Abstract
pVHL, the protein product of the von Hippel-Lindau (VHL) tumor suppressor gene, is a ubiquitin ligase that targets hypoxia-inducible factor α (HIF-α) for proteasomal degradation. Although HIF-α activation is necessary for VHL disease pathogenesis, constitutive activation of HIF-α alone did not induce renal clear cell carcinomas and pheochromocytomas in mice, suggesting the involvement of an HIF-α-independent pathway in VHL pathogenesis. Here, we show that the transcription factor B-Myb is a pVHL substrate that is degraded via the ubiquitin-proteasome pathway and that vascular endothelial growth factor (VEGF)- and/or platelet-derived growth factor (PDGF)-dependent tyrosine 15 phosphorylation of B-Myb prevents its degradation. Mice injected with B-Myb knockdown 786-O cells developed dramatically larger tumors than those bearing control cell tumors. Microarray screening of B-Myb-regulated genes showed that the expression of HIF-α-dependent genes was not affected by B-Myb knockdown, indicating that B-Myb prevents HIF-α-dependent tumorigenesis through an HIF-α-independent pathway. These data indicate that the regulation of B-Myb by pVHL plays a critical role in VHL disease.
INTRODUCTION
Ubiquitin-mediated proteolysis by the 26S proteasome plays an important role in the elimination of short-lived proteins (1), including those involved in cell cycle progression, cellular signaling in response to environmental stress or extracellular ligands, morphogenesis, secretion, DNA repair, and organelle biogenesis (2, 3). The pathway consists of two key steps, namely, the covalent attachment of multiple ubiquitin molecules to a target protein and the degradation of the ubiquitinated protein by the 26S proteasome complex. Several components act in concert to attach ubiquitin to a target protein, including a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin-protein isopeptide ligase (E3). E3 is directly responsible for substrate recognition. On the basis of structural similarity, E3 enzymes are classified into three families: the HECT (homologous to E6-AP COOH terminus) family, the U-box family, and the RING finger-containing protein family.
The elongin B and C-Cul2 or Cul5-SOCS box protein (ECS) family belongs to the cullin RING ligase (CRL) superfamily (4). pVHL, the protein product of the von Hippel-Lindau (VHL) tumor suppressor gene, is a member of the ECS family. pVHL forms a complex with elongins B and C, Cul2, and the RING finger protein Rbx1 (5, 6). The CRL2pVHL complex has ubiquitin ligase activity and targets the hypoxia-inducible factor α (HIF-α) family of transcription factors (HIF-1 to -3α) for proteasomal degradation (7). At normal oxygen levels, proline residues in the LXXLAP sequence motif of HIF-α proteins are hydroxylated by three prolyl hydroxylases (PHD1 to -3), and an in-depth study revealed that PHD2 is a critical enzyme for the hydroxylation of HIF-1α (8, 9). Hydroxylated HIF-α is targeted by pVHL for polyubiquitination and proteasomal degradation (10–12). Under conditions of hypoxia (low oxygen level), HIF-α is not hydroxylated by PHDs and is therefore not recognized or targeted for degradation by pVHL. The unhydroxylated HIF-α dimerizes with constitutively expressed HIF-1β, also known as an aryl hydrocarbon receptor nuclear translocator (ARNT), and translocates to the nucleus, where it induces the transcription of downstream target genes, including the genes coding for vascular endothelial growth factor A (VEGFA), solute carrier family 2 member 1 (SLC2A1; also known as GLUT1), and platelet-derived growth factor (PDGF) (13). Loss of functional pVHL protein prevents the O2-dependent degradation of HIF-α, resulting in constitutive expression of HIF-dependent genes and VHL disease, which is characterized by a variety of lesions, including hemangioblastomas, renal clear cell carcinomas, pheochromocytomas, pancreatic islet cell tumors, endolymphatic sac tumors, and papillary cystadenomas of the broad ligament (females) and epididymis (males) (13).
Studies showing that heterozygous pVHL+/− mice are phenotypically normal and VHL−/− mice die at embryonic day 10.5 (E10.5) to E12.5 (14), together with the existence of many pVHL-interacting proteins (13), and the fact that constitutive activation of HIF-α alone is not sufficient for the development of renal clear cell carcinomas and pheochromocytomas in mice (15) suggest the involvement of an HIF-α-independent pathway in VHL pathogenesis.
The v-Myb myeloblastosis viral oncogene homolog (avian)-like 2 gene (MYBL2 [B-Myb]) encodes a transcription factor involved in the regulation of cell cycle progression, differentiation, and apoptosis (16). The Myb gene family consists of three members, namely, A-Myb, B-Myb, and c-Myb, which share a highly conserved DNA-binding domain and bind to the consensus sequence PyAACNG, although they have different biological roles (17). Mammalian A-Myb and c-Myb are expressed in restricted cell types and stages of development; however, B-Myb was found to be expressed in all proliferating cells analyzed (18). A-Myb plays a critical role in spermatogenesis and mammary gland development (19). c-Myb regulates the proliferation and differentiation of hematopoietic stem and progenitor cells (20). B-Myb is a cell cycle-regulated protein that is maximally induced at the G1/S boundary and during S phase (16). Cyclin A/cdk2 phosphorylates and activates B-Myb (21), and phosphorylated B-Myb is polyubiquitinated and degraded by ectopically overexpressed ubiquitin ligase Skp2 (22). Although B-Myb is highly expressed at the G1/S boundary and during S phase, it regulates the G2/M transition (23). Further studies showed that B-Myb is involved in chromosomal condensation and stability (24). B-Myb forms a complex with clathrin and filamin, and this complex is required for the normal localization of clathrin, which stabilizes kinetochore fibers, at the mitotic spindle (25). Among Myb family members, only B-Myb is expressed in embryonic stem (ES) cells, and B-Myb-deficient mice die at an early stage of development, at approximately E4.5 to E6.5 (26). Knockdown of B-Myb in ES cells results in delayed G2/M transition, severe mitotic spindle and centrosome defects, and aneuploidy (27). Furthermore, knockdown of B-Myb results in the downregulation of OCT3/4 and thereby the differentiation of ES cells (27).
In the present study, B-Myb was identified as a substrate of the pVHL ubiquitin ligase complex, which targets it for degradation via the ubiquitin-proteasome pathway. VEGF-dependent tyrosine 15 phosphorylation of B-Myb prevented its degradation, and B-Myb knockdown 786-O cells generated larger tumors in vivo than control cells, indicating that B-Myb prevents tumorigenesis. Microarray screening showed that the expression of HIF-α-dependent genes was not affected by B-Myb knockdown. These data suggested that B-Myb functions via an HIF-α-independent pathway to regulate VHL pathogenesis.
MATERIALS AND METHODS
Plasmid construction.
Human A-Myb (GenBank/EBI accession no. NM_001144755.2), B-Myb (GenBank/EBI accession no. NM_002466.2), and pVHL (GenBank/EBI accession no. NM_000551.3) cDNAs were amplified by PCR from a 293T cDNA library and introduced into pcDNA3-puro or pMX-puro. c-Myb (GenBank/EBI accession no. NM_005375.3) was amplified by PCR from a human thymus cDNA library and contains a deletion of 48 to 71 amino acids, which is considered an isoform.
Knockdown.
Knockdown of B-Myb and Skp2 was performed as described previously (28). The target sequences for B-Myb#1 and B-Myb#2 were 5′-GACAATGCTGTGAAGAATCAC-3′ and 5′-GTCTGTCCTTCCTGGATTCCT-3′, respectively, and those for Skp2#1 and Skp2#2 were 5′-AAGGGAGTGACAAAGACTTTG-3′ and 5′-GCATGTACAGGTGGCTGTT-3′, respectively. Knockdown of pVHL or HIF-1α was performed as described previously (29, 30).
Reagents.
CoCl2, cycloheximide, anti-FLAG-M2–agarose, and FLAG peptide were purchased from Sigma (St. Louis, MO). Protein A-Sepharose was purchased from GE Healthcare Bioscience (Piscataway, NJ) and MG132 from Calbiochem (San Diego, CA). The receptor tyrosine kinase inhibitor set (PKI-RTKSET-250) was purchased from Biaffin GmbH & Co. KG (Kassel, Germany). Recombinant human VEGF165 was purchased from Humanzyme (Chicago, IL). Bafilomycin A1 was purchased from Wako (Tokyo, Japan).
Cell culture and transfection.
The 293T and 786-O cell lines were cultured as described previously (31); 293T cells were transfected using polyethyleneimine. Retrovirus infection was performed as described previously (31).
Cell fractionation.
Harvested cells were washed with phosphate-buffered saline (PBS) and resuspended in 0.1 ml of swelling buffer (10 mM Tris-HCl, 2 mM MgCl2, pH 7.4) and incubated for 5 min on ice. Then, 0.1 ml of lysis buffer (10 mM Tris-HCl, 2 mM MgCl2 1% Triton X-100 and protease inhibitor cocktail [Roche, Indianapolis, IN], pH 7.4) was added, and the buffer was mixed gently, followed by incubation on ice for 5 min. After being pipetted three times, samples were centrifuged for 10 min at 4,000 rpm. The supernatant was collected as the cytosolic fraction. Pellets were washed with 0.1 ml of swelling buffer twice and resuspended in 0.1 ml of extraction buffer (20 mM HEPES-KOH, 150 mM NaCl, 2 mM EDTA, 1 mM dithiothreitol [DTT], 10% glycerol, 1% Triton X-100 and protease inhibitor cocktail [Roche], pH 7.6) and incubated on ice for 5 min, followed by centrifugation for 10 min at 15,000 rpm. The supernatant was collected as the nuclear fraction.
IP and IB analyses.
Immunoprecipitation (IP) and immunoblot (IB) analyses were performed as reported previously (31).
qPCR analysis.
Quantitative PCR (qPCR) analysis was performed with a StepOne Plus instrument (Applied Biosystems, Foster City, CA) and KAPA SYBR Fast qPCR kit (Kapa Biosystems, Woburn, MA). The primer sequences were as follows: human B-Myb, 5′-AGTCTCTGGCTCTTGACATTG-3′ and 5′-GGGTGAGGCTGGAAGAGTTTG-3′; human complement factor B (CFB), 5′-AAAGCTCTGTTTGTGTCTGAG-3′ and 5′-ATGTCCTTGACTTTGTCATAG-3′; human FK506 binding protein 1B (FKBP1B), 5′-GGTTGCAGATTGAAGCATTTC-3′ and 5′-GGCAGTGTAGATTGTGCGAAC-3′; human septin 6 (SEPT6), 5′-TGTCAGCAACGGAGTCCAGAT-3′ and 5′-GTGCTGCCAATGACAGCAAAC-3′; and human GAPDH (glyceraldehyde-3-phosphate dehydrogenase), 5′-GCAAATTCCATGGCACCGT-3′ and 5′-TCGCCCCACTTGATTTTGG-3′.
Antibodies.
Antibodies against FLAG (M2; 1 μg/ml [Sigma]), hemagglutinin (HA) (12CA5; 1 μg/ml), B-Myb (sc-81192; 1 μg/ml [Santa Cruz Biotechnology, Santa Cruz, CA]), HIF-2α (sc-13596; 1 μg/ml [Santa Cruz Biotechnology]), HIF-1β (sc-17812; 1 μg/ml [Santa Cruz Biotechnology]), Skp2 (32-3400; 1 μg/ml [Invitrogen, Carlsbad, CA]), phosphotyrosine (610000; 1 μg/ml [BD Transduction Laboratories, MD]), phospho-histone H3.1 (GTX50231; 1 μg/ml; [GeneTex, Irvine, CA]), VEGF receptor 1 (VEGFR1) (sc-316; 1 μg/ml [Santa Cruz Biotechnology]), PDGFRα (D13C6; 1,000 dilution [Cell Signaling Technology, Chiyoda, Japan]), β-actin (AM4302; 1 μg/ml [Applied Biosystems]), Hsp70 (610607; 0.2 μg/ml [BD Transduction Laboratories]), and Hsp90 (610419; 0.2 μg/ml [BD Transduction Laboratories]) were used. Rabbit anti-pVHL antibody was generated using recombinant pVHL, which was purified from Escherichia coli by Ni-nitrilotriacetic acid (Ni-NTA)–agarose (Invitrogen). Anti-pVHL antibody was further purified by glutathione S-transferase (GST)-fused pVHL. All antibodies were diluted with PBS, except anti-VEGFR1, which was diluted with Can Get Signal Immunoreaction Enhancer Solution 1 (Toyobo, Tokyo, Japan).
Isolation and identification of pVHL-interacting proteins.
The substrates of the pVHL ubiquitin ligase were identified as described previously (32).
In vitro ubiquitination.
In vitro ubiquitination was performed as described previously with slight modifications (28). Recombinant CRL2pVHL complex was purified as described previously (5). Recombinant 3×HA–B-Myb was immunopurified from the lysates of transiently transduced 293T cells using anti-HA antibody and protein A-Sepharose.
Tumor growth in vivo.
786-O cells were cultured to confluence in 15-cm cell culture dishes for 1 week. The medium was replaced every day. The cells were harvested using a cell scraper, rinsed with PBS, and suspended in 600 μl PBS. A volume of 150 μl of suspension containing 2 × 107 cells was injected subcutaneously into 8-week-old female nude mice individually housed under sterile conditions. After 4 weeks, the longest tumor diameter was measured, and tumor volume was calculated using the following formula: volume = [(shorter diameter)2 × longer diameter]/2.
Phosphorylation of B-Myb.
786-O cells (2.6 × 106) expressing 3×HA–B-Myb (wild type [WT]or Y15A mutant) or control cells were cultured in either one 10-cm dish (confluent condition) or two 15-cm dishes (sparse condition) for 2 days with one change of culture medium. For tyrosine kinase inhibition, the following inhibitors were added to the culture medium for 1 day before harvesting the cells: 0.1 μM epidermal growth factor receptor (EGFR) inhibitor PD153035, 5 μM ErbB2 inhibitor AG825, 0.1 μM insulin-like growth factor receptor (IGFR) inhibitor picropodophyllin, 1 μM mesenchymal epithelial transition factor (MET) inhibitor SU11274, and 5 μM vascular endothelial growth factor receptor (VEGFR) and platelet-derived growth factor receptor (PDGFR) inhibitor SU4312. Phosphatase treatment was performed as follows: 3×HA–B-Myb was immunoprecipitated and washed with phosphatase buffer supplied by New England BioLabs (NEB; Tokyo, Japan). Immunoprecipitates were incubated with 10 U of Antarctic phosphatase (M0289; NEB) for 30 min at 37°C.
FACS analysis.
Trypsinized cells were fixed in 70% ethanol in PBS at −20°C for 1 day, followed by RNase treatment (500 μg/ml for 30 min at 37°C [Sigma]) and staining with propidium iodide (20 μg/ml [Sigma]). Then, 20,000 cells were analyzed with a FACSCalibur (Becton Dickinson, San Jose, CA) fluorescence-activated cell sorter (FACS).
Identification of B-Myb-regulated genes by microarray.
Total RNA was purified from control 786-O cells or two independent B-Myb knockdown (B-Myb#1 and B-Myb#2) cell lines, which were confluently cultured for 1 week with one daily medium change by TriPure isolation reagent (Roche, Indianapolis, IN). Synthesis of cDNA and cRNA was performed by Hokkaido System Science Co., Ltd. (Hokkaido, Japan), using an Agilent low-input Quick Amp labeling kit (Agilent Technologies, Santa Clara, CA). Fragmented cRNA was hybridized to SurePrint G3 human GE 8x60K version 2.0 (Agilent Technologies).
Statistical analysis.
The Student t test was used to determine the statistical significance of the experimental data.
RESULTS
pVHL interacts with B-Myb.
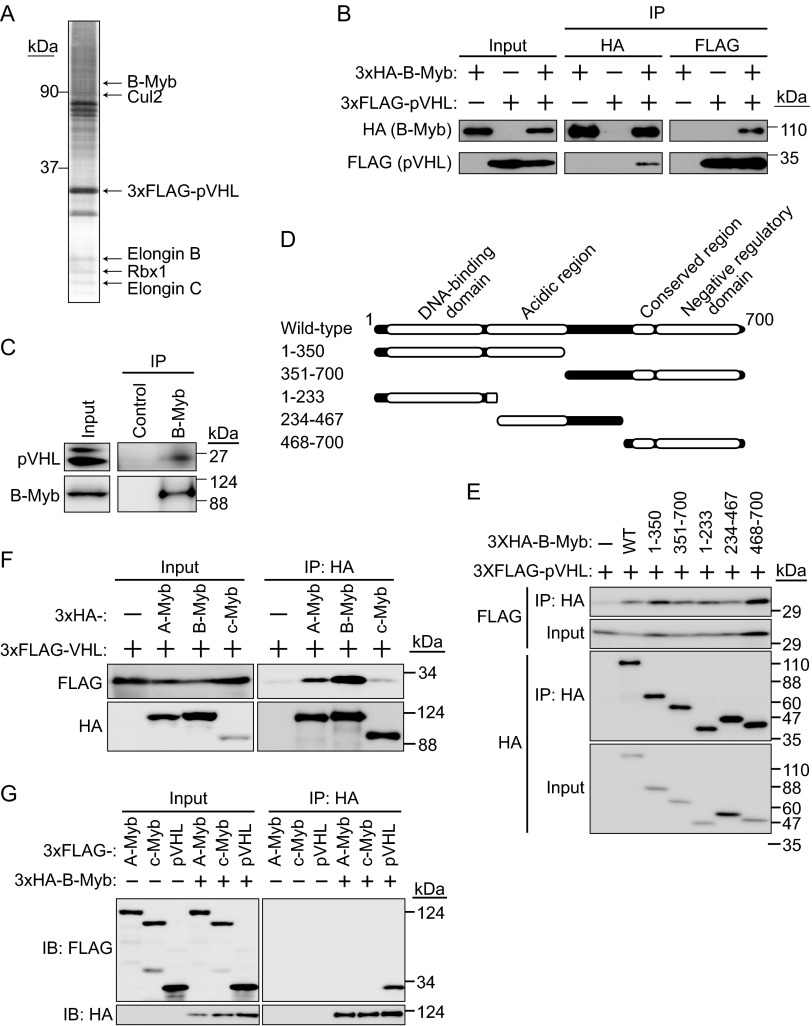
Evidence that constitutive activation of HIF-α is not sufficient for the development of renal clear cell carcinomas and pheochromocytomas in mice (15) suggested the involvement of an HIF-α-independent pathway in VHL pathogenesis. To identify novel substrates targeted by pVHL, 3×FLAG-tagged pVHL was purified from 293T cell lysates, and potential pVHL-interacting proteins were analyzed by mass spectrometry. In addition to molecules previously known to interact with pVHL, such as Cul2, elongin B, elongin C, and Rbx1, B-Myb was identified as a novel pVHL-interacting protein from an excised SDS-PAGE gel slice of approximately 100 kDa (Fig. 1A). As shown in Fig. 1A, several bands overlapped, and some proteins were identified from the same gel slice as that containing B-Myb (data not shown). Since similar experiments were performed using other E3s and B-Myb was identified by pVHL pulldown but not by pulldown with other E3s, the interaction between pVHL and B-Myb was examined further. To confirm the interaction between B-Myb and pVHL, 3×HA-tagged B-Myb and 3×FLAG-tagged pVHL were expressed in HEK293T cells. The cell lysates were subjected to immunoprecipitation (IP) with anti-HA or anti-FLAG antibody, and the resulting immunoprecipitates were subjected to SDS-PAGE and Western blotting with anti-HA or anti-FLAG antibody. The interaction between 3×HA–B-Myb and 3×FLAG-pVHL was detected by reciprocal IP (Fig. 1B). HEK293T cells express two biologically active VHL gene products of approximately 19 and 30 kDa, similar to other cell lines (Fig. 1C) (33). HEK293T cells were cultured in the presence of the proteasome inhibitor MG132 for 6 h, and cell lysates were prepared and subjected to IP with anti-B-Myb antibody. Immunoblot analysis of the immunoprecipitates showed that endogenous B-Myb interacted with endogenous 19-kDa pVHL (Fig. 1C); however, whether B-Myb interacted with the 30-kDa pVHL remained unclear. To determine the binding domain of B-Myb for pVHL, several truncated mutants were generated (Fig. 1D). IP analysis showed all of the truncated mutants interacted with 3×FLAG-pVHL (Fig. 1E), suggesting the presence of several binding sites. It might also be possible that these truncated mutants bind pVHL because they were overexpressed. Since Myb family proteins have highly conserved domains (34), we next examined the interaction between pVHL and A-Myb or c-Myb. Our results showed that all Myb family proteins were able to interact with pVHL, although the interaction between c-Myb and pVHL was relatively weak (Fig. 1F). Further examination of the interaction between B-Myb and A-Myb or c-Myb did not yield clear results showing that A-Myb or c-Myb interacted with B-Myb (Fig. 1G). Mammalian A-Myb and c-Myb are expressed in restricted cell types and at specific stages of development; for example, A-Myb plays a critical role in spermatogenesis and mammary gland development (19), and c-Myb regulates the proliferation and differentiation of hematopoietic stem and progenitor cells (20); therefore, the physiological roles of pVHL with regard to A-Myb or c-Myb should be investigated under the appropriate conditions in the future. The present results indicated that pVHL interacts with Myb family proteins, especially B-Myb, under physiological conditions.
FIG 1.
Interaction between pVHL and B-Myb. (A) Purification of the CRL2pVHL complex. The 3×FLAG-pVHL expressed in 293T cells was purified using anti-FLAG antibody and resolved by SDS-PAGE. In-gel digestion by trypsin was performed for mass spectrometry analysis. A silver-stained SDS-PAGE gel is shown, and the protein bands identified are indicated. B-Myb was identified as a novel protein interacting with pVHL. (B) Interaction between 3×HA–B-Myb and 3×FLAG-pVHL. HEK293T cells expressing 3×HA–B-Myb or 3×FLAG-pVHL (as indicated) were cultured in the presence of the proteasome inhibitor MG132 (10 μM for 6 h), immunoprecipitated (IP) with anti-HA or anti-FLAG antibody, and immunoblotted (IB) with anti-HA or anti-FLAG antibody. (C) Interaction between endogenous pVHL and B-Myb. HEK293T cells were cultured in the presence of MG132 (10 μM for 6 h), immunoprecipitated with anti-B-Myb antibody, and immunoblotted with anti-pVHL or anti-B-Myb antibody. (D) Schematic representation of B-Myb mutants used in this study. (E) Interaction between pVHL and B-Myb via several binding sites. The wild type (WT) or a deletion mutant of 3×HA–B-Myb as indicated was coexpressed with 3×FLAG-pVHL in HEK293T cells. Lysates were immunoprecipitated with anti-HA antibody and immunoblotted with anti-FLAG or anti-HA antibody. (F) Interaction between Myb family proteins and pVHL. HEK293T cells expressing 3×HA–A-Myb, 3×HA–B-Myb, 3×HA–c-Myb, or 3×FLAG-pVHL (as indicated) were cultured in the presence of MG132 (10 μM for 6 h), immunoprecipitated with anti-HA antibody, and immunoblotted with anti-HA or anti-FLAG antibody. (G) Assessment of the interaction between B-Myb and A-Myb or c-Myb. HEK293T cells expressing 3×FLAG–A-Myb, 3×FLAG–c-Myb, 3×FLAG-pVHL, or 3×HA–B-Myb (as indicated) were cultured in the presence of MG132 (10 μM for 6 h), immunoprecipitated with anti-HA antibody, and immunoblotted with anti-HA or anti-FLAG antibody.
pVHL targets B-Myb for proteasomal degradation.
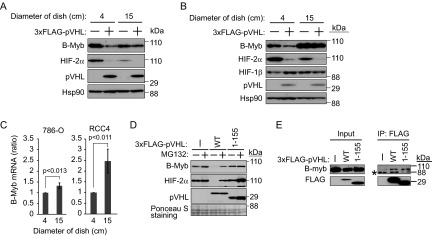
In preliminary experiments using pVHL-deficient 786-O cells, which were established from a renal clear cell carcinoma, B-Myb was downregulated by coexpression of pVHL when cells were cultured to confluence (1.3 × 106 cells grown in a 4-cm culture dish for 1 day) (Fig. 2A). This was confirmed in a different pVHL-deficient renal clear cell carcinoma cell line, RCC4 (Fig. 2B). Control or 3×FLAG-pVHL was expressed in RCC4 cells (1.3 × 106), which were cultured under confluent (4-cm dish) or sparse (15-cm dish) conditions for 1 day, and the levels of B-Myb and HIF-2α were compared. B-Myb was downregulated in the presence of 3×FLAG-pVHL under confluent conditions (Fig. 2B). In contrast, HIF-2α was downregulated by coexpression of pVHL under both conditions (Fig. 2A and B). These data suggested that pVHL targets B-Myb in cells grown to confluence. Since rat pVHL stably expressed in normal rat kidney (NRK) epithelial cells or NIH 3T3 cells predominantly localized to the cytoplasm under confluent conditions, whereas it localized to the nucleus under the sparse condition (35), we examined the localization of pVHL in 786-O cells or RCC4 cells (see Fig. S1 in the supplemental material). The fact that pVHL was mainly localized to the cytoplasm under both culture conditions and HIF-2α was downregulated by pVHL reintroduction in both conditions (Fig. 2A and B) suggests that cell culture conditions do not affect the activity of pVHL. B-Myb was slightly increased under the confluent condition in 786-O cells (Fig. 2A), whereas it was strongly upregulated under the sparse condition in RCC4 cells (Fig. 2B). To identify the molecular mechanism underlying this difference, we quantified the mRNA encoding B-Myb in these cell lines cultured under different conditions (Fig. 2C). Although B-Myb mRNA was increased under the sparse condition in both cell lines, its upregulation was greater in RCC4 cells than in 786-O cells. The strong upregulation of mRNA expression could be one of the reasons for the strong expression of the B-Myb protein in RCC4 cells. The inhibition of protein translation and/or enhanced protein degradation could explain the slight downregulation of the B-Myb protein in 786-O cells under the sparse condition.
FIG 2.
Destabilization of B-Myb by pVHL. (A) Regulation of B-Myb by cell density and pVHL. 786-O cells (1 × 106) were cultured for 1 day in culture dishes of different diameters as indicated. The resulting cell lysates were subjected to Western blotting with antibodies against B-Myb, HIF-2α, pVHL, or Hsp90. Hsp90 was used as a loading control. (B) The experiment described in panel A was performed using RCC4 cells. (C) Upregulation of B-Myb mRNA in 786-O cells and RCC4 cells cultured sparsely. Total RNA was purified and analyzed by quantitative RT-PCR. (D) Accumulation of B-Myb upon exposure to the proteasome inhibitor MG132. 786-O cells stably expressing wild-type 3×FLAG-pVHL or mutant 3×FLAG-pVHL(1–155) lacking the VHL box or control cells were cultured under confluent conditions for 1 day, followed by exposure to MG132 (10 μM) for 6 h and Western blotting with antibodies against B-Myb, HIF-2α, or pVHL. Ponceau S staining was used as the loading control. (E) 786-O cells were cultured under confluent conditions for 1 day, exposed to MG132 (10 μM) for 6 h, immunoprecipitated (IP) with anti-FLAG antibody, and immunoblotted with anti-B-Myb or anti-FLAG antibody. The asterisk represents a nonspecific band.
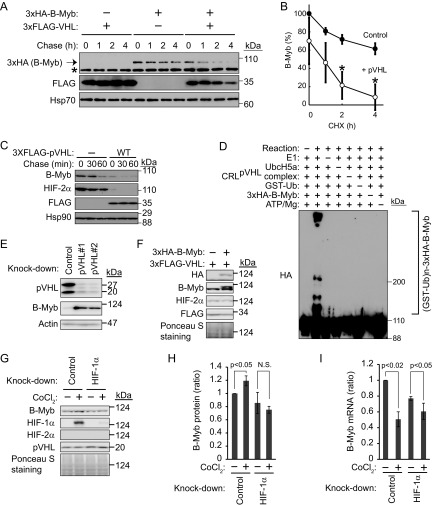
Reintroduction of wild-type pVHL downregulated endogenous B-Myb, but failed to do so in the presence of the proteasome inhibitor MG132 (Fig. 2D). Although VHL box-deficient functionally inactive pVHL(1–155) (32) retained the same B-Myb-binding activity as wild-type pVHL (Fig. 2E), pVHL(1–155) had no effect on the expression of B-Myb (Fig. 2D), supporting the idea that B-Myb is a substrate of the pVHL ubiquitin ligase. To examine the stability of B-Myb in the presence or absence of pVHL, 3×HA–B-Myb was expressed in 293T cells with or without 3×FLAG-pVHL, and the levels of 3×HA–B-Myb were measured after treatment with the protein translation inhibitor cycloheximide (Fig. 3A). The half-life of 3×HA–B-Myb was shortened upon overexpression of pVHL (Fig. 3A and B). Despite the fact that pVHL is the major ubiquitin ligase targeting B-Myb and HIF-2α, these proteins were gradually degraded even in the absence of pVHL in control 786-O cells, suggesting the involvement of another ubiquitin ligase (Fig. 3C). Because SCFSkp2 was suggested as a ubiquitin ligase targeting B-Myb (22), we investigated the role of SCFSkp2 in the regulation of B-Myb by knocking down Skp2 and analyzing the stability of B-Myb in the absence of pVHL (see Fig. S2 in the supplemental material). Knockdown of Skp2 did not stabilize B-Myb more than that in control cells, indicating that B-Myb is not a substrate of Skp2, at least in 786-O cells cultured under confluent conditions.
FIG 3.
Destabilization of B-Myb by pVHL. (A) HEK293T cells expressing 3×HA–B-Myb with or without 3×FLAG-pVHL were exposed to cycloheximide (CHX [50 μg/ml]) for 1, 2, or 4 h. The lysates were subjected to Western blotting with antibodies against HA, FLAG, or Hsp70. Hsp70 was used as a loading control. HA–B-Myb is indicated by an arrow. The asterisk represents a nonspecific band. (B) The intensities of HA–B-Myb bands in panel A were normalized to those of the corresponding Hsp70 bands and plotted as a percentage of the normalized value without pVHL expression at 0 h. Open circles, with pVHL expression; closed circles, mock. *, P < 0.02. Data represent means ± standard deviations (SD) from three independent experiments. (C) Destabilization of endogenous B-Myb by pVHL in 786-O cells. Control or pVHL-reintroduced 786-O cells were exposed to cycloheximide (50 μg/ml) for 30 or 60 min. The lysates were subjected to Western blotting with antibodies against B-Myb, HIF-2α, FLAG, or Hsp90. Hsp90 was used as a loading control. (D) pVHL-dependent polyubiquitination of B-Myb in vitro. Recombinant ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (UbcH5a), CRL2pVHL complex (pVHL, Cul2, elongin B, elongin C, and Rbx1), GST-ubiquitin, and immunopurified HA–B-Myb were mixed in vitro with ATP (in various combinations), incubated at 30°C for 3 h, and subjected to Western blotting with anti-HA antibody. (E) Accumulation of endogenous B-Myb by knockdown of pVHL. Two independent small interfering RNAs (siRNAs) targeting pVHL (pVHL#1 and pVHL#2) were transfected into HEK293T cells and cultured for 2 days (100% confluent on day 2), followed by Western blotting with anti-pVHL, anti-B-Myb, and antiactin antibodies. Actin was used as a loading control. (F) No change of HIF-2α expression by overexpression of B-Myb. 786-O cells stably expressing 3×FLAG-pVHL with or without 3×HA–B-Myb were cultured under confluent conditions for 1 day. The lysates were subjected to Western blotting with antibodies against HA, B-Myb, HIF-2α, or FLAG. Ponceau S staining was used as a loading control. (G) Upregulation of B-Myb by HIF-1α in 293T cells. siRNA targeting HIF1α or the control was transfected into HEK293T cells and cultured for 40 h (100% confluent). 293T cells were further cultured in the presence of CoCl2 (400 μM) for 6 h and subjected to Western blotting with antibodies against B-Myb, HIF-1α, HIF-2α, or pVHL. Ponceau S staining was used as the loading control. (H) The intensities of B-Myb bands in panel G were normalized to those of Ponceau S-stained bands. B-Myb expression level in cells without CoCl2 was set as 1. Data represent means ± SD from three independent experiments. (I) Downregulation of B-Myb mRNA by CoCl2 or knockdown of HIF-1α. Total RNA was purified from control or HIF-1α knockdown 786-O cells and analyzed by quantitative RT-PCR. Data represent means ± SD from three independent experiments.
We next performed in vitro ubiquitination assays (Fig. 3D). The recombinant CRL2pVHL complex, including elongin B/C, Rbx1, and Cul2, was expressed in insect SF21 cells, purified, and mixed with or without recombinant ubiquitin-activating enzyme E1, ubiquitin-conjugating enzyme UbcH5a, ubiquitin, ATP, and 3×HA–B-Myb immunopurified from HEK293T cell lysates. After incubation for 2 h at 30°C, the reaction mixtures were analyzed by Western blotting with anti-HA antibody to detect the polyubiquitination of B-Myb (Fig. 3D). Polyubiquitinated B-Myb was detected when all components were included in the reaction, supporting the notion that B-Myb is a substrate of the pVHL ubiquitin ligase. Furthermore, knockdown of pVHL resulted in the accumulation of endogenous B-Myb in HEK293T cells (Fig. 3E). To examine whether B-Myb and HIF-2α are competitively recognized by pVHL and targeted for proteasomal degradation, a 3×HA–B-Myb-overexpressing 786-O cell line was established (Fig. 3F). Increased expression of B-Myb did not affect the expression of HIF-2α, indicating that there was no competitive stabilization of HIF-2α by B-Myb overexpression, or that the expression level of 3×HA–B-Myb was not sufficient to competitively stabilize HIF-2α. We next examined B-Myb expression levels in 293T cells cultured under hypoxic conditions. HEK293T cells were cultured in the presence of the hypoxia mimetic agent CoCl2 for 7 h, and cell lysates were prepared and subjected to Western blotting. B-Myb was slightly but significantly increased under the hypoxia mimetic condition (Fig. 3G and H). Since HIF-1α but not HIF-2α was detected in 293T cells, the expression of B-Myb was examined in HIF-1α knockdown 293T cells. B-Myb was not upregulated under the hypoxia mimetic condition when HIF-1α was knocked down (Fig. 3G and H). The fact that the B-Myb mRNA did not increase under the hypoxia mimetic condition (Fig. 3I) indicates that HIF-1α may competitively block the degradation of B-Myb in 293T cells. HIF-1α would not be recognized by pVHL under hypoxia (13); therefore, HIF-1α may prevent the degradation of B-Myb mediated by another ubiquitin ligase but not pVHL. Further investigation of the molecular mechanisms by which CoCl2 treatment downregulates B-Myb mRNA is warranted based on the lack of correlation between HIF-1α protein expression and the mRNA encoding B-Myb (Fig. 3G to I). These data indicated that pVHL polyubiquitinates B-Myb and targets it for proteasomal degradation.
Phosphorylation of tyrosine 15 of B-Myb prevents its degradation.
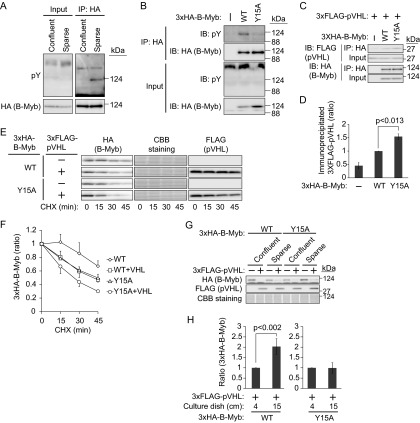
Given that the downregulation of B-Myb by pVHL was apparent when cells were cultured at high density (Fig. 2A and B) and that B-Myb has at least 10 different phosphorylation sites (36), we speculated that phosphorylation of B-Myb may regulate its stability. The phosphorylation statuses of 3×HA–B-Myb were compared between cells cultured under confluent (3.8 × 106 cells in two 6-cm dishes) and sparse (same cell number but cultured in three 15-cm dishes) conditions for 1 day by incubating cell lysates with or without a phosphatase to distinguish phosphorylated B-Myb (see Fig. S3 in the supplemental material). The results indicated that the phosphorylation statuses of B-Myb were similar in cells grown under both conditions. However, because B-Myb is poly-phosphorylated, minor differences in phosphorylation status, such as tyrosine phosphorylation, could have been missed. A search of the PhosphoSitePlus database (http://www.phosphosite.org) (37) revealed that tyrosine 15 of B-Myb was the only tyrosine residue that could be phosphorylated. To confirm the tyrosine phosphorylation of B-Myb, 786-O cells expressing 3×HA–B-Myb were cultured under confluent or sparse conditions, and 3×HA–B-Myb immunoprecipitates were blotted with an antiphosphotyrosine antibody (Fig. 4A). As expected, the tyrosine residues of 3×HA–B-Myb were phosphorylated when cells were cultured sparsely. To confirm that tyrosine 15 is the target residue for phosphorylation, 786-O cells stably expressing mutant 3×HA–B-Myb(Y15A) were generated. Culturing of 786-O cells stably expressing 3×HA–B-Myb (wild type or Y15A mutant) under sparse conditions and analysis of tyrosine phosphorylation status showed that the Y15A mutation suppressed the tyrosine phosphorylation of B-Myb, indicating that tyrosine 15 is the B-Myb phosphorylation site under sparse culture conditions (Fig. 4B). A residual tyrosine phosphorylation signal was observed in B-Myb(Y15A), suggesting that another tyrosine residue could be phosphorylated. To determine the cellular location of B-Myb tyrosine phosphorylation, 3×HA–B-Myb was immunoprecipitated from the cytosolic or nuclear fraction. Immunoblotting of the precipitates with antiphosphotyrosine antibody showed tyrosine phosphorylation in the cytosol but not in the nucleus (see Fig. S4 in the supplemental material). Because B-Myb was tyrosine phosphorylated when cells were cultured sparsely and its resistance to pVHL-dependent degradation was observed under the same conditions (Fig. 2A and B), we speculated that the tyrosine phosphorylation of B-Myb prevented its interaction with pVHL. To verify this hypothesis, the interaction between B-Myb (wild type or Y15A mutant) and pVHL was examined in cells cultured under sparse conditions (Fig. 4C and D). The results showed that the interaction of B-Myb(Y15A) with pVHL was slightly but reproducibly stronger than that of wild-type B-Myb, supporting the idea that unphosphorylated B-Myb was targeted by pVHL. Given that the tyrosine phosphorylation of wild-type B-Myb might not be complete even under sparse culture conditions, only a slight increase in affinity may be detected. Examination of the stability of B-Myb (wild type or Y15A mutant) in the presence of pVHL under sparse culture conditions showed that pVHL destabilized wild-type B-Myb (Fig. 4E and F). Furthermore, the half-life of B-Myb(Y15A) in the presence of pVHL was further shortened. Since the Y15A mutation itself destabilized B-Myb compared with wild-type B-Myb in the absence of pVHL, we further examined the effect of tyrosine phosphorylation on the stability of B-Myb. 786-O cells expressing 3×HA–B-Myb (wild type or Y15A mutant) with or without 3×FLAG-pVHL were cultured under sparse or confluent conditions. Because B-Myb was upregulated in cells cultured under confluent conditions in the absence of pVHL (Fig. 2A), the amount of 3×HA–B-Myb in the presence of pVHL was normalized to the amount in the absence of pVHL under the same culture conditions (Fig. 4G and H). Wild-type B-Myb was downregulated by coexpression of pVHL in 786-O cells, and it was destabilized further under confluent conditions. In contrast, although B-Myb(Y15A) was also downregulated by coexpression of pVHL, this process was not accelerated by culturing the cells under confluent conditions, as quantified in Fig. 4H. These data suggested that tyrosine 15 of B-Myb is a major phosphorylation site and that the phosphorylation of B-Myb may prevent its degradation.
FIG 4.
Regulation of B-Myb stability by phosphorylation of tyrosine 15. (A) Tyrosine phosphorylation of B-Myb under sparse culture conditions. 786-O cells (2.6 × 106) stably expressing 3×HA–B-Myb were cultured under confluent (in one 10-cm dish) or sparse (in two 15-cm dishes) conditions for 1 day. 3×HA–B-Myb was immunoprecipitated (IP) using an anti-HA antibody and immunoblotted (IB) with antiphosphotyrosine (anti-pY) or anti-HA antibody. (B) Phosphorylation of tyrosine 15 of B-Myb. 786-O cells (2.6 × 106) stably expressing 3×HA–B-Myb (WT or Y15A mutant) or control cells were cultured under sparse conditions for 1 day. 3×HA–B-Myb was immunoprecipitated with anti-HA antibody and immunoblotted with anti-pY or anti-HA antibody. (C) Enhancement of the interaction between 3×HA–B-Myb and 3×FLAG-pVHL by mutation of tyrosine 15 of B-Myb to alanine. 786-O cells stably expressing 3×HA–B-Myb (WT or Y15A) and 3×FLAG-pVHL or control cells were cultured under sparse conditions for 2 days. 3×HA–B-Myb was immunoprecipitated with anti-HA antibody and immunoblotted with anti-FLAG or anti-HA antibody. (D) The intensities of coimmunoprecipitated 3×FLAG-pVHL bands in panel C were normalized to those of input bands, and the amount of 3×FLAG-pVHL coimmunoprecipitated with 3×HA–B-Myb (WT) was set as 1. Data represent means ± SD from three independent experiments. (E) Destabilization of B-Myb by Y15A mutation. 786-O cells stably expressing 3×HA–B-Myb (WT or Y15A mutant) with or without 3×FLAG-pVHL were cultured under sparse conditions and exposed to CHX (50 μg/ml) for 15, 30, or 45 min. The lysates were subjected to Western blotting with antibodies against HA, or FLAG. Coomassie brilliant blue (CBB) staining was used as a loading control. (F) The intensities of 3×HA–B-Myb (WT or Y15A mutant) bands in panel E were normalized to those of CBB-stained bands. The amount of 3×HA–B-Myb (WT or Y15A mutant) at the beginning of the chase was set as 1. Data represent means ± SD from three independent experiments. (G) Cell density-dependent destabilization of wild-type B-Myb but not the Y15A mutant in the presence of pVHL. 786-O cells stably expressing 3×HA–B-Myb (WT or Y15A mutant) with or without 3×FLAG-pVHL were cultured under sparse or confluent conditions for 2 days. The lysates were subjected to Western blotting with antibodies against HA or FLAG. CBB staining was used as a loading control. (H) The intensities of 3×HA–B-Myb (WT or Y15A mutant) bands with 3×FLAG-pVHL coexpression in panel G were normalized to those without 3×FLAG-pVHL coexpression because the expression of B-Myb increased when cells were cultured under confluent conditions compared with when they were cultured under sparse culture conditions. The normalized intensity of 3×HA–B-Myb with 3×FLAG-pVHL coexpression under confluent culture conditions was set as 1. Data represent means ± SD from four independent experiments.
VEGF-dependent tyrosine phosphorylation of B-Myb prevents its degradation.
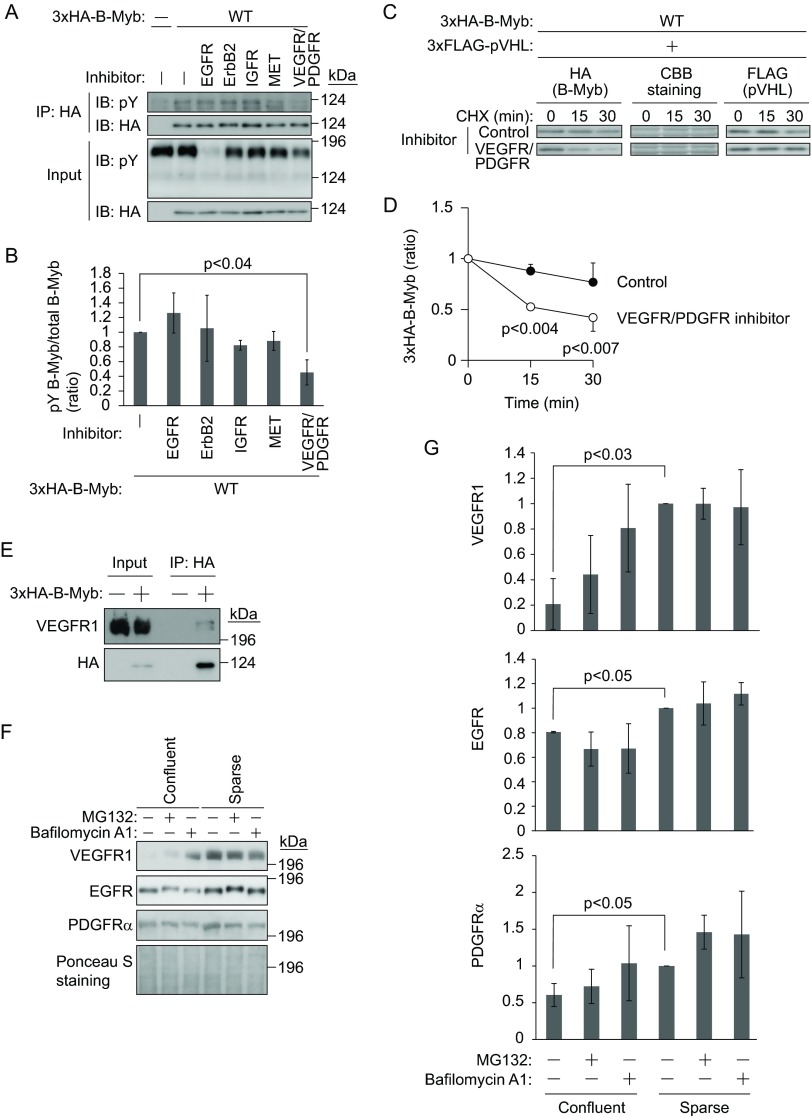
We next investigated the identity of the kinase responsible for B-Myb phosphorylation under sparse culture conditions. Because the growth factors in the culture medium may mediate the tyrosine phosphorylation of B-Myb through receptor tyrosine kinases (RTKs), the effect of RTK inhibitors was assessed (Fig. 5A). 3×HA–B-Myb-expressing 786-O cells were cultured sparsely in the presence of inhibitors against EGFR, erythroblastic leukemia viral oncogene 2 (ErbB2)/human EGFR-related 2 (HER2), IGFR, MET, or VEGFR and PDGFR. Immunoprecipitation of 3×HA–B-Myb and immunoblotting with antiphosphotyrosine antibody showed that the VEGFR/PDGFR inhibitor modestly prevented tyrosine phosphorylation of B-Myb (Fig. 5A and B). Then, the stability of 3×HA–B-Myb in the presence of 3×FLAG-pVHL was examined with or without VEGFR/PDGFR inhibitor (Fig. 5C and D). The presence of the VEGFR/PDGFR inhibitor shortened the half-life of 3×HA–B-Myb. The possible interaction of VEGFR with B-Myb was then examined. Because 786-O cells express VEGFR1 but not VEGFR2 (38), the interaction between VEGFR1 and B-Myb was assessed. Immunoprecipitation of 3×HA–B-Myb from 786-O cells cultured sparsely and immunoblotting with anti-VEGFR1 antibody showed that 3×HA–B-Myb interacted with VEGFR1 (Fig. 5E). However, VEGFR1 and B-Myb showed an unstable interaction, even when B-Myb was overexpressed. Because tyrosine phosphorylation of B-Myb was not detected in response to VEGF stimulation under confluent culture conditions, the expression of VEGFR1 and PDGFRα was assessed, which showed that they tended to be downregulated by confluent culture conditions (Fig. 5F and G). The downregulation of VEGFR1 and PDGFRα was partially prevented by the lysosome inhibitor bafilomycin A1, whereas the proteasome inhibitor MG132 had little effect, indicating that VEGFR1 and PDGFRα were degraded by several pathways, including the endosome-lysosome pathway and the ubiquitin-proteasome pathway (Fig. 5F and G). Although EGFR was also slightly downregulated under confluent culture conditions (Fig. 5F and G), its downregulation was not prevented by bafilomycin A1 or MG132, indicating that the expression of VEGFR1 and PDGFRα was selectively regulated by cell culture conditions. Since EGFR is ubiquitinated by CBL upon ligand stimulation (39), the shift in the migration of EGFR caused by MG132 treatment (Fig. 5F) indicates the phosphorylation and ubiquitination of EGFR as reported previously (40). These data indicate that VEGFR1-dependent tyrosine phosphorylation may directly or indirectly mediate the phosphorylation and stabilization of B-Myb.
FIG 5.
Regulation of B-Myb by VEGF and PDGF. (A) VEGFR and PDGFR inhibitor prevents tyrosine phosphorylation of B-Myb. 786-O cells stably expressing 3×HA–B-Myb or control cells were sparsely cultured for 1 day, followed by RTK inhibitor treatment for 1 day. 3×HA–B-Myb was immunoprecipitated (IP) and immunoblotted (IB) with anti-pY or anti-HA antibody. (B) The intensities of tyrosine-phosphorylated 3×HA–B-Myb bands in panel A were normalized to those of total immunoprecipitated 3×HA–B-Myb bands, and mock treatment was set as 1. Data represent means ± SD from three independent experiments. (C) VEGFR and PDGFR inhibitor destabilizes B-Myb. 786-O cells stably expressing 3×HA–B-Myb and 3×FLAG-pVHL were sparsely cultured for 1 day, followed by RTK inhibitor treatment for 1 day. Then, cells were exposed to CHX (50 μg/ml) for 15 or 30 min. The lysates were subjected to Western blotting with antibodies against HA or FLAG. CBB staining was used as a loading control. (D) The intensities of 3×HA–B-Myb bands in panel C were normalized to those of CBB-stained bands. The amount of 3×HA–B-Myb at the beginning of the chase was set as 1. Data represent means ± SD from three independent experiments. (E) B-Myb interacts with VEGFR1. 786-O cells stably expressing 3×HA–B-Myb or control cells were sparsely cultured for 2 days. 3×HA–B-Myb was immunoprecipitated and immunoblotted with antibodies against VEGFR1 or HA. (F) Downregulation of VEGFR1, PDGFRα, and EGFR by confluent culture conditions. pVHL-reintroduced 786-O cells were cultured under confluent or sparse conditions overnight with or without MG132 (10 μM) or bafilomycin A1 (0.5 μM). The lysates were subjected to Western blotting with antibodies against VEGFR1, PDGFRα, or EGFR. Ponceau S staining was used as a loading control. (G) The intensities of VEGFR1, PDGFRα, or EGFR bands in panel F were normalized to those of Ponceau S-stained bands. The amount of VEGFR1, PDGFRα, or EGFR expressed under sparsely cultured conditions was set as 1. Data represent means ± SD from three independent experiments.
Accelerated tumor development upon B-Myb knockdown.
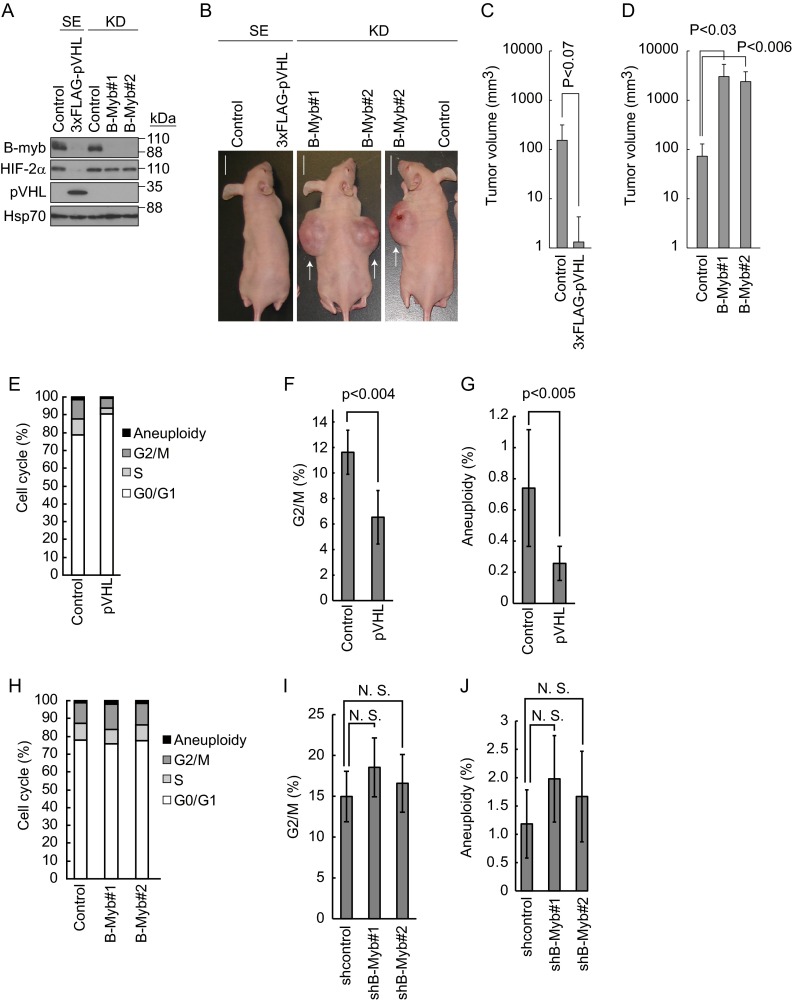
The above data prompted us to assess the biological significance of B-Myb in tumorigenesis in vivo. For this purpose, 786-O cells expressing 3×FLAG-pVHL or control cells, as well as control knockdown 786-O cells or two independent B-Myb knockdown (B-Myb#1 and B-Myb#2) 786-O cell lines, each targeting different sequences of B-Myb, were implanted subcutaneously into BALB/c athymic nude mice and allowed to form tumors (Fig. 6A and B). Tumor volumes were calculated as described in Materials and Methods. Reintroduction of pVHL in 786-O cells slowed down tumor development by downregulating HIF-2α as reported previously (41) (Fig. 6B and C). Tumor development in B-Myb knockdown cells was accelerated compared with that in control cells, indicating that B-Myb negatively regulates HIF-2α-dependent tumor development (Fig. 6B and D). Control and B-Myb knockdown cell lines formed a similar number of colonies in the in vitro anchorage-independent soft agar colony formation assay, and their growth rates were similar (data not shown). These data indicated that when the expression of B-Myb was downregulated genetically or epigenetically in the presence of pVHL mutation, tumor development was accelerated. A search of an integrated cancer microarray database provided by Oncomine (https://www.oncomine.org) showed that B-Myb expression was significantly downregulated in two independent statistical analyses of human clear cell renal cell carcinoma (42, 43) (see Fig. S5 in the supplemental material). These findings indicate that the downregulation of B-Myb and pVHL may be crucial for the development of renal cell carcinomas.
FIG 6.
Regulation of tumor development in vivo by B-Myb. (A) Knockdown of B-Myb in 786-O cells. 786-O cells stably expressing (SE) pVHL or short hairpin (sh) RNA to knockdown (KD) B-Myb under confluent conditions were lysed and subjected to Western blotting with antibodies against B-Myb, HIF-2α, pVHL, or Hsp70. Hsp70 was used as a loading control. (B) Representative images of mice implanted with 786-O cells in which pVHL function was reconstituted, cells in which B-Myb was knocked down, or the corresponding control cells. Cells were implanted, and photographs were taken after 1 month as indicated. Representative tumors are indicated by arrows. Bars, 1 cm. (C) The estimated volume (cubic centimeters) of tumors generated by control or pVHL-reconstituted cells is indicated. Data represent means ± SD (n = 5). (D) The estimated volume (cubic centimeters) of tumors generated by control or B-Myb knockdown cells is indicated. Data represent means ± SD (n = 5). (E) Accumulation of cells in G0/G1 phase by pVHL reintroduction. 786-O cells stably expressing pVHL or control cells cultured under confluent conditions were subjected to FACS analysis to analyze aneuploidy and the G2/M, S, and G0/G1 phases. (F) Decrease in the G2/M-phase population by pVHL. FACS analysis was performed as described in panel E. Data represent means ± SD from three independent experiments. (G) Decrease in aneuploidy by pVHL. Data represent means ± SD from three independent experiments. (H) Knockdown of B-Myb had no effect on cell cycle progression. Control 786-O cells or 786-O cells with B-Myb knockdown (targeting independent sequences B-Myb#1 and B-Myb#2) cultured under confluent conditions were subjected to FACS to analyze aneuploidy, G2/M, S, and G0/G1 phases. (I) Knockdown of B-Myb did not cause accumulation of cells in G2/M phase. Data represent means ± SD from three independent experiments. N.S., not significant. (J) No increase in aneuploidy was observed upon knockdown of B-Myb. Data represent means ± SD from three independent experiments.
B-Myb does not regulate the cell cycle in 786-O cells.
B-Myb transactivates genes required for the G2/M cell cycle transition by forming the dREAM/Myb–MuvB-like complex, which was originally identified in Drosophila (23, 44–47). Furthermore, mutation or downregulation of B-Myb results in genome instability (27, 48, 49). In contrast, pVHL induces cell cycle arrest at the G0/G1 phase (50), and knockdown of HIF-2α is sufficient to suppress aneuploidy (50). Based on these data, we hypothesized that pVHL might regulate the cell cycle and genomic instability by inducing the degradation of B-Myb. Reintroduction of pVHL into 786-O cells resulted in the accumulation of cells in the G0/G1 phase and a decrease in G2/M-phase cells compared with control 786-O cells (Fig. 6E and F; see Fig. S6 in the supplemental material). pVHL expression also reduced aneuploidy (Fig. 6G; see Fig. S6), suggesting that pVHL contributes to genomic stability. In contrast and unexpectedly, knockdown of B-Myb increased the number of cells in the G2/M phase and aneuploidy, although without statistical significance (Fig. 6H to J; see Fig. S6). These data suggested that B-Myb regulates tumor development through a process independent of the regulation of cell cycle and aneuploidy.
Regulation of HIF-α-independent genes by B-Myb.
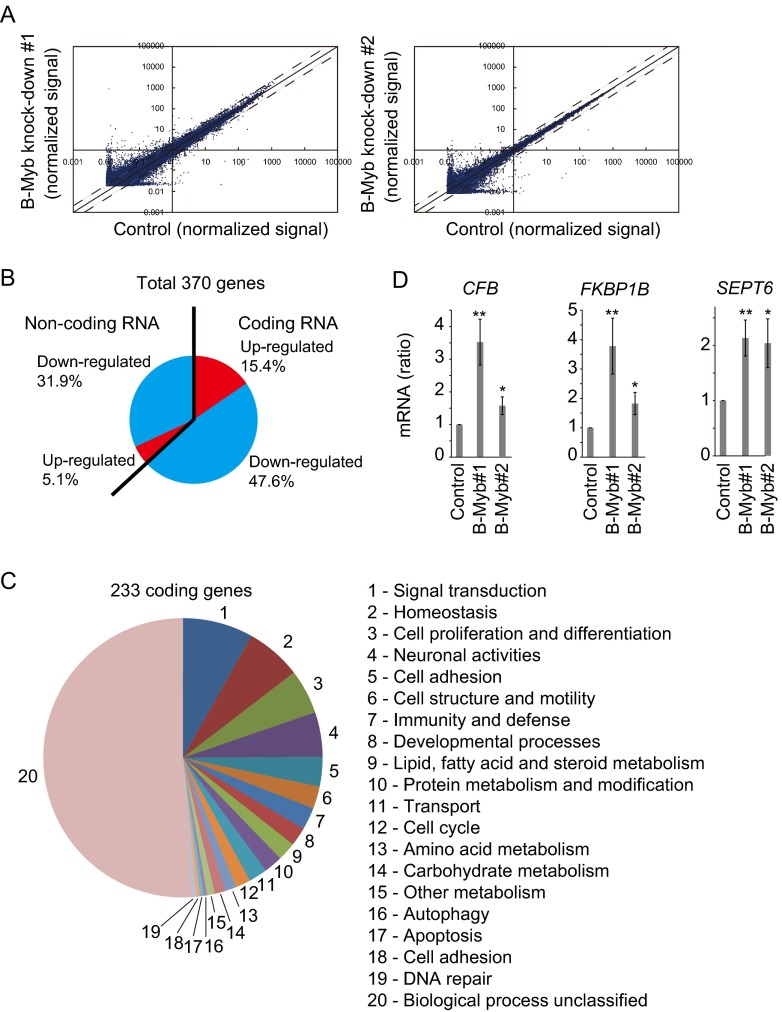
Given that knockdown of B-Myb significantly accelerated tumor development (Fig. 6), we assumed that the transcription factor B-Myb may inhibit the expression of HIF-α-dependent tumorigenic genes, such as VEGFA, GLUT1, and PDGF, and knockdown of B-Myb might result in the upregulation of those genes and tumor development. Assessment of the expression of HIF-α-dependent tumor-related genes in B-Myb knockdown 786-O cell lines by quantitative reverse transcription (RT)-PCR showed that B-Myb knockdown had no effect on their expression (data not shown). To identify B-Myb-regulated genes, total RNA was purified from control or B-Myb knockdown 786-O cells (two independent cell lines, B-Myb#1 and B-Myb#2) and analyzed by genome-wide microarray screening. Although the expression of most genes was not affected by B-Myb knockdown, 76 genes were significantly upregulated (>2-fold) and 294 genes were downregulated (<1/2-fold) in both B-Myb knockdown cell lines (Fig. 7A and B). Among the 370 genes, 63% were coding RNA genes, and the rest were noncoding RNA genes (Fig. 7B). Of these, 233 coding genes were classified by known or conceivable biological functions (Fig. 7C; see Tables S1 and S2 in the supplemental material). The results suggested that B-Myb regulates a broad spectrum of biological functions. HIF-2α-dependent genes such as VEGFA, GLUT1, and PDGF, were not affected by B-Myb knockdown, supporting the hypothesis that B-Myb regulates tumor development independently of HIF-α activity. Among the candidate genes regulated by B-Myb directly or indirectly, upregulation of those coding for complement factor B (CFB), FK506 binding protein 1B (FKBP1B), and septin 6 (SEPT6) was validated by quantitative RT-PCR (Fig. 7D). These data suggested that two independent parallel pathways mediated by HIF-α or B-Myb may finely regulate cell growth, and breakdown of this regulation might result in VHL disease.
FIG 7.
Regulation of HIF-α-independent genes by B-Myb. (A) Scatter plots of total probes examined by microarray. Total RNA purified from control or B-Myb knockdown 1 or 2 cells was used for microarray analysis to identify genes differentially expressed in response to B-Myb knockdown. The solid line indicates identical gene expression. The dashed lines indicate the threshold defined as 2 or 1/2 times expression. (B) The 370 B-Myb-regulated genes are grouped by coding or noncoding and by upregulated or downregulated. (C) The 233 B-Myb-regulated coding genes are grouped by conceivable biological functions. (D) Upregulation of CFB, FKBP1B, and SEPT6 by knockdown of B-Myb. Total RNA was purified from control or B-Myb knockdown (B-Myb#1 or B-Myb#2) cells and analyzed by quantitative RT-PCR. *, P < 0.05; **, P < 0.01.
DISCUSSION
In the present study, we identified B-Myb as a novel substrate of the ubiquitin ligase pVHL and as a novel molecule involved in VHL pathogenesis. Although previous studies showed that B-Myb is an oncogene and that knockdown of B-Myb causes G2/M-phase arrest and increased aneuploidy, we showed that B-Myb acts as a tumor suppressor gene, particularly in 786-O cells. This unexpected result could be explained by the upregulation of HIF-2α-regulated tumorigenic genes in 786-O cells caused by the possible constitutive activation of HIF-2α. Under these conditions, the oncogenic effect of B-Myb might be masked. The fact that constitutive activation of HIF-α alone is not sufficient for the development of renal clear cell carcinomas and pheochromocytomas in mice (15) suggests the involvement of an HIF-α-independent pathway in VHL pathogenesis. In our study, B-Myb was stabilized in pVHL-deficient tumor cells, and depletion of B-Myb in 786-O cells promoted tumor development (Fig. 6B); this suggested that B-Myb plays a tumor suppressor role by preventing tumor development caused by HIF-2α accumulation.
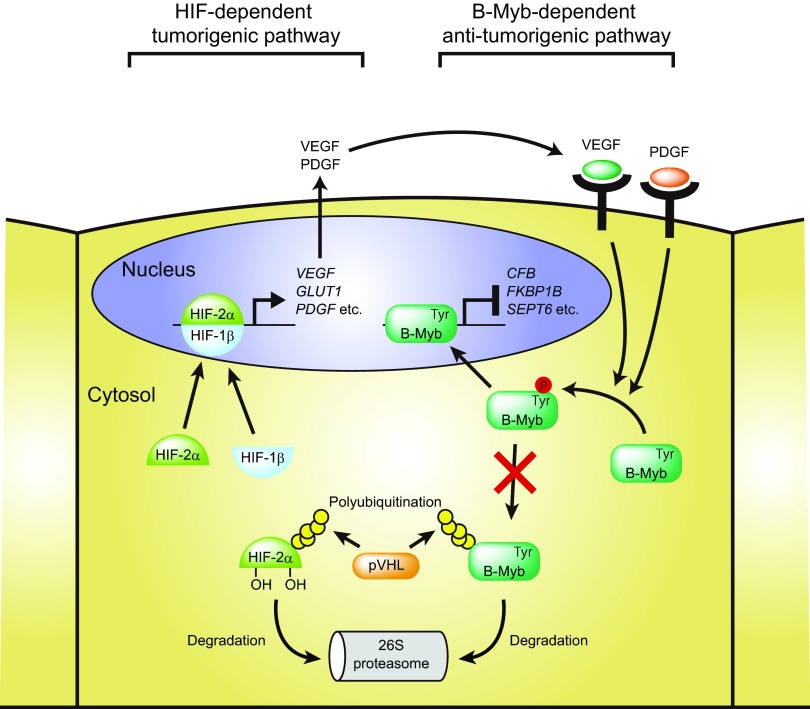
Downregulation of HIF-2α is sufficient to suppress pVHL-deficient tumor growth in some model systems (41), and high expression of HIF-1α alone is not sufficient to induce tumor growth. These facts together with the upregulation of VEGF in other model systems (51) suggest that both HIF-2α-dependent and B-Myb-dependent pathways play an important role in pVHL-deficient tumor development. We propose that the transcription factor B-Myb is a novel inhibitor of VHL disease, as illustrated in Fig. 8. Wild-type (pVHL-reconstituted 786-O) cells are able to regulate the amount of HIF-2α and B-Myb appropriately and are not tumorigenic. Loss of pVHL results in the accumulation of HIF-2α and downstream target molecules, including VEGF, GLUT1, and PDGF, leading to tumorigenesis (52). However, B-Myb, a transcription factor that regulates G2/M cell cycle transition, chromosomal condensation, and stability, is also upregulated and prevents tumor development. Downregulation of B-Myb and deficiency of pVHL result in the accumulation of cells in G2/M phase, abnormal mitosis, and increased aneuploidy in addition to accumulation of HIF-2α (27). Phosphorylation of tyrosine residue 15 of B-Myb by VEGFR1 and/or PDGFR stabilizes B-Myb (Fig. 4 and 5), suggesting the existence of a feedback loop to prevent HIF-α-dependent tumorigenesis. Deficiency of pVHL results in the constitutive activation of HIF-α/HIF-1β transcription factors and the extended expression of downstream target genes such as those coding for VEGF, GLUT1, and PDGF, which contribute to accelerated cell growth and metabolism. However, secreted VEGF and PDGF stimulate VEGFR and PDGFR, respectively, which induces tyrosine phosphorylation of B-Myb, allowing B-Myb to escape pVHL-dependent ubiquitination and degradation and regulate the expression of downstream target genes.
FIG 8.
Putative model of pVHL-, B-Myb-, and HIF-2α-mediated regulation of tumorigenesis. HIF-2α is regulated by pVHL and stabilized under certain conditions (for example, hypoxia), thereby preventing tumorigenesis. pVHL deficiency causes accumulation of HIF-2α, resulting in tumorigenesis by constitutive induction of downstream target molecules, including the VEGF, GLUT1, and PDGF genes. B-Myb inhibits HIF-2α-dependent tumorigenesis by regulating the expression of specific genes, including the CFB, FKBP1B, and SEPT6 genes. VEGF- and PDGF-dependent tyrosine phosphorylation of B-Myb prevents its degradation mediated by pVHL, suggesting the existence of a feedback loop to stabilize B-Myb when the HIF pathway is activated. B-Myb deficiency causes the upregulation of tumorigenesis-related proteins. These proteins cooperate with the constitutively active HIF pathway and accelerate tumor development.
Despite the identification of many B-Myb-regulated genes and noncoding RNAs, their potential involvement in gene regulatory functions or pathways associated with VHL disease related processes remains unclear because most of them are of unknown function. As the HIF pathway directly regulates more than 60 genes to resolve and counteract hypoxic conditions (53), the simultaneous regulation of B-Myb-targeted genes could be important to prevent HIF-dependent tumor development. Constitutive activation of the HIF pathway upregulates interleukin-1β (IL-1β), NF-κB, and cyclooxygenase 2 (COX2), which are related to inflammatory responses, and cytokines such as IL-1β and tumor necrosis factor alpha (TNF-α) induce HIF transactivation activity (53, 54). Acute inflammation mediated by complement proteins prevents tumor development, whereas chronic inflammation facilitates tumorigenesis and promotes tumor invasiveness (55, 56). In particular, complement factor B accelerates tumor invasion and migration (55). Complement activation supports chronic inflammation, the establishment of an immunosuppressive microenvironment, angiogenesis, and cancer-related signaling pathways (56). Therefore, the upregulation of complement factor B by B-Myb knockdown could contribute to tumor development.
FKBP1B is a cis-trans prolyl isomerase and regulates protein folding (57). The role of FKBP1B in the regulation of tumor development is not clear. However, FKBP1B-immunosuppressant drug complexes inhibit calcineurin (58, 59) and bind mTOR (60, 61), thereby inhibiting cytokine production and cytotoxic T-cell proliferation (58, 60–62); therefore, upregulation of FKBP1B in renal cells may also be involved in inflammation. FKBP1A, which shares approximately 85% similarity and shows almost identical structure to FKBP1B (63), is a component of the EGFR/FKBP1A/HIF-2α pathway, which plays a role in childhood high-grade astrocytomas (64), suggesting that increased FKBP1B may also cause astrocytomas.
Septins are GTP-binding proteins, and there are 13 septins (SEPT1 to -12 and -14) in humans, which exist in 6- to 8-subunit complexes (65). SEPT6 forms a complex with SEPT2 and SEPT7 (SEPT7-SEPT6-SEPT2-SEPT2-SEPT6-SEPT7), resulting in the formation of the septin filament (66). Septins also form ring-like structures (septin rings) that regulate cell division, ciliogenesis, and neuronal synapses (65). SEPT2 is upregulated in renal cell carcinomas (67), which suggests that the upregulation of SEPT6 increases the SEPT2-SEPT6-SEPT7 filament and may play a role in tumorigenesis and/or metastasis (68–70), especially in VHL disease. However, the underlying molecular mechanism remains to be identified. Simultaneous regulation of B-Myb-targeted genes, including those coding for CFB, SEPT6, and FKBP1B, may prevent tumorigenesis driven by constitutively active HIF-α. Future work should aim to shed light on any crucial hidden pathways that may exist.
VEGFR tyrosine kinase inhibitors are used in the treatment of VHL disease. However, we suggested that inhibition of VEGFR tyrosine kinase blocked the phosphorylation of B-Myb and destabilized B-Myb in patients with a functional pVHL. Certain patients have wild-type pVHL and mutant HIF-α, which cannot be polyubiquitinated by pVHL. In this case, the use of VEGFR tyrosine kinase inhibitors as a treatment for VHL disease could actually have detrimental effects. We also found that VEGFR was downregulated by confluent culture conditions. Therefore, future work will include the assessment of VEGFR expression in VHL disease patients. If VEGFR is indeed downregulated, VEGFR tyrosine kinase inhibitors would have a minor effect on patients.
In conclusion, the findings of the present study indicate that the stability and balance between HIF-2α and B-Myb regulated by pVHL are responsible for the broad spectrum of tumors associated with VHL disease.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yoji Andrew Minamishima (Keio University, Japan) for providing the RCC4 cells (control and pVHL-expressing cells).
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00067-16.
REFERENCES
- 1.Peters JM. 1998. SCF and APC: the Yin and Yang of cell cycle regulated proteolysis. Curr Opin Cell Biol 10:759–768. doi: 10.1016/S0955-0674(98)80119-1. [DOI] [PubMed] [Google Scholar]
- 2.Hershko A, Ciechanover A. 1998. The ubiquitin system. Annu Rev Biochem 67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 3.Weissman AM. 1997. Regulating protein degradation by ubiquitination. Immunol Today 18:189–198. doi: 10.1016/S0167-5699(97)84666-X. [DOI] [PubMed] [Google Scholar]
- 4.Lipkowitz S, Weissman AM. 2011. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer 11:629–643. doi: 10.1038/nrc3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG Jr, Elledge SJ, Conaway RC, Harper JW, Conaway JW. 1999. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 6.Kibel A, Iliopoulos O, DeCaprio JA, Kaelin WG Jr. 1995. Binding of the von Hippel-Lindau tumor suppressor protein to Elongin B and C. Science 269:1444–1446. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- 7.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 8.Masson N, Willam C, Maxwell PH, Pugh CW, Ratcliffe PJ. 2001. Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J 20:5197–5206. doi: 10.1093/emboj/20.18.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian YM, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ, Ratcliffe PJ. 2001. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107:43–54. doi: 10.1016/S0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 10.Hon WC, Wilson MI, Harlos K, Claridge TD, Schofield CJ, Pugh CW, Maxwell PH, Ratcliffe PJ, Stuart DI, Jones EY. 2002. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature 417:975–978. doi: 10.1038/nature00767. [DOI] [PubMed] [Google Scholar]
- 11.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG Jr. 2001. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 12.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. 2001. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 13.Kaelin WG. 2007. von Hippel-Lindau disease. Annu Rev Pathol 2:145–173. doi: 10.1146/annurev.pathol.2.010506.092049. [DOI] [PubMed] [Google Scholar]
- 14.Gnarra JR, Ward JM, Porter FD, Wagner JR, Devor DE, Grinberg A, Emmert-Buck MR, Westphal H, Klausner RD, Linehan WM. 1997. Defective placental vasculogenesis causes embryonic lethality in VHL-deficient mice. Proc Natl Acad Sci U S A 94:9102–9107. doi: 10.1073/pnas.94.17.9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim WY, Safran M, Buckley MR, Ebert BL, Glickman J, Bosenberg M, Regan M, Kaelin WG Jr. 2006. Failure to prolyl hydroxylate hypoxia-inducible factor alpha phenocopies VHL inactivation in vivo. EMBO J 25:4650–4662. doi: 10.1038/sj.emboj.7601300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sala A. 2005. B-MYB, a transcription factor implicated in regulating cell cycle, apoptosis and cancer. Eur J Cancer 41:2479–2484. doi: 10.1016/j.ejca.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Ness SA. 2003. Myb protein specificity: evidence of a context-specific transcription factor code. Blood Cells Mol Dis 31:192–200. doi: 10.1016/S1079-9796(03)00151-7. [DOI] [PubMed] [Google Scholar]
- 18.Nomura N, Takahashi M, Matsui M, Ishii S, Date T, Sasamoto S, Ishizaki R. 1988. Isolation of human cDNA clones of myb-related genes, A-myb and B-myb Nucleic Acids Res 16:11075–11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toscani A, Mettus RV, Coupland R, Simpkins H, Litvin J, Orth J, Hatton KS, Reddy EP. 1997. Arrest of spermatogenesis and defective breast development in mice lacking A-myb. Nature 386:713–717. doi: 10.1038/386713a0. [DOI] [PubMed] [Google Scholar]
- 20.Sandberg ML, Sutton SE, Pletcher MT, Wiltshire T, Tarantino LM, Hogenesch JB, Cooke MP. 2005. c-Myb and p300 regulate hematopoietic stem cell proliferation and differentiation. Dev Cell 8:153–166. doi: 10.1016/j.devcel.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 21.Bessa M, Saville MK, Watson RJ. 2001. Inhibition of cyclin A/Cdk2 phosphorylation impairs B-Myb transactivation function without affecting interactions with DNA or the CBP coactivator. Oncogene 20:3376–3386. doi: 10.1038/sj.onc.1204439. [DOI] [PubMed] [Google Scholar]
- 22.Charrasse S, Carena I, Brondani V, Klempnauer KH, Ferrari S. 2000. Degradation of B-Myb by ubiquitin-mediated proteolysis: involvement of the Cdc34-SCF(p45Skp2) pathway. Oncogene 19:2986–2995. doi: 10.1038/sj.onc.1203618. [DOI] [PubMed] [Google Scholar]
- 23.Katzen AL, Jackson J, Harmon BP, Fung SM, Ramsay G, Bishop JM. 1998. Drosophila myb is required for the G2/M transition and maintenance of diploidy. Genes Dev 12:831–843. doi: 10.1101/gad.12.6.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manak JR, Wen H, Van T, Andrejka L, Lipsick JS. 2007. Loss of Drosophila Myb interrupts the progression of chromosome condensation. Nat Cell Biol 9:581–587. doi: 10.1038/ncb1580. [DOI] [PubMed] [Google Scholar]
- 25.Yamauchi T, Ishidao T, Nomura T, Shinagawa T, Tanaka Y, Yonemura S, Ishii S. 2008. A B-Myb complex containing clathrin and filamin is required for mitotic spindle function. EMBO J 27:1852–1862. doi: 10.1038/emboj.2008.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka Y, Patestos NP, Maekawa T, Ishii S. 1999. B-myb is required for inner cell mass formation at an early stage of development. J Biol Chem 274:28067–28070. doi: 10.1074/jbc.274.40.28067. [DOI] [PubMed] [Google Scholar]
- 27.Tarasov KV, Tarasova YS, Tam WL, Riordon DR, Elliott ST, Kania G, Li J, Yamanaka S, Crider DG, Testa G, Li RA, Lim B, Stewart CL, Liu Y, Van Eyk JE, Wersto RP, Wobus AM, Boheler KR. 2008. B-MYB is essential for normal cell cycle progression and chromosomal stability of embryonic stem cells. PLoS One 3:e2478. doi: 10.1371/journal.pone.0002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamura T, Hara T, Matsumoto M, Ishida N, Okumura F, Hatakeyama S, Yoshida M, Nakayama K, Nakayama KI. 2004. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27(Kip1) at G1 phase. Nat Cell Biol 6:1229–1235. doi: 10.1038/ncb1194. [DOI] [PubMed] [Google Scholar]
- 29.Zhao WT, Zhou CF, Li XB, Zhang YF, Fan L, Pelletier J, Fang J. 2013. The von Hippel-Lindau protein pVHL inhibits ribosome biogenesis and protein synthesis. J Biol Chem 288:16588–16597. doi: 10.1074/jbc.M113.455121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warnecke C, Zaborowska Z, Kurreck J, Erdmann VA, Frei U, Wiesener M, Eckardt KU. 2004. Differentiating the functional role of hypoxia-inducible factor (HIF)-1 alpha and HIF-2 alpha (EPAS-1) by the use of RNA interference: erythropoietin is a HIF-2 alpha target gene in Hep3B and Kelly cells. FASEB J 18:1462–1464. [DOI] [PubMed] [Google Scholar]
- 31.Okumura F, Zou W, Zhang DE. 2007. ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP. Genes Dev 21:255–260. doi: 10.1101/gad.1521607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI. 2004. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev 18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iliopoulos O, Ohh M, Kaelin WG Jr. 1998. pVHL19 is a biologically active product of the von Hippel-Lindau gene arising from internal translation initiation. Proc Natl Acad Sci U S A 95:11661–11666. doi: 10.1073/pnas.95.20.11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh IH, Reddy EP. 1999. The myb gene family in cell growth, differentiation and apoptosis. Oncogene 18:3017–3033. doi: 10.1038/sj.onc.1202839. [DOI] [PubMed] [Google Scholar]
- 35.Lee S, Chen DYT, Humphrey JS, Gnarra JR, Linehan WM, Klausner RD. 1996. Nuclear cytoplasmic localization of the von Hippel-Lindau tumor suppressor gene product is determined by cell density. Proc Natl Acad Sci U S A 93:1770–1775. doi: 10.1073/pnas.93.5.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson TK, Schweppe RE, Septer J, Lewis RE. 1999. Phosphorylation of B-Myb regulates its transactivation potential and DNA binding. J Biol Chem 274:36741–36749. doi: 10.1074/jbc.274.51.36741. [DOI] [PubMed] [Google Scholar]
- 37.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. 2012. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res 40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang D, Ding Y, Li Y, Luo WM, Zhang ZF, Snider J, Vandenbeldt K, Qian CN, Teh BT. 2010. Sunitinib acts primarily on tumor endothelium rather than tumor cells to inhibit the growth of renal cell carcinoma. Cancer Res 70:1053–1062. doi: 10.1158/0008-5472.CAN-09-3722. [DOI] [PubMed] [Google Scholar]
- 39.Haglund K, Dikic I. 2012. The role of ubiquitylation in receptor endocytosis and endosomal sorting. J Cell Sci 125:265–275. doi: 10.1242/jcs.091280. [DOI] [PubMed] [Google Scholar]
- 40.Alwan HAJ, van Zoelen EJJ, van Leeuwen JEM. 2003. Ligand-induced lysosomal epidermal growth factor receptor (EGFR) degradation is preceded by proteasome-dependent EGFR de-ubiquitination. J Biol Chem 278:35781–35790. doi: 10.1074/jbc.M301326200. [DOI] [PubMed] [Google Scholar]
- 41.Kondo K, Kim WY, Lechpammer M, Kaelin WG Jr. 2003. Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol 1:E83. doi: 10.1371/journal.pbio.0000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lenburg ME, Liou LS, Gerry NP, Frampton GM, Cohen HT, Christman MF. 2003. Previously unidentified changes in renal cell carcinoma gene expression identified by parametric analysis of microarray data. BMC Cancer 3:31. doi: 10.1186/1471-2407-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beroukhim R, Brunet JP, Di Napoli A, Mertz KD, Seeley A, Pires MM, Linhart D, Worrell RA, Moch H, Rubin MA, Sellers WR, Meyerson M, Linehan WM, Kaelin WG Jr, Signoretti S. 2009. Patterns of gene expression and copy-number alterations in von-Hippel Lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res 69:4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okada M, Akimaru H, Hou DX, Takahashi T, Ishii S. 2002. Myb controls G(2)/M progression by inducing cyclin B expression in the Drosophila eye imaginal disc. EMBO J 21:675–684. doi: 10.1093/emboj/21.4.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beall EL, Manak JR, Zhou S, Bell M, Lipsick JS, Botchan MR. 2002. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 420:833–837. doi: 10.1038/nature01228. [DOI] [PubMed] [Google Scholar]
- 46.Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS, DeCaprio JA. 2007. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell 26:539–551. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 47.Wen H, Andrejka L, Ashton J, Karess R, Lipsick JS. 2008. Epigenetic regulation of gene expression by Drosophila Myb and E2F2-RBF via the Myb-MuvB/dREAM complex. Genes Dev 22:601–614. doi: 10.1101/gad.1626308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fung SM, Ramsay G, Katzen AL. 2002. Mutations in Drosophila myb lead to centrosome amplification and genomic instability. Development 129:347–359. [DOI] [PubMed] [Google Scholar]
- 49.Garcia P, Frampton J. 2006. The transcription factor B-Myb is essential for S-phase progression and genomic stability in diploid and polyploid megakaryocytes. J Cell Sci 119:1483–1493. doi: 10.1242/jcs.02870. [DOI] [PubMed] [Google Scholar]
- 50.Hughes MD, Kapllani E, Alexander AE, Burk RD, Schoenfeld AR. 2007. HIF-2alpha downregulation in the absence of functional VHL is not sufficient for renal cell differentiation. Cancer Cell Int 7:13. doi: 10.1186/1475-2867-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maranchie JK, Vasselli JR, Riss J, Bonifacino JS, Linehan WM, Klausner RD. 2002. The contribution of VHL substrate binding and HIF1-alpha to the phenotype of VHL loss in renal cell carcinoma. Cancer Cell 1:247–255. doi: 10.1016/S1535-6108(02)00044-2. [DOI] [PubMed] [Google Scholar]
- 52.Li M, Kim WY. 2011. Two sides to every story: the HIF-dependent and HIF-independent functions of pVHL. J Cell Mol Med 15:187–195. doi: 10.1111/j.1582-4934.2010.01238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaber T, Dziurla R, Tripmacher R, Burmester GR, Buttgereit F. 2005. Hypoxia inducible factor (HIF) in rheumatology: low O2! See what HIF can do! Ann Rheum Dis 64:971–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szade A, Grochot-Przeczek A, Florczyk U, Jozkowicz A, Dulak J. 2015. Cellular and molecular mechanisms of inflammation-induced angiogenesis. IUBMB Life 67:145–159. doi: 10.1002/iub.1358. [DOI] [PubMed] [Google Scholar]
- 55.Rutkowski MJ, Sughrue ME, Kane AJ, Mills SA, Parsa AT. 2010. Cancer and the complement cascade. Mol Cancer Res 8:1453–1465. doi: 10.1158/1541-7786.MCR-10-0225. [DOI] [PubMed] [Google Scholar]
- 56.Pio R, Ajona D, Lambris JD. 2013. Complement inhibition in cancer therapy. Semin Immunol 25:54–64. doi: 10.1016/j.smim.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang CB, Hong Y, Dhe-Paganon S, Yoon HS. 2008. FKBP family proteins: immunophilins with versatile biological functions. Neurosignals 16:318–325. doi: 10.1159/000123041. [DOI] [PubMed] [Google Scholar]
- 58.Fruman DA, Klee CB, Bierer BE, Burakoff SJ. 1992. Calcineurin phosphatase activity in T lymphocytes is inhibited by FK 506 and cyclosporin A. Proc Natl Acad Sci U S A 89:3686–3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sewell TJ, Lam E, Martin MM, Leszyk J, Weidner J, Calaycay J, Griffin P, Williams H, Hung S, Cryan J, Sigal NH, Wiederrecht GJ. 1994. Inhibition of calcineurin by a novel FK-506-binding protein. J Biol Chem 269:21094–21102. [PubMed] [Google Scholar]
- 60.Brown EJ, Beal PA, Keith CT, Chen J, Shin TB, Schreiber SL. 1995. Control of p70 s6 kinase by kinase activity of FRAP in vivo. Nature 377:441–446. doi: 10.1038/377441a0. [DOI] [PubMed] [Google Scholar]
- 61.Heitman J, Movva NR, Hall MN. 1991. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 62.Perrucci GL, Gowran A, Zanobini M, Capogrossi MC, Pompilio G, Nigro P. 2015. Peptidyl-prolyl isomerases: a full cast of critical actors in cardiovascular diseases. Cardiovasc Res 106:353–364. doi: 10.1093/cvr/cvv096. [DOI] [PubMed] [Google Scholar]
- 63.Deivanayagam CC, Carson M, Thotakura A, Narayana SV, Chodavarapu RS. 2000. Structure of FKBP12.6 in complex with rapamycin. Acta Crystallogr D Biol Crystallogr 56:266–271. doi: 10.1107/S0907444999016571. [DOI] [PubMed] [Google Scholar]
- 64.Khatua S, Peterson KM, Brown KM, Lawlor C, Santi MR, LaFleur B, Dressman D, Stephan DA, MacDonald TJ. 2003. Overexpression of the EGFR/FKBP12/HIF-2alpha pathway identified in childhood astrocytomas by angiogenesis gene profiling. Cancer Res 63:1865–1870. [PubMed] [Google Scholar]
- 65.Saarikangas J, Barral Y. 2011. The emerging functions of septins in metazoans. EMBO Rep 12:1118–1126. doi: 10.1038/embor.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weirich CS, Erzberger JP, Barral Y. 2008. The septin family of GTPases: architecture and dynamics. Nat Rev Mol Cell Biol 9:478–489. doi: 10.1038/nrm2407. [DOI] [PubMed] [Google Scholar]
- 67.Craven RA, Hanrahan S, Totty N, Harnden P, Stanley AJ, Maher ER, Harris AL, Trimble WS, Selby PJ, Banks RE. 2006. Proteomic identification of a role for the von Hippel Lindau tumour suppressor in changes in the expression of mitochondrial proteins and septin 2 in renal cell carcinoma. Proteomics 6:3880–3893. doi: 10.1002/pmic.200500811. [DOI] [PubMed] [Google Scholar]
- 68.Gilden JK, Peck S, Chen YC, Krummel MF. 2012. The septin cytoskeleton facilitates membrane retraction during motility and blebbing. J Cell Biol 196:103–114. doi: 10.1083/jcb.201105127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Osaka M, Rowley JD, Zeleznik-Le NJ. 1999. MSF (MLL septin-like fusion), a fusion partner gene of MLL, in a therapy-related acute myeloid leukemia with a t(11;17)(q23;q25). Proc Natl Acad Sci U S A 96:6428–6433. doi: 10.1073/pnas.96.11.6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jaeger J, Koczan D, Thiesen HJ, Ibrahim SM, Gross G, Spang R, Kunz M. 2007. Gene expression signatures for tumor progression, tumor subtype, and tumor thickness in laser-microdissected melanoma tissues. Clin Cancer Res 13:806–815. doi: 10.1158/1078-0432.CCR-06-1820. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.