ABSTRACT
Multiple essential small GTPases are involved in the assembly of the ribosome or in the control of its activity. Among them, ObgE (CgtA) has been shown recently to act as a ribosome antiassociation factor that binds to ppGpp, a regulator whose best-known target is RNA polymerase. The present study was aimed at elucidating the expression of obgE in Escherichia coli. We show that obgE is cotranscribed with ribosomal protein genes rplU and rpmA and with a gene of unknown function, yhbE. We show here that about 75% of the transcripts terminate before obgE, because there is a transcriptional terminator between rpmA and yhbE. As expected for ribosomal protein operons, expression was highest during exponential growth, decreased during entry into stationary phase, and became almost undetectable thereafter. Expression of the operon was derepressed in mutants lacking ppGpp or DksA. However, regulation by these factors appears to occur post-transcription initiation, since no effects of ppGpp and DksA on rplU promoter activity were observed in vitro.
IMPORTANCE The conserved and essential ObgE GTPase binds to the ribosome and affects its assembly. ObgE has also been reported to impact chromosome segregation, cell division, resistance to DNA damage, and, perhaps most interestingly, persister formation and antibiotic tolerance. However, it is unclear whether these effects are related to its role in ribosome formation. Despite its importance, no studies on ObgE expression have been reported. We demonstrate here that obgE is expressed from an operon encoding two ribosomal proteins, that the operon's expression varies with the growth phase, and that it is dependent on the transcription regulators ppGpp and DksA. Our results thus demonstrate that obgE expression is coupled to ribosomal gene expression.
INTRODUCTION
Ribosome biogenesis is a complex, hierarchical, and highly regulated process. The number of ribosomes and the synthesis rates of their components are constantly adapted to the cell's growth rate. In bacteria, the stringent response is mediated by guanosine tetra- and pentaphosphate (both referred to here as ppGpp), which control the expression of rRNA operons in response to changes in the availability of nutrients (1, 2). In Escherichia coli, ppGpp binds directly to RNA polymerase (RNAP) to regulate transcription from rRNA promoters (3–6) and requires the protein cofactor DksA to exert its full effect (7). As a consequence of the inhibition of rRNA synthesis by ppGpp and DksA, specific ribosomal proteins transiently accumulate in excess of the rRNAs, and these excess ribosomal proteins inhibit the expression of their own operons, in most cases by binding to their mRNAs and repressing translation (translational feedback control) (8).
It has become clear in recent years that transcription from many, but not all, ribosomal protein promoters is also inhibited directly by ppGpp/DksA (9). However, effects of ppGpp/DksA on transcription and/or translational feedback have not been investigated for all of the r-protein operons, and further information is still required to understand this fundamental gene regulatory network. For example, the regulation of the S2, L25, and S6 r-protein operons was elucidated only recently (10–12). In the present study, we investigated the regulation of a previously neglected operon, rplU-rpmA, that codes for 50S subunit r-proteins L21 and L27, respectively.
Ribosome biogenesis has been studied extensively, both in vitro and in vivo (13). The rRNAs are chemically modified and acted on by helicases; in addition, small GTPases are required for correct ribosome assembly (14). One of the small, essential GTPases, ObgE (CgtA), contains a typical GTPase domain and an N-terminal Obg domain that defines this family (for reviews, see references 15, 16, and 17). Obg proteins have been implicated in ribosome biogenesis repeatedly, and they have also been implicated in regulation by ppGpp during the stringent response (17). A cocrystal of Bacillus subtilis Obg was solved with ppGpp, suggesting that ppGpp might act on it directly (18). It was recently proposed that ObgE prevents the association of the 30S and 50S subunits by mimicking a tRNA in the peptidyltransferase center (19). These findings might explain some of the pleotropic phenotypes of obg mutants, which have led to proposals of ObgE involvement in central functions as diverse as chromosome segregation, cell division, resistance to DNA damage, and persistence (20).
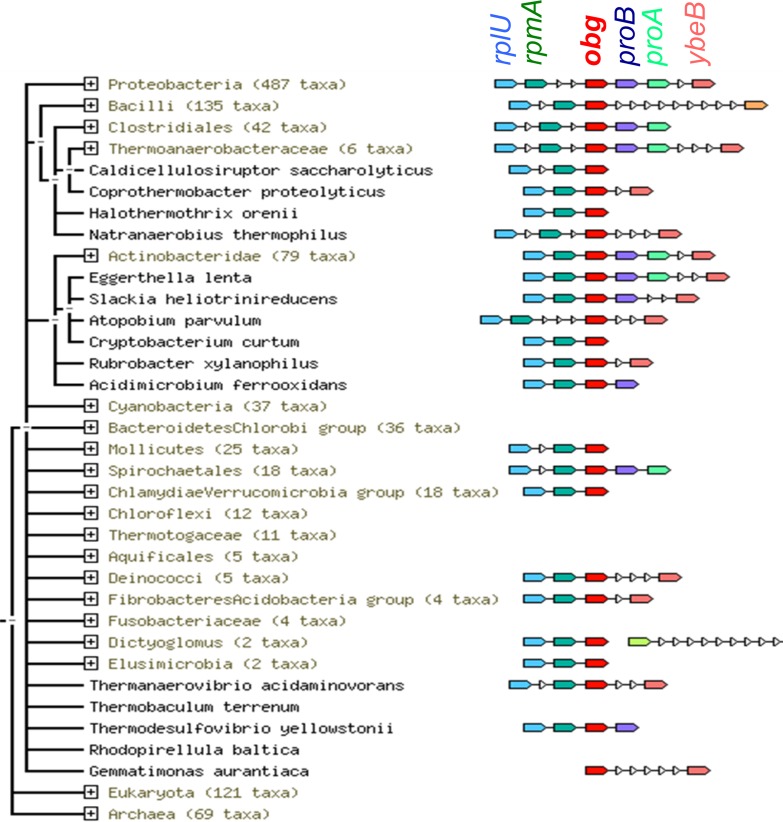
The obgE gene is located downstream from rplU, rpmA, and yhbE in E. coli (Fig. 1). yhbE has been annotated as coding for an integral inner membrane protein of unknown function. The obgE, rplU, and rpmA genetic neighborhood is conserved in the majority of bacteria, resulting in a score of 0.99 in String prediction (21) (Fig. 1). Regulation of the rplU and rpmA genes was not examined in the era when most other r-protein operon regulation studies were undertaken (8). Few studies have focused on these genes since that time, perhaps because the central role of the neighboring ObgE protein in ribosome biogenesis was not appreciated.
FIG 1.
Conservation of the rplU-rpmA-obg genetic association. The data represent a summary of the neighborhood of the obg gene in bacteria, obtained with the String tool (21), which shows the high conservation of the rplU-rpmA-obg association among bacteria (string score, >0.99). The triangles represent genes that do not show conserved genetic association with obg.
Transcriptome analysis has shown that rpmA, yhbE, and obgE are inhibited during the stringent response and that the inhibition is dependent on RelA and SpoT, the ppGpp synthases (22). These data are consistent with the organization of the rplU, rpmA, yhbE, and obgE genes as an operon and with the possibility that the operon's promoter(s), like many other r-protein promoters, is regulated by ppGpp/DksA (9). However, regulation of this promoter(s) has not been characterized and little or no information is available about whether expression of the operon is regulated posttranscriptionally by translational feedback.
Here we characterize the expression of obgE and its associated genes, rplU, rpmA, and yhbE. We found that obgE and yhbE are cotranscribed with rplU and rpmA and that the operon is regulated by nutritional conditions, consistent with an integral role of ObgE in ribosome assembly. However, unlike the majority of r-protein promoters, the rplU promoter does not appear to be regulated directly by ppGpp/DksA.
MATERIALS AND METHODS
Media and chemicals.
Cells were grown in lysogeny broth medium (LB; bactotryptone [10 g/liter], yeast extract [5 g/liter], NaCl [10 g/liter], pH 7.5) unless stated otherwise. The plasmids were maintained with ampicillin (Amp; 100 μg/ml), chloramphenicol (Cam; 50 μg/ml), or kanamycin (Kana; 50 μg/ml).
Plasmid constructions. (i) Transcriptional fusions with gfp.
Gene expression was monitored using transcriptional fusions with gfpmut2 using the pUA66 and pUA139 plasmids (23) or with gfpasv using the pZE-gfp plasmid (24). The GFPmut2 variant is highly stable, while the half-life of the GFPasv variant is about 100 min (25). The transcriptional fusion with the ispB upstream region was available in the E. coli promoter library (23). The other intergenic regions were amplified by PCR with different primer pairs (see Table S1 in the supplemental material) using purified genomic DNA of strain MG1655 as the template. PCR products were then digested by BamHI/XhoI restriction enzymes and cloned into pUA139, pUA66, or pZE-gfp (Table 1).
TABLE 1.
Plasmidsa
| Laboratory codeb | Name | Description | Transcriptional fusion limits (nt) | Reference or source |
|---|---|---|---|---|
| pEB266 | pCP20 | TS; Camr Ampr; FLP recombinase gene | 34 | |
| pEB232 | pKO3 | TS; Camr sacB | 27 | |
| pEB1449 | pKO3-U.A.ΔT.yhbE | Ebm892/893 terminator deletion | This work | |
| pEB267 | pKD46 | 31 | ||
| pEB794 | pJL148 | 32 | ||
| pEB898 | pUA66 | Kanar p15A ori; gfp transcriptional fusion | 23 | |
| pEB987 | pUA139 | Kanar p15A ori; gfp transcriptional fusion | 23 | |
| pUA66-ispB | 23 | |||
| pEB1364 | pUA139-rplU | XhoI/BamHI fragment from pUA66-ispB reversed in pUA139 | −289/+145 | This work |
| pEB1403 | pUA-rplUΔbox | Ebm860/861 PCR (XhoI/BamHI) product in pUA66 | −58/+94 | This work |
| pEB1485 | pUA-rplUΔbox2 | Ebm1005/861 PCR (XhoI/BamHI) product in pUA66 | −89/+94 | This work |
| pEB1412 | pUA-rplUbox* | Ebm886/887 mutagenesis on pEB1364 | −289/+145 | This work |
| pEB1197 | pUA-UPrpmA | Ebm555/556 PCR (XhoI/BamHI) product in pUA66 | This work | |
| pEB1383 | pUA-UPyhbE | Ebm848/849 PCR (XhoI/BamHI) product in pUA66 | This work | |
| pEB1198 | pUA-UPobgE | Ebm557/558 PCR (XhoI/BamHI) product in pUA66 | This work | |
| pEB1431 | pUA-csgD | Ebm917/918 PCR (XhoI/BamHI) product in pUA66 | −221/+83 | This work |
| pEB1323 | pZE-gfp | Ampr; gfpasv transcriptional fusion | 24 | |
| pEB1382 | pZE-rplU | Ebm846/847 PCR (XhoI/BamHI) product in pZE-gfp | −178/+94 | This work |
| pEB1242 | pASK-IBA37plus | Ampr; TetR promoter | IBA GmbH | |
| pEB1466 | pIBA-U.A.yhbE-SPA | Ebm852/968 PCR (EcoRI/XhoI) product from EB114 in pASK-IBA37plus | This work | |
| pEB1467 | pIBA-U.A.Δ.yhbE-SPA | Ebm892/893 mutagenesis on pEB1466 | This work | |
| pEB227 | pBAD24 | Ampr; ColE1 replication origin; PBAD promoter | 26 | |
| pEB1366 | pBAD-mlrA | Ebm770/771 (EcoRI/XhoI) product in pBAD24 (EcoRI/SalI) | This work | |
| pRLG770 | Transcription vector | 28 | ||
| pRLG11395 | rplU P | UPrplU in pRLG770 | −112/+48 | This work |
| pRLG13065 | rrnB P1 | Plasmid RLG589 maintained in XL1-Blue (Stratagene) | −88/+50 | 28 |
| pRLG11394 | PlacUV5 | Plasmid RLG593 maintained in XL1-Blue (Stratagene) | −59/+38 | 28 |
TS, thermosensitive; Ampr, Cmr, and Kanar, ampicillin, chloramphenicol, and kanamycin resistance, respectively; ori, origin of replication; FLP, FRT site flippase; UPrplU, DNA fragment upstream of rplU. Characteristics are given only for the vectors and the reference plasmids (indicated in bold).
The laboratory codes correspond to our stock numbering. Transcriptional fusions from the E. coli promoter library (23) do not have laboratory codes.
(ii) Plasmids for protein expression.
The DNA sequence of mlrA was amplified by PCR on genomic DNA using the oligonucleotides Ebm770/Ebm771, digested by EcoRI/XhoI, and cloned into the EcoRI/SalI sites of pBAD24 (26) to obtain plasmid pBAD-MlrA (pEB1366). The DNA sequences of rplU, rpmA, and sequential peptide affinity (SPA)-tagged yhbE (yhbE-SPA) were amplified by PCR of genomic DNA from strain EB114 using the oligonucleotides Ebm852/Ebm968, digested with EcoRI/XhoI, and cloned into pIBA37+ (IBA) to obtain plasmid pIBA-U.A.yhbE-SPA (pEB1466). The pIBA-U.A.Δ.yhbE-SPA plasmid (pEB1467) was obtained by mutagenesis of pEB1466 using oligonucleotides Ebm892/Ebm893.
(iii) Plasmids for recombination and terminator deletion.
A sequence of 1,940 bp encompassing the rplU-rpmA-yhbE region was amplified using oligonucleotides Ebm846/Ebm558 and cloned between the BamHI and SalI restriction sites of pKO3 (27). The terminator deletion was then introduced by directed mutagenesis using oligonucleotides Ebm892/Ebm893 in the pKO3-rplU-rpmA-yhbE plasmid, in order to obtain the pKO3-U.A.ΔT.yhbE plasmid (pEB1449).
(iv) Plasmids for in vitro transcription reaction.
The promoter for rplU was PCR amplified from the E. coli chromosome using primers RLG6565/RLG6566 and cloned into the EcoRI/HindIII site of the transcription vector pRLG770 (28).
Strain constructions.
Deletion mutant strains were obtained from the Keio collection (29). The tandem affinity purification (TAP)- or SPA-tagged strains were obtained from the collection of strains described in reference 30, obtained from Open Biosystems, or constructed as described previously (31) using plasmid pJL72 or plasmid pJL148 (32). For both types of strains, the recombinant genes were transferred to the desired strain background by P1 transduction (Table 2) (33). When required, the kanamycin resistance gene was removed using the pCP20 plasmid (34). The terminator deletion was introduced into the chromosome of MG1655 yhbE-SPA and MG1655 obgE-3Flag strains using the pEB1449 plasmid by the method described previously (27).
TABLE 2.
E. coli K-12 strainsa
| Laboratory code | Name | Description | Reference or source |
|---|---|---|---|
| EB698 | BW25113 ΔmlrA::Kanar | Keio collection; Kanar | 29 |
| EB132 | BW25113 ΔdksA::Kanar | Keio collection; Kanar | 29 |
| EB021 | CF4943 | MG1655 galK2 relA251 spoT203 zib563::Tn10 | 49 |
| EB149 | BW25113 | Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− rph-1 Δ(rhaD-rhaB)568 hsdR514 | 31 |
| MG1655 | Wild-type E. coli K-12; F− λ−ilvG rfb-50 rph-1 | 50 | |
| EB421 | MG1655 ΔrelA° | In-frame relA ORF deletion | 51 |
| EB425 | MG1655 ΔrelA° ΔspoT207 | ΔrelA spoT207::Camr | 51 |
| EB544 | MG1655 ΔrelA° spoT203 | Tetr; P1 transduction from CF4943 to EB421 | This work |
| EB559 | MG1655 ΔdksA° | In-frame dksA ORF deletion | 51 |
| EB708 | MG1655 ΔmlrA° | P1 transduction from EB698 to MG1655; kanamycin cassette removed | This work |
| EB113 | MG1655 L27-3Flag | PCR Ebm913/914 product on pEB794 recombined in MG1655 | This work |
| EB114 | MG1655 yhbE-SPA | PCR Ebm559/560 product on pEB794 recombined in MG1655 | This work |
| EB818 | MG1655 obgE-3Flag | PCR Ebm596/562 product on pEB794 recombined in MG1655 | This work |
| EB568 | MG1655 Δterm yhbE-SPA | Recombination of pEB1449 in EB114 | This work |
| EB565 | MG1655 Δterm obgE-3Flag | Recombination of pEB1449 in EB818 | This work |
The ° character after a strain name means that the kanamycin cassette was removed using the pCP20 plasmid. Tetr, Camr, and Kanar, tetracycline, chloramphenicol, and kanamycin resistance, respectively; Δterm, terminator deletion. Boldface indicates the wild-type strain.
RNA preparation, RT-PCR, and mapping of the TSS.
Total RNA was prepared from 10 ml of MG1655 cells grown to an optical density at 600 nm (OD600) of 1.3 using a PureYields RNA Midiprep system from Promega. RT-PCRs were performed using a Promega Access reverse transcriptase PCR (RT-PCR) system. The transcription start site (TSS) was determined using an RLM-RACE kit from Ambion.
Transcriptional fusion with gfp.
Wild-type E. coli strain MG1655 or isogenic mutant strains were transformed with plasmids carrying transcriptional fusions of gfpmut2 or gfpasv and were maintained with kanamycin or ampicillin, respectively. For cotransformation with pUA plasmids, compatible plasmids (pBAD24 and derivatives) were used and ampicillin was added. Selection plates were incubated at 37°C for 16 h. A 600-μl volume of LB medium supplemented with required antibiotics, and with arabinose (0.01% or 0.05%) when necessary for PBAD-driven expression, was inoculated (3 to 6 replicates per assay), and the cells were grown for 16 h at 30°C in 96-well polypropylene plates (2.2-ml wells) under conditions of aeration and agitation. A 150-μl volume was transferred from each well to a black Greiner 96-well plate for reading absorbance at 600 nm and fluorescence (excitation, 485 nm; emission, 530 nm) in a Tecan Infinite M200 plate reader. The expression levels were calculated by dividing the intensity of fluorescence by the absorbance at 600 nm. These results are given in arbitrary units because the intensity of fluorescence is acquired with variable gain and hence varies from one experiment to the other.
SDS-PAGE and Western blotting.
SDS-PAGE, electrotransfer onto nitrocellulose membranes, and Western blot analyses were performed as previously described (35). Total cell extracts were prepared by homogenizing cell pellets directly in 1× Laemmli buffer and heating 10 min at 96°C. Volumes corresponding to 1.5 · 108 cells were loaded in each lane. As an additional control, we verified that equivalent protein amounts were loaded by staining all proteins on the membrane with Ponceau red dye prior blocking. In order to run the YhbE-SPA-tagged protein at its expected molecular weight, samples in denaturing buffer were left unheated before loading on the gel. Monoclonal anti-Flag M2 used to detect the 3Flag and SPA tags was purchased from Sigma. Anti-mouse IgG–Alexa Fluor 680 secondary antibodies were purchased from Invitrogen. Western blots were imaged and quantified using an Odyssey Fc fluorescent reader from Li-Cor Biosciences.
RESULTS
The rplU, rpmA, yhbE, and obgE genes form an operon.
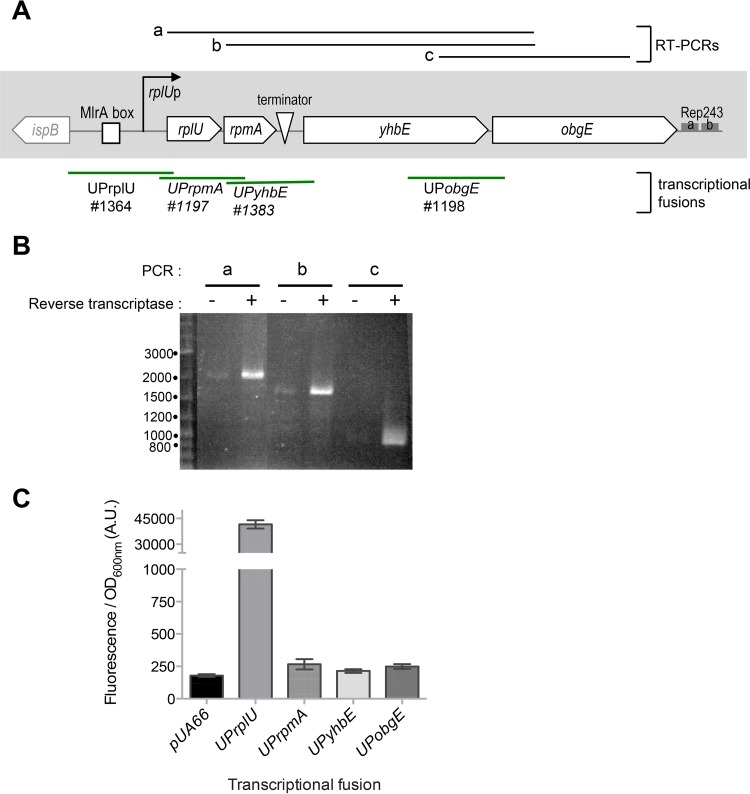
Because the genetic association between rplU, rpmA, and obgE is strongly conserved in bacteria (Fig. 1), and because these genes are regulated together during the stringent response (22), we hypothesized that they are expressed as an operon. To test this hypothesis, we performed RT-PCR analysis using three different pairs of primers; one primer in each pair was within obgE, and the other was within rplU, rpmA, or yhbE (Fig. 2A). Amplified DNA fragments of the expected sizes, based on the reported genome DNA sequence, were obtained for each set of oligonucleotides (Fig. 2B). The longest fragment (labeled “a”) corresponded to the size predicted for a product containing all 4 open reading frames (ORFs), strongly suggesting that the 4 genes were cotranscribed under the control of a promoter located upstream of rplU.
FIG 2.
The rplU, rpmA, yhbE, and obgE genes are cotranscribed. (A) obgE locus in Escherichia coli. The positions of the RT-PCR fragments shown in panel B (a, b, and c) are indicated at the top. A box indicates the binding region of MlrA, and a triangle indicates the terminator sequence between rpmA and yhbE. The transcription start site of rplU is indicated by an arrow. The limits of the regions cloned in transcriptional fusions with gfpmut2 (in the pUA66 or pUA139 plasmid) are shown below the scheme by green lines, and the laboratory codes of the corresponding plasmids are given. Two Rep elements downstream of obgE are indicated by gray boxes. (B) RT-PCR was performed on total RNAs prepared from cells in exponential phase with the oligonucleotides indicated in Table S1 in the supplemental material. The reactions were performed in parallel with (+) or without (−) reverse transcriptase in the PCR mix. The sizes (in base pairs) of the fragments in the DNA ladder are indicated to the left of the agarose gel. (C) Screen for promoter activity using transcriptional fusions with gfpmut2. The MG1655 strain was transformed using the indicated transcriptional fusion plasmids (pEB898, pEB1364, pEB1197, pEB1383, and pEB1198) (Table 1). After growth in LB at 30°C, fluorescence was measured at exponential phase, with the OD600 between 0.8 and 1. Error bars show the standard deviations calculated from the results determined for 4 replicates. UPrplU, UPrpmA, UPyhbE, and UPobgE refer to DNA fragments upstream of rplU, rpmA, yhbE, and obgE, respectively, cloned in the pUA66 or pUA139 plasmid.
To test for the presence of additional promoters inside the locus, we constructed a series of fusions to green fluorescent protein (GFP) containing the upstream regions from the rplU, rpmA, yhbE, and obgE genes (23) (Fig. 2A). Only the transcriptional fusion containing the DNA region upstream of rplU drove expression of GFP. Fluorescence from the other constructs was comparable to that from the pUA66 control plasmid (Fig. 2C). These results suggest that the 4 genes are transcribed as a polycistronic message from a single promoter located upstream from rplU.
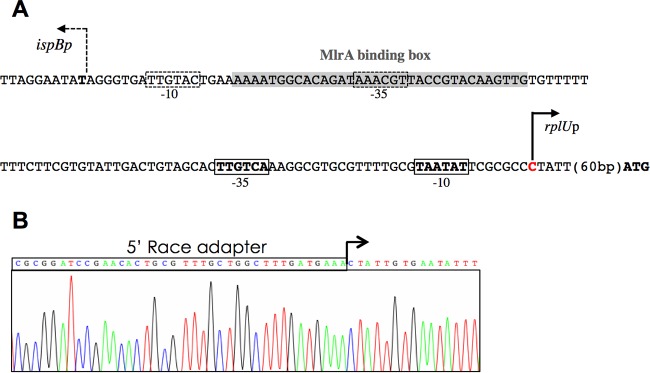
A promoter located upstream from rplU was also identified recently in a global RNA sequencing (RNA-seq) study (36) (Fig. 3A). This promoter contained close matches to the consensus −10 and −35 elements for sigma 70 promoters, separated by a consensus 17-bp spacer length (37). We confirmed the exact position of the transcription start site (TSS) using 5′ rapid amplification of cDNA ends (5′-RACE) (Fig. 3B). The mapped TSS is 8 nucleotides (nt) downstream of the predicted −10 hexamer, a common position for a TSS in E. coli, consistent with the predicted position of the rplU promoter. Taken together, the results suggest that rplU, rpmA, yhbE, and obgE are expressed as an operon from the rplU promoter.
FIG 3.
Characterization of the rplU promoter. (A) Sequence of the ispB-rplU intergenic region. The predicted promoter for ispB is depicted with dashed lines, while the rplU promoter is depicted with solid lines. The MlrA binding region is indicated by a gray rectangle. The transcription start site determined by 5′-RACE experiments is indicated in red. (B) Mapping of the transcription start site of rplU. The result of sequencing of one representative clone is shown.
Expression of the operon varies with growth phase.
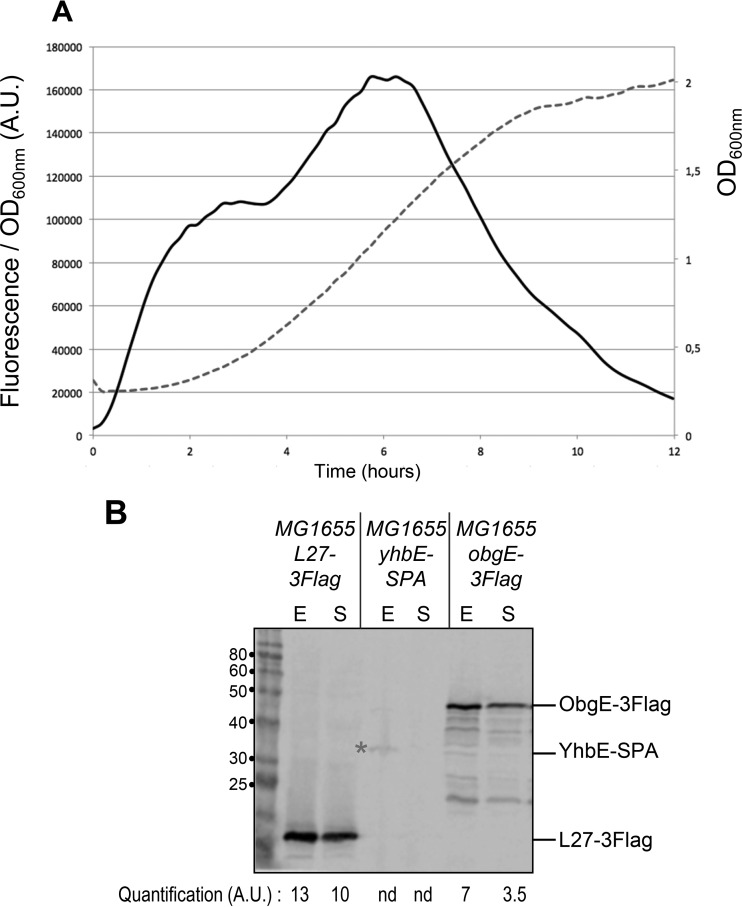
We also studied expression of the rplU operon at different growth phases using a transcriptional fusion to a less stable GFP variant, GFPasv (24). The GFPasv variant allows better estimation of relative expression levels during a typical growth curve analysis than the GFPmut2 used as described above. The activity of the rplU fusion was maximal during exponential growth, declined during entry into stationary phase, and shut off almost completely after several hours in stationary phase, consistent with expectations for r-protein expression (Fig. 4A) (9).
FIG 4.
Expression of the operon during growth. (A) The MG1655 strain was transformed by the transcriptional fusion of the region upstream of rplU (UPrplU) with GFPasv (pEB1382). Cultures were grown in 150 μl of M9 minimal medium supplemented with 0.2% glucose, Casamino Acids, and ampicillin, at 32°C, in a Tecan M200 plate reader. Every 10 min, OD600 and fluorescence levels (excitation, 485 nm; emission, 530 nm) were measured. The graph shows the fluorescence/OD600 ratio in arbitrary units (solid line, left y axis) and the OD600 (gray dashed line, right y axis) over time in hours. (B) The indicated strains (MG1655 L27-3Flag, EB113; MG1655 yhbE-SPA, EB114; and MG1655 obgE-3Flag, EB118) (Table 2) were grown in LB at 37°C. In exponential phase (E) and after overnight growth (S), total extracts were prepared and analyzed by 12% SDS-PAGE and Western blotting as described in Materials and Methods. Quantification of the band intensities is indicated below the image in arbitrary units (A.U.). nd, the intensity was too low to be quantified.
To test whether the reduction in rplU promoter activity during stationary phase resulted in a decrease in the amounts of the L21 (RplU), L27 (RpmA), YhbE, and ObgE proteins, we created strains in which the L27, YhbE, and ObgE proteins were tagged at their C termini with a 3Flag tag or an SPA (CBP-Tev-3Flag) tag and performed Western blot analyses with anti-Flag monoclonal antibodies. Growth of the L21-tagged strain was severely affected (data not shown); therefore, we did not use this strain further. In contrast, the L27- and ObgE-tagged recombinant strains grew similarly to the parental wild-type strains (data not shown), suggesting that the L27-3Flag and ObgE-3Flag proteins were functional.
Levels of L27-3Flag, YhbE-SPA, and ObgE-3Flag proteins were higher during exponential growth than during stationary phase (Fig. 4B). This was especially obvious for YhbE-SPA, which was undetectable in stationary phase. There was a decrease of about 2-fold for ObgE-3Flag, while the decrease in L27-3Flag was less marked. Therefore, the reduction in transcription in stationary phase did result in a decrease in protein amounts, although the reduction was not as large as the decrease in expression, suggesting that the proteins were relatively stable.
Expression of the operon is affected indirectly by ppGpp/DksA.
The rpmA, yhbE, and obgE genes were downregulated in response to amino acid starvation in microarray experiments, and this downregulation was dependent on the presence of the relA and spoT genes (22), suggesting that ppGpp regulates expression of the operon. In addition, the majority of promoters for ribosomal proteins are inhibited directly by ppGpp and DksA (9). These results led us to test the effects of ppGpp and DksA on transcription from the rplU promoter.
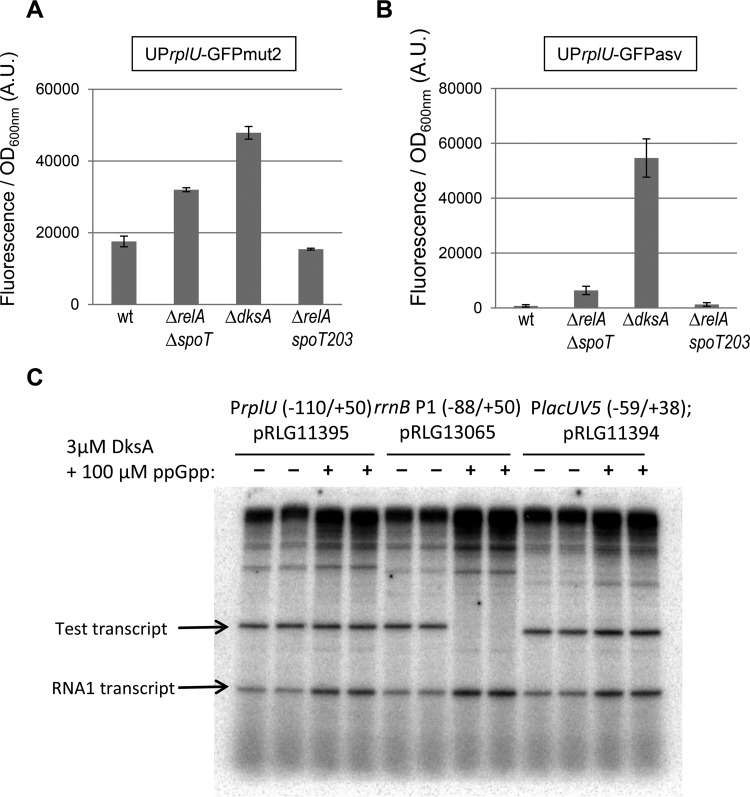
The activity of the rplU promoter was measured using constructs in which the promoter, mRNA leader, and ribosome binding site region from the rplU gene were fused to genes coding for two different GFP variants (see Materials and Methods). Fluorescence from these constructs was measured in stationary phase, when expression from r-protein promoters is relatively low in a wild-type strain (Fig. 4). rplU expression was significantly higher in the mutant that lacked ppGpp (ppGpp°; ΔrelA ΔspoT strain; EB425) than in the wild-type strain (Fig. 5A). The increase in the expression of the ΔdksA mutant (EB559) was even greater. rplU expression was lower in a strain defective for ppGpp degradation rather than synthesis (EB544; relA1 spoT203) than it was in a wild-type strain, again consistent with regulation of the promoter by ppGpp (Fig. 5A). When the less stable GFPasv protein was employed, the apparent differences between the wild-type and mutant strains were greater (Fig. 5B), consistent with the conclusion that ppGpp and DksA are required for inhibition of rplU expression during stationary phase and that the modest effects detailed in Fig. 5A resulted from the relative stability of the reporter protein.
FIG 5.
The rplU promoter is indirectly affected by the stringent response. (A and B) The MG1655 (wt) strain and the isogenic EB425 (ΔrelA ΔspoT; ppGpp°), EB559 (ΔdksA), and EB544 (ΔrelA spoT203) strains were transformed using UPrplU-GFPmut2 (A) or UPrplU-GFPasv (B) transcriptional fusions (with the pEB1364 and pEB1382 plasmids, respectively). Cells were grown overnight in LB at 30°C, and fluorescence and OD600 were measured. The data show the fluorescence/OD600 ratio in arbitrary units. Error bars indicate the standard deviations calculated from results determined for 3 replicates. (C) In vitro transcription assays. Single-round in vitro transcription assays were performed essentially as described previously (7) using Eσ70 RNAP (10 nM) and supercoiled plasmid templates (1 nM). The reaction mixtures were incubated at 30°C for 10 min and contained 10 mM Tris-HCl (pH 8), 60 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol (DTT), 200 μM (each) ATP, CTP, and GTP, 10 μM UTP, and ∼2 μCi [α-32P]UTP. Heparin (10 μg/ml) was added with the nucleoside triphosphates (NTPs) as a DNA competitor to prevent transcription reinitiation. Samples were electrophoresed on a 7 M urea–5.5% polyacrylamide gel and quantified by phosphorimaging. DksA (3 μM) and ppGpp (TriLink) (100 μM) were added to the transcription reaction mixtures where indicated.
Some r-protein promoters are regulated directly by effects of ppGpp/DksA on transcription initiation (9). The rplU promoter has a GC-rich discriminator region, like many r-protein and rRNA promoters that are regulated directly by ppGpp/DksA (2, 38, 39). However, even r-protein operons whose promoters contain G+C-rich discriminator sequences are also regulated by a translational inhibition mechanism in which an r-protein binds to the mRNA that encoded it and blocks translation when the repressor r-protein is in stoichiometric excess of rRNA (8). Thus, direct effects of ppGpp/DksA on rRNA transcription could also account for effects of ppGpp/DksA on r-protein synthesis.
Since it was not clear whether the observed inhibition of rplU by ppGpp/DksA was from direct inhibition of transcription initiation at the r-protein promoter complex or from mechanisms after transcription initiation generated by inhibition of rRNA transcription, we tested whether ppGpp/DksA inhibited rplU promoter activity directly in vitro. As a positive control, we measured the effects of ppGpp on transcription of the rrnB P1 promoter; as a negative control, we measured the effects of ppGpp/DksA on the lacUV5 promoter. rrnB P1 was strongly inhibited by ppGpp/DksA, but there was no inhibition of rplU or the lacUV5 promoter (Fig. 5C). We conclude that the observed inhibition of rplU expression by ppGpp/DksA in vivo must be indirect.
Effect of MlrA on rplU expression.
It was reported previously that MlrA is a transcription factor that acts as a positive regulator of σS-dependent curli and extracellular matrix production in E. coli and Salmonella enterica serovar Typhimurium (40). An MlrA binding motif was defined as AAAATTGTACA(12N)TGTACAATTTT (41), a sequence similar to one present ∼100 bp upstream of the rplU transcription start site (Fig. 3A). The MlrA binding motif also overlaps the predicted −35 element of the divergent ispB gene that codes for the essential octaprenyl diphosphate synthase involved in quinone synthesis (Fig. 3A). We therefore tested if MlrA affected the expression of rplU or ispB transcriptional fusions to GFPmut2 (Fig. 6A), using a series of reporter constructs with different rplU promoter upstream endpoints and/or with substitutions in the proposed recognition site for MlrA (Fig. 6A).
FIG 6.
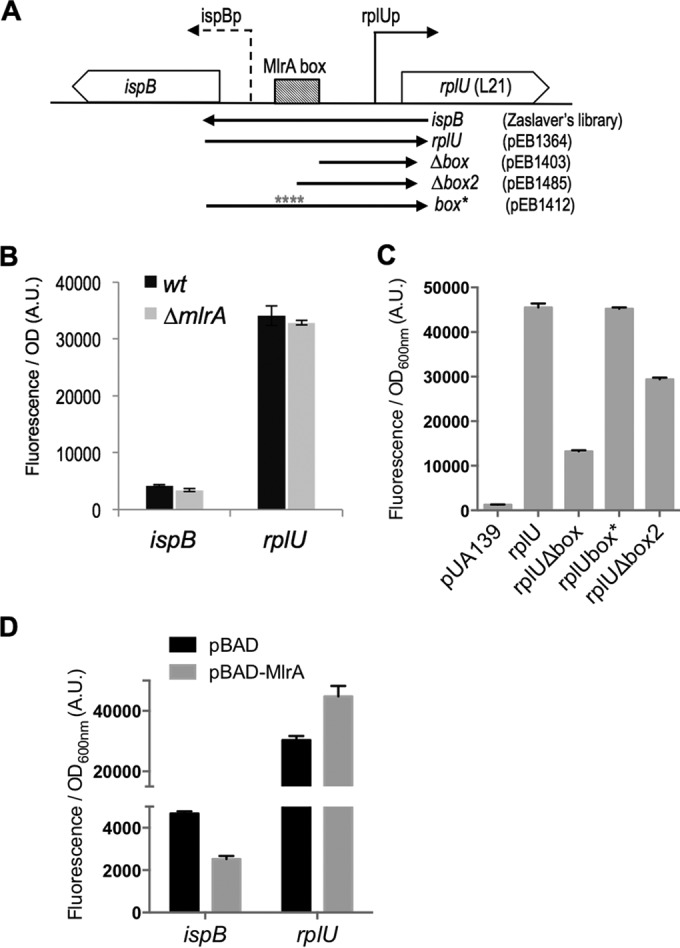
Impact of MlrA on the expression of ispB and rplU genes. (A) Limits of the different transcriptional fusion constructions are indicated below the scheme by arrows, and the laboratory codes of the corresponding plasmids are given. (The ispB plasmid comes from the library of A. Zaslaver [23].) (B) MG1655 (wt) and MG1655 ΔmlrA (EB708) strains were transformed by the indicated transcriptional fusions. Cells were grown overnight in LB at 30°C, and fluorescence and OD600 were measured. The data show the fluorescence/OD600 ratio in arbitrary units. Error bars indicate the standard errors of the means calculated from the results determined for 6 replicates. (C) The MG1665 wild-type strain was transformed using the indicated transcriptional fusions. Cultures were grown overnight at 30°C in LB supplemented with kanamycin. Error bars show the standard deviations calculated from results determined for 3 replicates. (D) The MG1665 ΔmlrA strain (EB708) was cotransformed using the indicated transcriptional fusions together with the pBAD24 or pBAD-MlrA (pEB1366) plasmid. Cultures were grown overnight at 30°C in LB supplemented with ampicillin, kanamycin, and 0.05% arabinose. Error bars show the standard errors of the means calculated from results determined for 3 replicates.
Deletion of mlrA had no effect on expression of either rplU or ispB (Fig. 6B). Although deletion of the upstream region, including the proposed MlrA recognition site (rplUΔbox), or part of the recognition site (rplUΔbox2), reduced expression, a 4-bp substitution in the MlrA recognition site (rplUbox*), without deletion of the upstream DNA, had no effect on activity (Fig. 6C). This suggested that the effect of the upstream region did not result from MlrA binding under these conditions.
Consistent with the conclusion that MlrA has only a minimal role or no role in rplU expression, overproduction of MlrA (from a pBAD-mlrA plasmid) increased expression of rplU promoter only very slightly (Fig. 6D). Overexpression of MlrA did inhibit expression of ispB about 2-fold (Fig. 6D), but understanding the physiological significance of this result will require quantitation of MlrA levels and comparison of the overexpressed levels to natural levels of MlrA. We also note that MlrA activity might not be maximal under the conditions tested, since it has been reported that another protein, YdaM, directly stimulates MlrA activity as part of the c-di-GMP signaling cascade (42).
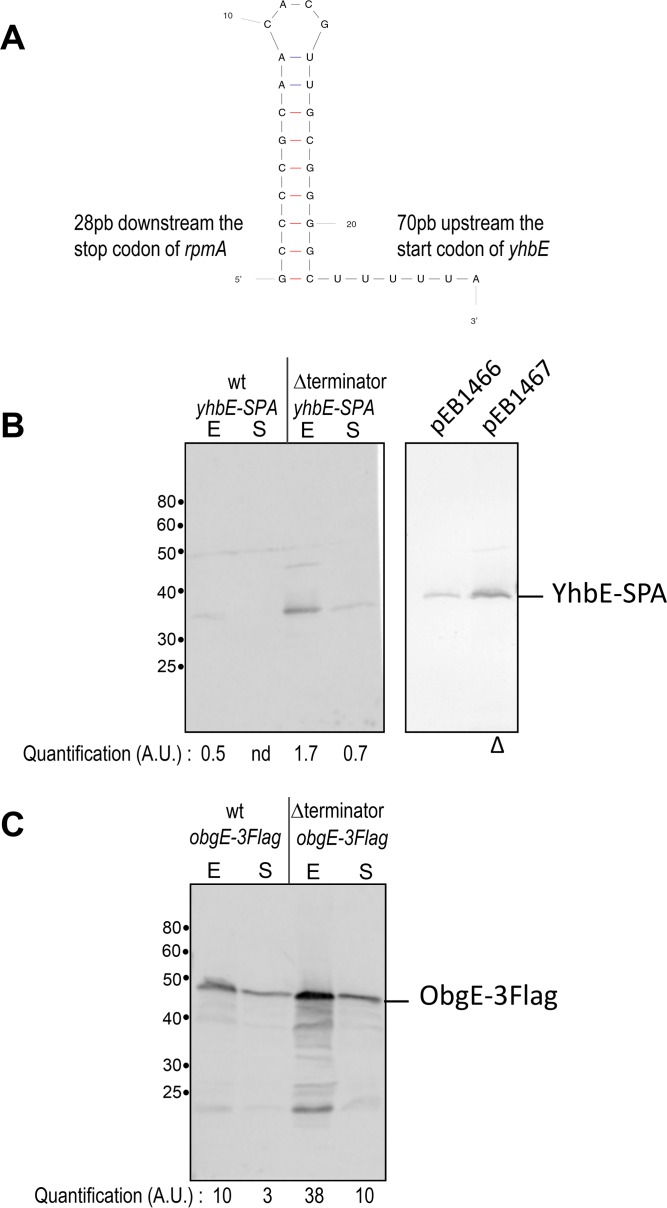
Role of the terminator between rpmA and yhbE.
Sequence analysis suggested that there might be an intrinsic terminator sequence between the rpmA and yhbE ORFs (RegulonDB [43, 44]) (Fig. 7A). To test the importance of the putative terminator, we deleted this sequence from the chromosome and then measured the effects on production of YhbE and ObgE. To measure the levels of YhbE and ObgE, we introduced the coding sequences for YhbE-SPA and ObgE-3Flag proteins into the chromosome, retaining control of their expression from their native regulatory regions. While YhbE-SPA was barely detectable in the wild-type strain, it was overproduced in the strain lacking the intrinsic terminator (Fig. 7B, left panel). Qualitatively similar effects were observed with plasmids expressing the rplU, rpmA, and yhbE-SPA genes as an operon under the control of the TetR repressor, with or without the termination sequence upstream of yhbE (Fig. 7B, right panel). Similarly, ObgE-3Flag amounts were strongly increased in the terminator deletion mutant compared to the wild type (Fig. 7C). Quantification of the band intensities indicated there was a 3-fold increase in the absence of the intrinsic terminator, suggesting that about 75% of the transcripts were attenuated in wild-type cells.
FIG 7.
The terminator downstream of rpmA affects yhbE and obgE expression. (A) Sequence, fold prediction, and position of the terminator located in the rpmA-yhbE intergenic region. The figure was obtained with the online MFold Web server (52). (B) Left panel: strains MG1655 yhbE-SPA (EB114) and MG1655 Δterm yhbE-SPA (EB568) were grown in LB at 37°C to exponential phase (E; OD600 = 0.5) and to stationary phase (S; 24 h). Right panel: strain MG1655, transformed using pIBA-U.A.yhbE-SPA (pEB1466) or pIBA-U.A.Δ.yhbE-SPA (pEB1467), was grown in LB, and expression was induced by the use of 200 ng/ml anhydrotetracycline. The Δ symbol below the image indicates that the terminator is deleted in the pEB1467 plasmid. (C) Strains MG1655 obgE-3Flag (EB818) and MG1655 Δterm obgE-3Flag (EB565) were grown in LB at 37°C to exponential phase (E; OD600 = 0.5) and to stationary phase (S; 24 h). In the experiments whose results are presented in panels B and C, total cell extracts were analyzed by 10% SDS-PAGE and Western blotting as described in Materials and Methods. Quantification of the band intensities is indicated in arbitrary units (A.U.) below the images. nd, the intensity was too low to be quantified.
The attenuator decreased expression of YhbE and ObgE in stationary phase as well as in exponential phase. The same 3-fold increase in protein amounts was observed for YhbE-SPA and ObgE-3Flag protein expression in the absence of the terminator (Fig. 7B and C). Thus, the terminator appears to set the amounts of the ObgE and YhbE proteins relative to those of the ribosomal proteins but not in the regulation of their synthesis with nutritional conditions.
DISCUSSION
ObgE is proposed to play a role in ribosome biogenesis. Here, we demonstrated that obgE is cotranscribed with rplU and rpmA, ensuring that its expression is linked to that of ribosomal proteins. Thus, consistent with its proposed function, obgE's genetic linkage and regulation are coordinated, like those of many other translation factors and small GTPases (8, 16).
A relationship between yhbE and the translation machinery is less clear. In some bacteria, rplU, rpmA, and obg are not genetically linked to yhbE, and another gene is inserted between rpmA and obg. In firmicutes, the sporulation-related gene spo0B, rather than yhbE, is located between rpmA and obg (45). In Listeria, genes coding for a glycerol kinase and an uptake facilitator are located between rpmA and obg. In alphaproteobacteria, a gene coding for an acetyl-transferase is sometimes found at this location. This evolutionary variation in gene position adds to the uncertainty regarding a relationship between yhbE and translation.
Despite the conserved genetic association between rplU, rpmA, and obgE, their coexpression and coregulation have not been demonstrated previously. We report here that an attenuator following rpmA reduces transcription of yhbE and obgE by ∼75%. Use of an attenuator to reduce expression of distal genes in E. coli r-protein operons has been reported previously (10, 46). For example, the intergenic attenuator that decreases readthrough of the rpsB-tsf operon, which codes for r-protein S2 and elongation factor Ts (EF-Ts), respectively, reduces transcription of tsf by ∼2/3 (10). It was reported previously that attenuation of obg expression in B. subtilis occurs at the translation level through an mRNA hairpin that sequesters the ribosome binding site (45).
Whereas the terminator upstream from yhbE sets the ratio of L21 and L27 to ObgE, this does not account for their regulation with nutritional conditions. It was reported previously that the level of ObgE is about 6-fold lower in stationary-phase E. coli than in exponential-phase E. coli (6,000 versus 30,000 molecules, respectively) (47), a finding qualitatively similar to what is reported here (Fig. 4B and 7C). As expected from studies on other r-protein operons (9), we found that ppGpp and DksA are required for this regulation in vivo (Fig. 5A and B). We tested whether ppGpp/DksA directly regulated the rplU promoter in vitro, as has been reported for a subset of other r-protein operons (9) (Fig. 5C). ppGpp/DksA had no effect on rplU promoter activity in vitro. Therefore, we propose that regulation of the operon is likely the consequence of direct regulation of rRNA transcription by ppGpp/DksA, which in turn affects expression of the operon by a posttranscriptional mechanism involving binding of either L21 or L27 to the rplU mRNA (translational feedback) (8). Because L21 is one of the proteins that binds directly to 23S rRNA during the early stages of 50S ribosomal subunit assembly (48), it is a good candidate for a translational repressor of the rplU operon. However, further studies will be needed to test this hypothesis.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Centre National de la Recherche Scientifique (CNRS), Aix-Marseille University; by the ANR (French National Research Agency) grant Lipid-Stress (ANR-09-JCJC-0018); and by the U.S. National Institutes of Health (R37 GM37048 to R.L.G.). L.M. was the recipient of a Medical Research Foundation (FRM) fellowship, and H.L.B. was supported by a Molecular Biosciences Training Grant from the NIH (grant no. T32GM007215) and by a fellowship from the University of Wisconsin—Madison.
We thank Patrice Moreau and Hans Geiselman for gifts of materials.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00159-16.
REFERENCES
- 1.Srivatsan A, Wang JD. 2008. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr Opin Microbiol 11:100–105. doi: 10.1016/j.mib.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu Rev Microbiol 62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 3.Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL. 2013. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell 50:420–429. doi: 10.1016/j.molcel.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zuo Y, Wang Y, Steitz TA. 2013. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol Cell 50:430–436. doi: 10.1016/j.molcel.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mechold U, Potrykus K, Murphy H, Murakami KS, Cashel M. 2013. Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res 41:6175–6189. doi: 10.1093/nar/gkt302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross W, Sanchez-Vazquez P, Chen AY, Lee J-H, Burgos HL, Gourse RL. ppGpp binding to a site at the RNAP-DksA interface accounts for its dramatic effects on transcription initiation during the stringent response. Mol Cell, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paul BJ, Ross W, Gaal T, Gourse RL. 2004. rRNA transcription in Escherichia coli. Annu Rev Genet 38:749–770. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- 8.Nomura M, Gourse R, Baughman G. 1984. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem 53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- 9.Lemke JJ, Sanchez-Vazquez P, Burgos HL, Hedberg G, Ross W, Gourse RL. 2011. Direct regulation of Escherichia coli ribosomal protein promoters by the transcription factors ppGpp and DksA. Proc Natl Acad Sci U S A 108:5712–5717. doi: 10.1073/pnas.1019383108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aseev LV, Levandovskaya AA, Tchufistova LS, Scaptsova NV, Boni IV. 2008. A new regulatory circuit in ribosomal protein operons: S2-mediated control of the rpsB-tsf expression in vivo. RNA 14:1882–1894. doi: 10.1261/rna.1099108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aseev LV, Bylinkina NS, Boni IV. 2015. Regulation of the rplY gene encoding 5S rRNA binding protein L25 in Escherichia coli and related bacteria. RNA 21:851–861. doi: 10.1261/rna.047381.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Babina AM, Soo MW, Fu Y, Meyer MM. 2015. An S6:S18 complex inhibits translation of E. coli rpsF. RNA 21:2039–2046. doi: 10.1261/rna.049544.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shajani Z, Sykes MT, Williamson JR. 2011. Assembly of bacterial ribosomes. Annu Rev Biochem 80:501–526. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- 14.Britton RA. 2009. Role of GTPases in bacterial ribosome assembly. Annu Rev Microbiol 63:155–176. doi: 10.1146/annurev.micro.091208.073225. [DOI] [PubMed] [Google Scholar]
- 15.Czyz A, Wegrzyn G. 2005. The Obg subfamily of bacterial GTP-binding proteins: essential proteins of largely unknown functions that are evolutionarily conserved from bacteria to humans. Acta Biochim Pol 52:35–43. [PubMed] [Google Scholar]
- 16.Verstraeten N, Fauvart M, Versees W, Michiels J. 2011. The universally conserved prokaryotic GTPases. Microbiol Mol Biol Rev 75:507–542, p 2 and 3 of table of contents. doi: 10.1128/MMBR.00009-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kint C, Verstraeten N, Hofkens J, Fauvart M, Michiels J. 2014. Bacterial Obg proteins: GTPases at the nexus of protein and DNA synthesis. Crit Rev Microbiol 40:207–224. doi: 10.3109/1040841X.2013.776510. [DOI] [PubMed] [Google Scholar]
- 18.Buglino J, Shen V, Hakimian P, Lima CD. 2002. Structural and biochemical analysis of the Obg GTP binding protein. Structure 10:1581–1592. doi: 10.1016/S0969-2126(02)00882-1. [DOI] [PubMed] [Google Scholar]
- 19.Feng B, Mandava CS, Guo Q, Wang J, Cao W, Li N, Zhang Y, Zhang Y, Wang Z, Wu J, Sanyal S, Lei J, Gao N. 2014. Structural and functional insights into the mode of action of a universally conserved Obg GTPase. PLoS Biol 12:e1001866. doi: 10.1371/journal.pbio.1001866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verstraeten N, Knapen WJ, Kint CI, Liebens V, Van den Bergh B, Dewachter L, Michiels JE, Fu Q, David CC, Fierro AC, Marchal K, Beirlant J, Versees W, Hofkens J, Jansen M, Fauvart M, Michiels J. 2015. Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol Cell 59:9–21. doi: 10.1016/j.molcel.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 21.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C. 2011. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T. 2008. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol 68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zaslaver A, Bren A, Ronen M, Itzkovitz S, Kikoin I, Shavit S, Liebermeister W, Surette MG, Alon U. 2006. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat Methods 3:623–628. doi: 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
- 24.de Jong H, Ranquet C, Ropers D, Pinel C, Geiselmann J. 2010. Experimental and computational validation of models of fluorescent and luminescent reporter genes in bacteria. BMC Syst Biol 4:55. doi: 10.1186/1752-0509-4-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andersen JB, Sternberg C, Poulsen LK, Bjorn SP, Givskov M, Molin S. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol 64:2240–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Link AJ, Phillips D, Church GM. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol 179:6228–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross W, Thompson JF, Newlands JT, Gourse RL. 1990. E. coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J 9:3733–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, Davey M, Parkinson J, Greenblatt J, Emili A. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 31.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeghouf M, Li J, Butland G, Borkowska A, Canadien V, Richards D, Beattie B, Emili A, Greenblatt JF. 2004. Sequential peptide affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J Proteome Res 3:463–468. doi: 10.1021/pr034084x. [DOI] [PubMed] [Google Scholar]
- 33.Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- 34.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-A. [DOI] [PubMed] [Google Scholar]
- 35.Gully D, Moinier D, Loiseau L, Bouveret E. 2003. New partners of acyl carrier protein detected in Escherichia coli by tandem affinity purification. FEBS Lett 548:90–96. doi: 10.1016/S0014-5793(03)00746-4. [DOI] [PubMed] [Google Scholar]
- 36.Thomason MK, Bischler T, Eisenbart SK, Forstner KU, Zhang A, Herbig A, Nieselt K, Sharma CM, Storz G. 2015. Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J Bacteriol 197:18–28. doi: 10.1128/JB.02096-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huerta AM, Collado-Vides J. 2003. Sigma70 promoters in Escherichia coli: specific transcription in dense regions of overlapping promoter-like signals. J Mol Biol 333:261–278. doi: 10.1016/j.jmb.2003.07.017. [DOI] [PubMed] [Google Scholar]
- 38.Travers AA. 1980. Promoter sequence for stringent control of bacterial ribonucleic acid synthesis. J Bacteriol 141:973–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Travers AA. 1984. Conserved features of coordinately regulated E. coli promoters. Nucleic Acids Res 12:2605–2618. doi: 10.1093/nar/12.6.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown PK, Dozois CM, Nickerson CA, Zuppardo A, Terlonge J, Curtiss R III. 2001. MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium. Mol Microbiol 41:349–363. doi: 10.1046/j.1365-2958.2001.02529.x. [DOI] [PubMed] [Google Scholar]
- 41.Ogasawara H, Yamamoto K, Ishihama A. 2010. Regulatory role of MlrA in transcription activation of csgD, the master regulator of biofilm formation in Escherichia coli. FEMS Microbiol Lett 312:160–168. doi: 10.1111/j.1574-6968.2010.02112.x. [DOI] [PubMed] [Google Scholar]
- 42.Lindenberg S, Klauck G, Pesavento C, Klauck E, Hengge R. 2013. The EAL domain protein YciR acts as a trigger enzyme in a c-di-GMP signalling cascade in E. coli biofilm control. EMBO J 32:2001–2014. doi: 10.1038/emboj.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gama-Castro S, Salgado H, Peralta-Gil M, Santos-Zavaleta A, Muñiz-Rascado L, Solano-Lira H, Jimenez-Jacinto V, Weiss V, García-Sotelo JS, López-Fuentes A, Porrón-Sotelo L, Alquicira-Hernández S, Medina-Rivera A, Martínez-Flores I, Alquicira-Hernández K, Martínez-Adame R, Bonavides-Martínez C, Miranda-Ríos J, Huerta AM, Mendoza-Vargas A, Collado-Torres L, Taboada B, Vega-Alvarado L, Olvera M, Olvera L, Grande R, Morett E, Collado-Vides J. 2011. RegulonDB version 7.0: transcriptional regulation of Escherichia coli K-12 integrated within genetic sensory response units (Gensor units). Nucleic Acids Res 39:D98–D105. doi: 10.1093/nar/gkq1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merino E, Yanofsky C. 2005. Transcription attenuation: a highly conserved regulatory strategy used by bacteria. Trends Genet 21:260–264. doi: 10.1016/j.tig.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 45.Asayama M, Saito K, Kobayashi Y. 1998. Translational attenuation of the Bacillus subtilis spo0B cistron by an RNA structure encompassing the initiation region. Nucleic Acids Res 26:824–830. doi: 10.1093/nar/26.3.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barry G, Squires C, Squires CL. 1980. Attenuation and processing of RNA from the rplJL–rpoBC transcription unit of Escherichia coli. Proc Natl Acad Sci U S A 77:3331–3335. doi: 10.1073/pnas.77.6.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kobayashi G, Moriya S, Wada C. 2001. Deficiency of essential GTP-binding protein ObgE in Escherichia coli inhibits chromosome partition. Mol Microbiol 41:1037–1051. [DOI] [PubMed] [Google Scholar]
- 48.Chen SS, Williamson JR. 2013. Characterization of the ribosome biogenesis landscape in E. coli using quantitative mass spectrometry. J Mol Biol 425:767–779. doi: 10.1016/j.jmb.2012.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gentry DR, Hernandez VJ, Nguyen LH, Jensen DB, Cashel M. 1993. Synthesis of the stationary-phase sigma factor sigma s is positively regulated by ppGpp. J Bacteriol 175:7982–7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bachmann BJ. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p 2460–2488. In Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. [Google Scholar]
- 51.Wahl A, My L, Dumoulin R, Sturgis JN, Bouveret E. 2011. Antagonistic regulation of dgkA and plsB genes of phospholipid synthesis by multiple stress responses in Escherichia coli. Mol Microbiol 80:1260–1275. doi: 10.1111/j.1365-2958.2011.07641.x. [DOI] [PubMed] [Google Scholar]
- 52.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.