Abstract
The Wnt/β-catenin signaling pathway controls embryonic development and adult stem cell maintenance through the regulation of transcription. Failure to downregulate Wnt signaling can result in embryonic malformations and cancer, highlighting the important role of negative regulators of the pathway. The Wnt pathway activates several negative feedback targets, including axin2 and Dkk1, that function at different levels of the signaling cascade; however, none have been identified that directly target active β-catenin/Tcf1 transcriptional complexes. We show that Zfp703 is a Wnt target gene that inhibits Wnt/β-catenin activity in Wnt reporter assays and in Wnt-dependent mesoderm differentiation in embryonic stem cells. Zfp703 binds directly to Tcf1 to inhibit β-catenin/Tcf1 complex formation and does so independently of the Groucho/Tle transcriptional corepressor. We propose that Zfp703 is a novel feedback suppressor of Wnt/β-catenin signaling that functions by inhibiting the association of β-catenin with Tcf1 on Wnt response elements in target gene enhancers.
INTRODUCTION
Wnt/β-catenin signaling plays a key role in promoting the growth and self-renewal of stem cell and progenitor populations during embryonic development and adult tissue homeostasis (1). Negative regulators of Wnt/β-catenin signaling play important roles in controlling growth and preventing the occurrence of cancer (1). This is best illustrated by the adenomatous polyposis coli (APC) tumor suppressor that functions in the β-catenin destruction complex to limit Wnt signaling and is commonly mutated in gastrointestinal cancers (2, 3). Additionally, some Wnt/β-catenin inhibitors are themselves transcriptional targets of Wnt and thus function as feedback suppressors (i.e., axin2, Nkd1, ΔN-LEF1, and Dkk1) (4–11). As these feedback suppressors keep Wnt/β-catenin signaling in check, loss-of-function (LOF) mutations or epigenetic silencing of these genes can result in embryonic malformation or cancers (10, 12–14). Thus, identifying feedback suppressors, and understanding where in the signaling cascade they function, may assist in the identification of drugs that affect cancer cell growth and survival.
Wnt signaling in vertebrates is regulated by a family of 20 conserved genes, several of which are capable of signaling through the β-catenin pathway (15). In the absence of a Wnt ligand, Wnt target gene expression is repressed by the binding of an inhibitory complex, which includes the HMG-box transcription factor Tcf3 (Tcf7l1) and Groucho/Tle corepressors, to Tcf/Lef binding sites on target gene regulatory elements (8, 16–19). With the stimulation of signaling, through the presence of Wnt ligand or mutations that activate signaling (20), stabilized β-catenin enters the nucleus and binds to Tcf1 (Tcf7), Tcf4 (Tcf7l2), or Lef1. Activating β-catenin/Tcf1 complexes subsequently bind to Tcf/Lef binding sites to displace inhibitory Tcf3/Groucho complexes (4, 8, 17, 21–24). β-Catenin/Tcf1 complexes directly activate target genes, including genes that encode components of the Wnt signaling pathway itself (e.g., Lef1).
During mouse axial development, Wnt3a/β-catenin signaling controls posterior progenitor maintenance and differentiation (25–28). Genetic removal of Wnt3a, Tcf1/Lef1, or β-catenin in posterior progenitors leads to downregulation of Wnt/β-catenin target genes (29–35) and disrupts paraxial mesoderm (PM) formation (25, 31, 36, 37). Here, we describe a functional screen to identify inhibitors of Wnt/β-catenin signaling from candidate genes arising from a transcriptome analysis of embryonic day 7.75 (E7.75) Wnt3a−/− embryos (29) and Lef1 and Sp5 gain-of-function (GOF) embryonic stem cells (ESCs). We show that the Nlz-like zinc finger transcription factor gene Zfp703 (also known as Csmn1, End2, Zeppo1, Zpo1, and NLZ1), the mixed paired-like homeobox gene Mixl1, and caudal-type homeobox genes (Cdx1, -2, and -4) all possess inhibitory activity in Wnt reporter assays. We characterize the embryonic expression of Zfp703 and describe a molecular mechanism by which it may antagonize the β-catenin/Tcf1 signaling complex.
MATERIALS AND METHODS
Microarray, RNA-Seq analysis, and candidate gene selection.
Microarray analysis data for control and Wnt3a−/− node and primitive streak (PS) were previously deposited in the GEO database (accession number GSE29995). To enhance the likelihood of identifying target genes that lie downstream of the Wnt3a signaling pathway, the list of differentially downregulated genes (P value, ≤0.05) identified in Wnt3a−/− embryos by microarray analysis was further refined by comparing it to the list of genes identified by transcriptome sequencing (RNA-Seq) that were upregulated in ESCs by the overexpression of the Wnt effectors Lef1 and/or Sp5 (false discovery rate [q value], ≤0.05) (GEO accession number GSE73084). This candidate list included 114 genes, of which 24 genes were tested for inhibitory activity in the Super TOPFlash (STF) luciferase assay (see Table S1 in the supplemental material).
Super TOPFlash screen and luciferase assays.
Selected candidate genes were PCR amplified from E7.5-to-E9.5 whole-embryo cDNA or mouse ESC cDNA and cloned into pcDNA3.1-V5-His TOPO TA expression vector (Life Technologies). Three hundred nanograms of pcDNA3.1-V5-His candidate constructs was cotransfected with 100 ng of Super TOPFlash reporter and 20 ng of thymidine kinase (TK)-Renilla luciferase using 3 μl of FuGene HD/1 μg of DNA (Promega). Empty pcDNA3.1-V5-His vector served as a negative control. Super FOPFlash (SFF) reporter constructs carrying mutated Tcf/Lef binding sites were also used as controls for specificity of Tcf/Lef activity in some experiments. Luciferase activity was assayed according to Dual-Luciferase assay kit instructions (Promega). For luciferase assays shown in Fig. 2F, 854 bp of the Zfp703 promoter, including 836 bp upstream of the transcription start site (TSS) and 19 bp of 5′ untranslated region upstream of the ATG, was cloned into pGL4.10 (luc2) vector and transfected into differentiating (day 2) A2lox.Wnt3a ESCs (see below) cultured in N2B27 serum-independent medium. Wnt3a expression was induced by the addition of 1 μg/ml doxycycline (Dox) to the culture medium. Alternatively, Wnt/β-catenin signaling was activated in A2lox.Wnt3a cells by treatment with 3 μM CHIR99021 (Stemgent).
FIG 2.
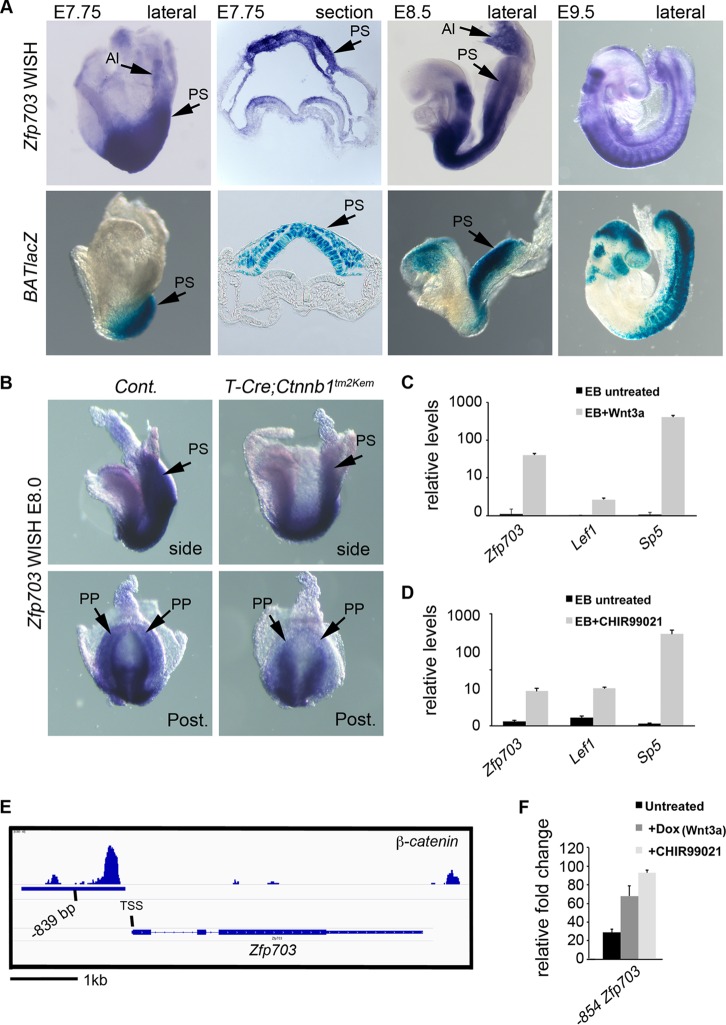
Zfp703 is a direct target gene of Wnt/β-catenin signaling. (A) Whole-mount in situ hybridization (WISH) detection of Zfp703 transcripts (top panels) and comparison to BATlacZ (Wnt/β-catenin/Tcf1-Lef1 reporter) β-galactosidase-stained mouse embryos (bottom panels) (stages E7.75 to E9.5). Sections (second panels from left) are through the PS region with the ingressed PS region marked with black arrows. (B) WISH analysis of Zfp703 expression in E8.0 control (left) and mutant (right) embryos lacking β-catenin function in the PS (T-Cretg/+; Ctnnb1Δ/flox). Posterior views (bottom panels) show that Zfp703 expression was reduced in posterior progenitors in the PS. (C) qPCR of Zfp703 expression in untreated and Wnt3a-treated mouse embryoid bodies, compared to Wnt target genes Lef1 and Sp5. (D) qPCR of Zfp703 expression in untreated and Wnt agonist CHIR99021-treated mouse embryoid bodies, compared to Wnt target genes Lef1 and Sp5. (E) Genomic map of β-catenin binding to the mouse Zfp703 locus with center of peak indicated as bp −839 from the transcriptional start site. (F) Luciferase assay of 854 bp upstream from ATG (includes 836-bp upstream putative regulatory region from transcription start site) of Zfp703 under untreated and β-catenin-stimulating conditions (Wnt3a and/or CHIR99021) relative to vector control. Abbreviations: Al, allantois; PS, primitive streak; PP, posterior progenitors; EB, embryoid bodies.
Quantitative reverse transcription-PCR (RT-qPCR)–ChIP-qPCR.
RNA was isolated using Tri reagent (Ambion) and converted to cDNA using the iScript cDNA synthesis kit (Bio-Rad). Real-time PCR was performed using universal SYBR green Supermix (Bio-Rad) with specific oligonucleotide primer pairs for each gene (see Table S2 in the supplemental material) using a cycle of 95°C for 10 s for denaturing and 60°C for 30 s for annealing and extension. Chromatin immunoprecipitation-qPCR (ChIP-qPCR) utilized primers encompassing Tcf/Lef binding sites on Msgn1, T, Axin2, and Cdx1 (38) and negative-control regions (see Table S2 in the supplemental material).
Embryos and gene expression analysis.
Embryos for detection of Zfp703 mRNA were dissected from pregnant NIH-Swiss females in phosphate-buffered saline (PBS) prior to fixation in 4% paraformaldehyde. Embryos from BATlacZ β-catenin/Tcf1-Lef1 lacZ reporter transgenic mice were used to localize Wnt signaling as described previously (27). Ctnnb1tm2Kem (floxed β-catenin loss-of-function [LOF] allele) mice were crossed to the T-Cre driver line to conditionally delete β-catenin in the PS of mutant embryos as described previously (25, 39, 40). Embryos were assayed by whole-mount in situ hybridization (WISH) as described previously (41) with antisense RNA probes synthesized from mouse Zfp703 using digoxigenin-UTP (Roche) and developed using nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate (NBT-BCIP) (Roche). Embryos were photographed in PBS plus 0.1% Tween 20 (PBT) using a Leica MZFLlII microscope and an AxioCam digital camera and AxioVision software (Zeiss). Paraffin sectioning was performed by standard methods to a thickness of 5 μm and imaged on a Zeiss Axioplan II microscope. This study was carried out in compliance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (Frederick National Laboratory Animal Care and Use Committee proposal 12-408). Rodents were euthanized by CO2 inhalation in accordance with the most recent AVMA Guidelines for the Euthanasia of Animals (61).
ESC lines, culture, and immunocytochemistry.
To generate Dox-inducible Wnt3a, epitope control (3×Flag), and epitope-tagged Zfp703 (3×Flag-Zfp703) and Cdx2 (3×Flag-Cdx2) GOF ESCs, cDNAs were first cloned into p2lox plasmid to generate targeting vectors suitable for inducible cassette exchange recombination (42). One day before electroporation, parental A2Lox.Cre ESCs that constitutively express rtTA from the ROSA26 locus were treated with 1 μg/ml Dox to induce Cre recombinase expression from a Tet-responsive element (TRE)-Cre-Δneo cassette inserted into the Hprt locus (42). These cells were then electroporated with the targeting constructs. Successful cassette exchange results in the replacement of Cre with the cDNA of interest downstream of the TRE and the restoration of the selectable PGK-neo cassette. Targeted cell lines were selected using resistance to 300 μg/ml G418 antibiotic for 1 to 2 weeks as previously described (43). At least 2 independent clones for each construct were characterized for targeted-gene expression and protein subcellular localization. ESC culture and differentiation protocols were performed as previously described (29). Briefly, ESCs were cultured on mitomycin C mitotically inactivated mouse embryonic fibroblasts (MEFs) in Dulbecco's modified Eagle medium (DMEM) plus 15% fetal bovine serum (FBS), penicillin-streptomycin (Pen-Strep), and 2-mercaptoethanol. For ESC differentiation, MEFs were removed from ESCs and differentiated as embryoid bodies (EBs) for 2 days on 10-cm petri dishes (Fisher Scientific) in ESC medium containing 10% FBS and 50 μg/ml l-ascorbic acid (Sigma). On day 2, EBs were transferred to ultralow-attachment dishes (Costar), and Dox (1 μg/ml) was added to the medium to induce gene expression. EBs were subsequently cultured for another 24 to 72 h.
For immunocytochemistry, EBs were transferred to Matrigel (Corning)-coated glass coverslips and cultured for another 24 h with or without Dox. Cells were fixed in 4% paraformaldehyde for 10 min at room temperature and permeabilized in 0.1% cold Triton X-100 for 5 min. Cells were blocked in 1% bovine serum albumin (BSA) in PBS for 30 min and incubated with primary antibody (anti-Flag M2, 1:1,000) followed by fluorescently labeled secondary antibodies. Images were acquired using an Axioplan II (Zeiss) microscope.
Immunoprecipitation and Western analysis.
Immunoprecipitations were performed on nuclear extracts using antibodies to Tcf1 (Cell Signaling Technology catalog no. 2203S), pan-Tle (Santa Cruz catalog no. sc-13373), β-catenin (Sigma catalog no. C7207), and Flag M2 (Sigma catalog no. F3165) using protein G-Sepharose beads (GE Healthcare) preblocked in 1% BSA. Extracts were separated on a 10% Bis-Tris gel using morpholinepropanesulfonic acid (MOPS) buffer (Invitrogen) and transferred to nitrocellulose membranes using standard procedures. Blots were blocked in 5% fat-free milk in Tris-buffered saline (TBS) and incubated with primary antibodies to Tcf1, pan-Tle, Flag M2, β-catenin (Santa Cruz catalog no. sc-7199), V5 (Thermo Scientific catalog no. R960-25, Sigma catalog no. V8137), and actin (Chemicon catalog no. MAB1501) followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies (Promega). SuperSignal West Pico chemiluminescent substrate (Thermo Scientific) was used to detect the protein.
Chromatin immunoprecipitation (ChIP).
Noninduced 3×Flag ESCs and 3×Flag-Zfp703 ESCs were differentiated as EBs for 2 days. Dox (1 μg/ml) was added on day 2 to induce expression of 3×Flag and 3×Flag-Zfp703. EBs were fixed for 15 min in 1% formaldehyde-HEPES buffer at room temperature as previously described and quenched in 0.125 M glycine solution (43). Cells were lysed in lysis buffer, and chromatin was sheared to 100 to 300 bp. The protein-DNA complexes were immunoprecipitated using anti-β-catenin (BD Transduction Laboratory; catalog no. 610154) and IgG (Cell Signaling; catalog no. 2729) as a negative control. The protein-DNA complexes were treated with proteinase K to digest proteins, and DNA was purified and suspended in TE buffer (43). Enrichment of immunoprecipitated DNA was assessed using Universal Sybr green (Bio-Rad)-based quantitative PCR.
RESULTS
Functional screen for Wnt feedback suppressors.
A multiparametric approach was taken to winnow a list of 379 potential Wnt3a target genes, i.e., genes that are downregulated in E8.5 Wnt3a−/− node and PS regions (29), down to a manageable size to screen for Wnt feedback suppressors. We compared the list of Wnt3a-dependent genes identified from mutant embryos (29) with the lists of genes upregulated in mouse ESCs by the overexpression of the Wnt effectors Lef1 and Sp5. Twenty-four genes that were dependent on Wnt3a for expression and upregulated by Lef1 and Sp5 and/or were previously associated with the Wnt/β-catenin pathway were selected for further analysis (see Table S1 in the supplemental material for the complete list of genes).
To test for inhibitory activity of candidates, we used the Super TOPFlash (STF) Wnt/β-catenin/Tcf luciferase reporter assays performed in 293T human embryonic kidney (HEK) cells (44). As the STF assay reports the activity of β-catenin/Tcf complexes, it is a direct method to identify potential transcriptional inhibitors; however, a caveat to this approach is that the candidate genes might indirectly regulate STF activity through transcriptional modulation of other Wnt component genes. We identified Cdx2, Zfp703, Mixl1, Cdx1, and Cdx4 as Wnt target genes that inhibit STF activity (in order of greatest to least inhibition) (Fig. 1B). Interestingly, Zfp703 and Cdx genes have previously been shown to lower Wnt/β-catenin signaling in cancer models, suggesting that our embryo screen successfully identified Wnt pathway inhibitors (45–47). We narrowed this list down to genes that consistently suppressed STF activity by at least 50% for further validation in mouse ESCs. Although Cdx2 and Zfp703 consistently inhibited STF activity in 293T cells (Fig. 1B; see also Fig. S1A and B in the supplemental material), Cdx2 inhibition of Wnt signaling in ESCs was inconsistent presumably because Cdx2 also activated Sp5, a prominent direct Wnt target gene in the mouse PS that functions to activate Wnt target genes (29, 48) (see Fig. S1E and F). We therefore focused on Zfp703, a zinc finger transcription factor gene with a known association with luminal breast cancer (49, 50) but an uncharacterized role in the mouse embryo.
FIG 1.
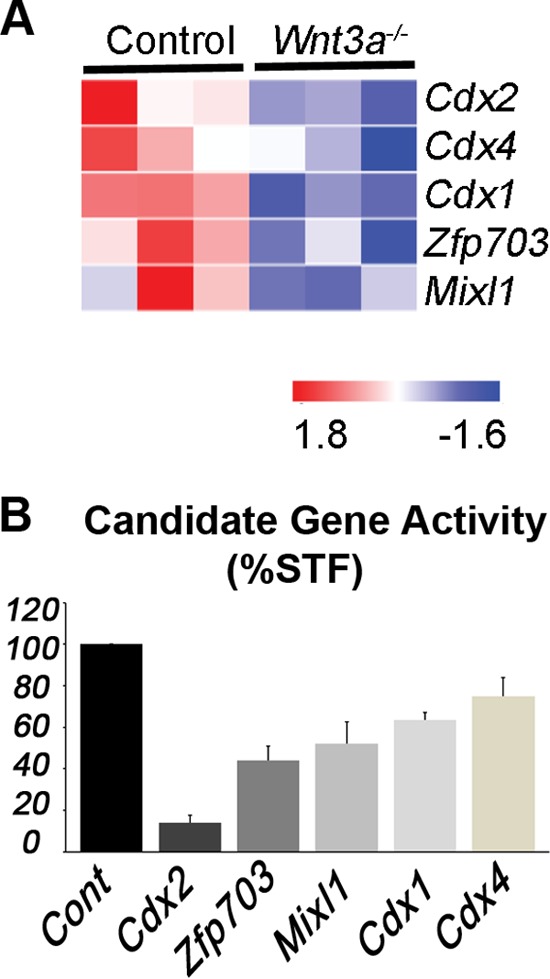
Functional screen for Wnt3a-regulated Wnt pathway antagonists. (A) Heat map of differentially expressed genes in wild-type control (n = 3, biological replicates) and Wnt3a−/− (n = 3, biological replicates) node/PS that showed inhibitory activity in STF reporter assays. Colors represent log intensity values (red, upregulated; blue, downregulated). (B) STF luciferase activity (stimulated by Wnt3a treatment) is reduced by the expression of candidate genes in HEK 293T cells (average plus standard deviation from 3 or more independent experiments).
Zfp703 is a Wnt/β-catenin target gene in developing embryos and differentiating ESCs.
To determine if Zfp703 was spatially and temporally expressed in the appropriate domain to regulate Wnt/β-catenin signaling in vivo, we compared Zfp703 expression by whole-mount in situ hybridization (WISH) to that of BATlacZ (a transgenic Wnt/β-catenin/Tcf β-galactosidase reporter) (27) in developing mouse embryos. We focused our analysis of Zfp703 transcript expression on the E7.75-to-E9.5 stages (Fig. 2A; see also Fig. S2 in the supplemental material), when Wnt3a is first required for PS development (25, 37). Further analysis of Zfp703 expression in mouse embryos is provided in the supplemental material (see Fig. S2). At E7.75 and E8.5, Zfp703 was detected in the posterior embryo in the caudal neural plate, PS, and allantois (Fig. 2A, top 3 left panels), including the posterior progenitors that require Wnt/β-catenin signaling (25). Comparison of Zfp703 expression to that of BATlacZ showed that Zfp703 is initially expressed at sites of Wnt activity, particularly at E7.75 (Fig. 2A, left 2 panels). At E8.5, Zfp703 and BATlacZ showed continued overlapping sites of expression in the PS region (Fig. 2A); however, Zfp703 expression often occurred in regions of the embryo with reduced BATlacZ activity. In particular, comparison of E8.5 and E9.5 Zfp703 and BATlacZ expression levels showed complementary expression patterns in the developing brain (Zfp703 was expressed in the hindbrain while BATlacZ was expressed in the adjacent midbrain) but considerable overlap in posterior regions of the embryo (Fig. 2A, right two panels). These data suggest that Zfp703 is expressed appropriately to be both a target of, and to potentially influence, Wnt signaling.
To determine if Zfp703 expression is regulated by Wnt/β-catenin signaling, we conditionally deleted β-catenin using a floxed LOF allele of β-catenin (Ctnnb1tm2Kem) excised under the control of T-Cre to inhibit all Wnt/β-catenin signaling in posterior progenitors (Fig. 2B) (25). Zfp703 expression was reduced in the PS in E8.0 T-Cretg/+; Ctnnb1Δ/flox mutants compared to controls (Fig. 2B), suggesting that Wnt regulates Zfp703 in the PS region but that additional pathways are also involved in the maintenance of expression. In addition, Zfp703 was upregulated in differentiating embryoid bodies (EBs) by the addition of Wnt3a or the Wnt pathway agonist CHIR99021 (51, 52) (Fig. 2C and D), indicating that Zfp703 is regulated by Wnt/β-catenin signaling. This relationship appears evolutionarily conserved, as similar results are observed with the zebrafish homolog (53).
Genome-wide analysis of β-catenin distribution by chromatin immunoprecipitation sequencing (ChIP-seq) (54) identified a β-catenin binding region at the Zfp703 locus that is centered at bp −839 from the transcriptional start site. In silico analysis shows a high level of sequence conservation in this putative regulatory region in mammals (Fig. 2E; see also Fig. S3A in the supplemental material). This β-catenin peak contains three consensus Tcf/Lef binding sites (see Fig. S3B); however, only the most proximal Tcf/Lef site is present in the conserved putative upstream regulatory region. To test if this conserved region is responsive to Wnt/β-catenin signaling, we transfected a luciferase reporter construct containing this putative DNA regulatory element into ESCs and found that it was indeed activated by Wnt3a and CHIR99021 (Fig. 2F). Taken together, these results are consistent with the Wnt/β-catenin signaling pathway directly regulating Zfp703 expression.
Zfp703 inhibits Wnt/β-catenin signaling in 293T cells and differentiating mouse embryonic stem cells.
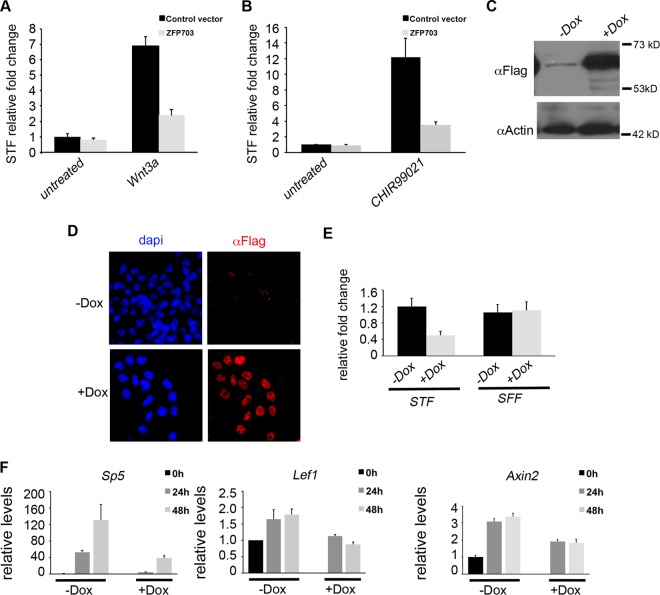
Our independent demonstration that Zfp703 can inhibit STF Wnt reporter activity stimulated by Wnt3a (Fig. 3A), or CHIR99021 (Fig. 3B), confirms similar previously published results (45, 53). The differentiation of pluripotent epiblast stem cells into the embryo germ layers is an early and important developmental event that is controlled by Wnt signaling and is closely modeled in vitro by the differentiation of ESCs (34). To determine whether Zfp703 has inhibitory activity in differentiating ESCs, an N-terminus triple-Flag-tagged Zfp703 (3F-Zfp703) cDNA was targeted to the Hprt locus of A2lox.Cre ESCs for doxycycline (Dox)-inducible expression as previously described (42, 43). Addition of Dox to the culture medium induced the expression of 3F-Zfp703 (Fig. 3C; see also Fig. S4A in the supplemental material). 3F-Zfp703 protein was localized to the nucleus (Fig. 3D) consistent with its function as a regulator of gene expression. Dox-induced Zfp703 blocked STF activity in differentiating ESCs by 67.2% compared to untreated cells, whereas it had negligible effects on the SFF control (Fig. 3E). Similarly, induction of Zfp703 also strongly inhibited the endogenous expression of the Wnt-responsive target genes Sp5, Lef1, and Axin2 (Fig. 3F) but had no effect on the expression of the housekeeping gene Gapdh (see Fig. S4C). These data demonstrate that Zfp703 can function as an inhibitor of the Wnt/β-catenin signaling pathway in differentiating ESCs.
FIG 3.
Zfp703 inhibits Wnt signaling in HEK 293T kidney cells and mouse embryonic stem cells. (A and B) Comparisons of transfected Zfp703 and control vector activity in Super TOPFlash (STF) Tcf/Lef luciferase reporter assays in untreated, Wnt3a (A)-, and CHIR99021 (B)-treated 293T cells. (C) Immunoblotting assay of A2lox.3F-Zfp703 mouse ESCs (with [+] or without [−] Dox]) detected 3F-Zfp703 at the expected molecular mass (62 kDa). (D) Immunocytochemistry of 3F-Zfp703 expression in A2lox.3F-Zfp703 mouse ESCs (with or without Dox) using anti-Flag antibodies and 4′,6-diamidino-2-phenylindole (DAPI) as a nuclear marker. (E) Luciferase assay of STF and Tcf/Lef-mutated Super FOPFlash (SFF) control reporter in A2lox.3F-Zfp703 mouse ESCs with or without Dox. (F) qPCR of Wnt target genes in differentiating A2lox.3F-Zfp703 mouse ESCs with or without Dox.
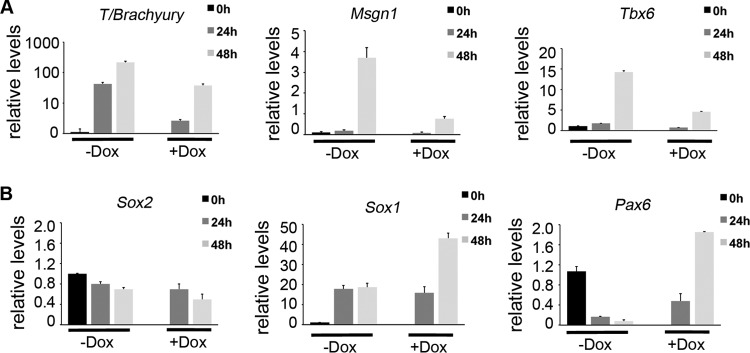
A biological readout of Wnt signaling in differentiating ESCs is the induction of mesoderm determination genes (34, 55, 56). Zfp703 overexpression inhibited the transcription of the Wnt-regulated mesodermal genes Brachyury (T), Msgn1, and Tbx6 (Fig. 4A) (31, 43, 57). In posterior embryonic development, inhibiting Wnt signaling promotes neural/spinal cord differentiation over paraxial mesodermal differentiation (36, 58). Examination of neural progenitor markers Sox2, Sox1, and Pax6 showed a marked increase in Sox1 and Pax6 but not Sox2, indicating that expression of Zfp703 was promoting some neural gene expression (Fig. 4B). Taken together, the data suggest that Zfp703 inhibits Wnt signaling and Wnt-dependent developmental processes such as mesoderm formation.
FIG 4.
Zfp703 inhibits mesoderm determination genes and activates neural markers. (A) qPCR of Wnt-dependent mesoderm determination genes in differentiating A2lox.3F-Zfp703 mouse ESCs with or without Dox. (B) qPCR of neural progenitor genes in differentiating A2lox.3F-Zfp703 mouse ESCs with or without Dox.
Zfp703 interacts with Tcf1 to destabilize Tcf1/β-catenin complex formation.
In light of our findings that Zfp703 inhibits both Wnt/β-catenin/Tcf reporter activation and Wnt/β-catenin/Tcf target gene expression, we asked if Zfp703 could do so by destabilizing the β-catenin/Tcf1 transcriptional complex. Coimmunoprecipitation (co-IP) analysis results for endogenous Tcf1 and β-catenin protein complexes in HEK 293T cells transfected with control and those transfected with Zfp703-V5-His-expressing vector were compared. Remarkably, the expression of exogenous Zfp703-V5-His resulted in a substantial reduction in Tcf1 bound to β-catenin (Fig. 5A). A possible explanation for reduced Tcf1/β-catenin complex formation is a Zfp703-mediated reduction in the nuclear levels of either β-catenin or Tcf1. Comparisons of the input levels of nuclear β-catenin and Tcf1 showed that they were unaffected by Zfp703 expression (Fig. 5A). These data suggest that Zfp703 expression is blocking Wnt signaling by disrupting the Tcf1 and β-catenin interactions in HEK 293T cells.
FIG 5.
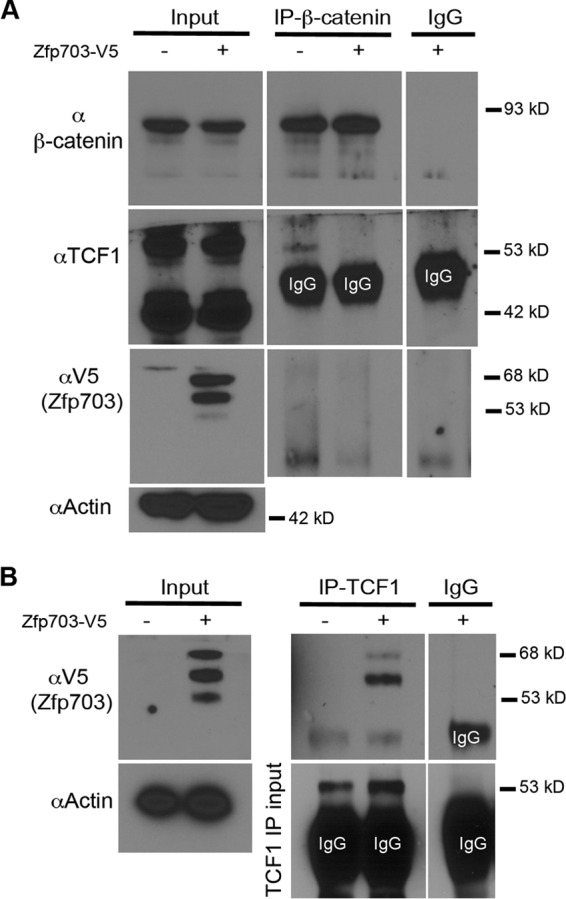
Zfp703 binds to Tcf1 and disrupts binding of β-catenin in 293T kidney cells. (A) Co-IP of endogenous β-catenin with endogenous Tcf1 in HEK 293T cells transfected with empty vector or Zfp703-V5-His. Equal amounts of lysate were used in co-IP experiments as illustrated by β-catenin, Tcf1, and actin input immunoblot assays. IgG precipitates were used as control. (B) Immunoblot assays with a V5 antibody to detect transfected Zfp703-V5-His in co-IPs of endogenous Tcf1 in HEK 293T cells. IgG precipitates were used as controls.
As Zfp703 can block β-catenin/Tcf1-mediated transcription stimulated by CHIR99021, we considered whether Zfp703 could associate with Tcf1. To test this, we performed co-IP from nuclear extracts isolated from Zfp703-V5-His-transfected HEK 293T cells. Co-IP experiments showed an interaction between endogenous TCF1 and exogenous Zfp703-V5-His (Fig. 5B). However, we were unable to detect interaction between endogenous β-catenin and exogenous Zfp703-V5-His (Fig. 5A), although the same samples showed an interaction between β-catenin and TCF1 (Fig. 5A). This result suggests that the ZFP703 interaction with TCF1 could be displacing β-catenin and thereby inhibiting Wnt signaling in HEK 293T cells.
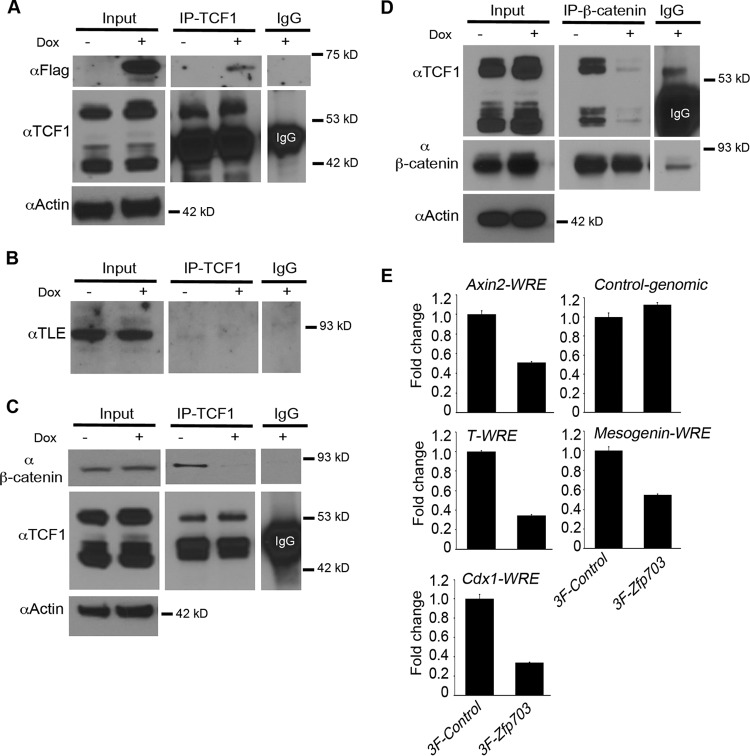
Since Zfp703 inhibited Wnt signaling and mesodermal gene induction in differentiating ESCs, we further explored the molecular mechanism of Zfp703 activity in ESCs. We performed co-IP from nuclear extracts isolated from 3F-Zfp703 ESCs with or without Dox treatments. Co-IP experiments support an interaction between endogenous Tcf1 and 3F-Zfp703 (Fig. 6A). Zfp703 is a known transcriptional corepressor that is thought to function through its interaction with Groucho/Tle proteins (45). To examine whether Groucho/Tle proteins could be identified in the Zfp703-Tcf1 immunoprecipitates, immunoblotting assays were performed; however, Groucho/Tle protein was not identified (Fig. 6B). Similarly, Tcf1 was not identified when Tle was immunoprecipitated (see Fig. S5 in the supplemental material), even in the absence of Zfp703. This observation is consistent with the function of Tcf1 in the active β-catenin/Tcf1 transcriptional complex, i.e., the Wnt ”ON” state, and not in association with Groucho/Tle inhibitory complexes (8, 24). These data suggest that Zfp703 binds to components of the active β-catenin/Tcf1 complex, independently of Groucho/Tle.
FIG 6.
Zfp703 binds to Tcf1 and disrupts binding of β-catenin. (A) 3F-Zfp703 was coimmunoprecipitated (co-IP) with endogenous Tcf1 in A2lox.3F-Zfp703 mouse ESCs (with or without Dox). Equal amounts of lysate were used in co-IP experiments as illustrated by Tcf1 and actin immunoblot assays. IgG precipitates were used as control. (B) Co-IPs of endogenous Tcf1 and Groucho/Tle using a pan-Tle antibody in A2lox.3F-Zfp703 mouse ESCs (with or without Dox). IgG precipitates were used as controls. (C) Co-IPs of endogenous Tcf1 and endogenous β-catenin in A2lox.3F-Zfp703 mouse ESCs (with or without Dox). Equal loadings used in co-IP experiments are evident from the actin immunoblot assay. Equal amounts of immunoprecipitated Tcf1 are shown by the anti-Tcf1 immunoblot assay. IgG precipitates were used as controls. (D) The reciprocal experiment of panel C showing co-IPs of endogenous β-catenin and endogenous Tcf1 in A2lox.3F-Zfp703 mouse ESCs (with or without Dox). Equal loadings used in co-IP experiments are evident from the actin immunoblot assay. Equal amounts of immunoprecipitated β-catenin are shown by anti-β-catenin immunoblot assay. (E) ChIP-qPCR of endogenous β-catenin-precipitated samples in A2lox.3F and A2lox.3F-Zfp703 mouse ESCs (with Dox) at Wnt response elements (WREs) of Wnt target genes. Levels of background β-catenin binding to control genomic regions were similar between 3F-Control cells and 3F-Zfp703 cells.
To characterize the Wnt-inhibiting activity of Zfp703 in ESCs, we compared co-IPs of endogenous Tcf1 and β-catenin from Dox− and Dox+ cells. As in the HEK 293T cells (Fig. 5), the expression of exogenous 3F-Zfp703 in ESCs resulted in a substantial reduction in β-catenin bound to Tcf1, regardless of whether Tcf1 or β-catenin was immunoprecipitated (Fig. 6C and D). Additionally, we observed no difference in the input levels of β-catenin and Tcf1 by Zfp703 expression (Fig. 6C and D; see also Fig. S4B in the supplemental material). Interestingly, levels of Lef1 mRNA (Fig. 3F) and protein (see Fig. S4B) were decreased by Zfp703, presumably because Lef1 is a Wnt target gene (11). Nevertheless, lower Lef1 levels might exacerbate the Zfp703-mediated inhibition of Wnt signaling. Reduced Tcf1 and Lef1 signaling is notable since the simultaneous loss of both genes results in a severe posterior axis truncation phenotype (36) that is remarkably similar to the Wnt3a−/− and T-Cre; Ctnnb1Δ/flox mutant phenotypes (25, 37).
If Zfp703 disrupts binding of β-catenin to Tcf1, then it follows that induction of Zfp703 in differentiating ESCs should inhibit the association of β-catenin with Tcf/Lef binding sites at Wnt target gene enhancers. To directly test this hypothesis, we performed ChIP for endogenous β-catenin on Wnt target genes in Dox-treated 3F-Zfp703 ESCs and 3×Flag control ESCs. ChIP-qPCR showed a reduced association of β-catenin on Wnt response elements (WREs) of several Wnt target genes in the presence of exogenous Zfp703 (Fig. 6E). The WREs assayed at the Axin2 (5), T/Brachyury (31), Cdx1 (35), and Msgn1 (43) loci are well characterized and contain previously validated functional Tcf/Lef binding sites. These data suggest that Zfp703 inhibits binding of β-catenin with Tcf1 at Tcf/Lef binding sites of Wnt-regulated genes.
DISCUSSION
We utilized HEK 293T cells, which have all the necessary components for Wnt/β-catenin signaling (59), and the well-characterized Super TOPFlash Wnt/β-catenin/Tcf reporter assay to screen for Wnt pathway suppressors. We hypothesized that testing putative Wnt target genes, initially identified in mouse embryos, in a heterologous system (human embryonic kidney cells) might best identify genes with a propensity to function as general Wnt feedback suppressors. Members of the Cdx transcription factor family were identified as strong inhibitors of Wnt signaling, at least in 293T cells. These results are consistent with the Wnt-inhibiting activity of Cdx2 observed in some tumor cells (46, 47). However, in the context of heterogeneous, differentiating mouse ESCs, Cdx2 both promoted the expression of the direct Wnt target gene Sp5 and modestly suppressed the Wnt target gene T/Brachyury, suggesting that Cdx2 activity is cell context dependent. Given the abundant studies of Cdx genes (60), we chose to focus on the little-studied transcription factor Zfp703, which appeared to unequivocally function as a Wnt pathway suppressor.
We show that Zfp703 can function as a feedback suppressor of Wnt/β-catenin signaling through the inhibition of the active nuclear β-catenin/Tcf complex. Feedback suppressors characterized to date typically target the pathway at or near the membrane, affecting ligand-receptor interactions and β-catenin stabilization (4). To the best of our knowledge, Zfp703 is unique, as it appears to target the activated β-catenin/Tcf complex directly. Truncated LEF1 protein (ΔN-LEF1), which is unable to bind to β-catenin, functions at a similar level as Zfp703, presumably occupying TCF/LEF binding sites at Wnt response elements to exclude the binding of active β-catenin/Tcf complexes (10). However, as Zfp703 directly, or indirectly, binds to Tcf1 to inhibit its association with β-catenin, it may have a more immediate effect on active Wnt target gene transcription, even in the presence of abundant ligand or high β-catenin levels. This activity could function to rapidly inhibit or dampen Wnt signaling in cellular microenvironments where Wnt signaling is actively occurring. Although it seems likely that Zfp703 binds to Tcf1 and not β-catenin, given that Zfp703 associates with Tcf1 even when β-catenin is displaced, it remains unclear how that binding leads to β-catenin displacement. Further studies are needed to characterize the binding targets of Zfp703 and to understand how Zfp703 disrupts the association of Tcf1 and β-catenin. Regardless, our data support a role for Zfp703 as an inhibitor of active Wnt signaling.
Interestingly, Zfp703 regulates cancer stem cell numbers in invasive luminal breast cancer (49, 50), wherein Zfp703 binds to Groucho/Tle proteins presumably functioning in the repressor complex to inhibit Wnt signaling (45). Zfp703 and its related homolog Zfp503 function similarly in breast cancer cells and may form a complex together (45), suggesting that these factors may show redundant activities in LOF studies. Ongoing genetic experiments will clarify a role for Zfp703 and Zfp503 in Wnt target gene repression and in posterior embryonic development. Factors like Zfp703 show great promise as major regulators of Wnt signaling, as Zfp703 can block signaling in two different ways: in conjunction with repressor Groucho/Tle complexes and through the inhibition of Tcf1/Lef1 active transcription complexes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alan Perantoni for providing materials and support and Ruth Wolfe for assistance with mouse colony management.
M.W.K. designed the screen. A.K. and R.B.C. performed the majority of experiments with assistance from M.W.K., R.J.G., S.T., and S.N.I. R.J.G. and T.P.Y. wrote the manuscript.
Funding Statement
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01010-15.
REFERENCES
- 1.Clevers H, Nusse R. 2012. Wnt/beta-catenin signaling and disease. Cell 149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 2.Fodde R, Brabletz T. 2007. Wnt/beta-catenin signaling in cancer stemness and malignant behavior. Curr Opin Cell Biol 19:150–158. doi: 10.1016/j.ceb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Network. 2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu F, van den Broek O, Destree O, Hoppler S. 2005. Distinct roles for Xenopus Tcf/Lef genes in mediating specific responses to Wnt/beta-catenin signalling in mesoderm development. Development 132:5375–5385. doi: 10.1242/dev.02152. [DOI] [PubMed] [Google Scholar]
- 5.Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. 2002. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol 22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niida A, Hiroko T, Kasai M, Furukawa Y, Nakamura Y, Suzuki Y, Sugano S, Akiyama T. 2004. DKK1, a negative regulator of Wnt signaling, is a target of the beta-catenin/TCF pathway. Oncogene 23:8520–8526. doi: 10.1038/sj.onc.1207892. [DOI] [PubMed] [Google Scholar]
- 7.Yan D, Wallingford JB, Sun TQ, Nelson AM, Sakanaka C, Reinhard C, Harland RM, Fantl WJ, Williams LT. 2001. Cell autonomous regulation of multiple Dishevelled-dependent pathways by mammalian Nkd. Proc Natl Acad Sci U S A 98:3802–3807. doi: 10.1073/pnas.071041898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cadigan KM, Waterman ML. 2012. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol 4(11):a007906. doi: 10.1101/cshperspect.a007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng W, Wharton KA Jr, Mack JA, Wang K, Gadbaw M, Suyama K, Klein PS, Scott MP. 2000. naked cuticle encodes an inducible antagonist of Wnt signalling. Nature 403:789–795. doi: 10.1038/35001615. [DOI] [PubMed] [Google Scholar]
- 10.Hovanes K, Li TW, Munguia JE, Truong T, Milovanovic T, Lawrence Marsh J, Holcombe RF, Waterman ML. 2001. Beta-catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nat Genet 28:53–57. doi: 10.1038/ng0501-53. [DOI] [PubMed] [Google Scholar]
- 11.Filali M, Cheng N, Abbott D, Leontiev V, Engelhardt JF. 2002. Wnt-3A/beta-catenin signaling induces transcription from the LEF-1 promoter. J Biol Chem 277:33398–33410. doi: 10.1074/jbc.M107977200. [DOI] [PubMed] [Google Scholar]
- 12.Aguilera O, Fraga MF, Ballestar E, Paz MF, Herranz M, Espada J, Garcia JM, Munoz A, Esteller M, Gonzalez-Sancho JM. 2006. Epigenetic inactivation of the Wnt antagonist DICKKOPF-1 (DKK-1) gene in human colorectal cancer. Oncogene 25:4116–4121. doi: 10.1038/sj.onc.1209439. [DOI] [PubMed] [Google Scholar]
- 13.Guo J, Cagatay T, Zhou G, Chan CC, Blythe S, Suyama K, Zheng L, Pan K, Qian C, Hamelin R, Thibodeau SN, Klein PS, Wharton KA, Liu W. 2009. Mutations in the human naked cuticle homolog NKD1 found in colorectal cancer alter Wnt/Dvl/beta-catenin signaling. PLoS One 4:e7982. doi: 10.1371/journal.pone.0007982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salahshor S, Woodgett JR. 2005. The links between axin and carcinogenesis. J Clin Pathol 58:225–236. doi: 10.1136/jcp.2003.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garriock RJ, Warkman AS, Meadows SM, D'Agostino S, Krieg PA. 2007. Census of vertebrate Wnt genes: isolation and developmental expression of Xenopus Wnt2, Wnt3, Wnt9a, Wnt9b, Wnt10a, and Wnt16. Dev Dyn 236:1249–1258. doi: 10.1002/dvdy.21156. [DOI] [PubMed] [Google Scholar]
- 16.Yi F, Pereira L, Hoffman JA, Shy BR, Yuen CM, Liu DR, Merrill BJ. 2011. Opposing effects of Tcf3 and Tcf1 control Wnt stimulation of embryonic stem cell self-renewal. Nat Cell Biol 13:762–770. doi: 10.1038/ncb2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hikasa H, Ezan J, Itoh K, Li X, Klymkowsky MW, Sokol SY. 2010. Regulation of TCF3 by Wnt-dependent phosphorylation during vertebrate axis specification. Dev Cell 19:521–532. doi: 10.1016/j.devcel.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A. 1998. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature 395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- 19.Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, van de Wetering M, Destree O, Clevers H. 1998. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature 395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- 20.Polakis P. 2012. Wnt signaling in cancer. Cold Spring Harb Perspect Biol 4(5):a008052. doi: 10.1101/cshperspect.a008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanson AJ, Wallace HA, Freeman TJ, Beauchamp RD, Lee LA, Lee E. 2012. XIAP monoubiquitylates Groucho/TLE to promote canonical Wnt signaling. Mol Cell 45:619–628. doi: 10.1016/j.molcel.2011.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. 1996. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature 382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 23.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. 1996. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell 86:391–399. doi: 10.1016/S0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 24.Chodaparambil JV, Pate KT, Hepler MR, Tsai BP, Muthurajan UM, Luger K, Waterman ML, Weis WI. 2014. Molecular functions of the TLE tetramerization domain in Wnt target gene repression. EMBO J 33:719–731. doi: 10.1002/embj.201387188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunty WC Jr, Biris KK, Chalamalasetty RB, Taketo MM, Lewandoski M, Yamaguchi TP. 2008. Wnt3a/beta-catenin signaling controls posterior body development by coordinating mesoderm formation and segmentation. Development 135:85–94. [DOI] [PubMed] [Google Scholar]
- 26.Garriock RJ, Chalamalasetty RB, Kennedy MW, Canizales LC, Lewandoski M, Yamaguchi TP. 2015. Lineage tracing of neuromesodermal progenitors reveals novel Wnt-dependent roles in trunk progenitor cell maintenance and differentiation. Development 142:1628–1638. doi: 10.1242/dev.111922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakaya MA, Biris K, Tsukiyama T, Jaime S, Rawls JA, Yamaguchi TP. 2005. Wnt3a links left-right determination with segmentation and anteroposterior axis elongation. Development 132:5425–5436. doi: 10.1242/dev.02149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB, Volpin D, Bressan GM, Piccolo S. 2003. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci U S A 100:3299–3304. doi: 10.1073/pnas.0434590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunty WC Jr, Kennedy MW, Chalamalasetty RB, Campbell K, Yamaguchi TP. 2014. Transcriptional profiling of Wnt3a mutants identifies Sp transcription factors as essential effectors of the Wnt/beta-catenin pathway in neuromesodermal stem cells. PLoS One 9:e87018. doi: 10.1371/journal.pone.0087018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chalamalasetty RB, Garriock RJ, Dunty WC Jr, Kennedy MW, Jailwala P, Si H, Yamaguchi TP. 2014. Mesogenin 1 is a master regulator of paraxial presomitic mesoderm differentiation. Development 141:4285–4297. doi: 10.1242/dev.110908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamaguchi TP, Takada S, Yoshikawa Y, Wu N, McMahon AP. 1999. T (Brachyury) is a direct target of Wnt3a during paraxial mesoderm specification. Genes Dev 13:3185–3190. doi: 10.1101/gad.13.24.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galceran J, Sustmann C, Hsu SC, Folberth S, Grosschedl R. 2004. LEF1-mediated regulation of Delta-like1 links Wnt and Notch signaling in somitogenesis. Genes Dev 18:2718–2723. doi: 10.1101/gad.1249504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hofmann M, Schuster-Gossler K, Watabe-Rudolph M, Aulehla A, Herrmann BG, Gossler A. 2004. WNT signaling, in synergy with T/TBX6, controls Notch signaling by regulating Dll1 expression in the presomitic mesoderm of mouse embryos. Genes Dev 18:2712–2717. doi: 10.1101/gad.1248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.ten Berge D, Koole W, Fuerer C, Fish M, Eroglu E, Nusse R. 2008. Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell Stem Cell 3:508–518. doi: 10.1016/j.stem.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pilon N, Oh K, Sylvestre JR, Savory JG, Lohnes D. 2007. Wnt signaling is a key mediator of Cdx1 expression in vivo. Development 134:2315–2323. doi: 10.1242/dev.001206. [DOI] [PubMed] [Google Scholar]
- 36.Galceran J, Farinas I, Depew MJ, Clevers H, Grosschedl R. 1999. Wnt3a−/−-like phenotype and limb deficiency in Lef1−/− Tcf1−/− mice. Genes Dev 13:709–717. doi: 10.1101/gad.13.6.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takada S, Stark KL, Shea MJ, Vassileva G, McMahon JA, McMahon AP. 1994. Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev 8:174–189. doi: 10.1101/gad.8.2.174. [DOI] [PubMed] [Google Scholar]
- 38.Wu CI, Hoffman JA, Shy BR, Ford EM, Fuchs E, Nguyen H, Merrill BJ. 2012. Function of Wnt/beta-catenin in counteracting Tcf3 repression through the Tcf3-beta-catenin interaction. Development 139:2118–2129. doi: 10.1242/dev.076067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R. 2001. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128:1253–1264. [DOI] [PubMed] [Google Scholar]
- 40.Perantoni AO, Timofeeva O, Naillat F, Richman C, Pajni-Underwood S, Wilson C, Vainio S, Dove LF, Lewandoski M. 2005. Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development 132:3859–3871. doi: 10.1242/dev.01945. [DOI] [PubMed] [Google Scholar]
- 41.Biris KK, Dunty WC Jr, Yamaguchi TP. 2007. Mouse Ripply2 is downstream of Wnt3a and is dynamically expressed during somitogenesis. Dev Dyn 236:3167–3172. doi: 10.1002/dvdy.21342. [DOI] [PubMed] [Google Scholar]
- 42.Iacovino M, Bosnakovski D, Fey H, Rux D, Bajwa G, Mahen E, Mitanoska A, Xu Z, Kyba M. 2011. Inducible cassette exchange: a rapid and efficient system enabling conditional gene expression in embryonic stem and primary cells. Stem Cells 29:1580–1588. doi: 10.1002/stem.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chalamalasetty RB, Dunty WC Jr, Biris KK, Ajima R, Iacovino M, Beisaw A, Feigenbaum L, Chapman DL, Yoon JK, Kyba M, Yamaguchi TP. 2011. The Wnt3a/beta-catenin target gene Mesogenin1 controls the segmentation clock by activating a Notch signalling program. Nat Commun 2:390. doi: 10.1038/ncomms1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. 2003. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol 13:680–685. doi: 10.1016/S0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 45.Slorach EM, Chou J, Werb Z. 2011. Zeppo1 is a novel metastasis promoter that represses E-cadherin expression and regulates p120-catenin isoform expression and localization. Genes Dev 25:471–484. doi: 10.1101/gad.1998111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu X, Zhang X, Zhan Q, Brock MV, Herman JG, Guo M. 2012. CDX2 serves as a Wnt signaling inhibitor and is frequently methylated in lung cancer. Cancer Biol Ther 13:1152–1157. doi: 10.4161/cbt.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hryniuk A, Grainger S, Savory JG, Lohnes D. 2014. Cdx1 and Cdx2 function as tumor suppressors. J Biol Chem 289:33343–33354. doi: 10.1074/jbc.M114.583823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weidinger G, Thorpe CJ, Wuennenberg-Stapleton K, Ngai J, Moon RT. 2005. The Sp1-related transcription factors sp5 and sp5-like act downstream of Wnt/beta-catenin signaling in mesoderm and neuroectoderm patterning. Curr Biol 15:489–500. doi: 10.1016/j.cub.2005.01.041. [DOI] [PubMed] [Google Scholar]
- 49.Ginestier C, Sircoulomb F, Charafe-Jauffret E, Chaffanet M, Birnbaum D. 2011. ZNF703: a novel oncogene involved in breast cancer. Med Sci (Paris) 27:357–359. (In French.) doi: 10.1051/medsci/2011274008. [DOI] [PubMed] [Google Scholar]
- 50.Sircoulomb F, Nicolas N, Ferrari A, Finetti P, Bekhouche I, Rousselet E, Lonigro A, Adelaide J, Baudelet E, Esteyries S, Wicinski J, Audebert S, Charafe-Jauffret E, Jacquemier J, Lopez M, Borg JP, Sotiriou C, Popovici C, Bertucci F, Birnbaum D, Chaffanet M, Ginestier C. 2011. ZNF703 gene amplification at 8p12 specifies luminal B breast cancer. EMBO Mol Med 3:153–166. doi: 10.1002/emmm.201100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ring DB, Johnson KW, Henriksen EJ, Nuss JM, Goff D, Kinnick TR, Ma ST, Reeder JW, Samuels I, Slabiak T, Wagman AS, Hammond ME, Harrison SD. 2003. Selective glycogen synthase kinase 3 inhibitors potentiate insulin activation of glucose transport and utilization in vitro and in vivo. Diabetes 52:588–595. doi: 10.2337/diabetes.52.3.588. [DOI] [PubMed] [Google Scholar]
- 52.Bennett CN, Ross SE, Longo KA, Bajnok L, Hemati N, Johnson KW, Harrison SD, MacDougald OA. 2002. Regulation of Wnt signaling during adipogenesis. J Biol Chem 277:30998–31004. doi: 10.1074/jbc.M204527200. [DOI] [PubMed] [Google Scholar]
- 53.Dutta S, Sriskanda S, Boobalan E, Alur RP, Elkahloun A, Brooks BP. 2015. nlz1 is required for cilia formation in zebrafish embryogenesis. Dev Biol 406:203–211. doi: 10.1016/j.ydbio.2015.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X, Peterson KA, Liu XS, McMahon AP, Ohba S. 2013. Gene regulatory networks mediating canonical Wnt signal-directed control of pluripotency and differentiation in embryo stem cells. Stem Cells 31:2667–2679. doi: 10.1002/stem.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gadue P, Huber TL, Paddison PJ, Keller GM. 2006. Wnt and TGF-beta signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc Natl Acad Sci U S A 103:16806–16811. doi: 10.1073/pnas.0603916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lindsley RC, Gill JG, Murphy TL, Langer EM, Cai M, Mashayekhi M, Wang W, Niwa N, Nerbonne JM, Kyba M, Murphy KM. 2008. Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem Cell 3:55–68. doi: 10.1016/j.stem.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szeto DP, Kimelman D. 2004. Combinatorial gene regulation by Bmp and Wnt in zebrafish posterior mesoderm formation. Development 131:3751–3760. doi: 10.1242/dev.01236. [DOI] [PubMed] [Google Scholar]
- 58.Yoshikawa Y, Fujimori T, McMahon AP, Takada S. 1997. Evidence that absence of Wnt-3a signaling promotes neuralization instead of paraxial mesoderm development in the mouse. Dev Biol 183:234–242. doi: 10.1006/dbio.1997.8502. [DOI] [PubMed] [Google Scholar]
- 59.Gujral TS, MacBeath G. 2010. A system-wide investigation of the dynamics of Wnt signaling reveals novel phases of transcriptional regulation. PLoS One 5:e10024. doi: 10.1371/journal.pone.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mallo M, Wellik DM, Deschamps J. 2010. Hox genes and regional patterning of the vertebrate body plan. Dev Biol 344:7–15. doi: 10.1016/j.ydbio.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.American Veterinary Medical Association. 2013. AVMA guidelines for the euthanasia of animals. American Veterinary Medical Association, Schaumburg, IL: https://www.avma.org/KB/Policies/Pages/Euthanasia-Guidelines.aspx. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.