Abstract
Long noncoding RNAs (lncRNAs) can regulate gene expression in a cell-specific fashion during development. Here, we identify a novel lncRNA from the X chromosome that we named lncRHOXF1 and which is abundantly expressed in trophectoderm and primitive endoderm cells of human blastocyst-stage embryos. lncRHOXF1 is a spliced and polyadenylated lncRNA about 1 kb in length that is found in both the nuclear and cytoplasmic compartments of in vitro differentiated human trophectoderm progenitor cells. Gain-of-function experiments in human embryonic stem cells, which normally lack lncRHOXF1 RNA, revealed that lncRHOXF1 reduced proliferation and favored cellular differentiation. lncRHOXF1 knockdown using small interfering RNAs (siRNAs) in human trophectoderm progenitors increased expression of viral response genes, including type I interferon. Sendai virus infection of human trophectoderm progenitor cells increased lncRHOXF1 RNA levels, and siRNA-mediated disruption of lncRHOXF1 during infection reduced the expression of viral response genes leading to higher virus replication. Thus, lncRHOXF1 RNA is the first example of a lncRNA that regulates the host response to viral infections in human placental progenitor cells, and we propose that it functions as a repressor of the viral response during early human development.
INTRODUCTION
The mammalian genome contains thousands of long noncoding RNAs (lncRNAs) that are transcribed in a cell- and tissue-specific fashion. While only a few of these lncRNAs have been functionally characterized, some are known to play important roles during development. X chromosome inactivation and genomic imprinting, classic epigenetic processes required for the development of the early embryo and placenta, are regulated by lncRNAs (1). Relative to other somatic tissues, many lncRNAs are exclusively or predominantly expressed in the placenta (2). Recent studies suggest that the formation of the placenta likely involves lncRNAs and that that some of these lncRNAs become differentially expressed during complicated pregnancies (2). However, the function for the majority of these placental lncRNAs is still unknown.
The development of the mammalian early embryo is regulated by epigenetic mechanisms that coordinate gene expression changes required to transition from totipotency to more-differentiated states. The placenta is formed about 1 week postconception and is a transient organ derived from the embryo, which supports its growth and development. The placenta originates in the preimplantation blastocyst, from the outer trophectoderm (TE) cells that surround the inner cell mass (ICM) and blastocoel cavity. During implantation, the TE progenitor cells differentiate into cytotrophoblasts (CTBs) and multinucleated syncytiotrophoblasts (SYNs) and begin to invade the endometrium (3). CTBs remodel the uterine spiral arterioles to sequester a maternal blood supply. SYNs are terminally differentiated cells that facilitate nutrient and gas exchange between the fetus and the mother and also produce hormones required to sustain the pregnancy.
The placenta is a physical barrier between the mother and fetus, and emerging data indicate that it is also an immunological barrier that prevents transmission of pathogens to the fetus (4). Recent studies indicate that the immune system is not suppressed during pregnancy but actually is actively engaged and carefully regulated at the implantation site (5). Placental trophoblasts and various immune cells (T cells, macrophages, natural killer cells, and dendritic cells) regulate immunity at the maternal-fetal interface, yet our understanding of the specific mechanisms by which the placenta protects the developing fetus from viral infections is not complete. The SYNs directly contact the maternal blood supply and are the first line of defense against invading pathogens. SYNs are resistant to infection by cytomegalovirus, herpes simplex viruses 1 and 2, human immunodeficiency virus, coxsackieviruses, and the nonviral prenatal pathogens Listeria monocytogenes and Toxoplasma gondii (4). In contrast, the CTBs, which reside between the SYNs and the fetal basement membrane, are susceptible to infection by viruses and nonviral pathogens that do not infect SYNs (4). However, human SYNs, which produce high levels of exosomes, transfer viral resistance to recipient cells through the generation of microRNA-containing exosomes (6).
Here, we identify a novel lncRNA, termed lncRHOXF1, that is robustly expressed from the X chromosome in TBs from preimplantation human embryos and in vitro-derived trophoblast progenitor cells. Strikingly, we found that lncRHOXF1 regulates expression of genes involved in the sensing and response to virus infection in these cells, providing the first example of a lncRNA that regulates the innate immune response in the early human placenta.
MATERIALS AND METHODS
Cell culture of in vitro-derived human TBs.
Human embryonic stem cells (hESCs; HUES-7 and HUES-9 cell lines) and human induced pluripotent stem cells (hiPSCs) (7) were cultured on mouse embryonic fibroblasts (MEFs) under 4% oxygen and passaged manually as described previously (7, 8). For bone marrow protein 4 (BMP4) with A83-01 and PD173074 (BMP4/A/P)-induced differentiation (here termed BMP4/A/P differentiation) to human trophectoderm progenitors, cells were passaged onto plates coated with Matrigel (R&D Biosystems) and then grown using MEF-conditioned medium containing β-fibroblast growth factor (β-FGF) (9). After 6 to 7 days, cells were passaged 1:3 using Dispase (Life Technologies) onto Matrigel-coated 6-well plates. To support differentiation, the medium, which lacked β-FGF and contained 50 ng/ml BMP4, 1 μM A83-01 (Tocris), and 0.1 μM PD173074 (Tocris) (BMP4/A/P), was added the next day and refreshed every 48 h for the course of the differentiation.
siRNA transfection and Sendai virus infection.
Three small interfering RNA (siRNA) target sequences for lncRHOXF1, a control scramble siRNA, and siRNA for hnRNP U were designed and manufactured by Dharmacon. BMP4/A/P-induced cells were transfected with siRNAs at a final concentration of 75 pmol using RNAi Max (Invitrogen). Transfections were performed twice at 24-h intervals during BMP4-induced differentiation before cells were collected. Knockdown was confirmed using reverse transcription-PCR (RT-PCR). Sendai virus strain Cantell was grown and characterized at the Lopez laboratory (10) and added at a multiplicity of infection of 1 to a 0.5-ml volume of cell medium following 48 h of knockdown. Cells were infected for 8 h and then harvested for RNA isolation. These experiments were repeated 8 times, yielding similar results (with varied knockdown efficiencies), and representative data from one experiment are shown.
RNA and DNA FISH.
Fluorescence in situ hybridization (FISH) experiments were performed as described previously (11). lncRHOXF1 RNA was detected using a Cy3-labeled probe of lncRHOXF1 cDNA (∼800 nucleotides [nt]) comprised of exons 1 to 4, which was labeled by nick translation (Roche). DNA FISH for the X chromosomes was performed using X-paints (Cambio, Cambridge, United Kingdom). For experiments combining RNA and DNA FISH, the RNA FISH was performed first. Images were captured and positions recorded, then slides were refixed in 4% paraformaldehyde, treated with RNase A (to degrade RNA signals), and then denatured for DNA FISH. After hybridization overnight at 37°C, the slides were reimaged at the recorded positions. For RNA FISH analyses preserving the cytoplasm, cells were either cytospun (hESCs and hiPSCs) or grown on chamber slides, then fixed for 10 min using 4% paraformaldehyde, washed, and permeabilized with 70% ethanol for 1 h (12). Each chamber was washed, then hybridization buffer containing denatured Cy3-labeled lncRHOXF1 probe was added, and the mixture was incubated overnight. Slides were washed twice and then mounted using 4′,6-diamidino-2-phenylindole–VectaShield before imaging.
Nuclear-cytoplasmic fractionation.
BMP4/A/P-differentiated cells (derived from HUES-9 cells) were washed twice with cold phosphate-buffered saline and pelleted at 200 × g for 5 min, then resuspended in fractionation buffer (10 mM Tris-HCl [pH 8.4], 1.5 mM MgCl2, 140 mM NaCl), and 5 μl of 5% NP-40 and 1 μl RNase inhibitor (Roche) were added. Samples of the fractionation reaction mixture were taken at intervals of 5, 10, 15, 20, and 30 min, with 5 μl of 5% NP-40 added to the remaining reaction mixture at each time point. The harvested samples were spun at 2,500 rpm for 2 min in a tabletop centrifuge at 4°C, and the supernatant was mixed with TRIzol-L/S (Thermo Fisher Scientific), while the pellet was resuspended in TRIzol (Thermo Fisher Scientific). Samples were stored at −80°C prior to RNA isolation. Cell fractionation experiments were repeated at least twice, demonstrating nuclear and cytoplasmic expression of lncRHOXF1, and results from one experiment (female cells) are shown below in Fig. 2.
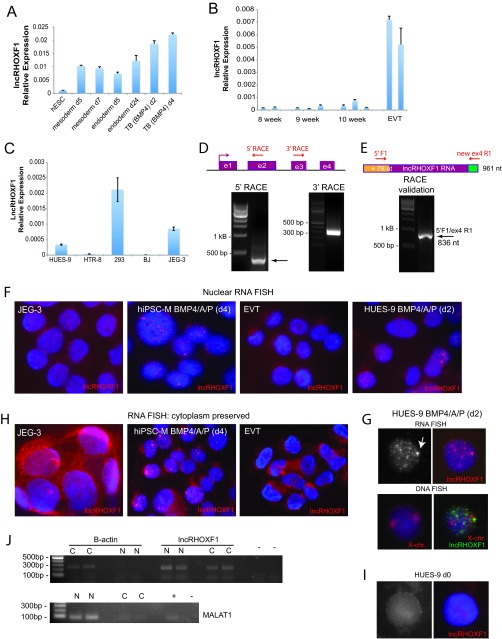
FIG 2.
lncRHOXF1 RNA isoforms are present in the nucleus and cytoplasm. (A) qRT-PCR analysis of lncRHOXF1 expression for in vitro-differentiated hESCS to mesoderm, endoderm, and trophectoderm lineages. Error bars denote standard deviations from the means. (B) qRT-PCR analysis of lncRHOXF1 expression in human placental samples from 8-, 9-, and 10-week conceptuses (three independent placentas for each time point) and primary extravillous trophoblast cells (from two different placentas). Error bars denote standard deviations from the means. (C) qRT-PCR analysis of lncRHOXF1 RNA in transformed epithelial fetal kidney cells (293T), placental choriocarcinoma cells (JEG3), transformed trophoblast cells (HTR-8), fibroblast foreskin cells (BJ), and BMP4/A/P-differentiated hESCs for comparison. Error bars denote standard deviations from the means. (D) Schematic of the lncRHOXF1 gene, with locations of the 5′- and 3′-RACE primers. PCR results for 5′- and 3′-RACE of lncRHOXF1. The arrow denotes the lncRHOXF1 transcript. (E) Validation of 5′- and 3′-RACE ends for lncRHOXF1, using primers 5′ F1 and new ex4 R1. (F) Nuclear RNA FISH analyses for lncRHOXF1 expression in primary and immortalized placental progenitor cells. Cells were disrupted to preserve the nucleus, then fixed prior to probe hybridization. (G) Sequential RNA-DNA FISH analyses of lncRHOXF1 transcripts in BMP4/A/P-differentiated (day 2) female hESCs. The arrow denotes nascent transcription (as a pinpoint) from the active X chromosome. Images are from the same nucleus. (H) Whole-cell (cytoplasm preserved) RNA FISH analysis of lncRHOXF1 expression in primary and immortalized placental progenitor cells. lncRHOXF1 RNA transcripts are present in both the cytoplasm and nucleus. (I) Nuclear RNA FISH analysis for undifferentiated hESCs, which lack detectable lncRHOXF1 transcripts. (J) Cell fractionation of BMP4/A/P cells (HUES-9) revealed that lncRHOXF1 is present in the nucleus and cytoplasm. MALAT1 RNA is exclusively nuclear, and β-actin RNA is cytoplasmic. +, inclusion of reverse transcriptase for cDNA synthesis.
Transgene constructs.
The lncRHOXF1 cDNA was amplified by PCR with NcoI and KpnI restriction sites, then cloned into the pTRE tight vector that contained homology arms for targeting the adeno-associated virus integration site (AAVS) locus on chromosome 19 (13).
Quantitative RT-PCR (qRT-PCR) experiments.
Total RNA was isolated using TRIzol (Invitrogen), and 500 ng was reverse transcribed with random primers using qScript cDNA master mix (Quanta Biosciences). Control reaction mixtures without reverse transcriptase were also prepared. Melting curve analyses showed a single peak for each primer pair, indicative of homogeneity of PCR products. Expression levels were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) levels, and relative expression levels were plotted using the ΔΔCT comparison. Error bars in the figures represent standard deviations among triplicate reactions, and each RT-PCR experiment was repeated at least three times. Primer sequences are available upon request.
Cell growth and cellular differentiation assays.
We tested five replicates for each condition (with or without doxycycline [DOX]) and followed the cells for over five passages. Statistical significance was determined using a two-tailed paired Student's t test. Cell growth experiments were repeated two times using both clone 23 and clone 28 (with 5 to 6 replicates for each clone and treatment) and yielded similar results. The results from one experiment using clone 28 are shown below in Fig. 3C. For embryoid body (EB) formation, equal numbers of HUES9-lncRHOXF1 cells (in triplicate) were transferred to low-retention tissue culture plates, using hESC medium lacking β-FGF. After 8 days, the total number of EB spheres was counted for each clone for each treatment (with or without doxycycline). Three independent EB differentiation experiments using clones 23 and 28 were performed and yielded similar results; results from one experiment are shown below in Fig. 3D. Statistical significance between treated and untreated EB groups was determined using a one-tailed paired Student t test.
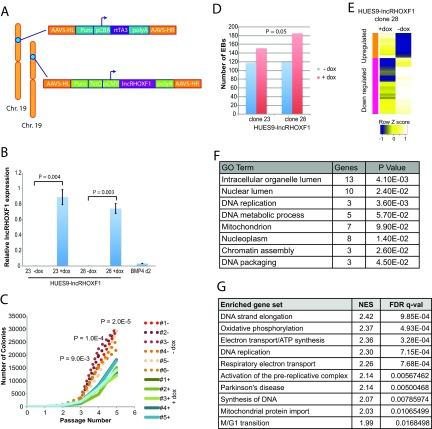
FIG 3.
Overexpression of lncRHOXF1 in undifferentiated hESCs promotes cellular differentiation by changing gene expression. (A) Targeting constructs for inducible expression of lncRHOXF1 RNA from the AAVS locus on chromosome 19 in hESCs. The reverse tetracycline transactivator (rtTA3) was constitutively expressed using the chicken β-actin (CBA) promoter. (B) qRT-PCR analysis of lncRHOXF1 expression in uninduced (−dox) and induced (+dox) hESCs, and BMP4/A/P-differentiated hESCs (for comparison). P values were determined using a two-tailed t test to compare untreated versus induced samples. Error bars denote standard deviations from the mean. (C) Overexpression of lncRHOXF1 RNA (clone 28) reduces cell growth, which was quantified based on the number of undifferentiated colonies during routine passaging. Five and six independent replicates for each condition (with or without doxycycline) were followed for 5 passages. Statistical significance was determined using a two-tailed paired t test comparing –dox and +dox samples. (D) Overexpression of lncRHOXF1 (clones 23 and 28) increases the number of embryoid bodies during differentiation. The P value was determined using a one-tailed t test to compare EB quantities between clones 23 and 28. (E) Microarray analyses of lncRHOXF1 overexpression in hESCs. The heat map indicates clustering of upregulated and downregulated genes for the 162 differentially expressed genes in HUES9-lncRHOXF1 cells (clone 28) treated or not with doxycycline. Biological replicates (triplicates for each sample) were averaged together to determine gene expression differences with lncRHOXF1 induction. (F) GO analysis for the 162 differentially expressed genes (red, upregulated; blue, downregulated) when lncRHOXF1 is overexpressed (+dox). P values were determined by using the DAVID online analysis tool. (G) GSEA of gene sets enriched in DOX-treated HUES9-lncRHOXF1 cells. NES, normalized enrichment score; FDR, false-discovery rate. FDR q values were calculated by using the GSEA tool.
Expression profiling using microarrays.
BMP4/A/P-differentiated hESCs (HUES7 and HUES9) were used for siRNA-mediated knockdown of lncRHOXF1. Microarray analyses were performed using male HUES7 cells. Undifferentiated HUES9 cells containing the lncRHOXF1 and rtTA3 cDNAs inserted into the AAVS locus of chromosome 19 were used in overexpression experiments. Total RNA was isolated using TRIzol (Invitrogen). RNA was processed using the NuGEN Ovation Pico system (for knockdown) and the TargetAmp Nano labeling kit (for overexpression). Labeled cDNA probes were hybridized onto the human Affymetrix U133 Plus 2.0 system for knockdown experiments, and for overexpression experiments we used Illumina Human HT12 V4 chips. Biological replicates (in triplicate) were analyzed, and results were averaged together to determine gene expression differences.
RESULTS
lncRHOXF1 encodes a lncRNA expressed in the human trophectoderm lineage.
We hypothesized that lncRNAs important for early placental development would be robustly expressed in extraembryonic cells and missing in the early epiblast. Therefore, we searched for lncRNAs that were specifically expressed in the trophectoderm but not the inner cell mass by using single-cell RNA sequencing data from blastocyst-stage human embryos (14). Using this criterion, we identified a gene (ENSG00000234493; RHOXF1P1; LNCRHOXF2B), which we named lncRHOXF1. It exhibited high numbers of reads in both the TE and primitive endoderm (PE) cells of human blastocysts, yet was absent in oocytes, zygotes, epiblast cells, and human embryonic stem cells (Fig. 1A). Reads were also detected at the 4-cell to morula stage of embryos, during the transition from totipotency to pluripotency, yet at lower levels than in TE and PE cells. We concluded that lncRHOXF1 RNA is one of the most abundant lncRNAs expressed in human trophectoderm cells.
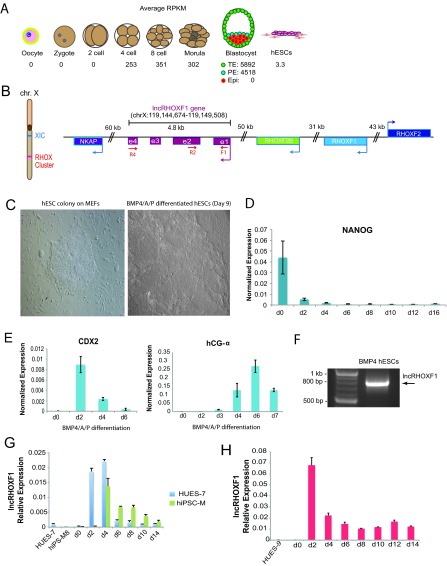
FIG 1.
lncRHOXF1 is robustly expressed during early human development. (A) Schematic showing RNA sequencing reads (RPKM) for lncRHOXF1 during early human development (14). (B) Genomic location of lncRHOXF1 on the X chromosome. XIC, X-inactivation center. Primer locations for transcript cloning (F1 and R4) and qPCR (F1 and R2) are shown. (C) Bright-field images of a human embryonic stem cell colony cultured using mouse embryonic fibroblasts (left) and BMP4/A/P-differentiated cells after 9 days (right). (D) qRT-PCR analysis of decreased NANOG expression during BMP4/A/P differentiation. Error bars denote standard deviations from the means. (E) qRT-PCR analysis of increased placentation markers CDX2 and hCG-α expression during BMP4/A/P differentiation. Error bars denote standard deviations from the means. (F) RT-PCR amplification of exons 1 to 4 of lncRHOXF1 by using primers F1 and R4 and RNA from BMP4/A/P-differentiated hESCs. (G) qRT-PCR analysis of lncRHOXF1 expression in male hESCs (blue) and hiPSCs (green) during BMP4/A/P differentiation. Error bars denote standard deviations from the mean. (H) qRT-PCR analysis of lncRHOXF1 expression during BMP4/A/P differentiation of human female pluripotent stem cells. Error bars denote standard deviations from the means.
The lncRHOXF1 gene is 4.8 kb in size and located within the reproductive homeobox (RHOX) gene locus on the X chromosome, in between the protein-coding genes RHOXF2B and NKAP (Fig. 1B). The human RHOX genes are not well characterized, but they are expressed in oocytes and male germ cells and have been associated with male infertility (15). Mouse RHOX genes are also genes that are expressed during early embryonic development, and some family members are essential for male and female reproduction (16). Although the lncRHOXF1 gene was predicted to be noncoding, we used in silico analyses to assess its coding potential. Using the NCBI open reading frame (ORF) finder, we detected short open reading frames that would be unlikely to produce a protein (data not shown). PhyloCSF analysis of the lncRHOXF1 gene yielded negative values (data not shown), supporting its noncoding status. Next, we searched for sequence homology using NCBI BLAST and PhyloP, and we found that rhesus monkeys, dogs, and elephants have significant matches across the human lncRHOXF1 gene (data not shown). The mouse genomic sequence has some homology to the exon 3 region of human lncRHOXF1. Thus, lncRHOXF1 encodes a recently evolved lncRNA from the X chromosome and is likely present in humans and higher placental mammals, excluding mice.
The lncRHOXF1 gene is also located near another long noncoding RNA, RHOXF1-AS1 (NR_131238; ENST0000553843; RP4-755D9.1). According to the NONCODE database, RHOXF1-AS1 is predominantly expressed in breast and thyroid tissues, and its expression is quite low in placenta and ovary. RHOXF1-AS1 reads were also absent in the single-cell RNA sequencing data sets used to identify lncRHOXF1 RNA (14). Because lncRHOXF1 RNA and RHOXF1-AS1 are expressed in different cells, we concluded that they are not functionally linked and are likely regulated independently of each other. Examination of the lncRHOXF1 region using the UCSC genome browser revealed the presence of a CpG island in the promoter region of this gene. We examined the expression level for lncRHOXF1 by using available ENCODE RNA sequencing data for five different cell lines (including H1 hESCs), and we found that lncRHOXF1 was not expressed in any of them (data not shown). DNA methylation profiles for the CpG island of lncRHOXF1 across these different cell lines varied from being unmethylated (for the primary cell H1 hESCs and the B lymphocyte line GM12878) to hypermethylated for the three carcinoma cell lines (A549, HepG2, and HeLa), consistent with the frequently observed hypermethylation of cancer cells. We conclude that DNA methylation of this CpG island is unlikely to regulate expression of lncRHOXF1.
Because of ethical and technical limitations of working with human embryos, we validated the presence of lncRHOXF1 RNA during early human preimplantation development by using an in vitro model for human trophoblast progenitors. We differentiated hESCs by removing β-FGF from the culture medium and adding three factors: BMP4 and the inhibitors A83-01 and PD173074, which block the SMAD2/3 and MEK1/2 signaling pathways, respectively (17). These compounds induce differentiation of pluripotent stem cells into early human trophectoderm-like progenitors, mainly SYNs and extravillous cytotrophoblasts (EVTs), without extensive generation of mesoderm, endoderm, or ectoderm cells (17–20). Using this method, the cells acquired a flattened epithelial appearance (Fig. 1C), and expression of the pluripotency marker NANOG decreased within 48 h of treatment (Fig. 1D). Importantly, the BMP4/A/P culture conditions stimulate expression of various trophoblast markers and placental hormones (9, 17, 18), and we validated expression of HLA-G, CDX2, human chorionic gonadotropin, and placental growth factor transcripts by using RT-PCR (Fig. 1E and data not shown). To determine whether the full-length lncRHOXF1 RNA transcript was expressed in this model system, we isolated RNA and amplified one transcript using primers specific for exons 1 and 4 (Fig. 1F). Sanger sequencing confirmed the identity of lncRHOXF1.
Next, we used the BMP4/A/P model of in vitro trophoblast development to examine when lncRHOXF1 is expressed. Undifferentiated male and female hESCs and hiPSCs express very little lncRHOXF1 RNA, in agreement with the single-cell RNA sequencing data (Fig. 1G and H) (14). Similar to CDX2 expression (Fig. 1E), which specifies the trophectoderm lineage in mice (21), we found that lncRHOXF1 expression was highest during days 2 to 4 of BMP4/A/P differentiation (Fig. 1G and H). Female BMP4/A/P-differentiated trophoblast progenitors expressed greater levels of lncRHOXF1 transcript than did differentiated male cells (Fig. 1H). Next, we examined lncRHOXF1 expression in in vitro-differentiated early mesoderm and endoderm progenitor cells (22), and we found that this transcript was present, although at lower levels than in trophoblast progenitors (Fig. 2A). lncRHOXF1 was expressed at low levels in first-trimester human placentas (8-, 9-, and 10-week gestational age) and at higher levels in primary EVTs (Fig. 2B), although levels were still not as high as in BMP4/A/P-differentiated cells. In contrast, lncRHOXF1 was expressed at very low (but detectable) levels in placenta-derived transformed cell lines (HTR-8, JEG-3) that are used to model trophoblast-related events (Fig. 2C). The transformed human female fetal kidney cell line 293T had higher lncRHOXF1 expression than the transformed trophoblast cell lines, likely a result of its aneuploidy and multiple X chromosomes (Fig. 2C). In conclusion, lncRHOXF1 is expressed in early progenitor cells that have recently exited the pluripotent state, suggesting that this transcript may function during the initial stages of trophoblast lineage development.
lncRHOXF1 RNA is a spliced and polyadenylated cytoplasmic transcript in human placental progenitor cells.
Using RNA isolated from BMP4/A/P-differentiated hESCs, we performed rapid amplification of cDNA ends (RACE) to determine the 5′ and 3′ ends of the lncRHOXF1 transcript (Fig. 2D). We found that lncRHOXF1 RNA had a different 5′ sequence in BMP4/A/P-differentiated hESCs than found in the NCBI and ENSEMBL database records for this transcript, with an additional 78-nt sequence immediately upstream of exon 1. Using 3′-RACE, we found that lncRHOXF1 RNA is polyadenylated with 12 or 13 A's, and we observed that exon 4 has a longer and slightly different nucleotide sequence than the NCBI sequence. We confirmed our 5′- and 3′-RACE results by using PCR primers located in the novel upstream 5′ and novel 3′ ends of exon 4 (Fig. 2E). Thus, lncRHOXF1 is 961 nucleotides in length and is spliced and polyadenylated.
Next, we examined the nuclear localization pattern of lncRHOXF1 by using RNA FISH. We designed a probe specific for exons 1 to 4 of lncRHOXF1 (774 bp), and we found that this transcript is abundant and easily detectible in nuclei from BMP4/A/P-differentiated hESCs, hiPSCs, JEG-3 cells, and primary EVTs (Fig. 2F). lncRHOXF1 transcripts localized to multiple small bright foci widely distributed throughout the nucleus, similar to the NEAT1/Neat1 RNA localization pattern, also determined using RNA FISH (23). Next, we performed sequential RNA/DNA FISH using X-paint probes, and we confirmed that the brightest pinpoint observed in all nuclei corresponded to the nascent transcript that overlapped with one X chromosome. Female BMP4/A/P cells had just one pinpoint overlapping the X chromosome, which indicates that lncRHOXF1 is subject to X chromosome inactivation in female cells (Fig. 2G). Consistent with this idea, RNA FISH using male cells, which have only one active X chromosome, routinely displayed one bright pinpoint, corresponding to the nascent lncRHOXF1 transcript (Fig. 2F). We conclude that lncRHOXF1 RNA is abundant in the nucleus of human trophoblast progenitor cells.
We next determined the cellular distribution of lncRHOXF1 by using RNA FISH and cell fractionation experiments. Using fixation conditions that preserve the cytoplasm for RNA-FISH, we detected abundant lncRHOXF1 transcripts in the cytoplasm of BMP4/A/P cells, JEG-3 cells, and primary EVTs (Fig. 2H). We confirmed lncRHOXF1 probe specificity by using undifferentiated hESCs as a negative control, which express very low levels of lncRHOXF1 RNA, and we found that these cells lacked lncRHOXF1 foci (Fig. 2I). Next, we fractionated BMP4/A/P -differentiated cells into nuclear and cytoplasmic extracts, and we detected lncRHOXF1 expression in both compartments (Fig. 2J). The presence of lncRHOXF1 transcripts in cytoplasm suggests that this transcript has the potential to regulate gene expression in trans, acting at the RNA level and not through transcriptional interference.
Overexpression of lncRHOXF1 RNA in hESCs induces differentiation and reduces cell growth.
To determine the functional relevance of the lncRHOXF1 RNA during early human development, we used a gain-of-function approach to induce expression in undifferentiated hESCs that lack this transcript. To this end, we cloned exons 1 to 4 of lncRHOXF1 into a targeting construct for DOX-inducible expression from the AAVS.1 “safe harbor” locus on chromosome 19 (Fig. 3A) (13). We targeted female hESCs (HUES-9 cells) with lncRHOXF1 and the reverse tetracycline transactivator protein (rtTA3) constructs simultaneously, and we obtained two correctly targeted clones (23 and 28). Addition of DOX for 24 h to HUES9-lncRHOXF1 clones 23 and 28 significantly increased lncRHOXF1 transcript levels relative to untreated cells (Fig. 3B). HUES9-lncRHOXF1 clones treated with DOX expressed more lncRHOXF1 than did BMP4/A/P-differentiated trophoblast progenitors. We observed that DOX-treated HUES9-lncRHOXF1 cells required less frequent passaging than did wild-type hESCs, suggesting reduced proliferation rates. We performed cell growth assays by following the number of hESC colonies originating from two colonies at each passage, using clone 28 HUES9-lncRHOXF1 cells treated with DOX. The DOX-treated HUES9-lncRHOXF1 cells had significantly fewer colonies at passages 3, 4, and 5 (P = 0.009, P = 0.0001, and P = 2.0 × 10−5, respectively) relative to untreated cells (Fig. 3C), indicating that lncRHOXF1 expression in hESCs reduced cellular growth rates. Next, we investigated whether lncRHOXF1 expression in hESCs affected cellular differentiation by performing an EB formation assay. HUES9-lncRHOXF1 clones 23 and 28 treated with DOX had greater numbers of EBs than did untreated cells (P = 0.05) (Fig. 3D), supporting our hypothesis that inappropriate expression of lncRHOXF1 in pluripotent stem cells favors cellular differentiation.
We used microarray profiling to determine how exogenous lncRHOXF1 expression in hESCs affects global gene expression patterns. DOX treatment of HUES9-lncRHOXF1 cells altered the expression of over 150 genes (1.5-fold or greater), and the majority were downregulated. Altered genes clustered into two groups, depending on the direction of expression change (upregulated or downregulated) (Fig. 3E). Gene ontology (GO) analysis of differentially expressed genes showed they belonged to categories pertaining to DNA replication, cell adhesion, cell migration, and development of bone and blood (Fig. 3F). Gene set enrichment analysis (GSEA) demonstrated that DOX-treated HUES9-lncRHOXF1 cells were significantly enriched for genes involved in DNA synthesis and elongation, DNA replication, and oxidative phosphorylation (Fig. 3G). These gene expression changes are consistent with our observations that lncRHOXF1 expression in hESCs affects cellular growth and differentiation, and the changes suggest that this transcript functions in trans to regulate gene expression.
Disruption of lncRHOXF1 RNA in human placental progenitors increases viral response gene expression.
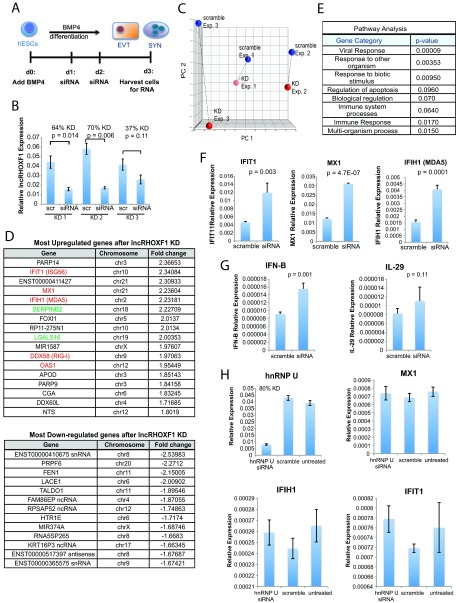
To determine the function of lncRHOXF1 in human trophectoderm progenitor cells, we used siRNAs to disrupt lncRHOXF1 RNA expression during in vitro BMP4/A/P differentiation of hESCs (Fig. 4A). We optimized transfection and differentiation conditions and achieved a maximum of 60 to 70% lncRHOXF1 knockdown efficiency via BMP4/A/P differentiation on day 3 (Fig. 4B). We examined global gene expression changes resulting from siRNA-mediated lncRHOXF1 disruption in the in vitro human trophectoderm progenitors by using microarray data from three separate knockdown experiments, comparing lncRHOXF1 knockdown to scrambled siRNA controls (Fig. 4C). We identified 109 genes that were upregulated and 100 genes downregulated 1.5-fold or more when lncRHOXF1 expression was disrupted. Downregulated genes included noncoding RNAs with unknown function and PRPF6, which is involved in pre-mRNA splicing (Fig. 4D). Intriguingly, we observed that various virus sensing genes exhibited the highest upregulation with lncRHOXF1 disruption (Fig. 4D). IFIH1 (MDA5) and DDX58 (RIG-I), two pattern recognition receptors (PRR) with RNA helicase activity that sense intracellular viral RNA (24), were upregulated 2- to 2.2-fold. MX1, a member of the interferon (IFN)-stimulated gene (ISG) family that is induced by type I and III IFNs upon viral infection (25), was also elevated. In addition, OAS1, another ISG with antiviral activity that detects cytosolic double-stranded RNA and activates RNase L (26), IFIT1, an antiviral molecule upregulated by type I IFN signaling and virus infection (27, 28), and LGALS16, which encodes a placenta-specific galectin protein expressed by SYNs that is involved in regulating immune responses, were also elevated 2-fold following lncRHOXF1 knockdown. Galectins induce T cell apoptosis and are thought to minimize the maternal immune response against fetal semiallograft (29). GO analysis confirmed the enrichment of genes involved in viral and immune responses (P = 9.0 × 10−5 and P = 0.02) when lncRHOXF1 expression was reduced in our in vitro model of early human placental progenitors (Fig. 4E). We confirmed that lncRHOXF1 disruption increased the expression of the three viral sensing genes IFIT1, MX1, and IFIH1 in day 3 BMP4/A/P-differentiated human cells by using qRT-PCR (Fig. 4F).
FIG 4.
lncRHOXF1 RNA disruption increases viral response gene expression in human trophoblast progenitors. (A) Schematic of siRNA-mediated reduction of lncRHOXF1 RNA during in vitro BMP4/A/P differentiation. (B) qRT-PCR analysis verifying lncRHOXF1 expression and knockdown for three independent siRNA transfections used for microarray analyses. Statistical significance was calculated using a two-tailed Student's t test. Error bars denote standard deviations from the mean. (C) Principal component analysis for the three independent knockdown experiments used for microarray analyses. (D) Genes exhibiting the greatest upregulation and downregulation after lncRHOXF1 RNA disruption in BMP4/A/P-differentiated HUES-7 cells was determined by microarray profiling. Genes in red are viral signature response genes; green denotes immune response genes. (E) GO analysis of differentially expressed genes following lncRHOXF1 RNA knockdown. P values were determined by using the DAVID online tool. (F) qRT-PCR analysis for expression of viral response genes, normalized to results with GAPDH. Representative results from one experiment are shown, and similar results were obtained from 5 independent experiments. Statistical significance was calculated using a one-tailed t test. Error bars denote standard deviations from the means. (G) qRT-PCR analysis of IFN-β and IL-29, downstream targets of viral response genes. Statistical significance was calculated using a one-tailed t test. (H) siRNA-mediated knockdown of hnRNP U in human trophoblast progenitors does not affect viral response genes. qRT-PCR analyses for expression of viral response genes are shown and were normalized to results with GAPDH. Representative results from one experiment are shown, and similar results were obtained from two independent experiments. Error bars denote standard deviations from the means.
Because IFIT1 (MDA5) and DDX58 (RIG-I) expression increased with lncRHOXF1 disruption, we examined expression levels of type I and III IFNs, IFN-β, and interleukin-29 (IL-29; IFN-λ1), which become enhanced in response to PRR-mediated activation (24, 30). We observed that IFN-β expression increased with lncRHOXF1 knockdown (P = 0.001), while IL-29 expression remained unchanged (Fig. 4G). siRNA transfection can have off-target effects and activate the type I IFN response, resulting in induction of ISGs (31, 32). To eliminate the possibility of siRNA-mediated activation of the IFN response, we used siRNAs to disrupt expression of hnRNP U, a nuclear matrix protein with RNA binding activity expressed at similar levels as lncRHOXF1 in human trophectoderm cells. We did not observe any changes in expression of MX1, IFIH1, or IFIT1 following hnRNP U RNA disruption using siRNAs, supporting the specificity of lncRHOXF1 disruption for affecting viral response gene expression in differentiated hESCs (Fig. 4H). We conclude that lncRHOXF1 RNA functions in trans to suppress the expression of genes involved in the innate immune response to virus infection in human placental progenitor cells.
lncRHOXF1 RNA increases following viral infection, and knockdown affects virus infectivity.
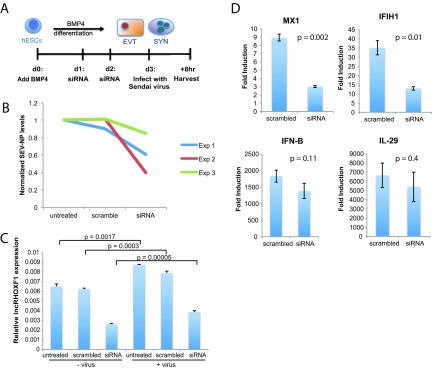
To determine the impact of lncRHOXF1 in the antiviral response of human trophoblast progenitors, we used Sendai virus to infect BMP4/A/P-differentiated cells with disrupted lncRHOXF1 expression (Fig. 5A) and compared their response to control cells. We found that infected cells treated with siRNAs against lncRHOXF1 consistently expressed less Sendai viral protein mRNA (SEV-NP) than did scramble siRNA-treated or untreated cells (Fig. 5B). These results confirmed our hypothesized enhanced antiviral state of cells with reduced lncRHOXF1 expression. Interestingly, we observed that Sendai virus infection increased the expression of lncRHOXF1 in BMP4/A/P-differentiated cells, even when cells were transfected with either scrambled siRNAs or siRNAs specific for lncRHOXF1 (Fig. 5C). Therefore, we asked whether viral infection in combination with lncRHOXF1 RNA disruption would affect the expression of viral response genes. As expected due to the reduced virus infectivity, the levels of MX1 and IFIH1 were significantly reduced (Fig. 5D). We did not observe changes with IFN-β and IL-29 induction. We conclude that lncRHOXF1 RNA is responsive to viral infections and that reducing lncRHOXF1 levels protected early human trophectoderm progenitor cells from Sendai virus infection.
FIG 5.
Sendai virus infection increases lncRHOXF1 expression in human trophophoblast progenitors. (A) Schematic of Sendai virus infection following siRNA-mediated knockdown of lncRHOXF1 RNA during in vitro BMP4/A/P differentiation. (B) qRT-PCR analysis of Sendai virus protein SEV-NP transcript levels in BMP4/A/P-differentiated cells transfected with scramble or lncRHOXF1 siRNAs. Results were normalized to those for untreated controls; results from three independent experiments are shown. (C) qRT-PCR analysis of lncRHOXF1 RNA levels in infected and uninfected BMP4/A/P-differentiated HUES-9 cells. Statistical analysis was determined using a one-tailed t test to compare triplicate measurements of infected versus uninfected cells. Error bars denote standard deviations from the means. (D) Relative expression of each gene was determined in cells treated with mock siRNA (scrambled) or siRNA for lncRHOXF1 RNA by quantifying RNA abundance in cells treated with and without Sendai virus; results were normalized to GAPDH expression results. Relative expression was compared using a two-tailed Welch's t test, and a representative example of 4 independent experiments is shown, as the mean relative expression level, 2(ΔCT [virus] − ΔCT [no virus]) ± the standard deviation.
DISCUSSION
Here we have described the identification and characterization of a novel lncRNA, which we named lncRHOXF1, that functions in the innate immune response in early human placental progenitor cells. We found that lncRHOXF1 is one of the most abundantly expressed lncRNAs in trophectoderm and primitive endoderm cells from human blastocyst-stage embryos, and it is also robustly expressed in an in vitro model of placental trophoblast progenitors differentiated from human pluripotent stem cells. Using this in vitro model of human trophoblasts, we discovered that siRNA-mediated disruption of lncRHOXF1 expression increased expression of viral sensor genes. To our knowledge, this is the first example of a lncRNA that regulates viral sensor genes by repressing their expression during development of human trophoblast cells.
lncRHOXF1 transcripts were most abundant in TE and primitive endoderm cells, yet absent in epiblast cells of human blastocyst-stage embryos. Our study found that lncRHOXF1 RNA is not expressed in hPSCs, in agreement with its absence in the inner cell mass of the human embryo (14). lncRNAs typically exhibit cell- and tissue-specific expression patterns (33), and lncRHOXF1 RNA has a defined expression pattern during early human development. The in vitro differentiation of hPSCs to the trophectoderm, mesoderm, or endoderm lineages resulted in upregulation of lncRHOXF1 expression. We speculate that lncRHOXF1 RNA primarily functions in early human development during differentiation from pluripotency, because this RNA was almost undetectable in 8- to 10-week placentas and fetal somatic cell lines (Fig. 2B and C). It is possible that lncRHOXF1 is expressed in just one of the many cell types present in 8- to 10-week placentas, and perhaps these cells were not present in sufficient quantity in the tissue sample that we examined in our study. Primary EVT cells taken from late-term placentas had low levels of lncRHOXF1 RNA transcripts. This suggests that lncRHOXF1 RNA is not functional in mature EVT cells but instead is important at an earlier developmental stage, perhaps in a progenitor cell population. Our study found that lncRHOXF1 RNA is abundant in mesoderm and endoderm progenitor cells, and we hypothesize that it may also regulate the host response to viral infections in the human epiblast during development. We conclude that lncRHOXF1 RNA is an abundant transcript expressed during early human development.
In this study, we observed that overexpression of lncRHOXF1 RNA in hESCs, which normally express very low levels of this lncRNA, reduced cell growth rates of pluripotent stem cells. Consistent with this observation, we detected altered expression of genes involved in DNA replication, DNA synthesis, and the M/G1 transition when lncRHOXF1 RNA was introduced into hESCs. Expression of lncRHOXF1 RNA also increased the number of EBs formed during differentiation, which suggests that elevated levels of lncRHOXF1 RNA in pluripotent stem cells favor cellular differentiation instead of the pluripotent state. hESCs with elevated lncRHOXF1 RNA had more morphological evidence of differentiated cells despite the presence of β-FGF in the medium. Differentiated cells have different metabolic requirements than pluripotent stem cells (34), and we observed that lncRHOXF1 RNA expression altered expression of genes involved in oxidative phosphorylation, electron transport/ATP synthesis, respiratory electron transport, and mitochondrial protein import. We did not observe altered expression of viral response genes when lncRHOXF1 was overexpressed in hESCs. We speculate that the reason for this could be that lncRHOXF1 RNA regulates viral response gene expression specifically in trophoblast progenitors, which are the cells that have the highest expression of this transcript. At this time, it is unclear whether overexpression of lncRHOXF1 in trophoblast progenitors will further repress the expression of these viral response genes and whether the endogenous levels of this transcript are sufficient for full repression.
Our gain- and loss-of-function experiments indicated that lncRHOXF1 is a functional lncRNA transcript that can alter gene expression in trans. We did not observe any altered gene expression of neighboring genes on the X chromosome following siRNA-mediated knockdown, thus eliminating a cis-acting mechanism. Consistent with a functional role in trans, we found that lncRHOXF1 is abundant in both the nucleus and cytoplasm in BMP4/A/P-differentiated cells, primary EVTs, and the placental cell line JEG-3. The nuclear and cytoplasmic signals appeared evenly distributed between both cellular compartments. Cellular fractionation experiments using BMP4/A/P-differentiated cells confirmed our RNA FISH results, where lncRHOXF1 RNA was detected in both the nucleus and cytoplasm. The nuclear lncRHOXF1 RNA signal is specific because we observed similar speckled patterns in all cell types where we detected lncRHOXF1 RNA expression. We did not detect lncRHOXF1 RNA transcripts in undifferentiated hESCs, which have very low steady-state levels of lncRHOXF1 RNA (Fig. 1G and H), indicating that the probe is specific for lncRHOXF1 and that this transcript is found in both nuclear and cytoplasmic compartments.
Additionally, we found that lncRHOXF1 RNA levels could be reduced using siRNAs, further supporting the hypothesis that this transcript mediates gene expression changes in the cytoplasm. One possibility is that lncRHOXF1 RNA in the cytoplasm inhibits the translation of viral sensing genes, perhaps by initiating mRNA decay of these transcripts. The lncRNAs ½-sbsRNAs, which bind to the 3′ untranslated regions of some mRNAs, form a duplex that is recognized by Staufen 1 and then targeted for degradation (35). However, siRNAs specific for nuclear lncRNAs, such as XIST or HOTAIR RNA, can also disrupt their expression (36, 37), so it is also possible that lncRHOXF1 RNA plays a regulatory role in the nucleus. lncRHOXF1 RNA may also function as a molecular scaffold in the nucleus. Examples of lncRNAs that act as molecular scaffolds include XIST and HOTAIR, which recruit the Polycomb repressive complex 2 to specific genes or chromosomes for transcriptional silencing (37, 38). It is possible that the secondary structure of lncRHOXF1 RNA recruits repressive histone modifier complexes to certain viral sensor genes to regulate their expression. Additional experiments are required to identify the lncRHOXF1-interacting protein(s) and the chromatin modification changes, if any, at viral sensor genes in early human placental progenitor cells.
The innate immune system responds to viral infection via PRRs that sense cytoplasmic virus-derived RNAs, which then signal for a type I IFN-α/β response that induces transcription of ISGs. Both IFIH1 (MDA5) and DDX58 (RIG-I) are PRRs that are induced when lncRHOXF1 expression is reduced. In this study, we found that siRNA-mediated reduction of lncRHOXF1 RNA increased the expression of some ISGs (IFIT, OAS1, and MX1) and the type I IFN gene IFN-β. At this time, it is unclear how reduced lncRHOXF1 RNA levels might directly activate ISGs and IFN-β. One possibility is that these genes become upregulated as a response to IFIH1 and DDX58 activation when lncRHOXF1 transcript levels decrease. In this study, we found that Sendai virus infection increased lncRHOXF1 RNA levels in human trophoblast progenitors, suggesting that factors induced by the viral response may directly regulate expression of this lncRNA. Alternatively, viral infection may stabilize lncRHOXF1 RNA transcripts within the cytoplasm, which would explain the increased steady-state levels (Fig. 5B). Studies have found that viral infection with severe acute respiratory syndrome coronavirus or influenza virus changes the expression of hundreds of lncRNAs, many of which are regulated downstream of type I IFN signals (39, 40). Additional work is necessary to determine how lncRHOXF1 expression is regulated during early human development and how viral infections influence lncRHOXF1 transcriptional expression and RNA stability.
Some lncRNAs can regulate the innate immune response during pathogen infection outside the context of pregnancy and early development. The lncRNAs NeST, THRIL, NEAT1, and NRAV regulate expression of IFN-γ, tumor necrosis factor alpha, IL-8, and interferon-stimulated genes, respectively (41–44). NEAT1 and 7SL RNA can also modulate viral infection by affecting virion packaging and posttranscriptional expression. NEAT1 RNA levels increase after infection with herpes simplex or influenza viruses, which elevates levels of the antiviral cytokine gene interleukin-8 in mice (42). Similar to NEAT1, we found that viral infection stimulated lncRHOXF1 RNA levels, which supports the idea that lncRNAs play important roles as gene expression regulators for innate immunity. Our study also found that human trophoblast progenitors were less susceptible to Sendai virus infection when lncRHOXF1 RNA levels were reduced. This reduction of viral infectivity could result from the increased expression of PRRs and ISGs when lncRHOXF1 expression is disrupted (Fig. 4D and F). Another possibility is that lncRHOXF1 RNA directly regulates the expression of the Sendai viral SEV-NP gene, similar to NEAT1 RNA interference with virion packaging (42). As a result of impaired viral infection, we observed reductions in steady-state levels of IFIH and MX1 in lncRHOXF1-disrupted cells following acute Sendai virus infection, yet downstream type I and III IFN genes were unchanged. Our results suggest that human trophoblasts continue to sense and respond to the presence of Sendai virus following lncRHOXF1 RNA disruption. Additional work is necessary to examine whether other viruses increase lncRHOXF1 expression in human trophectoderm progenitors and how recurring viral infections affect lncRHOXF1 RNA levels.
ACKNOWLEDGMENTS
We thank L. King, and M. Bartolomei for reading the manuscript and the members of the Lengner lab for many helpful suggestions. We also thank Yan Sun for help with SeV infections. We are grateful for bioinformatics help from D. Beiting and J. Tobias and statistical consultations with C. Berry. AAVS and rtTA3 targeting constructs were a gift from P. Gadue. RNA samples of in vitro-differentiated human endoderm and mesoderm cells were from P. Gadue. The primary EVTs and JEG-3 cells were a gift from M. Elovitz. Placental samples (8- to 10-week conceptus) were collected by L. Goetzl in collaboration with Temple University.
We do not have any conflicts of interest.
Funding Statement
This work was partially supported by grants from the Pennsylvania Health Research Formula Fund (Project 13, Long noncoding RNAs required for early human development) and the Abramson Cancer Center ACS pilot award to M.C.A. C.M.S. was supported by training grant T32 GM-07229.
REFERENCES
- 1.Lee JT, Bartolomei MS. 2013. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 2.Buckberry S, Bianco-Miotto T, Roberts CT. 2014. Imprinted and X-linked non-coding RNAs as potential regulators of human placental function. Epigenetics 9:81–89. doi: 10.4161/epi.26197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossant J, Cross JC. 2001. Placental development: lessons from mouse mutants. Nat Rev Genet 2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- 4.Delorme-Axford E, Sadovsky Y, Coyne CB. 2014. The placenta as a barrier to viral infections. Annu Rev Virol 1:133–146. doi: 10.1146/annurev-virology-031413-085524. [DOI] [PubMed] [Google Scholar]
- 5.Mor G, Cardenas I. 2010. The immune system in pregnancy: a unique complexity. Am J Reprod Immunol 63:425–433. doi: 10.1111/j.1600-0897.2010.00836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delorme-Axford E, Donker RB, Mouillet JF, Chu T, Bayer A, Ouyang Y, Wang T, Stolz DB, Sarkar SN, Morelli AE, Sadovsky Y, Coyne CB. 2013. Human placental trophoblasts confer viral resistance to recipient cells. Proc Natl Acad Sci U S A 110:12048–12053. doi: 10.1073/pnas.1304718110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anguera MC, Sadreyev R, Zhang Z, Szanto A, Payer B, Sheridan SD, Kwok S, Haggarty SJ, Sur M, Alvarez J, Gimelbrant A, Mitalipova M, Kirby JE, Lee JT. 2012. Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. Cell Stem Cell 11:75–90. doi: 10.1016/j.stem.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, Guenther MG, Welstead GG, Alagappan R, Frampton GM, Xu P, Muffat J, Santagata S, Powers D, Barrett CB, Young RA, Lee JT, Jaenisch R, Mitalipova M. 2010. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell 141:872–883. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 9.Marchand M, Horcajadas JA, Esteban FJ, McElroy SL, Fisher SJ, Giudice LC. 2011. Transcriptomic signature of trophoblast differentiation in a human embryonic stem cell model. Biol Reprod 84:1258–1271. doi: 10.1095/biolreprod.110.086413. [DOI] [PubMed] [Google Scholar]
- 10.Tapia K, Kim WK, Sun Y, Mercado-Lopez X, Dunay E, Wise M, Adu M, Lopez CB. 2013. Defective viral genomes arising in vivo provide critical danger signals for the triggering of lung antiviral immunity. PLoS Pathog 9:e1003703. doi: 10.1371/journal.ppat.1003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang LF, Huynh KD, Lee JT. 2007. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell 129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- 12.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. 2008. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods 5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sim X, Cardenas-Diaz FL, French DL, Gadue P. 2016. A doxycycline-inducible system for genetic correction of iPSC disease models. Methods Mol Biol 1353:13–23. doi: 10.1007/7651_2014_179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan L, Yang M, Guo H, Yang L, Wu J, Li R, Liu P, Lian Y, Zheng X, Yan J, Huang J, Li M, Wu X, Wen L, Lao K, Qiao J, Tang F. 2013. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol 20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- 15.Richardson ME, Bleiziffer A, Tuttelmann F, Gromoll J, Wilkinson MF. 2014. Epigenetic regulation of the RHOX homeobox gene cluster and its association with human male infertility. Hum Mol Genet 23:12–23. doi: 10.1093/hmg/ddt392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacLean JA II, Wilkinson MF. 2010. The Rhox genes. Reproduction 140:195–213. doi: 10.1530/REP-10-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amita M, Adachi K, Alexenko AP, Sinha S, Schust DJ, Schulz LC, Roberts RM, Ezashi T. 2013. Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc Natl Acad Sci U S A 110:E1212–E1221. doi: 10.1073/pnas.1303094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarkar P, Randall SM, Collier TS, Nero A, Russell TA, Muddiman DC, Rao BM. 2015. Activin/nodal signaling switches the terminal fate of human embryonic stem cell-derived trophoblasts. J Biol Chem 290:8834–8848. doi: 10.1074/jbc.M114.620641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das P, Ezashi T, Schulz LC, Westfall SD, Livingston KA, Roberts RM. 2007. Effects of FGF2 and oxygen in the BMP4-driven differentiation of trophoblast from human embryonic stem cells. Stem Cell Res 1:61–74. doi: 10.1016/j.scr.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu RH, Chen X, Li DS, Li R, Addicks GC, Glennon C, Zwaka TP, Thomson JA. 2002. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol 20:1261–1264. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- 21.Jedrusik A, Parfitt DE, Guo G, Skamagki M, Grabarek JB, Johnson MH, Robson P, Zernicka-Goetz M. 2008. Role of Cdx2 and cell polarity in cell allocation and specification of trophectoderm and inner cell mass in the mouse embryo. Genes Dev 22:2692–2706. doi: 10.1101/gad.486108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng X, Ying L, Lu L, Galvao AM, Mills JA, Lin HC, Kotton DN, Shen SS, Nostro MC, Choi JK, Weiss MJ, French DL, Gadue P. 2012. Self-renewing endodermal progenitor lines generated from human pluripotent stem cells. Cell Stem Cell 10:371–384. doi: 10.1016/j.stem.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. 2007. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dixit E, Kagan JC. 2013. Intracellular pathogen detection by RIG-I-like receptors. Adv Immunol 117:99–125. doi: 10.1016/B978-0-12-410524-9.00004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haller O, Kochs G, Weber F. 2007. Interferon, Mx, and viral countermeasures. Cytokine Growth Factor Rev 18:425–433. doi: 10.1016/j.cytogfr.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hornung V, Hartmann R, Ablasser A, Hopfner KP. 2014. OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nat Rev Immunol 14:521–528. doi: 10.1038/nri3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vladimer GI, Gorna MW, Superti-Furga G. 2014. IFITs: emerging roles as key anti-viral proteins. Front Immunol 5:94. doi: 10.3389/fimmu.2014.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reynaud JM, Kim DY, Atasheva S, Rasalouskaya A, White JP, Diamond MS, Weaver SC, Frolova EI, Frolov I. 2015. IFIT1 differentially interferes with translation and replication of alphavirus genomes and promotes induction of type I interferon. PLoS Pathog 11:e1004863. doi: 10.1371/journal.ppat.1004863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Than NG, Romero R, Goodman M, Weckle A, Xing J, Dong Z, Xu Y, Tarquini F, Szilagyi A, Gal P, Hou Z, Tarca AL, Kim CJ, Kim JS, Haidarian S, Uddin M, Bohn H, Benirschke K, Santolaya-Forgas J, Grossman LI, Erez O, Hassan SS, Zavodszky P, Papp Z, Wildman DE. 2009. A primate subfamily of galectins expressed at the maternal-fetal interface that promote immune cell death. Proc Natl Acad Sci U S A 106:9731–9736. doi: 10.1073/pnas.0903568106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. 2003. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol 4:69–77. doi: 10.1038/ni8753. [DOI] [PubMed] [Google Scholar]
- 31.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. 2003. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol 5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 32.Marques JT, Williams BR. 2005. Activation of the mammalian immune system by siRNAs. Nat Biotechnol 23:1399–1405. doi: 10.1038/nbt1161. [DOI] [PubMed] [Google Scholar]
- 33.Lee JT. 2012. Epigenetic regulation by long noncoding RNAs. Science 338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 34.Xu X, Duan S, Yi F, Ocampo A, Liu GH, Izpisua Belmonte JC. 2013. Mitochondrial regulation in pluripotent stem cells. Cell Metab 18:325–332. doi: 10.1016/j.cmet.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Gong C, Maquat LE. 2011. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nozawa RS, Nagao K, Igami KT, Shibata S, Shirai N, Nozaki N, Sado T, Kimura H, Obuse C. 2013. Human inactive X chromosome is compacted through a PRC2-independent SMCHD1-HBiX1 pathway. Nat Struct Mol Biol 20:566–573. doi: 10.1038/nsmb.2532. [DOI] [PubMed] [Google Scholar]
- 37.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng X, Gralinski L, Armour CD, Ferris MT, Thomas MJ, Proll S, Bradel-Tretheway BG, Korth MJ, Castle JC, Biery MC, Bouzek HK, Haynor DR, Frieman MB, Heise M, Raymond CK, Baric RS, Katze MG. 2010. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. mBio 1(5):e00206-10. doi: 10.1128/mBio.00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Q, Jeang KT. 2013. Long non-coding RNAs (lncRNAs) and viral infections. Biomed Pharmacother 3:34–42. doi: 10.1016/j.biomed.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. 2013. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell 152:743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, Kato A, Kawaguchi Y, Sato H, Yoneda M, Kai C, Yada T, Suzuki Y, Yamada T, Ozawa T, Kaneki K, Inoue T, Kobayashi M, Kodama T, Wada Y, Sekimizu K, Akimitsu N. 2014. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell 53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 43.Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, Head SR, Burns JC, Rana TM. 2014. The long noncoding RNA THRIL regulates TNF alpha expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A 111:1002–1007. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ouyang J, Zhu X, Chen Y, Wei H, Chen Q, Chi X, Qi B, Zhang L, Zhao Y, Gao GF, Wang G, Chen JL. 2014. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe 16:616–626. doi: 10.1016/j.chom.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]