Abstract
Multiple molecular markers contribute to the pathogenesis of thyroid cancer and can provide valuable information to improve disease diagnosis and patient management. We performed a comprehensive evaluation of miRNA gene expression in diverse thyroid lesions (n = 534) and developed predictive models for the classification of thyroid nodules, alone or in combination with genotyping. Expression profiling by reverse transcription‐quantitative polymerase chain reaction in surgical specimens (n = 257) identified specific miRNAs differentially expressed in 17 histopathological categories. Eight supervised machine learning algorithms were trained to discriminate benign from malignant lesions and evaluated for accuracy and robustness. The selected models showed invariant area under the receiver operating characteristic curve (AUC) in cross‐validation (0.89), optimal AUC (0.94) in an independent set of preoperative thyroid nodule aspirates (n = 235), and classified 92% of benign lesions as low risk/negative and 92% of malignant lesions as high risk/positive. Surgical and preoperative specimens were further tested for the presence of 17 validated oncogenic gene alterations in the BRAF, RAS, RET or PAX8 genes. The miRNA‐based classifiers complemented and significantly improved the diagnostic performance of the 17‐mutation panel (p < 0.001 for McNemar's tests). In a subset of resected tissues (n = 54) and in an independent set of thyroid nodules with indeterminate cytology (n = 42), the optimized ThyraMIR Thyroid miRNA Classifier increased diagnostic sensitivity by 30–39% and correctly classified 100% of benign nodules negative by the 17‐mutation panel. In contrast, testing with broad targeted next‐generation sequencing panels decreased diagnostic specificity by detecting additional mutations of unknown clinical significance in 19–39% of benign lesions. Our results demonstrate that, independent of mutational status, miRNA expression profiles are strongly associated with altered molecular pathways underlying thyroid tumorigenesis. Combined testing for miRNA gene expression and well‐established somatic gene alterations is a novel diagnostic strategy that can improve the preoperative diagnosis and surgical management of patients with indeterminate thyroid nodules.
Keywords: thyroid cancer, thyroid nodule, diagnosis, miRNA, gene mutation, gene rearrangement
Introduction
Much progress has been made over the past 30 years in understanding the molecular pathogenesis of thyroid cancer. The majority of thyroid carcinomas involve activating somatic mutations in genes encoding effectors of the MAPK and PI3K/AKT signaling pathways 1, 2. Point mutations in the BRAF gene are relatively frequent in papillary thyroid carcinomas (PTC) while RAS mutations are typically found in follicular variants of PTC (FvPTC), follicular thyroid carcinomas (FTC) and follicular adenomas (FA). Chromosomal rearrangements resulting in oncogenic gene fusions, such as the RET/CCDC6 inversion in PTC or the PAX8/PPARG translocation in FvPTC and follicular neoplasms, are another hallmark of thyroid cancer 1, 2. Gene mutations occurring at lower frequency in the PI3K/AKT pathway (eg AKT1, PIK3CA, PTEN) and other signaling pathways (eg CTNNB1, TP53, TSHR) as well as accumulation of alterations with synergistic oncogenic effects leading to cancer progression have also been reported 1, 2. Recent advances further suggest that reactivation of telomerase activity through TERT promoter mutations is another cooperative driver of progression and aggressiveness in thyroid cancer 3, 4.
Molecular testing for somatic gene alterations involved in tumorigenesis has considerably improved the preoperative diagnosis and surgical management of thyroid nodules 5, 6, 7, 8. Although nodules are frequent in adults, in particular in patients older than 60 years of age, only 5% of cases are ultimately diagnosed as malignant after surgical resection 9. Approximately 50% of thyroid cancers are initially identified by imaging and cytological evaluation of fine‐needle aspiration (FNA) biopsies 10. Nodules with an indeterminate cytological diagnosis have a highly variable residual risk of cancer and require additional risk stratification to rule in or rule out cancer 10, 11. Because oncogenic gene alterations are in most cases highly specific for thyroid cancer, a nodule with a positive molecular result is associated with an increased risk of malignancy 5, 6. Such tests with high specificity and positive predictive value (PPV) are referred to as ‘rule in’ tests.
Altered signaling pathways ultimately lead to changes in the expression of genes regulating thyroid cell growth, proliferation and survival, as well as angiogenesis, invasion and metastasis 2. These changes may be used as surrogate signatures to identify tumors with activated pathways 12. Based on microarray expression data in 220 surgical specimens and 234 preoperative nodule aspirates, a proprietary diagnostic algorithm involving 167 genes and seven classification signatures has been developed for the preoperative diagnosis of indeterminate thyroid nodules 13, 14. This gene expression classifier was optimized for high sensitivity at the detriment of specificity and therefore cannot be used to rule in cancer. It can, however, identify and rule out approximately 50% of benign nodules with high negative predictive value (NPV) 13.
Another class of gene expression marker with diagnostic potential is microRNA (miRNA). miRNAs are small, non‐coding RNAs that regulate fundamental cellular processes underlying cancer development and progression 15. A large number of studies have shown that the expression of specific oncomiRs is deregulated in the most common types of differentiated thyroid cancer such as PTC 16, 17, 18, 19, FvPTC 20, 21, 22, 23 and FTC 22, 23, 24, 25, 26. Similar to mRNA signatures, changes in miRNA expression can be assessed in thyroid nodule aspirates 27, 28, 29, 30 and may be associated with the presence of activating gene mutations 18, 20, 22, 30. Although a systematic large‐scale evaluation has not been performed to date, the bulk of these data suggest that classification algorithms based on miRNA expression may have clinical utility to rule in or rule out cancer in preoperative thyroid nodules with indeterminate cytology.
Recent integrated genomic studies have shown that thyroid cancer is more genetically diverse than previously recognized, with multiple layers of biomarkers contributing to the pathological genotypes and phenotypes 31. The logical extension of this recognition is that a molecular test assessing this complexity should improve current tests based on a single molecular dimension. In this study, we performed a comprehensive evaluation of the diagnostic potential of known miRNA biomarker candidates, developed supervised machine learning classification algorithms and assessed their performance, alone or in combination with genotyping for known gene alterations.
Materials and methods
Thyroid specimens
Surgical specimens were obtained from the University of Michigan pathology archives under a research protocol approved by the university's institutional review board. Thyroid tissues were histologically reviewed by a single expert pathologist (TJG) and classified according to current World Health Organization schemes. Remnant nucleic acids samples from preoperative thyroid nodule FNAs from 14 endocrinology centres across the United States were archived by Asuragen's clinical laboratory under a research protocol approved by a central investigational review board. The initial set of 235 aspirates consisted of 118 nodules with benign cytology, including 13 with surgical outcome (12 benign, 1 malignant), 73 with malignant cytology, including 51 with surgical outcome (1 benign, 50 malignant), and 44 with indeterminate cytology, all with available surgical outcome. The second set of aspirates consisted of 42 distinct nodules with indeterminate cytology and surgical outcome.
Molecular assays
Total nucleic acids were extracted and tested for 17 gene alterations using the Luminex 200 System (Luminex, Austin, TX) and for miRNA expression using custom Pick‐&‐Mix microRNA PCR Panels (Exiqon, Vedbaek, Denmark) as previously described 5, 32, 33. Reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) for each target miRNA were performed in duplicate. The 31 miRNA candidates were selected based on literature review and on differential expression analyses performed using GeneChip miRNA Arrays v2 (Affymetrix, Santa Clara, CA), 122 benign or malignant resected thyroid lesions (Asterand, Detroit, MI) and blood samples from healthy donors. Genotyping of over 1500 unique gene alterations was performed using a custom sequencing assay 34, 35.
Data analyses
Graphic and statistical analyses were performed in Excel (Microsoft Corp, Redmond, WA) or R (http://www.r-project.org/). miRNA expression data (cycle threshold or Ct) were normalized by mean‐centering. For miRNA evaluated in a given sample, the mean miRNA expression level ( ) was subtracted from each individual raw Ct. For hypothesis testing, p values were calculated using Welch's t‐test and adjusted using the Benjamini‐Hochberg procedure. The tests were considered significant when the adjusted p values were smaller than a false discovery rate set at 5% (Q value <0.05). The predictive performance of the algorithms trained by supervised machine learning was assessed using the area under the receiver operating characteristic curve (AUC). The probability scores generated by classifiers A and B were calculated using raw Ct results. Given an intercept , coefficients and raw Ct values , the score of each classifier with miRNA interrogated was calculated as with . Diagnostic sensitivity and specificity were calculated by comparing qualitative binary molecular test results (positive/high risk or negative/low risk) to the reference pathology diagnoses (benign or malignant). PPV and NPV were estimated using sensitivity, specificity, thyroid cancer prevalence and the Bayes theorem. Additional information is available in the Supplemental Materials and Methods.
Results
miRNA expression profiling in thyroid tissues
The expression levels of 31 target miRNAs were determined by RT‐qPCR in 257 unique surgical specimens representing 17 histopathological diagnostic categories. Unsupervised hierarchical clustering and principal component analyses confirmed highly variable miRNA expression profiles with distinct expression patterns in various benign or malignant lesions (Figure 1A and 1B). Almost all PTC separated from the other samples along the first principal component with FvPTC clustering between the PTC with true papillary architecture (cPTC and TcPTC) and the follicular neoplasms (FA, oFA, FTC and oFTC). There was no obvious segregation among the benign and malignant follicular tumours, reflecting their shared histopathological features. The anaplastic thyroid carcinomas (ATC) clearly separated from the other samples along the second component, consistent with their highly distinct histology. The chronic lymphocytic thyroiditis (CLT) cases formed a very tight cluster between the follicular cell‐derived tumours and the ATC.
Figure 1.
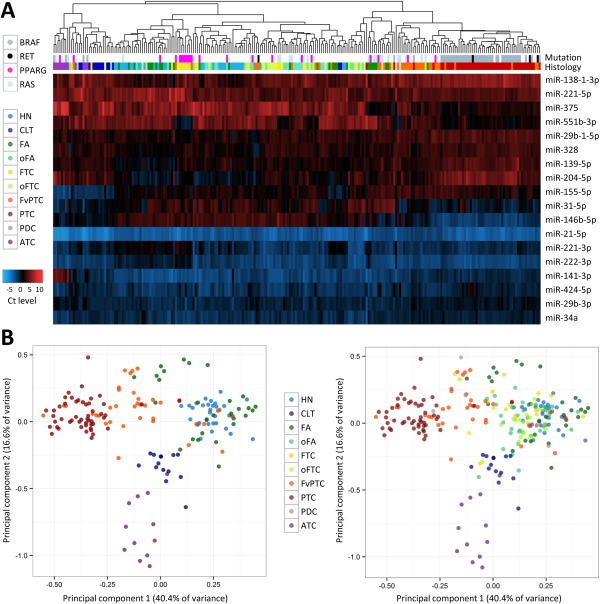
miRNA expression profiling in resected thyroid tissues. (A) Unsupervised hierarchical clustering for histopathological categories represented by at least five independent specimens (n = 248 specimens total). The expression levels of 18 miRNAs as determined by RT‐qPCR are colour‐coded from low expression (high Ct, red) to high expression (low Ct, blue) after mean‐centering normalization. The presence of gene alterations in the BRAF, RAS, PAX8 or RET genes detected with the Luminex platform is colour‐coded at the top of the figure. (B) Principal component (PC) analysis using the same miRNAs and sample set. For clarity, the colour‐coded markers corresponding to oFA, FTC, oFTC and PDC specimens are not displayed in the left panel. Abbreviations: HN, hyperplastic nodule (includes diffuse hyperplasia); CLT, chronic lymphocytic thyroiditis; FA, follicular adenoma; oFA, oncocytic FA; FTC, follicular thyroid carcinoma; oFTC, oncocytic FTC; PTC, papillary thyroid carcinoma (includes conventional and tall cell variants); FvPTC, follicular variant of PTC; PDC, poorly differentiated carcinoma; ATC, anaplastic thyroid carcinoma.
Differential miRNA expression in thyroid tissues
Statistical hypothesis testing identified specific miRNAs differentially expressed in malignant versus benign samples (Table 1). There were 13 miRNAs overexpressed in thyroid carcinoma by more than one Ct on average (effect size >1) and with a significant adjusted p value (Q value <0.05), mainly miR‐31‐5p, ‐146b‐3p/‐5p, ‐221‐3p, ‐222‐3p and ‐551‐3p. Conversely, there were only three miRNAs significantly over expressed in benign thyroid lesions, miR‐7‐5p, ‐204‐5p and ‐138‐1‐3p. Similar analyses using various combinations of histopathological categories showed that specific biomarker candidates such as miR‐221‐3p/‐5p, ‐222‐3p and ‐551‐3p were differentially expressed in both PTC and FTC relative to benign lesions (Supplemental Table 1). Other, such as miR‐31‐5p, ‐146b‐3p/‐5p and ‐375, were statistically significant only in PTC, including FvPTC. miR‐21‐5p was highly expressed in all thyroid specimens, in particular in cPTC/TcPTC and ATC (see also Figure 1A), but was not significant in FvPTC (Q = 0.69) and was barely significant in FTC (Q = 0.025) when compared to benign lesions. ATC samples had miRNA expression patterns mostly similar to other carcinomas but were characterized by high expression levels of miR‐155‐5p and low levels of miR‐141‐3p and ‐375 (see also Figure 1A). The five poorly differentiated carcinomas (PDC) were distributed along the first principal component (Figure 1B) and out of 11 miRNAs with an effect size >1 Ct, only miR‐96‐5p and miR‐139‐5p were statistically significant relative to benign lesions (Q = 0.024 and 0.0028, respectively). Interestingly, all miRNAs were expressed at lower levels in CLT samples relative to all other samples combined, with the exception of miR‐155‐5p that was over expressed by 1.7 Ct on average (Q = 3.1 × 10−10).
Table 1.
Differential miRNA expression in benign versus malignant resected tissues
| Effect size | p value | Q value | AUC | |
|---|---|---|---|---|
| miR‐138‐1‐3p | −0.77 | 1.1 E ‐03 | 1.7 E ‐03 | 0.64 |
| miR‐139‐5p | −0.40 | 3.8 E ‐02 | 0.051 | 0.58 |
| miR‐141‐3p | 0.04 | 0.904 | 0.904 | 0.53 |
| miR‐144‐5p | 0.29 | 0.288 | 0.308 | 0.51 |
| miR‐146b‐3p | 3.75 | 5.4 E ‐28 | 1.7 E ‐26 | 0.83 |
| miR‐146b‐5p | 3.78 | 3.8 E ‐24 | 5.9 E ‐23 | 0.80 |
| miR‐151‐3p | 0.51 | 6.5 E ‐03 | 9.6 E ‐03 | 0.57 |
| miR‐155‐5p | 0.39 | 0.069 | 0.088 | 0.55 |
| miR‐15a‐5p | 0.73 | 2.7 E ‐05 | 5.9 E ‐05 | 0.64 |
| miR‐181a‐2‐3p | 1.14 | 4.3 E ‐07 | 1.1 E ‐06 | 0.68 |
| miR‐187 | 1.73 | 2.8 E ‐08 | 7.9 E ‐08 | 0.72 |
| miR‐197‐3p | 0.58 | 1.0 E ‐04 | 1.8 E ‐04 | 0.62 |
| miR‐19b | 0.47 | 8.3 E ‐03 | 1.2 E ‐02 | 0.57 |
| miR‐204‐5p | −1.14 | 9.6 E ‐05 | 1.8 E ‐04 | 0.65 |
| miR‐21‐5p | 1.22 | 2.6 E ‐09 | 1.0 E ‐08 | 0.72 |
| miR‐221‐3p | 1.90 | 8.8 E ‐13 | 4.5 E ‐12 | 0.75 |
| miR‐221‐5p | 1.69 | 9.9 E ‐10 | 4.4 E ‐09 | 0.73 |
| miR‐222‐3p | 1.98 | 2.4 E ‐16 | 1.8 E ‐15 | 0.78 |
| miR‐222‐5p | 1.20 | 2.8 E ‐06 | 6.7 E ‐06 | 0.68 |
| miR‐224‐5p | 1.12 | 1.2 E ‐08 | 3.8 E ‐08 | 0.70 |
| miR‐29b‐1‐5p | 0.17 | 0.300 | 0.309 | 0.51 |
| miR‐29b‐3p | 0.55 | 4.9 E ‐03 | 7.6 E ‐03 | 0.61 |
| miR‐31‐5p | 2.29 | 4.0 E ‐14 | 2.5 E ‐13 | 0.75 |
| miR‐328 | 0.30 | 0.100 | 0.119 | 0.54 |
| miR‐34a | 0.69 | 7.0 E ‐05 | 1.5 E ‐04 | 0.64 |
| miR‐375 | 2.34 | 1.0 E ‐08 | 3.4 E ‐08 | 0.70 |
| miR‐424‐5p | 0.36 | 0.125 | 0.143 | 0.54 |
| miR‐551b‐3p | 3.32 | 8.8 E ‐24 | 9.1 E ‐23 | 0.82 |
| miR‐7‐5p | −1.42 | 1.5 E ‐04 | 2.6 E ‐04 | 0.65 |
| miR‐885‐5p | 0.45 | 0.071 | 0.088 | 0.57 |
| miR‐96‐5p | 0.37 | 0.164 | 0.181 | 0.55 |
AUC, area under the receiver operating characteristic curve.
miRNA classifiers development
Predictive models to discriminate benign from malignant thyroid lesions were trained on surgical specimens (Supplemental Figure 1) and further tested in an independent set of 235 representative thyroid nodule FNAs. Principal component analyses confirmed that miRNA expression profiles were similar in surgical and preoperative specimens for both benign and malignant lesions (Supplemental Figure 2). miR‐187, ‐222‐5p, ‐224‐5p and ‐885‐5p were excluded from model training because of the lack of reliable detection (Ct > 35) in various histopathology categories and nodule FNAs. In addition, eight miRNAs over expressed in blood relative to thyroid tissue, that is, for which potential blood contamination in a nodule aspirate could interfere with small but diagnostically relevant variations in miRNA expression, were excluded from model training (Figure 2; miR‐7‐5p, ‐15a‐5p, ‐19b, ‐96‐5p, ‐151‐3p, ‐144‐5p, ‐197‐3p and ‐424‐5p). Classification models were built using the 19 remaining miRNAs and eight supervised learning methodologies (Table 2). Diagonal linear discriminant analysis (DLDA) proved very robust with invariant AUC calculated on specimen scores predicted under leave‐one‐out cross‐validation in the training set and with optimal performance in the nodule FNA set. In addition, DLDA resulted in the highest correlation between duplicates, indicating that this classification model was less sensitive to small‐to‐moderate variations in Ct signal.
Figure 2.
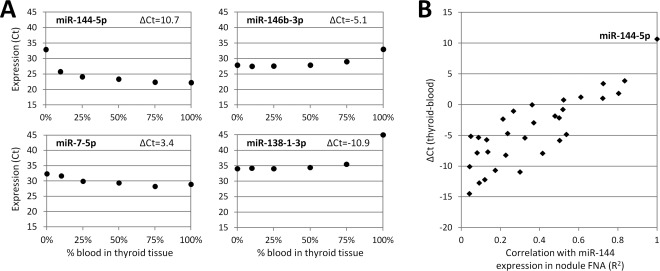
Impact of blood contamination on miRNA expression signatures. (A) Representative examples of miRNA expression levels (Ct) according to the percentage of blood contamination. Nucleic acids from resected thyroid samples (0%) were mixed with nucleic acids from blood samples (100%) to mimic clinical samples contaminated with 10, 25, 50 or 75% of blood. miR‐144‐5p was highly over expressed in blood relative to resected thyroid tissue (ΔCt=Ct 0% − Ct 100% >10), which was confirmed by microarray differential analyses (data not shown). (B) Relationship between miRNA expression difference in whole blood relative to resected thyroid tissues determined in Figure 2A (ΔCt, y axis) and the correlation between the Ct of each miRNA with the Ct of miR‐144‐5p measured in 235 preoperative thyroid nodule aspirates (R 2, x axis). In this experiment, the correlation of a given miRNA with miR‐144‐5p in nodule aspirates is taken as a proxy for the degree to which the miRNA's expression measurement is influenced by blood contamination. Abbreviation: FNA, fine‐needle aspiration.
Table 2.
Average performance of eight supervised machine learning algorithms
| Area under the ROC curve | Replicate correlation | |||
|---|---|---|---|---|
| RS | CV | FNA | ||
| Diagonal linear discriminant analysis | 0.89 | 0.89 | 0.94 | 0.98 |
| k‐nearest neighbours | 0.95 | 0.89 | 0.88 | 0.97 |
| Logistic regression | 0.92 | 0.90 | 0.91 | 0.97 |
| Neural network | 0.94 | 0.86 | 0.85 | 0.92 |
| Quadratic discriminant analysis | 0.93 | 0.88 | 0.87 | 0.96 |
| Random forest | 1.00 | 0.89 | 0.89 | 0.95 |
| Support vector machine, linear kernel | 0.91 | 0.89 | 0.89 | 0.96 |
| Support vector machine, radial kernel | 0.97 | 0.88 | 0.85 | 0.95 |
ROC, receiver operating characteristic; RS, resubstitution in training set; CV, cross‐validation in training set; FNA, fine‐needle aspiration test set.
Classification of benign and malignant lesions
Based on the above results, two DLDA classifiers were selected for further evaluation. Each classifier generated a probability score between 0 and 1 that could be further interpreted as high risk for malignancy (score ≥ 0.5) or low risk for malignancy (score < 0.5). There was 100% qualitative agreement between the resubstituted and cross‐validated scores in the training set with an overall score variability of ±1.37% (Supplemental Figure 3). The classifier A, consisting of miR‐138‐1‐3p, ‐146b‐5p, ‐155, ‐204‐5p and ‐375, correctly identified 81 of 130 malignant cases (62%) as high risk and 103 of 110 benign cases (94%) as low risk (Table 3). The classifier B interrogated the same miRNAs plus miR‐29b‐1‐5p, ‐31‐5p, ‐139‐5p, ‐222‐3p, and ‐551b‐3p. It classified only 64 benign cases (58%) in the low risk category but correctly identified 118 malignant cases (91%) as high risk. Similar distribution of molecular results was observed in the independent set of thyroid nodule FNAs (Table 3). Overall, the classifier A efficiently identified 92% (233/252) of benign lesions as low risk while the classifier B efficiently identified 92% (205/223) of malignant lesions as high risk.
Table 3.
Distribution of molecular results in resected tissues and nodules aspirates
| Resected tissues (n = 240) | Nodule aspirates (n = 235) | |||||||
|---|---|---|---|---|---|---|---|---|
| Malignant (n = 130) | Benign (n = 110) | Malignant (n = 93) | Benign (n = 142) | |||||
| HR/Pos | LR/Neg | HR/Pos | LR/Neg | HR/Pos | LR/Neg | HR/Pos | LR/Neg | |
| miRNA classifier A | 81 | 49 | 7 | 103 | 71 | 22 | 12 | 130 |
| miRNA classifier B | 118 | 12 | 46 | 64 | 87 | 6 | 42 | 100 |
| 17‐mutation panel | 71 | 59 | 15 | 95 | 65 | 28 | 10 | 132 |
| Mutation + classifier A | 97 | 33 | 18 | 92 | 78 | 15 | 15 | 127 |
| Mutation + classifier B | 122 | 8 | 51 | 59 | 88 | 5 | 45 | 97 |
HR/Pos, high risk by miRNA testing or positive by mutation testing; LR/Neg, low risk by miRNA testing or negative by mutation testing (negative by both tests for combined testing).
Combination with mutation testing
All specimens were also tested for the presence of 17 well‐characterized oncogenic gene alterations in the BRAF, RAS, RET or PAX8 genes using the Luminex platform 5, 33. A correlation between genotype, miRNA expression and histology was observed in the resected specimen set (Figure 1A). Gene alterations were distributed as expected, with BRAF and RET‐PTC detected mainly in carcinomas with true papillary architecture (cPTC and TcPTC) and RAS and PAX8‐PPARG detected mainly in FvPTC and follicular neoplasms. In the training set, 55% (71/130) of the malignant cases were mutation‐positive and 86% (95/110) of the benign cases were negative (Table 3). Mutation and miRNA testing identified overlapping but distinct subsets of thyroid lesions. Among the 59 mutation‐negative malignant specimens, classifier A identified 26 cases as high risk (44%) and classifier B identified 51 cases as high risk (86%). The cancer detection rate was significantly increased from 55% for mutation testing alone to 75% for mutation testing combined with classifier A (97/130; p = 9.4E‐07 for McNemar test) or 94% for mutation testing combined with classifier B (122/130; p = 2.5E‐12). Similar results were obtained with preoperative thyroid nodule FNAs (Table 3). Mutation testing correctly identified 70% (65/93) of malignant nodules and the detection rate was increased to 95% (88/93, p = 4.5 E ‐06) for mutation testing combined with classifier B. In both the resected and FNA sample sets, the majority of mutation‐negative benign specimens were correctly identified as low risk by classifier A (219/227 or 96%), that is, the benign detection rate for combined testing was only slightly reduced compared to that of mutation testing alone (87% [219/252] versus 90% [227/252], p = 0.013).
Comparison with high‐throughput targeted sequencing
An optimized miRNA classification algorithm, commercially available as the ThyraMIR Thyroid miRNA Classifier (Interpace Diagnostics Group Inc., Parsippany, NJ) 32, was next evaluated in parallel to a targeted next‐generation sequencing (NGS) assay interrogating mutation hotspots in 20 genes and 46 aberrant transcripts (Supporting Information Table 2). In a subset of resected thyroid tissues negative by the 17‐mutation panel (n = 54), additional gene alterations were detected by NGS in 13 out of 36 malignant cases, corresponding to a potential increase in diagnostic sensitivity of 36% relative to the 17‐mutation panel (Figure 3A). With ThyraMIR, 14 of the malignant cases were classified as high risk/positive (39% increase in sensitivity relative to the 17‐mutation panel), including five specimens negative by NGS (Supplemental Figure 4). None of the 18 benign lesions was high risk by miRNA testing (100% specificity), while seven cases were positive by NGS, corresponding to a potential decrease in diagnostic specificity of 39%. The distribution of molecular results was similar in a set of 42 thyroid nodule FNAs with indeterminate cytology (Bethesda categories III or IV) that had not been used for classifier development (Supplemental Figure 4). Both NGS and miRNA testing improved the diagnostic sensitivity of the 17‐mutation panel but NGS decreased its specificity by detecting additional gene alterations in 19% of benign cases (Figure 3B). Among 30 nodules negative by the 17‐mutation panel, ThyraMIR correctly classified 67% of malignant cases as high risk/positive and 100% of benign cases as low risk/negative. Testing for miRNA expression combined with genotyping for 17 well‐characterized gene alterations resulted in optimal diagnostic sensitivity (85%) and specificity (95%).
Figure 3.
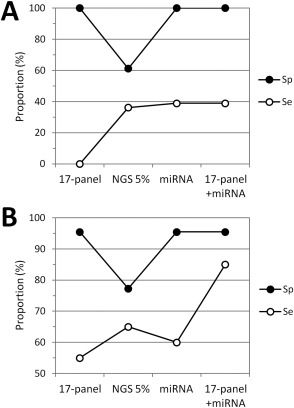
Comprehensive molecular characterization of thyroid lesions by miRNA gene expression and mutation testing. (A) Resected thyroid tissues. The graph shows the diagnostic specificity (Sp) and sensitivity (Se) for the panel of 17 gene alterations detected with the Luminex platform (17‐panel), next‐generation sequencing at 5% allele variant threshold (NGS 5%), the ThyraMIR test (miRNA), and for combination testing with the 17‐mutation panel and ThyraMIR in 54 resected tissues all negative with the 17‐mutation panel (100% specificity, 0% sensitivity). (B) Thyroid nodule aspirates. Same analysis for molecular testing performed in 42 aspirates from nodules with an indeterminate cytology diagnosis of atypia of undetermined significance/follicular lesion of undetermined significance (Bethesda category III) or follicular neoplasm/suspicious for a follicular neoplasm (Bethesda category IV).
Discussion
In the present study, we interrogated miRNA expression in a large and histologically diverse set of thyroid lesions and applied rigorous statistical analyses to the development of miRNA classification algorithms that discriminate between benign and malignant lesions. We show that multidimensional molecular analyses for miRNA expression, mRNA fusions and DNA mutations improve the predictive classification of thyroid lesions. Our data underline the relevance of miRNAs in the pathology of thyroid cancer and the value of combination testing in the preoperative risk‐based assessment of thyroid nodules with indeterminate cytology.
Ultrasound‐guided FNAs are the sample of choice for the diagnostic evaluation of thyroid nodules but present specific limitations for the development of a gene expression classifier. Those limitations include the imperfect traceability to the reference diagnosis, the unknown proportion of representative follicular cells in specimens dedicated to molecular testing and classifier training, and the potential contamination by other cellular components that may bias the expression levels of the biomarkers of interest. The latter is particularly significant for blood‐containing thyroid biopsies. For example, our data indicate that miR‐7‐5p and miR‐144‐5p, two miRNAs differentially expressed in FTC 22, 23, 24, 26, would not be dependable in a diagnostic FNA contaminated with a few percent of blood. Based on strict design requirements for test robustness and reliability, we elected to train our miRNA classification models using exclusively well‐characterized surgical specimens after exclusion of all the miRNAs that could interfere with the diagnostic signature (n = 8) or generate high signal variability or test failure (n = 4).
Following cross‐validation and evaluation of performance in an independent set of preoperative FNAs, this approach enabled us to select the most robust learning algorithm. Interestingly, algorithms with the highest initial AUC in the training set (random forest and support vector machine) showed the strongest drop in predictive performance after migration to FNA, suggesting that these statistical models overfitted the training data set. Overfitting is a well‐known problem in machine learning where overtraining complex algorithms on noisy features or training on a sample set too small or not diverse enough can lead to high training set performance that will not generalize well to a new data set 36. Significant decreases in predictive accuracy have previously been reported for miRNA classifiers that had optimal performance during training 27, 30. In contrast, we chose our DLDA classification model not based on the highest AUC during initial training but based on steady performance under leave‐one‐out cross‐validation and after testing in an independent set of nodule FNAs.
DLDA is a relatively simple linear classification model that assumes a common diagonal feature covariance matrix for the different classes to be predicted and is known to yield low error rates in situations where more complex predictors may suffer from overfitting 37. The parameters of the statistical model, estimated using the maximum likelihood method, can be easily constrained so that the sum of the model coefficients attributed to each miRNA predictor is equal to zero (built‐in normalization). The resulting classifier A contained two miRNAs down regulated in PTC, miR‐138‐1‐3p and miR‐204‐5p, and two miRNAs highly over expressed in PTC, miR‐146b‐5p and miR‐375 17, 18, 19, 20, 21. It also included the only miRNA overexpressed in CLT, miR‐155, which is known to control immune response and inflammation 38. The classifier B further interrogated two miRNAs with modest differential expression (miR‐139‐5p and miR‐29b‐1‐5p) and three miRNAs highly expressed in PTC (miR‐31‐5p) or in both PTC and FTC (miR‐222‐3p and miR‐551b‐3p) 16, 18, 19, 20, 21, 22, 25.
Noticeably, miR‐21 and miR‐221, two well‐known oncomiRs over expressed in many cancer types including thyroid cancer 15, 16, 19, 20, 21, 29, were not selected by the learning algorithms. Those miRNAs were outperformed by other candidates with similar expression patterns in various histopathological categories but better predictive performance, in particular miR‐146b‐5p and miR‐375 over miR‐21‐5p in PTC and miR‐222‐3p and miR‐551b‐3p over miR‐221‐3p/‐5p in FTC. However, our data did confirm that miR‐21 expression levels seem to correlate with aggressive histopathological features and may have prognostic value 31. Finally, several miRNAs such as miR‐7‐5p, ‐15a‐5p, ‐96‐5p, ‐187 or ‐885‐5p were valid candidates based on the literature and our data but were excluded based on our stringent design specifications. Previous work suggested that miR‐885 might be a good marker to discriminate oFTC from FTC and FA 25. Our data indicate that miR‐885‐5p is a general marker of oncocyticity over expressed in both oFTC and oFA, thus limiting its potential diagnostic utility at this time.
Importantly, predictive models interrogating the expression levels of a few miRNAs identified a subset of malignant lesions that lack oncogenic gene alterations. The sensitivity of mutation testing in thyroid tissues and nodule FNAs was 61% (136/223), in the range expected based from the literature 5, 6, and increased to 78% (175/223) when combined with classifier A (p = 1.2 E ‐09 for McNemar's test). Among the specimens positive by mutation and/or miRNA testing, the residual risk of cancer was high and unchanged relative to mutation testing alone (84%, p = 1.0). This performance would correspond to a calculated PPV of 60–80% in a given set of indeterminate thyroid nodules with a thyroid cancer prevalence of 20–40%. The sensitivity was further increased to 94% (210/223) when mutation testing was combined with classifier B (p = 2.1 E ‐17), leading to a low residual risk of cancer (8%) among specimens negative by mutation and miRNA testing. This performance would correspond to a calculated improvement from 78–90% NPV to 94–98% NPV at 20–40% cancer prevalence. These pivotal observations in our case‐control study suggest that miRNA testing could complement mutation testing and potentially improve the preoperative clinical management of patients with indeterminate thyroid nodules. Further optimization of the classifiers A and B resulted in the commercial ThyraMIR test that was recently evaluated in a multicenter cross‐sectional cohort study 32. As expected based on our data, mutation testing combined with ThyraMIR correctly ruled in 89% of malignant nodules with a PPV of 74% and correctly ruled out 85% of benign nodules with a NPV of 94% at 32% cancer prevalence 32.
An alternative method to improve the diagnostic performance of mutation testing is to interrogate additional genetic alterations associated with thyroid cancer. Single institution studies with unblinded surgical pathology review have previously shown that comprehensive genotyping by NGS can classify indeterminate nodules with high sensitivity and specificity 8, 39. Our data confirmed that expansive targeted NGS panels can increase test sensitivity in both resected tissues and indeterminate nodules. This higher sensitivity would correspond to a decrease in the number of false negative malignant nodules that are incorrectly ruled out, that is, an increase in NPV. However, NGS also uncovered additional gene alterations in specimens negative by ThyraMIR and classified as benign by independent pathologists from diverse endocrinology centres. Those cases included four mutations in TP53 (6–20% allele variant), two in TSHR (>30% allele variant), and four in HRAS, NRAS, PTEN or RET (5–8% allele variant) (Supplemental Table 3). TSHR mutations have previously been reported in benign nodules and low‐level mutations <10% of alleles may represent subclones within larger nodules that could be monitored for potential tumour growth by repeat FNA and molecular testing 8.
In term of diagnostic performance, this predictable decrease in specificity with NGS would correspond to an increase in the number of false positive benign nodules that are incorrectly ruled in and that may be referred for unnecessary surgery, that is, a decrease in PPV. In contrast, combination testing for miRNAs and a few well‐characterized oncogenic mutations provided a better balance between sensitivity and specificity. A key advantage of this strategy lies in its ability to rule in malignant nodules that lack oncogenic mutations and to rule out benign nodules with germline or low‐level somatic mutations of unknown clinical significance. Predictive performance could be further improved in the future by interrogating additional genetic alterations, provided they are highly specific for thyroid cancer. High test specificity is not only a requirement to efficiently rule in cancer with high PPV, it is also beneficial to increase the benign diagnostic yield, that is, the rate of benign nodules correctly ruled out by a test. For that reason, combination testing for oncogenic mutations and miRNA expression also represents a significant improvement over current mRNA‐based gene expression classification that can only rule out approximately 50% of benign nodules with indeterminate cytology 13, 32.
In summary, we have shown that miRNAs are practical surrogate markers to identify subsets of thyroid tumours with altered oncogenic pathways. This observation is consistent with their documented function in almost all aspects of cancer development and progression, and their strong association with distinct signaling changes, differentiation states and progression risks in thyroid cancer 15, 31. A direct application of our findings is that a molecular test combining an optimized miRNA classification algorithm with a validated panel of somatic driver mutations displays high diagnostic sensitivity and specificity. This novel approach can rule in cancer with high PPV and rule out cancer with high NPV, thus increasing the diagnostic yield of molecular cytology relative to current methods and further improving the preoperative risk‐based management of thyroid nodules with indeterminate cytology.
Author Contributions
DW designed the miRNAs classifiers, performed statistical analyses and generated figures and tables. SBH conceived the miRNA experiments, managed all specimens and collected the associated data. BCH conceived the next‐generation sequencing experiments and analyzed the results. TJG selected, reviewed and annotated the surgical specimens. EL designed the study, analyzed data and wrote the manuscript. All authors reviewed, revised and approved the manuscript before its submission.
Supporting information
The following supplementary material may be found in the online version of this article:
Supplemental Materials and Methods
Supplemental Table 1. Multiple hypothesis testing for differential miRNA expression in various histopathological categories
Supplemental Table 2. Genes targeted by next‐generation sequencing and number of PCR amplicons
Supplemental Table 3. Benign specimens negative by the ThyraMIR test and by the 17‐mutation panel but positive by targeted next‐generation sequencing
Supplemental Figure 1. Expression of 31 miRNAs in 240 resected tissue specimens used for classifier training. The strip plots show the expression levels (Ct) of each miRNA in specimens grouped and colour‐coded by histopathology category. Abbreviations: HN, hyperplastic nodule (includes diffuse hyperplasia); CLT, chronic lymphocytic thyroiditis; FA, follicular adenoma; oFA, oncocytic FA; FTC, follicular thyroid carcinoma; oFTC, oncocytic FTC; PTC, papillary thyroid carcinoma (includes conventional and tall cell variants); FvPTC, follicular variant of PTC.
Supplemental Figure 2. Comparison of miRNA expression profiles in resected thyroid tissues and preoperative thyroid nodule aspirates. Principal component (PC) analyses were performed for benign or malignant specimens using miRNA expression levels determined by RT‐qPCR.
Supplemental Figure 3. Comparison between resubstituted and cross‐validated scores in the 240 resected tissue specimens used for classifier training. The correlation plot (left) and the Bland and Altman bias plot (right) were built using n=960 scores generated with the classifiers A and B. The dashed lines in the bias plot represent the 95% limits of agreement or 1.96 times the standard deviation of the differences between resubstituted and cross‐validated scores (±0.0137).
Supplemental Figure 4. Sunburst plot showing the distribution of molecular results in a subset of resected tissues negative by the 17‐mutation panel (n = 54, left) and in an independent set of nodules aspirates indeterminate by cytology (n = 42, right). Each sample is represented by one spoke and the total numbers of samples within each category are indicated. From the inner‐most to the outer‐most circle, the chart shows the hierarchical relationships between surgical pathology (benign or malignant) and the molecular results obtained for the panel of 17 gene alterations detected with the Luminex platform (17‐panel), the ThyraMIR test (miRNA), and next‐generation sequencing (NGS) at 5% allele variant threshold.
Acknowledgements
The authors thank Sheila Puntnam and Michelle Vinco from the Department of Pathology at the University of Michigan Medical School for their assistance with the selection and annotation of surgical specimens as well as Mahima Bhatt, Andrew Hadd, Jeffrey Houghton, Julie Krosting, Laura Langfield and Rupali Shinde from Asuragen for the molecular characterization of surgical and preoperative specimens.
BCH is an employee of Asuragen Inc. EL, DW and SBH were employees of Asuragen Inc. at the time of the study. EL was a consultant to PDI, Inc. and TJG served as a scientific advisor for PDI Inc.
References
- 1. Hsiao SJ, Nikiforov YE. Molecular approaches to thyroid cancer diagnosis. Endocr Relat Cancer 2014; 21: T301–T313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer 2013; 13: 184–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xing M, Liu R, Liu X, et al BRAF V600E and TERT promoter mutations cooperatively identify the most aggressive papillary thyroid cancer with highest recurrence. J Clin Oncol 2014; 32: 2718–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Melo M, da Rocha AG, Vinagre J, et al TERT promoter mutations are a major indicator of poor outcome in differentiated thyroid carcinomas. J Clin Endocrinol Metab 2014; 99: E754–E765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beaudenon‐Huibregtse S, Alexander EK, Guttler RB, et al Centralized molecular testing for oncogenic gene mutations complements the local cytopathologic diagnosis of thyroid nodules. Thyroid 2014; 24: 1479–1487. [DOI] [PubMed] [Google Scholar]
- 6. Nikiforov YE, Ohori NP, Hodak SP, et al Impact of mutational testing on the diagnosis and management of patients with cytologically indeterminate thyroid nodules: a prospective analysis of 1056 FNA samples. J Clin Endocrinol Metab 2011; 96: 3390–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yip L, Wharry LI, Armstrong MJ, et al A clinical algorithm for fine‐needle aspiration molecular testing effectively guides the appropriate extent of initial thyroidectomy. Ann Surg 2014; 260: 163–168. [DOI] [PubMed] [Google Scholar]
- 8. Nikiforov YE, Carty SE, Chiosea SI, et al Highly accurate diagnosis of cancer in thyroid nodules with follicular neoplasm/suspicious for a follicular neoplasm cytology by ThyroSeq v2 next‐generation sequencing assay. Cancer 2014; 120: 3627–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brito JP, Yarur AJ, Prokop LJ, et al Prevalence of thyroid cancer in multinodular goiter versus single nodule: a systematic review and meta‐analysis. Thyroid 2013; 23: 449–455. [DOI] [PubMed] [Google Scholar]
- 10. Bongiovanni M, Spitale A, Faquin WC, et al The Bethesda System for Reporting Thyroid Cytopathology: a meta‐analysis. Acta Cytol 2012; 56: 333–339. [DOI] [PubMed] [Google Scholar]
- 11. Haugen BRM, Alexander EK, Bible KC, et al American Thyroid Association management guidelines for adult patients with thyroid nodules and differentiated thyroid cancer. Thyroid 2015; 26: 1–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Giordano TJ, Kuick R, Thomas DG, et al Molecular classification of papillary thyroid carcinoma: distinct BRAF, RAS, and RET/PTC mutation‐specific gene expression profiles discovered by DNA microarray analysis. Oncogene 2005; 24: 6646–6656. [DOI] [PubMed] [Google Scholar]
- 13. Alexander EK, Kennedy GC, Baloch ZW, et al Preoperative diagnosis of benign thyroid nodules with indeterminate cytology. N Engl J Med 2012; 367: 705–715. [DOI] [PubMed] [Google Scholar]
- 14. Chudova D, Wilde JI, Wang ET, et al Molecular classification of thyroid nodules using high‐dimensionality genomic data. J Clin Endocrinol Metab 2010; 95: 5296–5304. [DOI] [PubMed] [Google Scholar]
- 15. Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol 2014; 9: 287–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen YT, Kitabayashi N, Zhou XK, et al MicroRNA analysis as a potential diagnostic tool for papillary thyroid carcinoma. Mod Pathol 2008; 21: 1139–1146. [DOI] [PubMed] [Google Scholar]
- 17. Yip L, Kelly L, Shuai Y, et al MicroRNA signature distinguishes the degree of aggressiveness of papillary thyroid carcinoma. Ann Surg Oncol 2011; 18: 2035–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Swierniak M, Wojcicka A, Czetwertynska M, et al In‐depth characterization of the microRNA transcriptome in normal thyroid and papillary thyroid carcinoma. J Clin Endocrinol Metab 2013; 98: E1401–E1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. He H, Jazdzewski K, Li W, et al The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA 2005; 102: 19075–19080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sheu SY, Grabellus F, Schwertheim S, et al Differential miRNA expression profiles in variants of papillary thyroid carcinoma and encapsulated follicular thyroid tumours. Br J Cancer 2010; 102: 376–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dettmer M, Perren A, Moch H, et al Comprehensive MicroRNA expression profiling identifies novel markers in follicular variant of papillary thyroid carcinoma. Thyroid 2013; 23: 1383–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lassalle S, Hofman V, Ilie M, et al Can the microRNA signature distinguish between thyroid tumors of uncertain malignant potential and other well‐differentiated tumors of the thyroid gland? Endocr Relat Cancer 2011; 18: 579–594. [DOI] [PubMed] [Google Scholar]
- 23. Kitano M, Rahbari R, Patterson EE, et al Expression profiling of difficult‐to‐diagnose thyroid histologic subtypes shows distinct expression profiles and identify candidate diagnostic microRNAs. Ann Surg Oncol 2011; 18: 3443–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stokowy T, Wojtas B, Krajewska J, et al A two miRNA classifier differentiates follicular thyroid carcinomas from follicular thyroid adenomas. Mol Cell Endocrinol 2015; 399: 43–49. [DOI] [PubMed] [Google Scholar]
- 25. Dettmer M, Vogetseder A, Durso MB, et al MicroRNA expression array identifies novel diagnostic markers for conventional and oncocytic follicular thyroid carcinomas. J Clin Endocrinol Metab 2013; 98: E1–E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rossing M, Borup R, Henao R, et al Down‐regulation of microRNAs controlling tumourigenic factors in follicular thyroid carcinoma. J Mol Endocrinol 2012; 48: 11–23. [DOI] [PubMed] [Google Scholar]
- 27. Keutgen XM, Filicori F, Crowley MJ, et al A panel of four miRNAs accurately differentiates malignant from benign indeterminate thyroid lesions on fine needle aspiration. Clin Cancer Res 2012; 18: 2032–2038. [DOI] [PubMed] [Google Scholar]
- 28. Kitano M, Rahbari R, Patterson EE, et al Evaluation of candidate diagnostic microRNAs in thyroid fine‐needle aspiration biopsy samples. Thyroid 2012; 22: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nikiforova MN, Tseng GC, Steward D, et al MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab 2008; 93: 1600–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Agretti P, Ferrarini E, Rago T, et al MicroRNA expression profile helps to distinguish benign nodules from papillary thyroid carcinomas starting from cells of fine‐needle aspiration. Eur J Endocrinol 2012; 167: 393–400. [DOI] [PubMed] [Google Scholar]
- 31. Cancer Genome Atlas Research Network . Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014; 159: 676–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Labourier E, Shifrin A, Busseniers AE, et al Molecular Testing for miRNA, mRNA, and DNA on Fine‐Needle Aspiration Improves the Preoperative Diagnosis of Thyroid Nodules With Indeterminate Cytology. J Clin Endocrinol Metab 2015; 100: 2743–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Giordano TJ, Beaudenon‐Huibregtse S, Shinde R, et al Molecular testing for oncogenic gene mutations in thyroid lesions: a case‐control validation study in 413 postsurgical specimens. Hum Pathol 2014; 45: 1339–1347. [DOI] [PubMed] [Google Scholar]
- 34. Hadd AG, Houghton J, Choudhary A, et al Targeted, high‐depth, next‐generation sequencing of cancer genes in formalin‐fixed, paraffin‐embedded and fine‐needle aspiration tumor specimens. J Mol Diagn 2013; 15: 234–247. [DOI] [PubMed] [Google Scholar]
- 35. Sah S, Chen L, Houghton J, et al Functional DNA quantification guides accurate next‐generation sequencing mutation detection in formalin‐fixed, paraffin‐embedded tumor biopsies. Genome Med 2013; 5: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kourou K, Exarchos TP, Exarchos KP, et al Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J 2015; 13: 8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dudoit S, Fridlyand J, Speed TP. Comparison of Discrimination Methods for the Classification of Tumors Using Gene Expression Data. J Amer Statist Assoc 2002; 97: 77–87. [Google Scholar]
- 38. Seddiki N, Brezar V, Ruffin N, et al Role of miR‐155 in the regulation of lymphocyte immune function and disease. Immunology 2014; 142: 32–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nikiforov YE, Carty SE, Chiosea SI, et al Impact of the Multi‐Gene ThyroSeq Next‐Generation Sequencing Assay on Cancer Diagnosis in Thyroid Nodules with Atypia of Undetermined Significance/Follicular Lesion of Undetermined Significance Cytology. Thyroid 2015; 25: 1217–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following supplementary material may be found in the online version of this article:
Supplemental Materials and Methods
Supplemental Table 1. Multiple hypothesis testing for differential miRNA expression in various histopathological categories
Supplemental Table 2. Genes targeted by next‐generation sequencing and number of PCR amplicons
Supplemental Table 3. Benign specimens negative by the ThyraMIR test and by the 17‐mutation panel but positive by targeted next‐generation sequencing
Supplemental Figure 1. Expression of 31 miRNAs in 240 resected tissue specimens used for classifier training. The strip plots show the expression levels (Ct) of each miRNA in specimens grouped and colour‐coded by histopathology category. Abbreviations: HN, hyperplastic nodule (includes diffuse hyperplasia); CLT, chronic lymphocytic thyroiditis; FA, follicular adenoma; oFA, oncocytic FA; FTC, follicular thyroid carcinoma; oFTC, oncocytic FTC; PTC, papillary thyroid carcinoma (includes conventional and tall cell variants); FvPTC, follicular variant of PTC.
Supplemental Figure 2. Comparison of miRNA expression profiles in resected thyroid tissues and preoperative thyroid nodule aspirates. Principal component (PC) analyses were performed for benign or malignant specimens using miRNA expression levels determined by RT‐qPCR.
Supplemental Figure 3. Comparison between resubstituted and cross‐validated scores in the 240 resected tissue specimens used for classifier training. The correlation plot (left) and the Bland and Altman bias plot (right) were built using n=960 scores generated with the classifiers A and B. The dashed lines in the bias plot represent the 95% limits of agreement or 1.96 times the standard deviation of the differences between resubstituted and cross‐validated scores (±0.0137).
Supplemental Figure 4. Sunburst plot showing the distribution of molecular results in a subset of resected tissues negative by the 17‐mutation panel (n = 54, left) and in an independent set of nodules aspirates indeterminate by cytology (n = 42, right). Each sample is represented by one spoke and the total numbers of samples within each category are indicated. From the inner‐most to the outer‐most circle, the chart shows the hierarchical relationships between surgical pathology (benign or malignant) and the molecular results obtained for the panel of 17 gene alterations detected with the Luminex platform (17‐panel), the ThyraMIR test (miRNA), and next‐generation sequencing (NGS) at 5% allele variant threshold.


