Abstract
Molecular classification of hepatocellular carcinomas (HCC) could guide patient stratification for personalized therapies targeting subclass‐specific cancer ‘driver pathways’. Currently, there are several transcriptome‐based molecular classifications of HCC with different subclass numbers, ranging from two to six. They were established using resected tumours that introduce a selection bias towards patients without liver cirrhosis and with early stage HCCs. We generated and analyzed gene expression data from paired HCC and non‐cancerous liver tissue biopsies from 60 patients as well as five normal liver samples. Unbiased consensus clustering of HCC biopsy profiles identified 3 robust classes. Class membership correlated with survival, tumour size and with Edmondson and Barcelona Clinical Liver Cancer (BCLC) stage. When focusing only on the gene expression of the HCC biopsies, we could validate previously reported classifications of HCC based on expression patterns of signature genes. However, the subclass‐specific gene expression patterns were no longer preserved when the fold‐change relative to the normal tissue was used. The majority of genes believed to be subclass‐specific turned out to be cancer‐related genes differentially regulated in all HCC patients, with quantitative rather than qualitative differences between the molecular subclasses. With the exception of a subset of samples with a definitive β‐catenin gene signature, biological pathway analysis could not identify class‐specific pathways reflecting the activation of distinct oncogenic programs. In conclusion, we have found that gene expression profiling of HCC biopsies has limited potential to direct therapies that target specific driver pathways, but can identify subgroups of patients with different prognosis.
Keywords: hepatocellular carcinoma, molecular classification, transcriptome, gene expression
Introduction
Hepatocellular carcinoma (HCC) is one of the most frequent and deadliest cancers worldwide, with very limited therapeutic options 1. The prognosis of HCC patients is dismal, with less than 30% qualifying for curative treatments such as tumour resection or liver transplantation 2. Median survival time of patients who cannot be treated surgically is below 1 year. The only approved systemic treatment is sorafenib, a multikinase inhibitor that prolongs median patient survival by about 3 months 3. There is a pressing need for new therapies to treat advanced HCC.
It is believed that the heterogeneity of human HCC can be described through a system of discrete subclasses that could guide patient stratification for personalized therapies targeting subclass‐specific cancer ‘driver genes’. Currently, there are four transcriptome‐based molecular classifications of HCC with different subclass numbers, ranging from two to six 4, 5, 6, 7. Despite considerable research efforts, none of them has found application in clinical practice.
The four published molecular classifications have been developed using surgically obtained tumour specimens and, therefore, rely on the subset of patients selected for surgical treatment. For patients with liver cirrhosis and HCC, surgery (tumour resection or liver transplantation) is generally recommended only for early stage tumours in Barcelona‐Clinic Liver Cancer (BCLC) classes 0 and A 2. Unfortunately, less than 30% of patients in Europe and the United States are diagnosed with early stage tumours. In this study, we set out to develop a molecular classification system not biased by the stage of the HCC and to evaluate the existing molecular classifications of HCC in an unselected patient population using tumour biopsies instead of resection specimens.
Unexpectedly, we found that the inclusion of a control group of five normal liver samples into the analysis changed the interpretation of the existing and newly defined HCC subclasses. Our results challenge the general assumption that the diversity in gene expression profiles of human HCC is best described through a system of discrete class labels.
Patients and methods
HCC patients and normal controls
From August 2002 to March 2012, patients undergoing a liver biopsy for suspected hepatocellular carcinoma at the University Hospital Basel were asked for written informed consent to donate additional biopsies for research purposes. Tumour and paired non‐tumour liver biopsies were performed under ultrasound guidance using a coaxial needle technique allowing repetitive sampling from the same part of a focal lesion with a full‐core biopsy instrument (BioPince®, Angitech, Stenlose, Denmark). One biopsy cylinder was used for routine diagnostic histopathology, Edmondson grading, immunostaining and to quantify the relative contribution of cancer cells, necrotic areas and non‐cancerous vital tissue in the samples. Two additional biopsy cylinders were shock‐frozen in liquid nitrogen and stored at −70°C. For the current studies, only samples composed of at least 50% vital malignant cells in the tumour biopsy and tumour‐free in the parenchyma biopsy were used. The patients were managed according to best clinical practice. Treatment modalities as well as overall survival times were recorded. Deaths due to non‐liver‐related causes (n = 3) were included in the analysis as censored events. At the time of the final analysis, 18% of the patients were alive and 12% were lost to follow‐up, with mean follow‐up time of 130 weeks for the censored events. The normal control biopsy samples were obtained using the same technique. The clinical information concerning the control samples is summarized in Table 2. The study was approved by the Ethics Committee of Basel.
Table 2.
Patient characteristics of control biopsy samples
| Age | Sex | Reason for liver biopsy | Evidence for absence of liver pathology |
|---|---|---|---|
| 55 | M | Isolated slight gamma‐glutamyl transpeptidase elevation | Normal histology, liver ultrasound and transient elastography |
| 22 | M | Slight elevation of transaminases | Normal liver values at time of the biopsy, normal histology, normal liver ultrasound and normal transient elastography |
| 47 | F | Slight alanine transaminase and gamma‐glutamyl transpeptidase elevation | Normal liver values at the time of biopsy, normal histology, normal liver ultrasound and normal transient elastography |
| 78 | F | Biopsy of normal liver in a patient undergoing an ultrasound guided biopsy of a focal lesion (metastasis of neuroendocrine tumour) | Normal histology of liver parenchyma, normal liver values |
| 63 | F | Biopsy of normal liver in a patient undergoing an ultrasound guided biopsy of a focal lesion (focal nodular hyperplasia) | Normal histology of liver parenchyma, normal liver values |
RNA extraction, quantitative real‐time polymerase chain reaction (qPCR) and microarray hybridization
Total RNA was extracted using Qiazol reagent and RNAeasy Mini Kit (Qiagen, Hombrechtikon, Switzerland) according to the manufacturer's instructions. Total RNA, 250 ng, was reverse transcribed and biotinylated with the whole‐transcript Expression Kit (Ambion, Zug, Switzerland) and whole‐transcript Terminal Labeling Kit (Affymetrix, Santa Clara, CA) according to the manufacturer's instructions. The Hybridization and Wash Kit (Affymetrix) was used to hybridize all samples to Human Gene ST 1.0 arrays (Affymetrix). The data are available in the GEO database under accession number GSE64041. The same RNA samples were used for qPCR classification of the tumours based on expression levels of 16 genes as described previously 5.
Immunohistochemistry
Immunohistochemical analysis was performed on liver biopsy sections using an avidin‐biotin complex method (ABC detection kit, Vector Laboratories, Petersborough, UK). Following pressure cooker‐mediated antigen retrieval in 0.001 M ethylenediaminetetraacetic acid, pH 8, the sections were incubated with 10% goat serum (Dako Cytomation, Baar, Switzerland) for 20min. Endogenous peroxidase activity was blocked using 0.5% H2O2. Slides were then incubated with an antibody against β‐catenin (BD Biosciences, Allschwil, Switzerland) at a dilution 1:200. The labeling index was calculated in ‘hot‐spot’ areas as the proportion of positive nuclei as a percentage of the total number of nuclei.
Statistical analysis
Preprocessing
The data were preprocessed using the robust multiarray normalization procedure as implemented in the oligo package of the Bioconductor/R statistical software. We excluded measurements without a gene annotation and those with signal intensity below 80 in the highest expressing sample. If multiple measurements mapped to the same gene ID, we kept the one with the highest standard deviation.
Sample clustering
We applied consensus clustering on a set of 7404 probe sets and 60 tumour samples using the ConsensusClusterPlus package (Bioconductor/R) 8. Probe sets were included in the clustering if they fulfilled the minimum variability criterion: fold change greater than 2 between the 10th and the 90th percentile. The consensus result was calculated based on 1000 resamplings with 80% of the samples and clustering with Ward's linkage method, using 1‐Pearson correlation as a distance measure. We analyzed consensus matrices for number of clusters k from 2 to 10 and found the most robust result with a 3‐cluster solution.
Differential expression
The differential gene expression analysis of HCC samples in clusters 1–3 compared to the five normal samples was carried out using the moderated t statistics implemented in the limma package (Bioconductor/R).
Subclass prediction
The preprocessed data were classified with the Nearest Template Prediction algorithm implementation in the Gene Pattern software (Broad Institute, Boston, MA) using published gene signatures obtained from Molecular Signatures Database 9.
Pathway analysis
For pathway enrichment analysis, we applied the GSEAPreranked algorithm from the javaGSEA software version 2.0.13 (Broad Institute)10. We used the gene set collection C2‐Canonical Pathway and a selection of 683 gene sets from the C2‐Chemical and Genetic Perturbations collection (both version 3.1). The input for GSEAPreranked was a list of all genes on the preprocessed array and the log2 fold change values between the signal intensity in each tumour sample and the mean signal intensity in the 5 normal samples. The output files from the GSEA were then read into R software for filtering of the gene sets with highly significant scores in at least 10% of patients.
Identification of gene clusters
In order to identify clusters of genes that are likely to be co‐regulated and specifically upregulated or downregulated in a subset of patients, we applied the following procedure: (i) we considered only the genes that are not differentially expressed in the majority of patients (less than twofold change compared to the mean of normal samples), but altered in the same direction in a subgroup of at least six HCC samples (10% of the study population), (ii) we applied mixed Gaussian model density estimation (package mclust Bioconductor/R) to assess if the gene expression was more likely to originate from a bimodal or unimodal distribution, and kept the genes identified as bimodally distributed with at least 10% of samples assigned to each mode, (iii) we calculated all pairwise correlations between the genes selected in step (i) and kept only those that passed step (ii) and had at least six highly correlated partners (>0.75 Pearson coefficient). For each of the 265 genes that fulfilled these criteria, we retrieved all their high‐correlation partners from step (iii) and iteratively merged the lists that shared >50% of genes (looking at the shorter list) until no more merging occurred, resulting in nine clusters.
Results
Patients' characteristics
The study included paired liver needle biopsy samples from 60 HCC patients and five normal liver tissue samples (Tables 1 and 2). The HCC patients were predominantly male and 90% had liver cirrhosis (Table 1). The underlying liver disease was related to alcohol abuse (72%), hepatitis C virus (HCV) infection (15%) or hepatitis B virus (HBV) infection (15%). 32% of the patients were in BCLC stages 0 or A (early HCC), 40% in stage B (intermediate), 21% in stage C (advanced) and 7% in stage D (terminal). 72% of the biopsies were classified as Edmondson grade I or II, 28% were grade III or IV.
Table 1.
Patient characteristics of HCC and paired parenchyma samples
| Number of patients | 60 |
| Mean age ± SD | 64 ± 12 |
| Female sex | 12% |
| Cirrhosis | 90% |
| Serum AFP > 100 ng/ml | 33% |
| Largest tumour diameter, mean ± SD, cm | 5.7 ± 3.6 |
| Aetiology | |
| HBV infection | 15% |
| HCV infection | 15% |
| alcohol abuse | 72% |
| Edmondson grade | |
| I | 2% |
| II | 70% |
| III | 25% |
| IV | 3% |
| Barcelona Clinic Liver Cancer stage | |
| 0 | 4% |
| A | 28% |
| B | 40% |
| C | 21% |
| D | 7% |
| Treatment modalities | |
| Local treatment (EtOH/RFTA/TACE/SIRT) | 33% |
| Local treatment + sorafenib | 8.3% |
| Systemic treatment (sorafenib/sunitinib) | 5% |
| Tumour resection | 15% |
| Liver transplantation | 8.3% |
| Supportive care | 22% |
| Other/no data | 8.3% |
With one exception, the biopsies were obtained from treatment‐naïve HCCs. The patients then underwent treatment modalities suitable for their disease stage. A total of 23% patients were treated surgically by resection or liver transplantation, 33% received local treatments such as transarterial chemoembolization (TACE), radiofrequency thermoablation (RFTA), selective internal radiation therapy (SIRT) or percutaneous ethanol injection, 13% were treated with sorafenib or sunitinib (alone or in combination with local treatment) and 22% received best supportive care (Table 1).
Consensus clustering of HCC biopsy gene expression profiles reveals three robust subclasses
We applied unsupervised consensus clustering approach in order to generate a molecular classification of human HCCs using diagnostic biopsy specimens from an unselected patient population. In this method, the final clustering result is based on multiple rounds of resampling using subsets of samples. This approach makes it possible to discover which number of clusters results in the most stable subclass structure. In our dataset, the most robust solution was achieved with a three‐cluster partitioning (Figure 1A, Supplementary Table 1). The resulting sample grouping correlated with clinically relevant variables such as Edmondson grade, tumour size and survival. Tumours with Edmondson grades 3 and 4 (poor differentiation phenotype) were found predominantly in cluster 3 (p < 0.0001, Fisher test) (Figure 1A). Tumours in cluster 2 were significantly larger than the rest (p = 0.015, Student's t‐test; Figure 1B). We also found a statistically significant association of cluster membership with the BCLC stage: patients with early stage tumours (0/A) were overrepresented in cluster 1, intermediate‐stage (B) HCCs grouped mostly in cluster 2 and the late stage tumours (C/D) in cluster 3 (Figure 1C). There was a non‐significant trend towards improved survival of patients in cluster 1 (p = 0.2, log‐rank test)(data not shown). The difference became significant (p = 0.017, log‐rank test) after exclusion of surgically treated patients (N = 14) who as a group had markedly longer survival compared to all other patients regardless of the cluster membership (Figure 1D).
Figure 1.
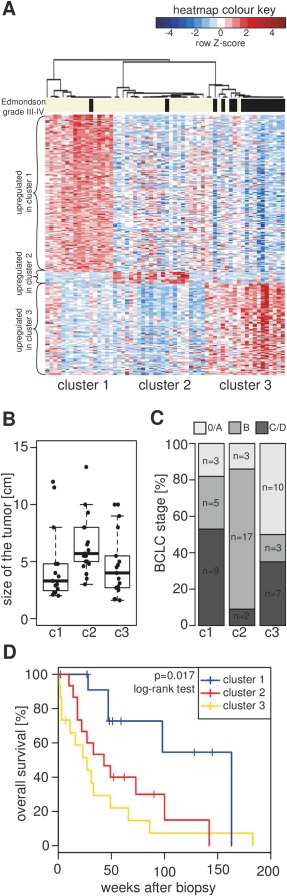
Consensus clustering of HCC biopsy gene expression profiles reveals three robust subclasses. (A) Heatmap representation of gene expression across the 60 tumour samples grouped into the three subclasses. Gene signatures, indicated by curly brackets to the left of the heatmap, consist of all genes that were twofold upregulated with a p‐value below 0.001 in one subclass of HCCs compared to the rest of the tumours (gene list provided in Supplementary Table 1). The heatmap colours represent scaled and centred expression values (shades of red indicate the genes with increased expression compared to the mean of the HCC samples for that gene, while blue indicates decreased expression). The bar above the heatmap identifies samples with Edmondson grade III or IV (black). The dendrogram is derived from hierarchical clustering of a consensus matrix resulting from 1000 rounds of clustering using a resampled dataset. (B) Boxplot showing the differences in the size of the largest tumour focus between the three HCC subclasses. (C) Distribution of BCLC stages between the three HCC subclasses. The numbers of patients in each stage are indicated in the bar plot. (D) Kaplan–Meier survival curves of non‐surgical patients in the three HCC clusters.
Paired study design does not reduce variability between the HCC gene expression profiles
The vast majority of HCCs arise in the context of chronic liver disease, and disease‐specific differences in gene expression profiles could potentially affect gene expression profiles of HCCs. Furthermore, human genetic diversity and host factors such as sex, age, race and comorbidities could also introduce variation of gene expression in HCCs. In an attempt to control for these inter‐individual differences in hepatic gene expression profiles, we performed microarray analysis of paired non‐cancerous parenchyma biopsies collected at the time of the tumour biopsy from all patients included in the study. We speculated that the inclusion of the non‐tumour samples in the analysis could filter out patient‐specific patterns from HCC expression profiles and thereby improve the discriminative power of the analysis. Contrary to our expectations, we observed that fold changes of gene expression generated by comparison of the paired parenchyma and HCC biopsies showed less cross‐correlation between the 60 patients than the tumour expression profiles normalized to the mean of normal controls (Figure 2). This means that the paired parenchyma samples added variability unrelated to HCC into the analysis. This was most likely caused by a significant diversity in gene expression profiles of non‐cancerous samples due to different grades of liver fibrosis and underlying aetiology (alcohol abuse, hepatitis B or C). In contrast, compared to non‐cancerous parenchyma, gene expression profiles of the five normal control samples showed very little variability despite differences in sex and age (Supplementary Figure 1).
Figure 2.
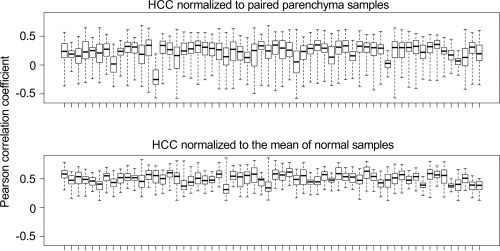
Paired study design does not reduce variability between the HCC gene expression profiles. Genome‐wide fold changes between 60 tumour samples and their corresponding paired controls (top panel), or between 60 tumour samples and the mean gene expression of the five normal samples (bottom panel) were correlated. Each box represents the 59 Pearson correlation coefficient values for one tumour sample calculated over all other samples in the dataset.
Consistent with these findings, we could not find a stable cluster structure that showed significant correlation with clinical variables or activation of specific oncogenic driver pathways in tumour expression profiles after normalization using paired parenchyma samples (data not shown). Therefore, we decided not to use the paired parenchyma samples in our subsequent analysis.
Including normal control samples changes the interpretation of gene expression‐based subclasses
The goal of targeted therapy is to correct abnormal and pathological activation status of cellular signaling pathways. Personalized medicine aims to identify specific patients who could benefit from these approaches. The challenge of molecular tumour classification is to identify both patient groups and their relevant therapeutic targets based on gene expression data.
Our clustering approach, similar to all previously published molecular classifications of human HCC, defines patient subclasses based on differences in gene expression among the tumour samples. In this context, upregulation means that expression of a set of genes is elevated in one tumour subclass compared to the rest of HCCs in the dataset (Figure 1A). This does not necessarily mean that this gene set is also upregulated with respect to the undiseased, baseline state of the cell. In order to interpret differential gene expression in HCC molecular subclasses against the appropriate biological baseline, we integrated a control set of five normal liver samples into our analysis (Figure 3A).
Figure 3.
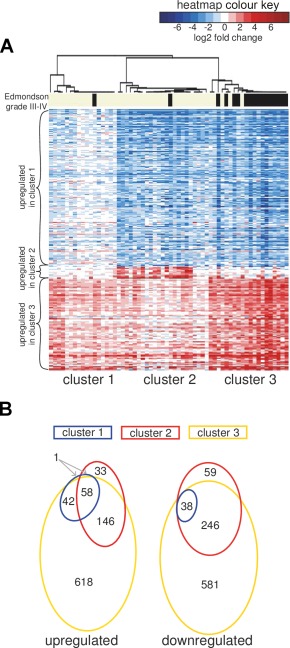
Including normal control samples changes the interpretation of gene expression‐based subclasses. (A) Heatmap representation of changes in gene expression as in Figure 1A, but using the expression data scaled with respect to the mean of normal liver tissue samples. The heatmap colours represent log2 fold change values in the tumour compared to the mean of 5 normal liver samples (shades of red indicate the genes upregulated with respect to the normal liver, while blue indicates downregulation). (B) Venn diagrams showing the overlap in the sets of genes significantly dysregulated in each HCC cluster compared to the reference set of normal liver tissue samples (>2‐fold, FDR < 0.01).
Intriguingly, we observed that the expression of subclass signature genes was generally altered in the same direction in the entire set of 60 tumour samples, and not only within specific clusters. For example, we realized that the gene signature ‘upregulated in cluster 1’ was in fact downregulated in all HCCs, with less‐pronounced regulation in cluster 1 tumours. Vice versa, genes downregulated in clusters 1 and 2 compared to 3 were in fact upregulated in all three clusters. Cluster 2 was an exception, since it was defined by a small set of genes that remained exclusively upregulated in cluster 2 samples and were unchanged in other samples after normalization of gene expression values to normal control liver samples.
In agreement with this result, we found that, apart from a single exception, all genes significantly differentially regulated relative to the normal tissue reference set (>2 fold, FDR < 0.01) in the samples of cluster 1 were also differentially regulated in the same direction in cluster 3. Cluster 2, on the other hand, was characterized by a small subset of differentially regulated genes that were not shared with clusters 1 or 3 (Figure 3B).
In conclusion, we observed that a large majority of genes whose expression was variable in HCC were cancer‐specific genes regulated to some extent across all tumour samples, and that the magnitude of this regulation influenced the outcome of unbiased sample clustering. This finding challenges the current paradigm of subclass‐specific differential gene expression and pathway activation as a general feature of human HCC.
HCC clusters are characterized by different extent of transcriptional regulation
We further observed that the molecular subclasses of HCC were characterized by the extent of transcriptional regulation, that is, the total number of differentially regulated genes, rather than just the identity of specific signature genes. The median number of genes upregulated or downregulated >2 fold in an HCC sample relative to the normal tissue increased from 582 and 397, respectively, in cluster 1 to 961 and 906 in cluster 2 and to 1528 and 1233 in cluster 3 (Figure 4A). Additionally, we found that the number of upregulated genes alone was a significant predictor of overall patient survival. In particular, patients with more than 1500 genes upregulated more than twofold compared to the mean of normal controls showed a substantial decrease in overall survival (p‐value 0.000004, log‐rank test, Figure 4B). These findings further imply that unbiased clustering of HCC expression profiles groups the samples according to the extent of differential expression of genes representing a universal ‘liver cancer phenotype’ rather than, as previously postulated, according to subclass‐specific upregulation or downregulation of a defined set of genes representing an abnormally activated signaling pathway.
Figure 4.
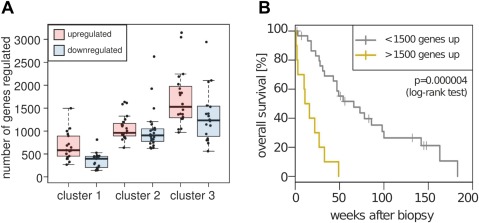
HCC clusters are characterized by different extent of transcriptional regulation. (A) Boxplot showing the number of genes up‐ and downregulated at least twofold in HCC subclasses compared to the mean of normal liver tissue. (B) Kaplan–Meier survival curves of non‐surgical HCC patients grouped based on the extent of gene upregulation (>2‐fold) compared to normal samples.
Integration of normal control samples changes the interpretation of the existing HCC classification systems
We next investigated whether the available classification systems that relied on resected HCC specimens were suitable to classify our biopsy samples that were derived from patients with all tumour stages. In order to do that, we used the available lists of subclass signature genes (Lee et al 4, Chiang et al 6, Hoshida et al 7) or qPCR classifier genes (Boyault et al 5) to predict membership of our biopsy samples in the subclasses of the existing classification systems.
All examined systems performed well in generating high confidence class membership predictions for our dataset, with 77 to 95% of samples assigned to molecular classes at the FDR < 0.05 (Table 3, Figure 4, Supplementary Figure 2). We could also validate several reported associations of cluster membership with clinical variables, including tumour size, differentiation grade and overall survival (Supplementary Figure 3). This finding demonstrates that gene expression patterns described in surgically obtained specimens are directly comparable with those derived from diagnostic tumour biopsies. Indeed, the majority of the published subclass signature genes showed the expected class‐specific expression pattern also in our dataset (Table 3). In agreement with previous studies 6, we observed considerable overlap between subclasses of the different classification systems in our dataset (Supplementary Figure 4 and 2).
Table 3.
Performance of resection‐derived molecular classifications in the biopsy dataset
| Molecular classification | % of biopsy samples classified with high confidence (FDR < 0.05) | % of signature genes showing differential expression (p < 0.05) |
|---|---|---|
| Lee et al | 77% | 81% |
| Boyault et al | NA (qPCR classifier used) | 56% |
| Chiang et al | 95% | 77% |
| Hoshida et al | 91% | 75% |
When the data were normalized to the normal liver reference set, it turned out that the “upregulated in good prognosis” gene signature of Lee et al 4 actually corresponds to genes with general downregulation in HCC, which are especially strongly suppressed in the set of ‘poor prognosis’ samples (Figure 5A). Also, compared to normal liver samples, the signatures reported in literature as ‘upregulated in unannotated’ (Chiang et al 6) and ‘upregulated in S3’ (Hoshida et al 7) were actually downregulated in all subclasses, but to a lower degree in the ‘unannotated’ and S3 groups. Importantly, regardless of the classification system, we found that the majority of subclass signature genes showed expression changes in the same direction in all tumours, with more pronounced upregulation or downregulation in a subset of samples (Figure 5).
Figure 5.
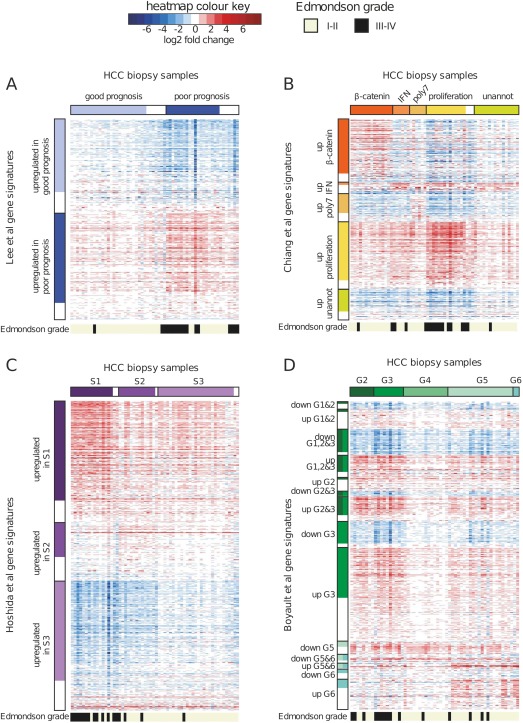
Integration of normal control samples changes the interpretation of existing HCC classification systems. Transcriptional profiles of HCC biopsies were classified into clusters described in the studies of Lee et al 4 (A), Chiang et al 6 (B) and Hoshida et al 7 (C) using the nearest template prediction (NTP) algorithm and the published gene signatures. For classification according to Boyault et al 5 study (D), a qPCR‐based algorithm was applied, as described previously. Heatmaps show log2 fold changes of gene expression for the published signature genes in HCC biopsies compared to the mean of five normal liver samples (shades of red indicate genes upregulated with respect to the normal liver while blue indicates downregulation). Bars above the heatmaps indicate class membership according to NTP/qPCR, with low‐confidence predictions (FDR > 0.05) represented by empty boxes, on the right side of the high confidence predictions for the given class. Bars to the left of the heatmaps indicate the published gene signatures. Genes described to be part of a signature, but without significant differential expression (p‐value > 0.05) in our dataset are shown as empty boxes below the significant genes. Bars below the heatmaps indicate the Edmondson grade, as explained in the colour key.
Unbiased pathway analysis of HCC transcriptomes identifies oncogenic driver pathways only in a set of samples with β‐catenin pathway activation
We next wanted to gain further insight into the biological processes and pathways involved in HCC, and possibly uncover a novel subclass structure related to activation of particular oncogenic signaling pathways in our dataset. To this end, we applied the Gene Set Enrichment Analysis (GSEA) method to each HCC sample. For every tumour sample, we generated a ranked gene list based on the fold‐change of expression compared to the mean of the reference set of normal samples. Subsequently, the gene set collections from the Molecular Signatures Database (MSigDB) were tested for enrichment in the ranked lists. The MSigDB collects lists of genes from several curated databases (KEGG, Reactome, Biocarta), as well as from high throughput measurements of gene expression after chemical or genetic perturbations.
Out of 1677 gene sets included in the analysis, we identified 163 with highly significant enrichment (FDR < 0.0001) in at least 10% of the patients. Consensus clustering of these gene sets based on their enrichment scores detected 4 clusters. Cluster A consisted of 58 gene sets related to cell proliferation, including pathways reported to play a role in HCC such as EGFR, MYC, MYB, WTAP, FOXP3, EZH2, E2F or RB1 11, 12, 13, 14, 15, 16. Cluster B was composed of 38 gene sets related to liver metabolism. These gene sets were almost universally downregulated relative to the normal controls, demonstrating that the loss of hepatocyte phenotype is a general feature of HCC and not limited to a subset of samples. The largest cluster (cluster C) consisted of 61 upregulated gene sets related to inflammation, cell adhesion and epithelial‐to‐mesenchymal transition, including pathways previously implicated in human HCC such as Notch, EGF and MYB 17. A small cluster (cluster D) grouped seven gene sets that all represented activation of interferon system.
This approach identified processes and pathways dysregulated in the majority of HCCs but, with the exception of a small interferon cluster, we did not find gene sets specifically altered in subgroups of patients, such as the genes specific for cluster 2 patients. In order to identify such gene sets we applied a filtering scheme using genes differentially expressed in 6–30 patients compared to normal liver samples (10–50% of the study population). The genes were further selected based on the distribution of expression values and the number of highly correlating genes, and subsequently grouped, generating nine gene sets of sizes ranging between 10 and 449 genes (see Statistical Analysis section). Of these nine gene sets, three were related to previously identified pathways and processes: cell proliferation, inflammation and interferon signaling, and five gene sets could not be assigned a meaningful biological interpretation. The remaining set was composed of 42 genes, the majority of which were known β‐catenin pathway targets or genes directly involved in β‐catenin signaling in the liver such as AXIN2, GLUL and LGR5 (Figure 6A). This β‐catenin pathway signature showed true class‐specific expression in that it was specifically upregulated in a subgroup of tumours, with expression levels equivalent to the normal liver in the remaining HCC specimens. Samples with high expression of genes belonging to this set constituted approximately one‐third of the HCC cases in our dataset. Activation of the β‐catenin pathway in these samples was confirmed by increased levels of nuclear β‐catenin observed by immunohistochemical staining (Figure 6B,C). We also noted that expression of the β‐catenin pathway signature was mostly upregulated in samples belonging to cluster 2 (16 out of the 23 samples classified in cluster 2 also showed high expression of β‐catenin signature genes).
Figure 6.
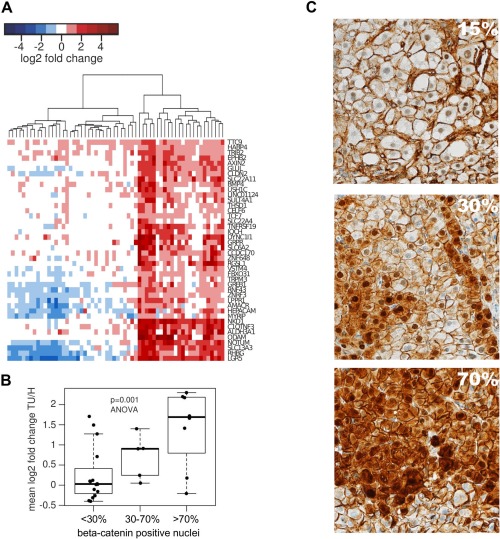
Unbiased pathway analysis of HCC transcriptomes identifies oncogenic driver pathways only in a set of samples with β‐catenin pathway activation. (A) Heatmap showing fold change of gene expression in HCC samples relative to the mean of the normal control samples. (B) HCC samples with increased nuclear β‐catenin staining show an increase in mean fold change of expression of β‐catenin related genes. (C) Representative images showing β‐catenin staining in HCC biopsies with 15, 30 and 70% of positive nuclei.
Discussion
Despite considerable efforts to increase therapeutic options for advanced hepatocellular carcinoma, sorafenib remains the only systemic treatment option. Several additional targeted therapeutics have been tested in large phase III trials, such as sunitinib 18, brivanib 19, linifanib 20, but none of them was found to be superior compared to sorafenib. HCC is a very heterogeneous cancer, and the failure of these trials has been attributed at least in part to the unselected inclusion of HCC patients with the entire spectrum of molecular subtypes of tumours. There is growing consensus in the field that progress in systemic therapies for HCC will depend on adequate selection of patients based on molecular classification of HCCs. The highly publicized phase‐II trial with tivantinib showed in a retrospective analysis significant survival efficacy in the subgroup of patients with c‐met overexpression, but no effect on median survival in patients with low expression of c‐met 21.
It is believed that the heterogeneity of human HCC can be described through a system of discrete subclasses that could guide patient stratification for personalized therapies targeting subclass‐specific cancer ‘driver genes’. Currently, there are four transcriptome‐based molecular classifications of HCC with different subclass numbers, ranging from two to six 4, 5, 6, 7. Despite considerable research efforts, none of them has found application in clinical practice. All of the published classification systems rely on surgically resected HCCs, and this bias towards early stage tumours or HCC arising in non‐cirrhotic livers might have been a reason for their limited usefulness for patients with advanced HCCs.
In the present work, we set out to establish a molecular classification of human HCC based on gene expression profiles from diagnostic biopsy specimens. We observed a robust 3‐subclass structure, which correlated with clinical variables and overall survival (Figure 1). We introduced two sets of control samples into our analysis in order to ensure correct biological interpretation of gene expression changes in HCC. Initially, a set of 60 paired non‐tumour liver tissue samples was included in an attempt to control for the inter‐individual differences in gene expression between the tumour samples. However, we found that this approach introduced additional variability into the dataset instead of removing irrelevant patient‐specific effects and exposing true differences between the tumour subclasses (Figure 2). Subsequently, we have resorted to using five normal liver biopsy gene expression profiles as the baseline for comparison of the tumour samples. Using this control set, we observed that the subclass‐defining ‘signature’ genes were mostly differentially regulated in the same direction across all tumours, regardless of their subclass membership (Figure 3). The same was true for the subclasses of previously published molecular classification systems (Figure 5).
We conclude that molecular classification systems of HCCs that are based on transcriptome analysis result largely from quantitative differences of expression levels of a set of tumour‐specific genes, instead of the upregulation or downregulation of class‐specific genes. A notable exception was a set of β‐catenin target genes (Figure 6) that defined cluster 2 in the current study as well as the ‘β‐catenin’ subclass of Chiang et al 6 and G5/G6 subclasses of Boyault et al 5 The β‐catenin pathway is central to liver development and often deregulated in liver cancer. Differential expression of genes relevant to this pathway is related to mutations in CTNNB1, which occur in about a third of HCC cases 22. Recent work points to the anti‐proliferative potential of β‐catenin pathway inhibitors in in vitro models of HCC, holding promise for future targeted therapies in this group of patients 23.
Ideally, a molecular classification of human HCC would highlight the differential activation of druggable targets, such as kinases or growth factor receptors, allowing identification of patients who could benefit from targeted therapies. So far, none of the existing molecular classifications of HCC has shown clear potential in this direction. The limited potential of gene expression profiling approaches might be caused by the convergence of multiple oncogenic driver pathways on a limited number of transcription factors that are central for the activation of cancer gene expression. Because of this convergence, the analysis of gene expression data sets is not very informative with regard to the activation of upstream signaling pathways. In order to classify HCC patients based on differential pathway activation it might be more promising to use phosphoproteomic assays, rather than transcriptome analysis. These approaches would directly measure the activation of the signaling molecules and attempt to resolve a set of HCC samples into clusters based on the similarities and differences in the identities of the signaling pathways involved. However, quantitative measurement of a large number of phosphoproteins in a sufficient number of tumour samples is currently a highly challenging task. In the future, progress in molecular classification of HCCs might result from further technical improvements in phosphoproteomic assays or from the integration of histological, immunohistochemical, genomic, transcriptomic and targeted biochemical analysis of oncogenic signaling pathways. The availability of tumour tissue from a large number of patients with all stages of HCC will be a prerequisite for the success of such an effort.
Author contributions
MHH designed the study; ZM, TB, LQ, MTD, MSM, LT collected the data; ZM, DA, JEV, VR performed the analysis; ZM and MHH wrote the manuscript. All authors read and approved the final manuscript.
Supporting information
The following supplementary material may be found in the online version of this article:
Supplementary Figure 1. Differential expression in gene expression profiles of paired parenchyma and tumour samples. The boxplot shows the number of genes up‐ or downregulated more than 2‐fold with respect to the mean of normal liver biopsies in the 5 normal liver samples (H), 60 non‐tumour samples from HCC patients (CT) and 60 HCC samples (TU). Lines connect paired samples from the same patient.
Supplementary Figure 2. Nearest template prediction and qPCR classify HCC biopsies into previously described subclasses. Transcriptional profiles of HCC biopsies were classified into clusters described in the studies of Lee et al (A), Chiang et al (B) and Hoshida et al (C) using the nearest template prediction (NTP) algorithm and the published gene signatures. For classification according to the Boyault et al study (D), qPCR‐based algorithm was applied, as described previously. Heatmaps show scaled and centred expression values (shades of red indicate the genes with increased expression compared to the mean of the HCC samples for that gene, while blue indicates decreased expression). Bars above the heatmaps indicate class membership according to NTP/qPCR, with low‐confidence predictions (FDR > 0.05) indicated by empty boxes on the right side of the high confidence predictions for the given class. Bars to the left of the heatmaps indicate the published gene signatures. Genes described to be part of a signature, but without significant differential expression (p value > 0.05) in our dataset are shown as empty boxes below the significant genes. Bars below the heatmaps indicate Edmonson grade, as explained in the colour key.
Supplementary Figure 3. Membership of published subclasses correlates with clinical variables in the biopsy dataset. (A) Membership of Lee et al classes is associated with overall survival in our set of HCC patients who were not treated surgically. Patients with low‐confidence predictions of class membership were excluded from the analysis. (B) Tumour differentiation grade is associated with overall survival in HCC patients who were not treated surgically. (C) Boxplot showing the size of the largest tumour focus in our 60 HCC patients classified into Chiang et al classes. (D) Boxplot showing log10 serum AFP in our 60 HCC patients classified into Chiang et al classes. (E) Boxplot showing the size of the largest tumour focus in our 60 HCC patients classified into Hoshida et al classes. (F) Boxplot showing log10 serum AFP in our 60 HCC patients classified into Hoshida et al classes.
Supplementary Figure 4. Comparison between the sample class assignments for the previously described molecular classifications. Sample classes were assigned to the HCC biopsy gene expression profiles using NTP or qPCR. Empty boxes indicate low‐confidence predictions. (A) Lee et al (NTP) and Boyault et al (qPCR) class memberships. (B) Lee et al (NTP) and Chiang et al (NTP) class memberships. (C) Lee et al (NTP) and Hoshida et al (NTP) class memberships. (D) Boyault et al (qPCR) and Chiang et al (NTP) class memberships. (E) Boyault et al (qPCR) and Hoshida et al (NTP) class memberships. (F) Chiang et al (NTP) and Hoshida et al (NTP) class memberships.
Supplementary Table S1
Supplementary Table S2
Acknowledgements
This work was funded by Swiss National Science Foundation grant 310030B_147089/1 and European Research Council Synergy grant 609883 (MERiC).
The authors declare no conflicts of interest.
Microarray data have been deposited in the GEO database.
References
- 1. Yang JD, Roberts LR. Hepatocellular carcinoma: A global view. Nat Rev Gastroenterol Hepatol 2010; 7: 448–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. European Association For The Study Of The Liver, European Organisation For Research And Treatment Of Cancer. EASL‐EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol 2012; 56: 908–943. [DOI] [PubMed] [Google Scholar]
- 3. Llovet JM, Ricci S, Mazzaferro V, et al Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008; 359: 378–390. [DOI] [PubMed] [Google Scholar]
- 4. Lee J‐S, Chu I‐S, Heo J, et al Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology 2004; 40: 667–676. [DOI] [PubMed] [Google Scholar]
- 5. Boyault S, Rickman DS, de Reyniès A, et al Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology 2007; 45: 42–52. [DOI] [PubMed] [Google Scholar]
- 6. Chiang DY, Villanueva A, Hoshida Y, et al Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res 2008;68:6779–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hoshida Y, Nijman SMB, Kobayashi M, et al Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res 2009; 69: 7385–7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 2010; 26: 1572–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hoshida Y. Nearest template prediction: a single‐sample‐based flexible class prediction with confidence assessment. PLoS One 2010; 5: e15543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Subramanian A, Tamayo P, Mootha VK, et al Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schiffer E, Housset C, Cacheux W, et al Gefitinib, an EGFR inhibitor, prevents hepatocellular carcinoma development in the rat liver with cirrhosis. Hepatology 2005; 41: 307–314. [DOI] [PubMed] [Google Scholar]
- 12. Nakajima T, Yasui K, Zen K, et al Activation of B‐Myb by E2F1 in hepatocellular carcinoma. Hepatol Res 2008; 38: 886–895. [DOI] [PubMed] [Google Scholar]
- 13. Sera T, Hiasa Y, Mashiba T, et al Wilms’ tumour 1 gene expression is increased in hepatocellular carcinoma and associated with poor prognosis. Eur J Cancer 2008; 44: 600–608. [DOI] [PubMed] [Google Scholar]
- 14. Huang Y, Wang F‐M, Wang T, et al Tumor‐infiltrating FoxP3+ Tregs and CD8+ T cells affect the prognosis of hepatocellular carcinoma patients. Digestion 2012; 86: 329–337. [DOI] [PubMed] [Google Scholar]
- 15. Xu L, Beckebaum S, Iacob S, et al MicroRNA‐101 inhibits human hepatocellular carcinoma progression through EZH2 downregulation and increased cytostatic drug sensitivity. J Hepatol 2014; 60: 590–598. [DOI] [PubMed] [Google Scholar]
- 16. Wang B, Hsu S, Wang X, et al Reciprocal regulation of microRNA‐122 and c‐Myc in hepatocellular cancer: role of E2F1 and transcription factor dimerization partner 2. Hepatology 2014; 59: 555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Villanueva A, Alsinet C, Yanger K, et al Notch signaling is activated in human hepatocellular carcinoma and induces tumor formation in mice. Gastroenterology 2012; 143: 1660–9.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheng A‐L, Kang Y‐K, Lin D‐Y, et al Sunitinib versus sorafenib in advanced hepatocellular cancer: results of a randomized phase III trial. J Clin Oncol 2013; 31: 4067–4075. [DOI] [PubMed] [Google Scholar]
- 19. Johnson PJ, Qin S, Park J‐W, et al Brivanib versus sorafenib as first‐line therapy in patients with unresectable, advanced hepatocellular carcinoma: results from the randomized phase III BRISK‐FL study. J Clin Oncol 2013; 31: 3517–3524. [DOI] [PubMed] [Google Scholar]
- 20. Cainap C, Qin S, Huang W‐T, et al Linifanib versus Sorafenib in patients with advanced hepatocellular carcinoma: results of a randomized phase III trial. J Clin Oncol 2015; 33: 172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Santoro A, Rimassa L, Borbath I, et al Tivantinib for second‐line treatment of advanced hepatocellular carcinoma: a randomised, placebo‐controlled phase 2 study. Lancet Oncol 2013; 14: 55–63. [DOI] [PubMed] [Google Scholar]
- 22. de La Coste A, Romagnolo B, Billuart P, et al Somatic mutations of the beta‐catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc Natl Acad Sci USA 1998; 95: 8847–8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gedaly R, Galuppo R, Daily MF, et al Targeting the Wnt/β‐catenin signaling pathway in liver cancer stem cells and hepatocellular carcinoma cell lines with FH535. PloS One 2014; 9: e99272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following supplementary material may be found in the online version of this article:
Supplementary Figure 1. Differential expression in gene expression profiles of paired parenchyma and tumour samples. The boxplot shows the number of genes up‐ or downregulated more than 2‐fold with respect to the mean of normal liver biopsies in the 5 normal liver samples (H), 60 non‐tumour samples from HCC patients (CT) and 60 HCC samples (TU). Lines connect paired samples from the same patient.
Supplementary Figure 2. Nearest template prediction and qPCR classify HCC biopsies into previously described subclasses. Transcriptional profiles of HCC biopsies were classified into clusters described in the studies of Lee et al (A), Chiang et al (B) and Hoshida et al (C) using the nearest template prediction (NTP) algorithm and the published gene signatures. For classification according to the Boyault et al study (D), qPCR‐based algorithm was applied, as described previously. Heatmaps show scaled and centred expression values (shades of red indicate the genes with increased expression compared to the mean of the HCC samples for that gene, while blue indicates decreased expression). Bars above the heatmaps indicate class membership according to NTP/qPCR, with low‐confidence predictions (FDR > 0.05) indicated by empty boxes on the right side of the high confidence predictions for the given class. Bars to the left of the heatmaps indicate the published gene signatures. Genes described to be part of a signature, but without significant differential expression (p value > 0.05) in our dataset are shown as empty boxes below the significant genes. Bars below the heatmaps indicate Edmonson grade, as explained in the colour key.
Supplementary Figure 3. Membership of published subclasses correlates with clinical variables in the biopsy dataset. (A) Membership of Lee et al classes is associated with overall survival in our set of HCC patients who were not treated surgically. Patients with low‐confidence predictions of class membership were excluded from the analysis. (B) Tumour differentiation grade is associated with overall survival in HCC patients who were not treated surgically. (C) Boxplot showing the size of the largest tumour focus in our 60 HCC patients classified into Chiang et al classes. (D) Boxplot showing log10 serum AFP in our 60 HCC patients classified into Chiang et al classes. (E) Boxplot showing the size of the largest tumour focus in our 60 HCC patients classified into Hoshida et al classes. (F) Boxplot showing log10 serum AFP in our 60 HCC patients classified into Hoshida et al classes.
Supplementary Figure 4. Comparison between the sample class assignments for the previously described molecular classifications. Sample classes were assigned to the HCC biopsy gene expression profiles using NTP or qPCR. Empty boxes indicate low‐confidence predictions. (A) Lee et al (NTP) and Boyault et al (qPCR) class memberships. (B) Lee et al (NTP) and Chiang et al (NTP) class memberships. (C) Lee et al (NTP) and Hoshida et al (NTP) class memberships. (D) Boyault et al (qPCR) and Chiang et al (NTP) class memberships. (E) Boyault et al (qPCR) and Hoshida et al (NTP) class memberships. (F) Chiang et al (NTP) and Hoshida et al (NTP) class memberships.
Supplementary Table S1
Supplementary Table S2


