ABSTRACT
Pseudomonas aeruginosa thrives in multiple environments and is capable of causing life-threatening infections in immunocompromised patients. RsmA is a posttranscriptional regulator that controls virulence factor production and biofilm formation. In this study, we investigated the expression and activity of rsmA and the protein that it encodes, RsmA, in P. aeruginosa mucA mutant strains, which are common in chronic infections. We determined that AlgU regulates a previously unknown rsmA promoter in P. aeruginosa. Western blot analysis confirmed that AlgU controls rsmA expression in both a laboratory strain and a clinical isolate. RNase protection assays confirmed the presence of two rsmA transcripts and suggest that RpoS and AlgU regulate rsmA expression. Due to the increased amounts of RsmA in mucA mutant strains, a translational leader fusion of the RsmA target, tssA1, was constructed and tested in mucA, algU, retS, gacA, and rsmA mutant backgrounds to examine posttranscriptional activity. From these studies, we determined that RsmA is active in mucA22 mutants, suggesting a role for RsmA in mucA mutant strains. Taken together, we have demonstrated that AlgU controls rsmA transcription and is responsible for RsmA activity in mucA mutant strains. We propose that RsmA is active in P. aeruginosa mucA mutant strains and that RsmA also plays a role in chronic infections.
IMPORTANCE P. aeruginosa causes severe infections in immunocompromised patients. The posttranscriptional regulator RsmA is known to control virulence and biofilm formation. We identify a new rsmA promoter and determine that AlgU is important in the control of rsmA expression. Mutant mucA strains that are considered mucoid were used to confirm increased rsmA expression from the AlgU promoter. We demonstrate, for the first time, that there is RsmA activity in mucoid P. aeruginosa strains. Our work suggests that RsmA may play a role during chronic infections as well as acute infections.
INTRODUCTION
Pseudomonas aeruginosa produces a myriad of virulence factors and can cause both acute and chronic infections (1, 2). To survive and persist, P. aeruginosa must coordinate gene expression in response to changing environmental conditions. Global regulatory networks respond to changing environmental conditions to allow the physiological changes necessary for survival to be made. The P. aeruginosa genome encodes many global regulatory systems, including posttranscriptional regulators, such as RsmA. RsmA is important in regulating several virulence factors involved in acute and chronic infections (3).
Acute infections are characterized by motility and expression of the type III secretion system (4). RsmA positively regulates both motility and type III secretion system expression (4–6). A defining characteristic of chronic infections is biofilm formation, which includes the production of exopolysaccharides, such as alginate, and the type VI secretion system (T6SS) (7, 8). RsmA is a negative regulator of the exopolysaccharide gene psl and the first hemolysin-coregulated secretion island type VI secretion system (HSI-I T6SS) (5, 8, 9). A characteristic of some P. aeruginosa isolates causing chronic infections, such as those found in cystic fibrosis patients, is alginate overproduction, which is the result of the extracytoplasmic sigma factor AlgU (7, 10).
AlgU is normally sequestered at the cytoplasmic membrane by the anti-sigma factor MucA (7, 10). MucA prevents AlgU from binding and activating target promoters (7, 10). Once liberated from MucA, AlgU activates a variety of genes, including those important for biosynthesizing alginate (7, 10–13). Strains overproducing alginate are termed mucoid. P. aeruginosa strains that initially colonize cystic fibrosis patients frequently evolve into strains that overproduce alginate, often because of mutations in the mucA gene (7, 10). A previous study found that a mucA mutant has increased rsmA expression (14). Because mucA mutations lead to hyperactive AlgU (7, 10), we hypothesized that AlgU might regulate rsmA transcription.
RsmA is a global posttranscriptional regulator that regulates several mRNAs involved in virulence (5, 6). RsmA positively regulates type III secretion and type IV pilus production, both of which are important virulence factors for acute infections (6, 14). In the case of biofilm formation, RsmA inhibits exopolysaccharide production (5, 9). Therefore, RsmA is important in determining what virulence factors are expressed by P. aeruginosa. RsmA activity is regulated by two small RNAs, RsmY and RsmZ, that bind RsmA and prevent RsmA from binding target mRNAs (15). RsmY and RsmZ sequester RsmA, allowing the production of exopolysaccharides and other RsmA target mRNAs (9, 16). Increased RsmY and RsmZ levels or decreased RsmA activity can allow biofilm formation to occur. The expression of rsmY and rsmZ is positively controlled by RsmA, GacA, and LadS and negatively controlled by RetS (16–18). RsmA binds to highly conserved GGA motifs. Usually, one of the GGA motifs is found in a stem-loop structure formed near the ribosome-binding site (19). Multiple copies of the same GGA motif are also found in the inhibitory small RNAs RsmY and RsmZ (20, 21).
Although RsmA is important for virulence and global gene expression (5, 22, 23), little is known regarding the genetic controls governing rsmA expression. Currently, only a single rsmA promoter region, which resembles a σ70 (RpoD) promoter, has been identified in P. aeruginosa (24–26). A recent chromatin immunoprecipitation sequencing study showed that both RpoD and RpoS likely control rsmA expression (27). However, another study found similar RsmA levels in both an rpoS mutant and the wild-type strain (9). Early studies found that optimal rsmA expression requires GacA and that rsmA expression increases throughout growth (28), findings which support a role for RpoS in controlling rsmA expression. These seemingly contradictory data have not yet been clarified.
Given the importance of RsmA in controlling P. aeruginosa virulence and biofilm formation, there is a critical need to understand rsmA expression. In this study, we determined the genetic elements controlling rsmA expression. We identified a new promoter region and determined that AlgU and RpoS contribute to rsmA expression. Importantly, we demonstrate that RsmA is active in P. aeruginosa mucA mutants. This work suggests that RsmA has a role in regulating gene expression in mucA mutants and that AlgU is an important activator of rsmA expression.
MATERIALS AND METHODS
Strains, plasmids, and media.
The strains used in this study are presented in Table 1. Escherichia coli strains were maintained on LB (Difco) plates or broth without or with antibiotics, as appropriate. Pseudomonas aeruginosa strains were grown on Pseudomonas isolation agar (PIA), LB medium, or Vogel-Bonner minimal medium (VBMM) supplemented with the appropriate concentrations of antibiotics. For E. coli, antibiotics were used at the following concentrations when appropriate: 10 μg/ml tetracycline, 15 μg/ml gentamicin, 100 μg/ml ampicillin, and 35 μg/ml kanamycin. For Pseudomonas strains, antibiotics were used at the following concentrations: 150 μg/ml gentamicin, 50 μg/ml tetracycline, and 300 μg/ml carbenicillin. For allelic exchange, sucrose was supplemented at 10% in YT (1% tryptone and 0.5% yeast extract) medium.
TABLE 1.
Strains, plasmids, and constructs used in this study
| Strain, plasmid, or construct | Genotype or relevant property(ies) | Reference or source |
|---|---|---|
| E. coli | ||
| NEB5α | fhuA2 Δ(argF-lacZ)U169 phoA glnV44 ϕ80dlacZΔM15 gyrA96 | New England BioLabs |
| SM10 | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu Kmr | 56 |
| pRK2013 strain | Helper strain | 31 |
| P. aeruginosa | ||
| PAO1 | Wild type | 58 |
| ΔalgU mutant | ΔalgU | 57 |
| mucA22 mutant | mucA22 | 59 |
| mucA22 ΔalgU strain | mucA22 ΔalgU | This study |
| 2192 | 45 | |
| 383 | 45 | |
| ΔrsmA mutant | rsmA nonpolar deletion | This study |
| mucA22 ΔrsmA mutant | rsmA nonpolar deletion in mucA22 strain | This study |
| PAO1 TF1-lacZ | rsmA transcriptional fusion strain | This study |
| ΔalgU TF1-lacZ strain | rsmA transcriptional fusion strain | This study |
| mucA22 TF1-lacZ strain | rsmA transcriptional fusion strain | This study |
| mucA22 ΔalgU TF1-lacZ strain | rsmA transcriptional fusion strain | This study |
| 2192 TF1-lacZ | rsmA transcriptional fusion strain | This study |
| 383 TF1-lacZ | rsmA transcriptional fusion strain | This study |
| 383-HA | Nonmucoid clinical isolate with rsmA-HA allele | This study |
| 2192-HA | Mucoid clinical isolate with rsmA-HA allele | This study |
| 2192-HA ΔalgU | 2192 with rsmA-HA allele and algU deletion | This study |
| 2192 ΔalgU TF3-lacZ | rsmA transcriptional fusion strain | This study |
| PAO1 TF3-lacZ | rsmA transcriptional fusion strain | This study |
| ΔalgU TF3-lacZ strain | rsmA transcriptional fusion strain | This study |
| mucA22 TF3-lacZ strain | rsmA transcriptional fusion strain | This study |
| mucA22 ΔalgU TF3-lacZ strain | rsmA transcriptional fusion strain | This study |
| PAO1 TF3USDM-lacZ | Mutant rsmA transcriptional fusion strain | This study |
| ΔalgU TF3USDM-lacZ strain | Mutant rsmA transcriptional fusion strain | This study |
| mucA22 TF3USDM-lacZ strain | Mutant rsmA transcriptional fusion strain | This study |
| mucA22 ΔalgU TF3USDM-lacZ strain | Mutant rsmA transcriptional fusion strain | This study |
| PAO1 TF2-lacZ | rsmA transcriptional fusion | This study |
| mucA22 TF2-lacZ strain | rsmA transcriptional fusion | This study |
| ΔalgU TF2-lacZ strain | rsmA transcriptional fusion | This study |
| mucA22 ΔalgU strain | rsmA transcriptional fusion | This study |
| PAO1-HA | PAO1 with epitope-tagged RsmA | This study |
| ΔalgU-HA mutant | ΔalgU mutant with epitope-tagged RsmA | This study |
| mucA22-HA mutant | mucA22 mutant with epitope-tagged RsmA | This study |
| mucA22 ΔalgU-HA mutant | mucA22 ΔalgU strain with epitope-tagged RsmA | This study |
| PAO1-HA/pTJ1 | PAO1 with rsmA-HA allele and vector pTJ1 alone | This study |
| PAO1-HA/pAlgU | PAO1 with rsmA-HA allele and algU under control of pBAD promoter | This study |
| PAO1 lacUV5-tssA1-lacZ | Leader/translational fusion | This study |
| PAO1 ΔalgU lacUV5-tssA1-lacZ | Leader/translational fusion | This study |
| PAO1 ΔgacA lacUV5-tssA1-lacZ | Leader/translational fusion | This study |
| PAO1 ΔretS lacUV5-tssA1-lacZ | Leader/translational fusion | This study |
| PAO1 ΔrsmA lacUV5-tssA1-lacZ | Leader/translational fusion | This study |
| PAO1 mucA22 lacUV5-tssA1-lacZ | Leader/translational fusion | This study |
| PAO1 mucA22 ΔgacA lacUV5-tssA1-lacZ | Leader/translational fusion | This study |
| PAO1 mucA22 ΔretS lacUV5-tssA1-lacZ | Leader/translational fusion | This study |
| PAO1 mucA22 ΔalgU lacUV5-tssA1-lacZ | Leader/translational fusion | This study |
| PAO1 mucA22 ΔrsmA lacUV5-tssA1-lacZ | Leader/translational fusion | This study |
| Plasmids and constructs | ||
| pEX18Tc | Allelic exchange vector | 30 |
| pEX18Gm | Allelic exchange vector | 30 |
| ΔrsmA pEX18Tc | Allelic exchange for rsmA nonpolar deletion | This study |
| ΔalgU pEX18Tc | Allelic exchange for algU nonpolar deletion | 57 |
| ΔrpoS pEX18Tc | Allelic exchange for rpoS nonpolar deletion | This study |
| rsmA-HA pEX18Gm | Allelic exchange for rsmA-HA allele | This study |
| miniCTXlacZ | Transcriptional fusion vector | 38 |
| TF1-lacZ miniCTX | Transcriptional fusion | This study |
| TF3-lacZ miniCTX | Transcriptional fusion | This study |
| TF3USDM-lacZ miniCTX | Site-directed mutagenesis transcriptional fusion | This study |
| TF2-lacZ | Transcriptional fusion | This study |
| pTJ1 | Integrating vector used for overexpression | 43 |
| algU pTJ1 (pAlgU) | algU complementation/overexpression | This study |
Strain construction.
All PCR products were amplified from P. aeruginosa PAO1, unless otherwise noted, using Q5 polymerase (New England BioLabs [ΝEB], Ipswich, MA) and primers listed in Table 2. Crossover PCR (splicing by overhang extension PCR) (29) was used to construct deletion mutations and to clone sequences into the suicide vector pEX18Tc or pEX18Gm (30). All cloned constructs were confirmed via sequencing. P. aeruginosa strains were conjugated with E. coli as a donor strain and the pRK2013-containing helper strain (31). Conjugations were performed overnight on LB plates at 37°C, and the conjugants were plated on the appropriate selective medium to obtain single-crossover mutants. Merodiploids were grown without selection and then screened for sucrose sensitivity on YT medium–10% sucrose plates. Mutations were confirmed using PCR with primers containing the suffix intF and intR in Table 2 and sequencing of the resulting PCR fragment. Hemagglutinin (HA) tagging of the proteins was accomplished using primers containing the HA tag at the 3′ end of the gene, and HA was introduced as described above using the suicide vector pEX18Gm.
TABLE 2.
Primers used in this study
| Primer name | Sequence | Usea |
|---|---|---|
| rsmARHindIII | GCGCAAGCTTCACGCGAATATTTCAGGACAAC | TF |
| rsmAFHindIII | GCGCAAGCTTCGGCAACATCACCACGCTGG | Mutant construction |
| rsmARXbaI | GCGCTCTAGAGCACGGTGATCCTGCAGACC | Mutant construction |
| rsmASOEF | GCGTGAGGAGAAAGGAATGCTGCAT TAATTTTTATCTAATTTTCCCTTTGC | Mutant construction |
| rsmASOER | GCAAAGGGAAAATTAGATAAAAATT AATGCAGCATTCCTTTCTCCTCACGC | Mutant construction |
| algUKOF | GCGCGAGCTCGACAGCAACTCGATGTTCGGTC | Mutant construction |
| algUKOR | GCGCTCTAGAGTCGTACCAGGAAGCCAGCTG | Mutant construction |
| algUTJ1F2 | GCGCCCATGGGAATGCTAACCCAGGAACAGGATCA | Complementation |
| algU HindIIIR | GCGCAAGCTTTCAGGCTTCTCGCAACAAAGGCTGCA | Complementation |
| algRTJ1F2 | GCGCCCATGGGAATGAATGTCCTGATTGTCGATGAC | Overexpression |
| rpoS EcoRIF | GCGCGAATTCGATGGCACTCAAAAAAGAAGGGCC | Mutant construction |
| rpoS HindIIIR | GCGCAAGCTTTCACTGGAACAGCGCGTCACT | Mutant construction |
| rpoS intF | CGTCGCTGACCGGGAGTTCG | PCR check |
| rpoS intR | CATTTATCTACTTAGGCTCACACG | PCR check |
| rsmASDMcheckF | GCCAAGGTTTCCATCGTCGG | PCR check |
| rsmAHAF | TACCCATACGATGTTCCAGATTACGCTTAATTTTTATCTAATTTTCCCTTTGC | HA allele construction |
| rsmAHAR | TTAAGCGTAATCTGGAACATCGTATGGGTAATGGTTTGGCTCTTGATCTTTCTC | HA allele construction |
| rsmASDMAlgUF | CAGAATTCTTCATTCCGGCGGGACTG | SDM |
| rsmASDMAlgUR | CCAAACGGTCAAAAACGACAAAAG | SDM |
| rsmAPE1F | GCGCGGTACCAAGGATCGCGCTCTTGATTTCTGCGGATCCGCCGCCATTTCTT | TF |
| rsmAPE3F | GCGCGGTACCGCTGACAGGCGAAAGGCG | TF/RPA |
| rsmAPE3R | GCGCGGATCCTCACCCAGTATTGACCAGTCC | TF |
| rsmAEcoRIR | GCGCGAATTCACGAGTCAGAATCAGCATTCCTTTC | TF |
| rsmAPE1 | GGCGCGTTGACGCCGATGCGC | Primer extension |
| rsmAPE3 | CACGCGAATATTTCAGGACAACAGTCTG | Primer extension |
| rsmAPE4 | GCGCGATCCTTCACCCAGTATTGACCAGTCC | Primer extension |
| gacAF | GCGCGAGCTCGAGGCGACCTTGCGTCATTC | Mutant construction |
| gacAR | GCGCTCTAGACGGCGATCGCCGAAACCAG | Mutant construction |
| gacASOEF-2 | GCGACGAGGTGCAGCGTGATTGTGAATTCCGCCAGCTAGATGAGCG | Mutant construction |
| gacASOER-2 | CGCTCATCTAGCTGGCGGAATTCACAATCACGCTGCACCTCGTCGC | Mutant construction |
| gacAintF | CCAGGGTGCTTGCGCTTTAC | PCR check |
| gacAintR | CCACGTAGAGAAGCTTGGCC | PCR check |
| retSFSacI | GCGCGAGCTCGAACGTCGCCATCGACGTGC | Mutant construction |
| PA0082FUV5 | GCGCAAGCTTTACACTTTATGCTTCCGGCTCGTATAATGTGTGGAGTCATCCAATATTCATCAATGGC | LF |
| retSRXbaI | GCGCTCTAGAGCAGGTCTGGCTATGCCGG | Mutant construction |
| retSSOEF | GACTTCGCCGTGGTACGGCTTTCCTGAGGGCAGCGACGTGCT | Mutant construction |
| retSSOER | AGCACGTCGCTGCCCTCAGGAAAGCCGTACCACGGCGAAGTC | Mutant construction |
| retSintF | CAAGGTCGAGGCGACCTGGG | PCR check |
| retSintR | GAATAACCGCGTGCGGTTATCC | PCR check |
| lacZScaIF | GCTATGACCATGATTAGTACTGATTCACTGGCCGTC | TLF vector |
| lacZRXhoI | CCCCTCGAGCAGACATGGCCTGCCCGGTTATTA | TLF vector |
| lacZRforTF | GATGTGCTGCAAGGCGATTAAG | SEQ |
| pHERDSF | ATCGCAACTCTCTACTGTTTCTC | SEQ |
| rsmASDMAlgRF | GAATTCGGCAGGAACTTTCATTCCGGC | RPA |
| rsmARPAR | TAATACGACTCACTATAGGGAGAGATAAAAATTAATGGTTTGGCTCTTG | RPA |
Abbreviations: TF, transcriptional fusion, SDM, site-directed mutagenesis; TLF, translational fusion, LF, leader fusion, SEQ, sequencing primer; RPA, RNase protection assay.
Transcriptional and translational leader fusion analysis.
Upstream DNA fragments containing promoter regions were generated by using the primers listed in Table 2 in conjunction with Q5 polymerase (NEB, Ipswich, MA). PAO1 genomic DNA was used as the template. PCR products were cloned into pCR2.1 and then subcloned into miniCTXlacZ using the restriction enzymes HindIII/BamHI, HindIII/EcoRI, or KpnI/BamHI (New England BioLabs, Ipswich, MA). To construct rsmA transcriptional fusions, the primer pair rsmAHindIII/rsmAEcoRIR was used and the product was inserted into the SalI/EcoRI sites of miniCTXlacZ using T4 DNA ligase (NEB, Ipswich, MA). The transcriptional fusion PE3-lacZ was constructed using the primers rsmAPE3F and rsmAPE3R and cloning into the KpnI/EcoRI site of miniCTXlacZ. The transcriptional fusion TF2-lacZ was constructed using primer pair rsmAPE1F/rsmAEcoRIR and cloned into miniCTXlacZ. The translational fusion vector CTXlacZCP was constructed by amplifying the lacZ gene using the primers listed in Table 2 in order to delete the lacZ ribosome-binding site and the translational initiation codon. The tssA1 leader fusion was constructed using the lacUV5 promoter in the 5′ primer (PA0082FUV5) and primer PA0082ScaIR to amplify the tssA1 untranslated region on the basis of the data from the study of Brencic and Lory (5) and cloned into CTXlacZCP. The fusion constructs were confirmed by sequencing and conjugated into P. aeruginosa strains by triparental conjugation. Strains were selected for tetracycline resistance and then conjugated with pFLP2 to remove vector sequences (30). Strains were selected for carbenicillin resistance, grown overnight without selection, and plated on YT medium with 10% sucrose to select for the loss of pFLP2. Individual colonies were patch plated onto VBMM supplemented with 300 µg/ml carbenicillin and PIA to ensure the loss of pFLP2. To confirm the presence of the fusion constructs, PCR was performed using the forward primer used to construct the fusion and the reverse primer lacZRforTF (Table 2). β-Galactosidase activity was determined by incubating cell extracts with o-nitrophenyl-β-d-galactopyranoside (4 mg/ml) as described by Miller (32). A strain carrying the empty vector, miniCTXlacZ, was assayed, and the value for this background (28 Miller units) was subtracted from all transcriptional fusions. All mucoid strains were confirmed to be mucoid at the end of each experiment by plating on PIA plates and making sure that all colonies were mucoid. Experiments were performed in triplicate at least three times.
Site-directed mutagenesis.
Phosphorylated primers rsmASDMAlgUF and rsmASDMAlgUR were used in an inverse PCR with the PE3-lacZ transcriptional fusions as the templates. The predicted AlgU-dependent promoter in the −35 consensus sequence was mutated from GAACTT to GAATTC. The successful inverse PCR product was self-ligated, transformed into NEB5α cells (NEB, Ipswich, MA), and screened by plasmid digestion with EcoRI digestion and sequencing. The constructs obtained by site-directed mutagenesis were conjugated into Pseudomonas strains as described above.
Western blot analysis.
P. aeruginosa strains were grown in LB broth at 37°C for various times. The bacteria were collected by centrifugation, resuspended in sterile phosphate-buffered saline, and lysed by sonication. Total protein concentrations were quantified by the Bradford protein assay (Bio-Rad, Carlsbad, CA). Cell extracts (5 μg) were separated by 12% SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Bio-Rad). The membranes were blocked and probed using a 1:2,500 dilution of anti-HA monoclonal antibody (Thermo Fisher, Pittsburgh, PA), followed by a 1:30,000 dilution of horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin antibody (33). Arbitrary units were determined by standardizing the Western blots to the Coomassie-stained SDS-polyacrylamide gel of each strain using total protein analysis as described previously (34, 35). Detection was performed using an ECL Plus kit (Thermo Fisher, Pittsburgh, PA) and chemiluminescence detection (ProteinSimple, Santa Clara, CA). The blots and Coomassie-stained SDS-polyacrylamide gels were analyzed with ImageJ software. All Western blot analyses were repeated at least three times.
Primer extension.
Total RNA (15 μg) from P. aeruginosa was isolated using a Pure-Link RNA minikit (Life Technologies, Grand Island, NY). Radiolabeled primers rsmAPE1 and rsmAPE3 (Table 2) were used in a reverse transcription reaction in a ThermoScript reverse transcription-PCR system (Life Technologies, Grand Island, NY). A temperature of 55°C was used for the extension. The same primer used for primer extension was also used in a sequencing reaction with the rsmA upstream sequence in pGEM-T Easy (Promega, Madison, WI) and a Sequenase 7-deaza-dGTP kit (Affymetrix, Cleveland, OH). The products of both the primer extensions and the sequencing reactions were run on a 6% acrylamide–8 M urea gel. The gel was dried, and the extension products were detected using autoradiography. Primer extension results were confirmed using two different primers a total of four times.
In vitro transcription and RPA.
The rsmA probe for the RNase protection assay (RPA) was synthesized using a PCR product generated with primers that incorporated a T7 promoter sequence at the 5′ end and a MAXIscript T7 kit (Life Technologies). The reverse primer containing the T7 promoter sequence contained a nonhomologous sequence to discriminate between the full-length probe and protected fragments. The primers rsmASDMAlgRF and rsmARPAR were used to produce the PCR product. The probes were labeled with UTP by the use of biotin-16-UTP (Life Technologies) at a ratio of 4:6 and were gel purified after in vitro transcription. Twenty micrograms of total RNA from each P. aeruginosa strain was precipitated with 800 pg of labeled probe and resuspended in hybridization buffer {80% formamide, 400 mM NaCl, 40 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid); pH 6.4], 1 mM EDTA}, heated at 95°C for 5 min, and incubated overnight at 42°C. Negative controls consisting of the probe incubated with 10 μg Saccharomyces cerevisiae yeast RNA were also included. Digestion of unhybridized RNA was performed using RNase A at 25 μg/ml and RNase T1 at 10 U/ml. A biotinylated RNA ladder (Kerafast) was also used to determine the size of the protected probe fragments. The products of the reactions were run on a 5% acrylamide–8 M urea gel, and the gels were transferred to a positively charged membrane (Hybond N+; Amersham Biosciences) using a semidry blotting system and 0.5× Tris-borate-EDTA buffer (Hoefer, Holliston, MA). Nucleic acid was UV cross-linked, and the probes were detected using a chemiluminescent nucleic acid detection module (Thermo Scientific). After washing, the blot was incubated for 5 min and developed using a FluoroChem M system (ProteinSimple). RNase protection assays were performed at least three times.
Overexpression/complementation studies.
In order to overexpress algU, the coding sequence was amplified (Table 2) and cloned into the integrating vector pTJ1 for complementation or overexpression (36). Trimethoprim-resistant colonies were picked, cultured overnight, and diluted 1,000-fold on the following day. The presence of the gene of interest was confirmed using primers pTJ1R and either algUTJ1F2 or algRTJ1F2. Cultures requiring induction used 1% arabinose and were grown for 8 h in broth or overnight on agar plates.
Statistical analyses.
Statistical analyses were performed using GraphPad Prism (version 6.0) software (GraphPad Software, La Jolla, CA). The results of transcriptional and translational fusions and densitometry analysis were compared using a one-way analysis of variance (ANOVA) with Tukey's posttest. ImageJ software was used for Western blot analysis.
RESULTS
AlgU hyperactivity significantly contributes to rsmA expression in mucA mutant strains.
Previous studies using an rsmA translational fusion and/or Western blot analyses found that RsmA levels increase throughout the growth phases (5, 21, 28), which suggests a role of the stationary-phase sigma factor RpoS (37). Another study using a transcriptional fusion indicated that a mucA strain has increased rsmA expression (14). Because the alternative sigma factor AlgU is available in strains lacking functional MucA, AlgU has increased activity in this background, and we postulated that AlgU might play a role in rsmA expression. To further understand rsmA regulation at a transcriptional level, an rsmA transcriptional fusion, TF1-lacZ, was constructed by cloning the entire intergenic region between rsmA and lysC in the transcriptional fusion vector miniCTXlacZ (Fig. 1) (38). Due to posttranscriptional rsmA autoregulation (39), it was important that only transcription be monitored. The miniCTXlacZ vector allows insertion of single-copy lacZ fusion integrants and contains an RNase III processing site to ensure that transcription is uncoupled from posttranscriptional effects (38, 40). In wild-type strain PAO1, the TF1-lacZ fusion had β-galactosidase activity of ∼380 Miller units at 8 h (Fig. 2A). This result indicates that rsmA was expressed at low levels at this time point. The fusion was also tested at additional time points, including 16 h, and there was little change in TF1-lacZ activity (data not shown). In a ΔalgU strain, the TF1-lacZ transcriptional fusion had no decrease in TF1-lacZ activity at 8 h (Fig. 2A). A ΔrpoS mutant was constructed and assayed for rsmA expression. There was no significant decrease in β-galactosidase activity at either 8 or 16 h (Fig. 2A). A ΔalgU ΔrpoS strain was constructed, and the TF1-lacZ transcriptional fusion was assayed. No change in TF1-lacZ activity was seen in the double mutant. A ΔalgR mutant had significantly increased rsmA expression at 8 h (Fig. 2A). A previous study indicated that AlgR activates rsmA expression (14), but the strain used, PA103, and the growth conditions differed from those used in our study. Our results suggest that rsmA is expressed at low levels in wild-type strain PAO1 and its mutant derivatives.
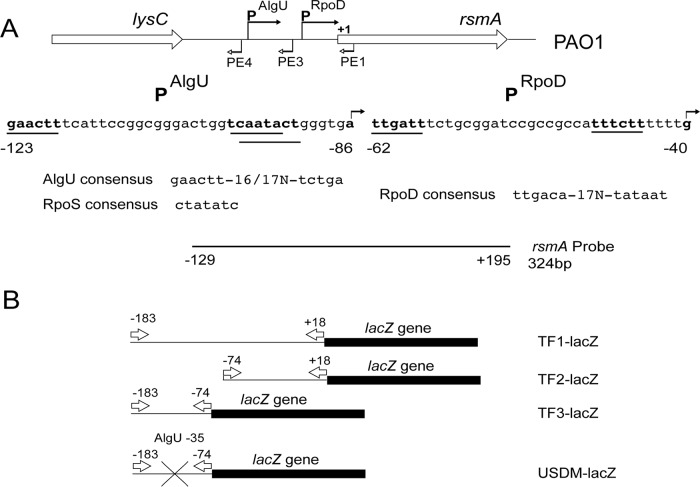
FIG 1.
Depiction of the rsmA genomic region, the promoters controlling rsmA expression, and the reagents used in this study. (A) Schematic representation of the rsmA genomic region. The sequences below the schematic represent those of the two potential promoters. The bent arrows above the sequence indicate the transcriptional start sites identified by primer extension analysis. The primers used in the primer extension experiments are identified by bent arrows below the genomic schematic. The potential promoters are indicated by a line underneath the sequence and are in bold. Potential sigma factor consensus sequences are indicated below the sequence. The probe used for RNase protection assays is indicated below, and position numbers are relative to the rsmA translational start site. (B) The rsmA-lacZ transcriptional fusions constructed are indicated. Arrows indicate the primers used in the construction of the transcriptional fusions. The position numbers above the arrows are in relation to the rsmA translational start codon. The X indicates the site-directed mutagenesis of the putative AlgU −35 consensus sequence.
FIG 2.
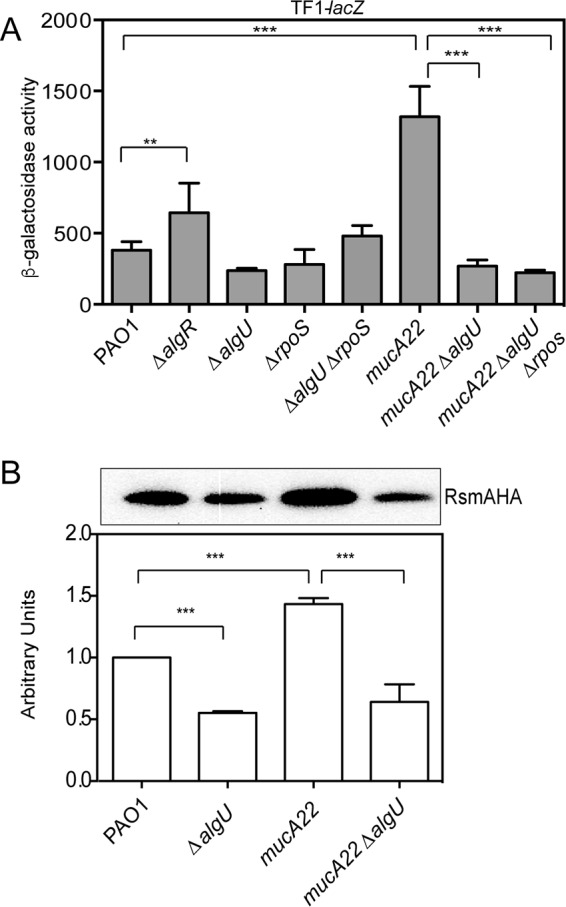
AlgU activates rsmA expression in mucA mutants. (A) Analysis of TF1-lacZ in mutant strains at 8 h of growth in LB broth. The TF1-lacZ transcriptional fusion was inserted into the attB site in the chromosomes of the indicated strains. Transcriptional fusion values are indicated as β-galactosidase activity. The values are the result of subtraction of the value for PAO1 containing the vector (miniCTXlacZ) alone (28 Miller units) from all test values. Fusion assays were all conducted at least three times. Determination of all significant differences from the results for the wild type was performed using a one-way analysis of variance and Tukey's posttest. **, P = 0.01; ***, P < 0.001. (B) (Top) Western analysis of RsmA in algU mutant strains. An rsmA-HA allele was constructed and introduced into various strains via homologous recombination, the strains were grown for 8 h, and whole-cell lysates were probed with anti-HA antibody. A single band was detected in all Western blots using the strains containing the rsmA-HA allele. A representative Western blot is shown. Western blot analysis was completed at least three times. The strains assayed are listed below the bottom panel. PAO1 without the rsmA-HA allele was run as a negative control (data not shown). (Bottom) Densitometry analysis was performed for each strain. Densitometry values, determined using ImageJ software, were standardized by dividing the intensity of the band in the Western blot by the intensity of the total protein in each lane stained with Coomassie. The result for PAO1 was set equal to 1, and the values for the other strains were normalized using the value for PAO1 as a reference. A one-way ANOVA with Tukey's posttest was used to determine statistical significance. ***, P < 0.0001.
To further investigate the role of AlgU in controlling rsmA expression, the level of TF1-lacZ fusion expression was measured in a mucA22 strain as well as the isogenic mucA22 ΔalgU nonmucoid derivative. As shown in Fig. 2A, a mucA22 strain had a >3-fold increase in rsmA expression compared to that in wild-type strain PAO1. In the mucA22 ΔalgU strain, rsmA transcriptional activity was reduced to the levels in wild-type strain PAO1 (Fig. 2A), confirming that AlgU does control rsmA expression. The analysis of the TF1-lacZ transcriptional fusion suggests that AlgU does play a role in regulating rsmA expression, but this effect is seen only when mucA is mutated and not able to sequester AlgU. In addition, RpoS does not regulate rsmA under these conditions.
To determine if RsmA protein levels mirrored the findings of rsmA expression analysis, we constructed a hemagglutinin (HA)-tagged rsmA allele that was introduced into strains by allelic exchange. As shown in Fig. 2B, the mucA22 mutant had significantly higher RsmA levels than wild-type strain PAO1. Densitometry analysis indicated that there was a significant decrease in RsmA levels in the ΔalgU mutant compared to those in wild-type strain PAO1 (Fig. 2B). The mucA22 ΔalgU mutant showed a drastic decrease in RsmA levels compared to those in the mucA22 strain (Fig. 2B). The results of Western blot analysis suggested that AlgU is required for increased rsmA expression in mucA mutant backgrounds.
Identification of a second rsmA promoter.
Because the sigma factor AlgU played a role in rsmA expression, the upstream region of rsmA was inspected for possible promoter sequences. A potential AlgU consensus sequence is located upstream of rsmA (Fig. 1A). To determine if this AlgU promoter is functional, we performed primer extension analysis. Total RNA was reverse transcribed using a radiolabeled primer (rsmAPE1) specific for a sequence located within the rsmA coding region, and extension products were detected on a denaturing polyacrylamide gel. Primer extension indicated that there were two transcriptional start sites (Fig. 3A), which suggests that rsmA has an additional promoter. The start site most distal to the coding region was identified as an A residue located 86 bp upstream of the predicted rsmA translational start codon. The first and closest transcriptional start site was a G residue located 40 bp upstream of the rsmA coding region. Primer extension analysis with an additional radiolabeled primer, rsmAPE3, produced a single extension product and confirmed the presence of the A residue identified to be a transcriptional start site 86 bp upstream of rsmA (data not shown). The second transcriptional start site was independently identified four times. Primer extension analysis with a third primer, rsmAPE4, whose sequence overlapped the most distal transcriptional start site, failed to produce an extension product, suggesting that rsmA has only two active promoters under the conditions tested.
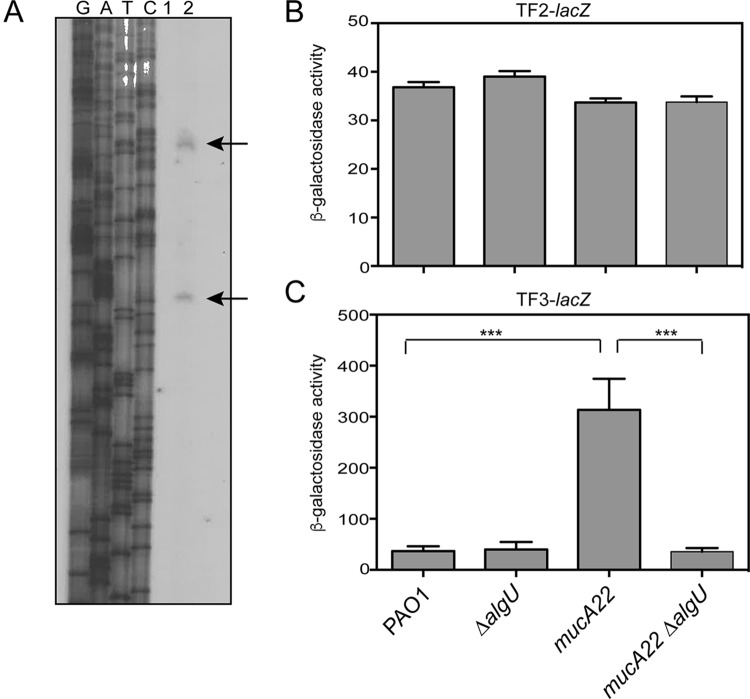
FIG 3.
Identification of a second rsmA promoter region controlled by AlgU. (A) Primer extension analysis was performed to identify the transcriptional start sites of rsmA. Total RNA was reverse transcribed using a radiolabeled primer, rsmAPE1 (Table 2), and run alongside the products of the sequencing reactions on a denaturing polyacrylamide gel. Lanes G, A, T, and C, the sequencing ladder generated using the same primer used in the reverse transcription reaction; lane 1, a control lane with total RNA without reverse transcriptase; lane 2, reverse-transcribed PAO1 RNA. Arrows, extension products. (B) β-Galactosidase activity of chromosomal transcriptional fusion TF2-lacZ in the indicated strains tested in triplicate after incubation in LB broth for 8 h at 37°C as described in the legend to Fig. 2A. The value for PAO1 carrying the vector alone was subtracted from the value for each strain. (C) β-Galactosidase activity of chromosomal transcriptional fusions of TF3-lacZ in the indicated strains tested in triplicate after incubation in LB broth for 8 h at 37°C. The value for PAO1 carrying the vector alone was subtracted from the value for each strain. Transcriptional fusion assays were performed at least three times. All significant differences from the wild type were determined using a one-way analysis of variance and Tukey's posttest. ***, P < 0.001.
Sequences upstream of the identified transcriptional start sites were examined for potential promoters (Fig. 1A). Three previous studies identified using transcriptome sequencing (25, 26) and 5′ rapid amplification of cDNA ends (24) a transcriptional start site that is 40 nucleotides upstream of the rsmA translational start codon. The sequence of the proximal promoter identified by our primer extension analysis resembled a σ70 (RpoD) promoter sequence. Therefore, the proximal promoter that we identified was the same promoter previously identified, validating the findings of our primer extension analysis.
The second transcriptional start site has two possible promoter consensus regions upstream. A potential AlgU consensus sequence GAACTT in the −35 region (Fig. 1A) (11) and a potential RpoS −10 region are located upstream of this transcriptional start site. The potential RpoS consensus sequence CAATACT differs from the RpoS consensus sequence CTATACT by 1 nucleotide (41). E. coli RpoS promoters frequently have an extended structure, denoted by a G/T residue directly upstream of the conserved −10 region (42). The potential RpoS promoter upstream of rsmA also follows this extended structure (Fig. 1A). The potential RpoS −10 region overlaps the potential AlgU −10 region (Fig. 1A). On the basis of the results of primer extension and sequence analyses, rsmA has a second promoter that is controlled by AlgU and/or RpoS.
AlgU controls the distal rsmA promoter.
Because two rsmA transcripts were detected by primer extension (Fig. 3A), this led us to test whether rsmA has two promoters. Two transcriptional fusions were constructed using the data generated by primer extension analysis (Fig. 1) to confirm the promoter activity of these regions. The activity of PAO1 carrying the vector alone (28 Miller units) was subtracted from the values obtained. A transcriptional fusion, TF2-lacZ, was constructed by deleting the putative distal promoter. TF2-lacZ has the 5′ end at position −74 in relation to the rsmA translational start site and extends 18 nucleotides into the rsmA coding region (Fig. 1B). The transcriptional fusion TF3-lacZ was constructed using the intergenic region from lysC to the distal transcriptional start site at position +1 as the 3′ end of the fusion and therefore contains only the putative distal promoter (Fig. 3B). When these fusions were analyzed in wild-type strain PAO1, both fusions produced β-galactosidase activity (Fig. 3B and C), but the level of expression was very low. The values for both the TF2-lacZ and TF3-lacZ fusions were much lower than the value for TF1-lacZ, suggesting that the entire region is necessary for optimal rsmA expression.
The TF2-lacZ transcriptional fusion was constructed and tested to determine the activity of the proximal promoter. The TF2-lacZ fusion did not have increased reporter activity in the mucA22 strain (Fig. 4B). When algU was deleted in either background, no decrease in reporter activity was detected, indicating that this promoter was constitutively active at low levels in the strains analyzed (Fig. 4C).
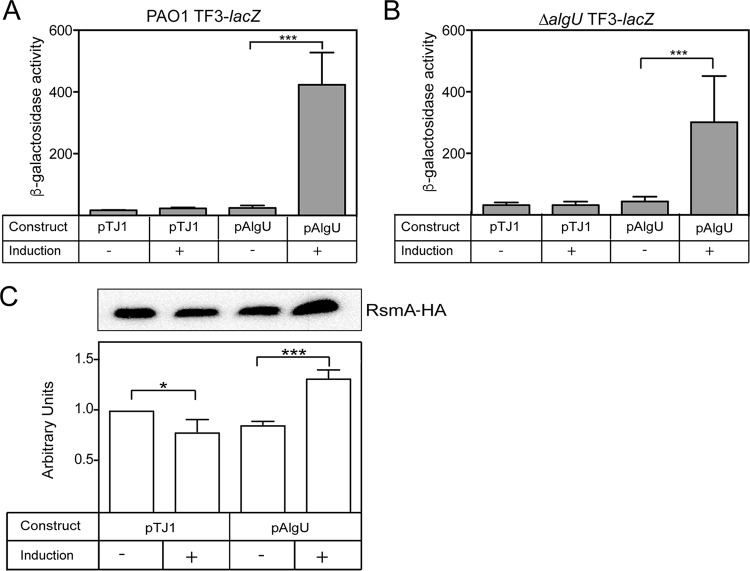
FIG 4.
AlgU overexpression and complementation confirm the AlgU regulation of rsmA. (A and B) β-Galactosidase activity of the TF3-lacZ (A) and ΔalgU TF3-lacZ (B) constructs. (A) The algU gene was cloned into the integrating vector pTJ1 and induced with 1% arabinose. Strains were grown in LB broth for 4 h and induced with arabinose for 4 h. PAO1 with the vector alone (pTJ1) was used as a negative control. Data from triplicate experiments were averaged after subtraction of the value for PAO1 carrying the vector alone (28 Miller units). (B) Complementation of the ΔalgU strain containing the transcriptional fusion TF3-lacZ was performed as described in the legend to panel A. (C) Overexpression of algU increased RsmA protein levels. PAO1-HA/pTJ1 was the PAO1 strain with the rsmA-HA allele and the vector alone. PAO1-HA/pAlgU contained the rsmA-HA allele and overexpressed algU when arabinose was added. ImageJ software was used for densitometry analysis, as described in the legend to Fig. 2B. (Top) A representative Western blot. Western blot analysis was performed at least three times. (Bottom) Densitometry results for the Western blots.
To determine if AlgU affected the distal promoter that we identified, the TF3-lacZ fusion was analyzed in wild-type strain PAO1 and a ΔalgU mutant. The activity of the TF3-lacZ fusion in PAO1 was low, and no difference in activity was seen when it was tested in a ΔalgU mutant at 8 h. However, when the activity of the TF3-lacZ fusion was tested in a mucA22 strain, there was an ∼8-fold increase (Fig. 3C). The activity of the TF3-lacZ transcriptional fusion in a mucA22 ΔalgU strain was significantly reduced ∼8-fold (Fig. 3C). These results show that the promoter contained in the TF3-lacZ fusion is AlgU dependent.
Overexpression and complementation of algU increase rsmA expression.
For overexpression of algU, we used an arabinose-inducible promoter to control algU expression (43). As shown in Fig. 4A, there was a statistically significant increase in TF3-lacZ activity when algU was induced with arabinose in strain PAO1. The results suggest a role for AlgU in activating rsmA expression.
Additionally, the ΔalgU mutant containing the TF3-lacZ transcriptional fusion was complemented with pAlgU. When 1% arabinose was used to induce algU expression, TF3-lacZ activity was also increased (Fig. 4B). Complementation and overexpression resulted in an increase in TF3-lacZ activity. From these results, we conclude that AlgU is necessary for increased rsmA expression.
To determine if RsmA protein levels matched those of the transcriptional fusions, we used the PAO1 rsmA-HA strain containing epitope-tagged rsmA. When algU expression was induced by the addition of arabinose, there was a significant increase in RsmA levels (Fig. 4C). On the basis of this result, it was determined that algU overexpression is able to increase rsmA transcription.
Site-directed mutagenesis of the putative AlgU-binding site in the distal promoter reduces rsmA expression.
Site-directed mutagenesis was performed on the TF3-lacZ transcriptional fusion to determine if the putative AlgU consensus sequence was required for increasing rsmA expression. The putative AlgU −35 sequence GAACTT was converted to GAATTC (underlined nucleotides indicate changes introduced) (Fig. 5). There was a >3-fold decrease in the level of transcription in the mucA22 strain with the mutagenized AlgU promoter (Fig. 5). The results of the site-directed mutagenesis experiment demonstrate the necessity of the conserved AlgU −35 sequence and strongly suggest that AlgU directly controls rsmA transcription by binding the distal rsmA promoter.
FIG 5.
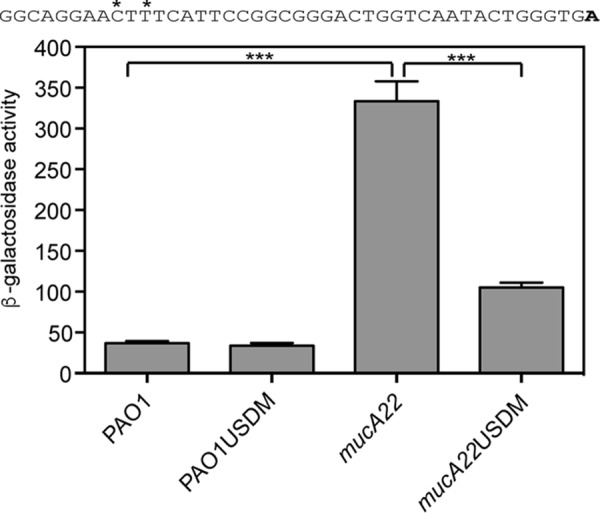
The potential AlgU-binding site is required for increased rsmA expression in a mucA22 mutant. The TF3-lacZ transcriptional fusion was engineered with two mutations in the potential AlgU −35 consensus sequence (indicated by asterisks above the indicated nucleotides in the sequence above the graph). The transcriptional start site (the 3′ end of the TF3-lacZ fusion) is indicated in bold. USDM indicates the fusion containing the mutation of the conserved AlgU −35 consensus sequence. β-Galactosidase activity was determined as described in Materials and Methods. Data from triplicate experiments were averaged after subtraction of the value for PAO1 carrying the vector (28 Miller units). A one-way ANOVA with Tukey's posttest was used to determine statistical significance. ***, P < 0.0001.
AlgU regulation of rsmA is conserved.
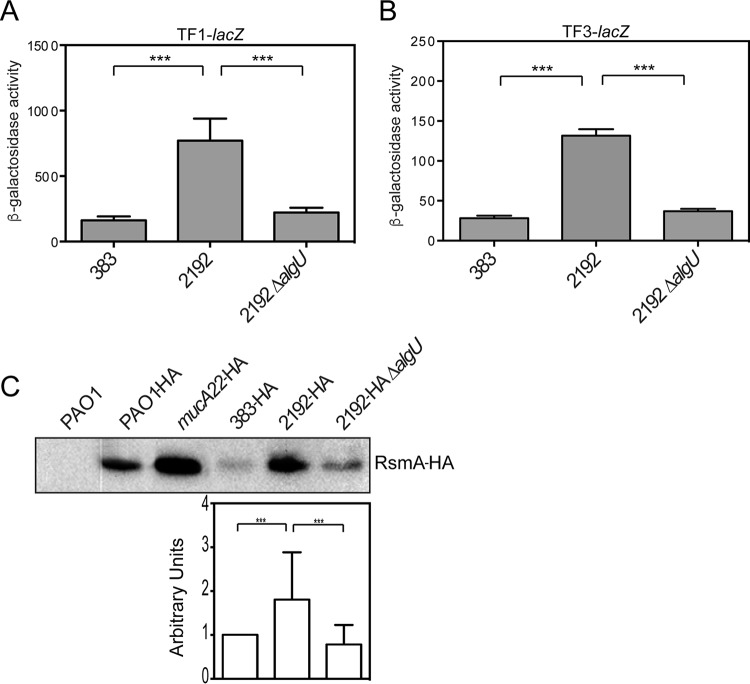
To determine if AlgU control over rsmA is a conserved mechanism in other P. aeruginosa strains, we investigated the role of AlgU in the clinical strains 2192 and 383. These strains were isolated from a longitudinal study (44) and have been determined to be isogenic (45). Strain 2192 is mucoid and has a mucA mutation (46). We took a genetic approach and assayed our transcriptional fusions TF1-lacZ and TF3-lacZ to determine the role of AlgU in activating rsmA expression in these strains (Fig. 6). Strain 2192 had an ∼3-fold increase in the amounts of both the TF1-lacZ fusion and the TF3-lacZ fusion compared to those in nonmucoid strain 383 (Fig. 6A and B). These results demonstrate that the AlgU-dependent promoter activity is increased in strain 2192 compared to that in its nonmucoid counterpart, strain 383. A 2192 ΔalgU mutant was constructed, and assays for the expression of the TF1-lacZ and TF3-lacZ fusions were performed to confirm a role for AlgU in other P. aeruginosa strains. As shown in Fig. 6A and B, there was a statistically significant decrease in the levels of TF1- and TF3-lacZ expression, supporting the role of AlgU in regulating the distal promoter. To confirm the transcriptional fusion results, the levels of RsmA in the clinical strains were determined using the rsmA-HA allele. RsmA levels were increased in strain 2192 compared to those in the nonmucoid 383 parent strain (Fig. 6C), suggesting that AlgU activated rsmA expression in strain 2192. The 2192 ΔalgU mutant had a significant decrease in the amount of RsmA compared to that in 2192 (Fig. 6C). On the basis of these results, it is likely that AlgU activates rsmA expression in multiple strains.
FIG 6.
Clinical mucA mutant strains have increased rsmA expression. (A and B) β-Galactosidase activities of the TF1-lacZ (A) and TF3-lacZ (B) transcriptional fusions. (A) The β-galactosidase activity of the TF1-lacZ transcriptional fusion was assayed in clinical isolates 383 and 2192 and a 2192 ΔalgU strain after 8 h, as described in the text. Data from triplicate experiments were averaged after subtraction of the value for PAO1 carrying the vector alone (28 Miller units). (B) The β-galactosidase activity of the TF3-lacZ transcriptional fusion was analyzed in strains 383, 2192, and 2192 ΔalgU at 8 h, as described in the text. Data from triplicate experiments were averaged after subtraction of the value for PAO1 carrying the vector alone (28 Miller units). A one-way ANOVA with Tukey's posttest was used to determine statistical significance. ***, P < 0.0001 (C) RsmA levels are also increased in clinical mucA mutants. The rsmA-HA allele was exchanged for the wild-type rsmA allele in clinical isolates 383 and 2192. Whole-cell lysates were produced and probed with anti-HA antibodies. PAO1 was used as a negative control (data not shown). The results for PAO1-HA and mucA22-HA are shown for reference. (Top) A representative Western blot. (Bottom) Results of densitometry analysis of the blot performed using ImageJ software. Data are in arbitrary units and were produced from three Western blots. Densitometry analysis was performed as described in the legend to Fig. 2B.
RpoS involvement in the control of rsmA.
There are conflicting data regarding RpoS control of rsmA expression (9, 27). The potential RpoS-binding site upstream of rsmA implicated RpoS in the control of rsmA (Fig. 1A). However, the TF1-lacZ transcriptional fusion analysis did not demonstrate a decrease in rsmA expression in a ΔrpoS mutant. Because sigma factor competition on the distal promoter could have influenced our ability to detect RpoS activity on the rsmA promoter (47), we tested a double mutant, the ΔalgU ΔrpoS mutant (Fig. 1A). No further decrease in TF1-lacZ activity in the ΔalgU ΔrpoS mutant was detected. To further define the role of RpoS in controlling rsmA expression, we tested the TF2- and TF3-lacZ fusions for activity in either ΔrpoS strains or ΔalgU ΔrpoS strains (Fig. 7). Interestingly, both a ΔrpoS mutant and a ΔalgU ΔrpoS mutant had significantly higher levels of activity when the TF2-lacZ promoter was used (Fig. 7A), suggesting that RpoS has a negative effect on transcription from the proximal rsmA promoter. No significant change was found when the TF3-lacZ transcriptional fusion was used in the ΔrpoS or ΔalgU ΔrpoS mutant strains (Fig. 7B). From these results, the role of RpoS in regulating rsmA is inconclusive.
FIG 7.
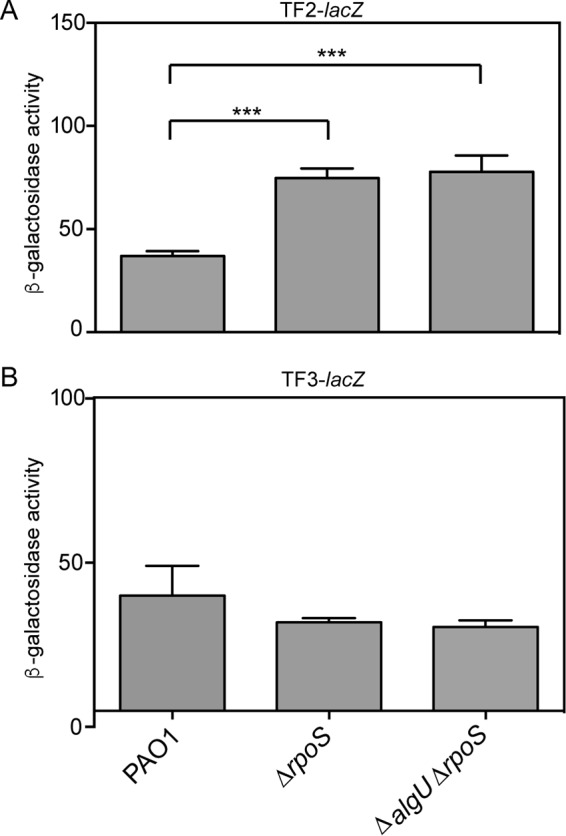
RpoS is not a significant regulator of rsmA expression. (A) β-Galactosidase activity of the TF2-lacZ transcriptional fusion in the denoted strains. Data from triplicate experiments were averaged after subtraction of the value for PAO1 carrying the vector alone (28 Miller units). (B) β-Galactosidase activity of the TF3-lacZ transcriptional fusion in the denoted strains. β-Galactosidase activity was determined as described in the text. Data from triplicate experiments were averaged after subtraction of the value for PAO1 carrying the vector alone (28 Miller units). Significant differences from the results for the wild type were determined using a one-way analysis of variance and Tukey's posttest. ***, P < 0.001.
RpoS and AlgU are necessary for rsmA expression from the distal promoter.
RNase protection assays (RPAs) were performed using an RNA probe specific for a region spanning the entire rsmA coding region and 129 bp upstream of the rsmA gene (Fig. 1A) to more directly analyze rsmA transcripts and better understand the role of AlgU and RpoS in controlling rsmA expression. The probe also includes non-template-encoded nucleotides to distinguish the full-length probe from protected fragments. As shown in Fig. 8, the RPAs confirmed the findings of our primer extension and transcriptional fusion analysis by detecting two rsmA transcripts in wild-type strain PAO1. The expected fragments were 281 bp and 235 bp, on the basis of the probe used. This matched what was seen in the RPAs (Fig. 8). Interestingly, the ΔalgU mutant contained the same two transcripts as wild-type strain PAO1. When the ΔrpoS mutant was analyzed, both transcripts were detected, with the amount of the first transcript resulting from the amount of the proximal promoter being increased. A ΔalgU ΔrpoS strain was also analyzed, and the larger transcript resulting from the distal promoter completely disappeared, demonstrating that both AlgU and RpoS contribute to transcription from the distal promoter (Fig. 8). The mucA22 strain had both transcripts as well, with both transcripts having greater intensities. The highest band in the ΔrpoS and mucA22 strains was likely undigested probe.
FIG 8.
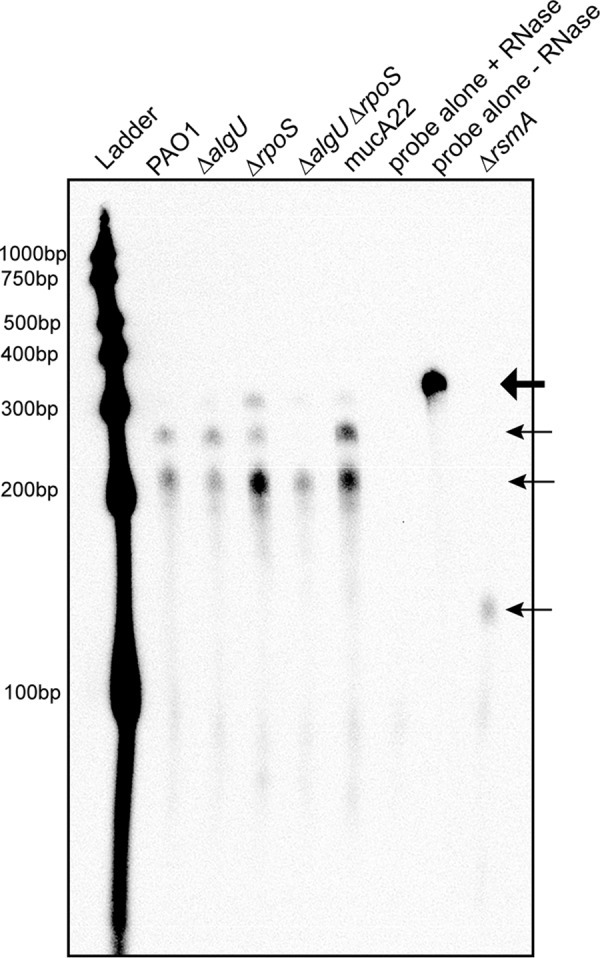
RPAs confirm the presence of two rsmA transcripts. RPAs of the indicated strains were performed after RNA was harvested from cultures grown in LB broth for 8 h at 37°C. Twenty micrograms of total RNA from the indicated strains was hybridized with 800 pg of biotinylated probe and subjected to RNase treatment. After RNase inactivation, RNA was precipitated from the samples and run on a 5% denaturing polyacrylamide gel, transferred to a nylon membrane, UV cross-linked, and developed using chemiluminescent detection. The undigested probe is indicated by the large arrow at the top. The protected rsmA probe is indicated by the three smaller arrows. Shown is a result representative of results from three RNase protection assays. Ladder, molecular size markers.
Three controls were included in the RPA. Two of the controls were the probe incubated with yeast RNA; one of these was treated with RNase, and the other was not. As shown in Fig. 8, RNase treatment completely degraded the probe, which indicates that the probe did not bind nonspecifically. The intact probe can be seen at ∼330 bp in the lane in Fig. 8 labeled “probe alone − RNase,” which is consistent with the additional nucleotides that are non-template encoded. A ΔrsmA mutant was constructed and used as an additional control. In the ΔrsmA mutant, there was only one discernible protected probe fragment at ∼150 bp. On the basis of how the ΔrsmA mutant was constructed, a protected fragment of 144 bp was expected. The results obtained with the ΔrsmA strain support the suggestion that there are only two rsmA transcripts, at least under the conditions used. Altogether, the results of the RPA suggested that there are two rsmA transcripts and that both AlgU and RpoS are involved in controlling the distal promoter region, suggesting overlapping promoters.
AlgU control of rsmA expression is responsible for RsmA activity in a P. aeruginosa mucA22 strain.
Because AlgU increased RsmA levels, we hypothesized that RsmA activity was also increased in a mucA22 strain. We analyzed a variety of mutant strains, including strains that should alter the activity of RsmA on the basis of the current RsmA paradigm (5). RetS interferes with GacA phosphorylation, and mutation of retS should lead to increased production of the small RNAs RsmY and RsmZ (18, 48). Therefore, the level of RsmA activity should be reduced. In contrast, mutation of gacA should lead to a reduction in the amounts of the small RNAs RsmY and RsmZ (49). In this case, increased RsmA activity should be observed. Mutants with mutations in the gacA and retS genes were constructed in both the PAO1 and mucA22 strain backgrounds.
RsmA has previously been shown to affect the production of the exopolysaccharide genes pel and psl (9, 16, 50). However, no studies have investigated the involvement of RsmA in alginate production. We hypothesized that if RsmA inhibits biofilm formation, then RsmA would inhibit alginate production. Interestingly, the mucA22 ΔgacA, mucA22 ΔretS, and mucA22 ΔrsmA mutants still overproduced alginate (data not shown), and we conclude that RsmA does not affect alginate production.
RsmA posttranscriptional activity was tested using a tssA1 leader/translational fusion containing the lacUV5 promoter and the tssA1 untranslated sequence, based on the study of Brencic and Lory (5) (Fig. 9A). Due to the low level of activity of this reporter, the data are presented as a percentage of the wild-type activity, where the values were normalized to those for the PAO1 strain (12 Miller units). The mucA22 strain had fusion activity decreased ∼3-fold compared to that of PAO1 (Fig. 9B). There was no difference in activity between PAO1 and a ΔalgU strain when the leader fusion was used (Fig. 9B). In the case of a mucA22 ΔalgU strain, there was a 5.5-fold increase in leader fusion activity compared to that in the mucA22 strain (Fig. 9B). These results suggest that RsmA is active in the mucA22 strain background and this is due to AlgU regulation of rsmA.
FIG 9.
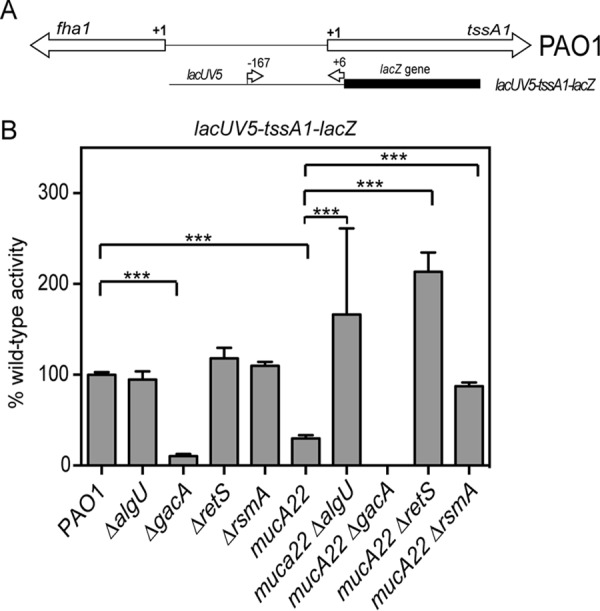
AlgU activation of rsmA leads to changes in tssA1 expression. (A) Schematic of the genomic region of the RsmA-controlled genes tssA1 and fha1. Below the genomic schematic is a depiction of the leader fusion that was constructed. The position numbers above the fusion construct indicate those of the primers used relative to the tssA1 translational start codon. (B) Analysis of a TssA1 leader translational fusion driven by the lacUV5 promoter in wild-type and mutant derivative strains. All assays were repeated at least three times in triplicate. Data were normalized to the mean for PAO1. PAO1 activity with the lacUV5-tssA1-lacZ fusion was 12 Miller units. A one-way ANOVA with Tukey's posttest was used to determine statistical significance. ***, P < 0.0001.
Further confirmation of RsmA activity in the mucA22 strain was carried out using mutants with mutations in gacA and retS, as described above. While the differences in activity were not significant by statistical analysis, there was no activity from a mucA22 ΔgacA strain, even after the reaction was allowed to proceed overnight. This result would be expected if RsmA activity were increased due to the absence of the small RNAs RsmY and RsmZ (49). As there was an increased amount of RsmA in the mucA22 strain due to AlgU activation of the distal promoter, the difference between the mucA22 strain and the mucA22 ΔgacA strain should not be so different if our hypothesis is correct (Fig. 10). The leader fusion also had increased activity in both the mucA22 ΔrsmA and the mucA22 ΔretS strains compared to that in the mucA22 strain (3-fold and 7-fold, respectively) (Fig. 9B). These results also indicate the RsmA activity in the mucA22 strain background. The increased activity of the leader fusion indicates that the absence of RsmA or an abundance of RsmY and RsmZ due to the absence of RetS allows greater expression from the leader fusion. Altogether, these data suggest that RsmA activity is present in a mucA22 strain.
FIG 10.
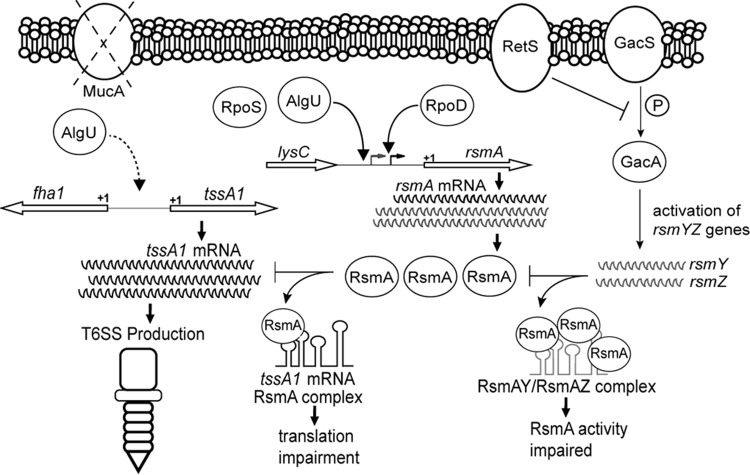
Model depicting the expression and activity of rsmA and the protein that it encodes, RsmA, in mucA mutant strains. Two transcripts of different sizes are produced. AlgU and RpoS control the longer transcript originating from the distal promoter. In the mucA strain background, AlgU increases the level of transcription from the distal promoter, but both transcripts are increased in amount either by transcription initiation at both promoters or by posttranscriptional processing of the longer message. Due to increased rsmA transcription, there is an increased amount of RsmA in mucA mutants. Transcriptional start sites are represented by bent arrows. The left and right bent arrows indicate the distal and proximal promoters, respectively. The +1 indicates the putative translational rsmA start site. RsmA levels increase, preventing translation of tssA1 and therefore production of the HSI-I T6SS. The dashed curved arrow on the left side indicates that AlgU either directly or indirectly leads to the activation of the tssA1 operon. RetS and GacA are depicted on the right to show how they are incorporated in the RsmA scenario. GacA, when phosphorylated by GacS, activates the expression of two small RNAs, RsmY and RsmZ, that bind and prevent RsmA from performing posttranscriptional regulation. RetS interferes with GacA phosphorylation by physically binding GacS and allows RsmA to regulate expression of mRNAs. The small RNAs RsmY and RsmZ bind and prevent RsmA activity.
DISCUSSION
RsmA is considered an important posttranscriptional regulator involved in the positive regulation of acute virulence factors, such as the type III secretion system and motility (5, 14). Little is known of the transcriptional regulation of rsmA. Initially, we undertook studies to determine how rsmA was regulated in P. aeruginosa. We demonstrate that AlgU activates rsmA expression and increases RsmA levels in a mucA22 mutant and a clinical isolate. Analysis of a leader fusion to assess the posttranscriptional activity of the RsmA target, tssA1 (5), led us to discover RsmA activity in the mucA22 strain. To our knowledge, this is the first demonstration of RsmA activity in a mucA mutant strain of P. aeruginosa. Our work demonstrates that increased rsmA expression due to AlgU activation leads to changes in the RsmA regulon. We hypothesize that RsmA plays an important role during P. aeruginosa chronic infection and that there are additional RsmA targets in mucA mutant strains.
One of the goals of this study was to determine how rsmA expression is regulated. Previous studies have raised questions about how rsmA is regulated in P. aeruginosa (9, 14, 24). In E. coli, five promoters control csrA (the orthologue of rsmA) expression (51). Only one rsmA promoter was previously identified in P. aeruginosa (24–26), and a previous study indicated that AlgR plays a role in regulating rsmA expression (14). Using primer extension, a new transcriptional start site was identified, indicating a second rsmA promoter (Fig. 2). Both transcriptional fusion analysis and RPA confirmed the presence of a second promoter (Fig. 3 and 8). Overexpression and Western blot analyses support a role of AlgU in controlling rsmA expression (Fig. 1E and 4). However, when MucA is functional, such as in PAO1, it is difficult to detect changes in rsmA expression. This explains the low levels of activity of the transcriptional reporters (Fig. 1A and 2). The role of RpoS in controlling rsmA expression is somewhat less clear. While we were not able to detect RpoS regulation of rsmA by transcriptional fusion analysis (Fig. 7), the results of RPA suggested that RpoS does play a role (Fig. 8). Further studies are needed to determine the significance of RpoS in controlling rsmA expression. A previous study identified AlgR to be a regulator of rsmA, rsmY, and rsmZ (14). It is possible that AlgR and AlgU work together in regulating rsmA in mucA mutant strains. It is clear that rsmA regulation is complex, but this work solidifies the role of AlgU as an rsmA activator.
Our finding that AlgU controls rsmA expression is bolstered by the analysis of rsmA in the clinical strains 2192 (mucoid) and 383 (nonmucoid) (45). These data indicate higher levels of RsmA in the mucA mutant strain 2192 (Fig. 6). The findings of our regulation studies are consistent with a previous observation that a mucA mutant had increased levels of rsmA expression (14). Thus, our work implicates RsmA to be necessary for the persistence of P. aeruginosa during chronic infections, because mucA mutants are found at later time points during infection. This is consistent with the requirement of Helicobacter pylori, which requires CsrA (the RsmA orthologue) for persistence in the stomach (52). Whether RsmA participates in regulating virulence factors in mucA mutant strains is currently not known and is an unexplored area.
After finding that AlgU, an important sigma factor in chronic infections (7), regulates rsmA expression, we asked if the increased RsmA levels have activity in mucA mutant strains. Our results support a model (Fig. 10) where RsmA activity in the mucA mutant background is due to AlgU. First, using the tssA1 leader fusion, the mucA22 strain showed decreased activity, indicating increased posttranscriptional regulation compared to that in PAO1. Second, the mucA22 ΔalgU strain had increased leader fusion activity compared to that of the mucA22 strain, which supports our conclusion that increased rsmA expression leads to increased RsmA activity. This result also demonstrates the importance of AlgU control of rsmA expression. Third, the increase in the fusion activity in the mucA22 ΔrsmA strain compared to that in the mucA22 strain was greater than the increase in the ΔrsmA mutant of PAO1. Further support for RsmA activity in mucA mutant strains comes from the increased leader fusion activity of the mucA22 ΔrsmA and mucA22 ΔretS strains compared to that of the mucA22 strain. An alternative explanation of our data is that some other form of posttranscriptional regulation occurs independently of RsmA. However, the gacA and retS mutants are known to regulate RsmA activity, even though rsmA is expressed in these strains (data not shown). We do not favor this alternative hypothesis, as previous studies have determined that GacA controls only rsmY and rsmZ expression (21, 49). Another posttranscriptional regulator, RsmF/RsmN, is also known to control tssA1 expression (24, 53). This could possibly have affected the fusion results found in the present study as well and may explain the increased expression of the tssA1 leader fusion in the mucA22 ΔretS strain. Regardless, the results still indicate increased posttranscriptional regulation of tssA1 in a mucA22 strain.
The results of a proteomic study of the isogenic clinical strains 383 and 2192 also support our findings. In this study, the mucoid strain 2192 has decreased amounts of HSI-I T6SS components, whereas the nonmucoid strain has increased HSI-I T6SS components (54). If the level of RsmA was increased due to AlgU in mucoid strain 2192, as presented here (Fig. 6), then the proteomic analysis suggests that RsmA is responsible for the decreased amounts of HSI-I T6SS components in mucoid strain 2192. We are currently examining RsmA activity in other clinical isolates.
However, a previous study did not find that RsmA posttranscriptionally regulates tssA1 in a mucA mutant (14). A possible reason for the discrepancy between the findings of that study and those of our study is that different strains and growth conditions were used. In addition, the previous study found that rsmY and rsmZ expression are increased in a PA103 mucA mutant (14). We analyzed rsmY and rsmZ expression using transcriptional fusions and also found the same result (data not shown). It is not known how RsmA functions if RsmY and RsmZ levels are increased, and this requires further investigation. One possibility is that the promoter fusions do not accurately monitor the levels of the small RNAs. We are directly measuring RsmY and RsmZ levels in a mucA mutant strain to determine if this is the case. If RsmY and RsmZ levels are increased and there is still RsmA activity, then we speculate the higher-affinity target mRNAs present compete with RsmY and RsmZ. Alternatively, other factors that appropriately direct RsmA to its targets, even in the presence of RsmY and RsmZ, exist.
The P. aeruginosa mucA mutant strains provide a snapshot of some of the bacteria found in the lungs of patients with cystic fibrosis during chronic infection. The use of mucA mutant strains may help provide an understanding of the factors required for persistence and adaptation in the lung environment. For instance, the overproduction of alginate by mucA mutant strains provides a competitive advantage by sequestering free radicals, preventing complement attack, and inhibiting phagocytosis (55). Our data suggest that RsmA also plays a role in chronic infections, such as those that occur in the lungs of cystic fibrosis patients. The RsmA regulon may have additional targets in mucA mutant backgrounds that have not yet been discovered. An understanding of RsmA in both acute and chronic infections may provide new ways to combat P. aeruginosa.
ACKNOWLEDGMENTS
We thank Joseph and Julia Albright for providing equipment and supplies for these studies. We thank Joanna Goldberg (Emory University) for plasmid construct pTJ1. We thank Laraine Powers and David Hurley (ETSU) for critical review of the manuscript. We thank Eric Jones for expert technical assistance.
This article is dedicated to Joseph Albright.
REFERENCES
- 1.Sadikot RT, Blackwell TS, Christman JW, Prince AS. 2005. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am J Respir Crit Care Med 171:1209–1223. doi: 10.1164/rccm.200408-1044SO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Driscoll JA, Brody SL, Kollef MH. 2007. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67:351–368. doi: 10.2165/00003495-200767030-00003. [DOI] [PubMed] [Google Scholar]
- 3.Vakulskas CA, Potts AH, Babitzke P, Ahmer BM, Romeo T. 2015. Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol Mol Biol Rev 79:193–224. doi: 10.1128/MMBR.00052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coggan KA, Wolfgang MC. 2012. Global regulatory pathways and cross-talk control Pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr Issues Mol Biol 14:47–70. [PubMed] [Google Scholar]
- 5.Brencic A, Lory S. 2009. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol 72:612–632. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burrowes E, Baysse C, Adams C, O'Gara F. 2006. Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology 152:405–418. doi: 10.1099/mic.0.28324-0. [DOI] [PubMed] [Google Scholar]
- 7.Govan JRW, Deretic V. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev 60:539–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. 2010. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol 78:158–172. doi: 10.1111/j.1365-2958.2010.07320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramsey DM, Wozniak DJ. 2005. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol Microbiol 56:309–322. doi: 10.1111/j.1365-2958.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- 11.Firoved AM, Boucher JC, Deretic V. 2002. Global genomic analysis of AlgU (sigma(E))-dependent promoters (sigmulon) in Pseudomonas aeruginosa and implications for inflammatory processes in cystic fibrosis. J Bacteriol 184:1057–1064. doi: 10.1128/jb.184.4.1057-1064.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Firoved AM, Deretic V. 2003. Microarray analysis of global gene expression in mucoid Pseudomonas aeruginosa. J Bacteriol 185:1071–1081. doi: 10.1128/JB.185.3.1071-1081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Firoved AM, Ornatowski W, Deretic V. 2004. Microarray analysis reveals induction of lipoprotein genes in mucoid Pseudomonas aeruginosa: implications for inflammation in cystic fibrosis. Infect Immun 72:5012–5018. doi: 10.1128/IAI.72.9.5012-5018.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Intile PJ, Diaz MR, Urbanowski ML, Wolfgang MC, Yahr TL. 2014. The AlgZR two-component system recalibrates the RsmAYZ posttranscriptional regulatory system to inhibit expression of the Pseudomonas aeruginosa type III secretion system. J Bacteriol 196:357–366. doi: 10.1128/JB.01199-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lapouge K, Schubert M, Allain FH, Haas D. 2008. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol 67:241–253. [DOI] [PubMed] [Google Scholar]
- 16.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell 7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 17.Heurlier K, Williams F, Heeb S, Dormond C, Pessi G, Singer D, Camara M, Williams P, Haas D. 2004. Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J Bacteriol 186:2936–2945. doi: 10.1128/JB.186.10.2936-2945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, Bleves S, Lazdunski A, Lory S, Filloux A. 2006. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci U S A 103:171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schubert M, Lapouge K, Duss O, Oberstrass FC, Jelesarov I, Haas D, Allain FH. 2007. Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat Struct Mol Biol 14:807–813. doi: 10.1038/nsmb1285. [DOI] [PubMed] [Google Scholar]
- 20.Valverde C, Lindell M, Wagner EG, Haas D. 2004. A repeated GGA motif is critical for the activity and stability of the riboregulator RsmY of Pseudomonas fluorescens. J Biol Chem 279:25066–25074. doi: 10.1074/jbc.M401870200. [DOI] [PubMed] [Google Scholar]
- 21.Kay E, Humair B, Denervaud V, Riedel K, Spahr S, Eberl L, Valverde C, Haas D. 2006. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J Bacteriol 188:6026–6033. doi: 10.1128/JB.00409-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulcahy H, O'Callaghan J, O'Grady EP, Adams C, O'Gara F. 2006. The posttranscriptional regulator RsmA plays a role in the interaction between Pseudomonas aeruginosa and human airway epithelial cells by positively regulating the type III secretion system. Infect Immun 74:3012–3015. doi: 10.1128/IAI.74.5.3012-3015.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mulcahy H, O'Callaghan J, O'Grady EP, Macia MD, Borrell N, Gomez C, Casey PG, Hill C, Adams C, Gahan CG, Oliver A, O'Gara F. 2008. Pseudomonas aeruginosa RsmA plays an important role during murine infection by influencing colonization, virulence, persistence, and pulmonary inflammation. Infect Immun 76:632–638. doi: 10.1128/IAI.01132-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marden JN, Diaz MR, Walton WG, Gode CJ, Betts L, Urbanowski ML, Redinbo MR, Yahr TL, Wolfgang MC. 2013. An unusual CsrA family member operates in series with RsmA to amplify posttranscriptional responses in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 110:15055–15060. doi: 10.1073/pnas.1307217110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dotsch A, Eckweiler D, Schniederjans M, Zimmermann A, Jensen V, Scharfe M, Geffers R, Haussler S. 2012. The Pseudomonas aeruginosa transcriptome in planktonic cultures and static biofilms using RNA sequencing. PLoS One 7:e31092. doi: 10.1371/journal.pone.0031092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wurtzel O, Yoder-Himes DR, Han K, Dandekar AA, Edelheit S, Greenberg EP, Sorek R, Lory S. 2012. The single-nucleotide resolution transcriptome of Pseudomonas aeruginosa grown in body temperature. PLoS Pathog 8:e1002945. doi: 10.1371/journal.ppat.1002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulz S, Eckweiler D, Bielecka A, Nicolai T, Franke R, Dotsch A, Hornischer K, Bruchmann S, Duvel J, Haussler S. 2015. Elucidation of sigma factor-associated networks in Pseudomonas aeruginosa reveals a modular architecture with limited and function-specific crosstalk. PLoS Pathog 11:e1004744. doi: 10.1371/journal.ppat.1004744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pessi G, Williams F, Hindle Z, Heurlier K, Holden MT, Camara M, Haas D, Williams P. 2001. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J Bacteriol 183:6676–6683. doi: 10.1128/JB.183.22.6676-6683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higuchi R. 1989. Using PCR to engineer DNA, p 61–70. In Erlich HA. (ed), PCR technology: principles and applications for DNA amplification. Stockton Press, New York, NY. [Google Scholar]
- 30.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 31.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller JF. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 33.Deretic V, Leveau JH, Mohr CD, Hibler NS. 1992. In vitro phosphorylation of AlgR, a regulator of mucoidy in Pseudomonas aeruginosa, by a histidine protein kinase and effects of small phospho-donor molecules. Mol Microbiol 6:2761–2767. doi: 10.1111/j.1365-2958.1992.tb01455.x. [DOI] [PubMed] [Google Scholar]
- 34.Aldridge GM, Podrebarac DM, Greenough WT, Weiler IJ. 2008. The use of total protein stains as loading controls: an alternative to high-abundance single-protein controls in semi-quantitative immunoblotting. J Neurosci Methods 172:250–254. doi: 10.1016/j.jneumeth.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eaton SL, Roche SL, Llavero Hurtado M, Oldknow KJ, Farquharson C, Gillingwater TH, Wishart TM. 2013. Total protein analysis as a reliable loading control for quantitative fluorescent Western blotting. PLoS One 8:e72457. doi: 10.1371/journal.pone.0072457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Damron FH, McKenney ES, Barbier M, Liechti GW, Schweizer HP, Goldberg JB. 2013. Construction of mobilizable mini-Tn7 vectors for bioluminescent detection of gram-negative bacteria and single-copy promoter lux reporter analysis. Appl Environ Microbiol 79:4149–4153. doi: 10.1128/AEM.00640-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Potvin E, Sanschagrin F, Levesque RC. 2008. Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol Rev 32:38–55. doi: 10.1111/j.1574-6976.2007.00092.x. [DOI] [PubMed] [Google Scholar]
- 38.Becher A, Schweizer HP. 2000. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques 29:948–950, 952. [DOI] [PubMed] [Google Scholar]
- 39.Jean-Pierre F, Perreault J, Deziel E. 2015. Complex autoregulation of the post-transcriptional regulator RsmA in Pseudomonas aeruginosa. Microbiology 161:1889–1896. doi: 10.1099/mic.0.000140. [DOI] [PubMed] [Google Scholar]
- 40.Linn T, St Pierre R. 1990. Improved vector system for constructing transcriptional fusions that ensures independent translation of lacZ. J Bacteriol 172:1077–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuster M, Hawkins AC, Harwood CS, Greenberg EP. 2004. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol Microbiol 51:973–985. doi: 10.1046/j.1365-2958.2003.03886.x. [DOI] [PubMed] [Google Scholar]
- 42.Typas A, Becker G, Hengge R. 2007. The molecular basis of selective promoter activation by the sigmaS subunit of RNA polymerase. Mol Microbiol 63:1296–1306. doi: 10.1111/j.1365-2958.2007.05601.x. [DOI] [PubMed] [Google Scholar]
- 43.Damron FH, McKenney ES, Schweizer HP, Goldberg JB. 2013. Construction of a broad-host-range Tn7-based vector for single-copy P(BAD)-controlled gene expression in gram-negative bacteria. Appl Environ Microbiol 79:718–721. doi: 10.1128/AEM.02926-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pier GB, Matthews WJ, Eardley DD. 1983. Immunochemical characterization of the mucoid exopolysaccharide of Pseudomonas aeruginosa. J Infect Dis 147:494–503. doi: 10.1093/infdis/147.3.494. [DOI] [PubMed] [Google Scholar]
- 45.Hanna SL, Sherman NE, Kinter MT, Goldberg JB. 2000. Comparison of proteins expressed by Pseudomonas aeruginosa strains representing initial and chronic isolates from a cystic fibrosis patient: an analysis by 2-D gel electrophoresis and capillary column liquid chromatography-tandem mass spectrometry. Microbiology 146(Pt 10):2495–2508. [DOI] [PubMed] [Google Scholar]
- 46.Mathee K, Narasimhan G, Valdes C, Qiu X, Matewish JM, Koehrsen M, Rokas A, Yandava CN, Engels R, Zeng E, Olavarietta R, Doud M, Smith RS, Montgomery P, White JR, Godfrey PA, Kodira C, Birren B, Galagan JE, Lory S. 2008. Dynamics of Pseudomonas aeruginosa genome evolution. Proc Natl Acad Sci U S A 105:3100–3105. doi: 10.1073/pnas.0711982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin Y, Withers TR, Wang X, Yu HD. 2013. Evidence for sigma factor competition in the regulation of alginate production by Pseudomonas aeruginosa. PLoS One 8:e72329. doi: 10.1371/journal.pone.0072329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goodman AL, Merighi M, Hyodo M, Ventre I, Filloux A, Lory S. 2009. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev 23:249–259. doi: 10.1101/gad.1739009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL, Lory S. 2009. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol Microbiol 73:434–445. doi: 10.1111/j.1365-2958.2009.06782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakuragi Y, Kolter R. 2007. Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J Bacteriol 189:5383–5386. doi: 10.1128/JB.00137-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yakhnin H, Yakhnin AV, Baker CS, Sineva E, Berezin I, Romeo T, Babitzke P. 2011. Complex regulation of the global regulatory gene csrA: CsrA-mediated translational repression, transcription from five promoters by Esigma(7)(0) and Esigma(S), and indirect transcriptional activation by CsrA. Mol Microbiol 81:689–704. doi: 10.1111/j.1365-2958.2011.07723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barnard FM, Loughlin MF, Fainberg HP, Messenger MP, Ussery DW, Williams P, Jenks PJ. 2004. Global regulation of virulence and the stress response by CsrA in the highly adapted human gastric pathogen Helicobacter pylori. Mol Microbiol 51:15–32. [DOI] [PubMed] [Google Scholar]
- 53.Morris ER, Hall G, Li C, Heeb S, Kulkarni RV, Lovelock L, Silistre H, Messina M, Camara M, Emsley J, Williams P, Searle MS. 2013. Structural rearrangement in an RsmA/CsrA ortholog of Pseudomonas aeruginosa creates a dimeric RNA-binding protein, RsmN. Structure 21:1659–1671. doi: 10.1016/j.str.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rao J, Damron FH, Basler M, Digiandomenico A, Sherman NE, Fox JW, Mekalanos JJ, Goldberg JB. 2011. Comparisons of two proteomic analyses of non-mucoid and mucoid Pseudomonas aeruginosa clinical isolates from a cystic fibrosis patient. Front Microbiol 2:162. doi: 10.3389/fmicb.2011.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pier GB, Coleman F, Grout M, Franklin M, Ohman DE. 2001. Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect Immun 69:1895–1901. doi: 10.1128/IAI.69.3.1895-1901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170:2575–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Damron FH, Qiu D, Yu HD. 2009. The Pseudomonas aeruginosa sensor kinase KinB negatively controls alginate production through AlgW-dependent MucA proteolysis. J Bacteriol 191:2285–2295. doi: 10.1128/JB.01490-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holloway BW, Krishnapillai V, Morgan AF. 1979. Chromosomal genetics of Pseudomonas. Microbiol Rev 43:73–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mathee K, Ciofu O, Sternberg C, Lindum PW, Campbell JI, Jensen P, Johnsen AH, Givskov M, Ohman DE, Molin S, Hoiby N, Kharazmi A. 1999. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology 145(Pt 6):1349–1357. doi: 10.1099/13500872-145-6-1349. [DOI] [PubMed] [Google Scholar]