi: 10.1038/nature08475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan S, Garg A, Camacho N, Van Rooyen J, Kumar Pole A, Belrhali H, Ribas de Pouplana L, Sharma V, Sharma A. Acta Crystallogr. Sect. D-Biol. Crystallogr. 2013;69:785. doi: 10.1107/S0907444913001923. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.