Abstract
Although domestic ruminants have long been recognized as the main source of human Q fever, little is known about the lifestyle that the obligate intracellular Gram-negative bacterium Coxiella burnetii adopts in its animal host. Because macrophages are considered natural target cells of the pathogen, we established primary bovine monocyte-derived macrophages (MDM) as an in vitro infection model to study reservoir host-pathogen interactions at the cellular level. In addition, bovine alveolar macrophages were included to take cell type peculiarities at a host entry site into account. Cell cultures were inoculated with the virulent strain Nine Mile I (NMI; phase I) or the avirulent strain Nine Mile II (NMII; phase II). Macrophages from both sources internalized NMI and NMII. MDM were particularly permissive for NMI internalization, but NMI and NMII replicated with similar kinetics in these cells. MDM responded to inoculation with a general upregulation of Th1-related cytokines such as interleukin-1β (IL-1β), IL-12, and tumor necrosis factor alpha (TNF-α) early on (3 h postinfection). However, inflammatory responses rapidly declined when C. burnetii replication started. C. burnetii infection inhibited translation and release of IL-1β and vastly failed to stimulate increased expression of activation markers, such as CD40, CD80, CD86, and major histocompatibility complex (MHC) molecules. Such capability of limiting proinflammatory responses may help Coxiella to protect itself from clearance by the host immune system. The findings provide the first detailed insight into C. burnetii-macrophage interactions in ruminants and may serve as a basis for assessing the virulence and the host adaptation of C. burnetii strains.
INTRODUCTION
Coxiella burnetii, a Gram-negative obligate intracellular bacterium, is the causative agent of Q fever, a widely distributed zooanthroponosis. After an incubation time of 2 weeks, Q fever can manifest as an acute, self-limiting flulike illness with complications such as pneumonia and hepatitis. Chronic disease occurs more rarely and presents as, e.g., endocarditis or fatigue syndrome (1). Pregnant women are at high risk for developing chronic Q fever, which can result in an adverse pregnancy outcome with, e.g., spontaneous abortion or fetal death (2).
The common sources for transmission of C. burnetii to humans are domestic ruminants. Infection in animals, called coxiellosis, is mostly inapparent, but chronic infection may lead to abortions with rates ranging from 5 to 91% in infected small-ruminant flocks (1). Placental membranes of infected animals contain up to 109 organisms/g of tissue (3). Humans become infected by aerosols derived from contaminated abortion material, birth products, urine, or feces. Many Q fever outbreaks can be traced back to C. burnetii-infected small ruminants (4). The potential risk arising from cattle has been discussed repeatedly because these animals excrete, more than do small ruminants, huge amounts of C. burnetii through milk over a long time (5). Oral transmission by ingestion of contaminated raw milk or dairy products could lead to seroconversion but reportedly has resulted so far only in a few cases of Q fever (5–7).
The threat to humans is primarily determined by the number of C. burnetii particles shed from infected ruminants. The magnitude of shedding eventually relies on the interaction with placental, intestinal, and mammary epithelia (8). Knowledge about tissue dissemination of C. burnetii in ruminants is sparse, but infection studies in pregnant goats revealed that C. burnetii has a strong tropism for placental trophoblasts (9). However, the initial contact of C. burnetii with the animal host at the entry site, e.g., the respiratory mucosa, and the characteristics of the subsequent host response are decisive for controlling pathogen replication. A C. burnetii infection initiates humoral and cellular immune responses (10). Peripheral blood mononuclear cells (PBMC) from infected goats respond to C. burnetii restimulation with proinflammatory signals, e.g., upregulation of tumor necrosis factor alpha (TNF-α) and gamma interferon (IFN-γ). Despite activation of innate immune effectors, the development of adaptive immune responses upon natural infection appears to be insufficient, as goats are not capable of clearing a C. burnetii infection (11).
In the lung and placenta of infected animals, C. burnetii antigens are often found in macrophages (12–14). Members of the mononuclear phagocyte system are generally regarded as the main target cells for C. burnetii. The ability of such cells to rapidly respond to internalization of pathogens is central for their antibacterial responses (15). In murine and human macrophages, C. burnetii replicates in parasitophorous vacuoles (PVs) with lysosomal acidic characteristics (16). Infection in these cells induces an early proinflammatory response characterized by increased expression of cytokines such as interleukin-12 (IL-12), TNF-α, and IL-1β (17, 18). These mediators are important for recruiting other immune cells to the infection site, promoting pathogen clearance, and development of adaptive immunity (15), essential for controlling C. burnetii infection in mice (19).
To develop novel measures for restricting the spread of C. burnetii in herds or their transmission to humans, alleged differences in the virulence of different C. burnetii strains must be considered. Different courses of natural and experimental infections in human patients and mice, respectively, imply that C. burnetii strains vary in their virulence properties, but an association with certain genotypes is weak at best. Gene mapping (20) and sequencing of the C. burnetii genome unveiled potential virulence genes based on sequence homologies. For example, the acute-disease antigen A (adaA) was described to be primarily present in strains isolated from acutely diseased patients (21). Despite an increasing number of effector proteins identified, their role as virulence factors during C. burnetii infection remains to be proven (22–25). Virulence of C. burnetii is strongly determined by phase variations similar to smooth-rough lipopolysaccharide (LPS) of enterobacteria (26). Phase I strains possess a full-length LPS and are regarded as the naturally occurring, highly virulent form of C. burnetii. Serial in vitro passage of phase I C. burnetii leads to a conversion into avirulent phase II variants with truncated LPS. Compared to phase I, phase II variants lack certain sugars as part of the LPS O chain. Avirulent phase II strains are more efficiently internalized by host cells and cleared faster by the immune system than virulent phase I strains (27).
Lacking appropriate test systems, it is difficult to determine to what extent virulence properties beyond LPS affect the capability of C. burnetii to multiply in and become shed by the reservoir host. Here we established an in vitro infection model with bovine monocyte-derived macrophages (MDM) and investigated bacterial replication and host cell cytokine responses after infection with virulent C. burnetii strain Nine Mile I (NMI) and avirulent strain NMII. For a better interpretation of key results, bovine alveolar macrophages were also infected. This is the first study to show that bovine MDM are a suitable model to explore the intracellular lifestyle of Coxiella in cells of the natural host and to provide insights into the immune response evoked by C. burnetii infections upon first host contact.
MATERIALS AND METHODS
Propagation, enumeration, and characterization of bacterial strains.
Chemicals were purchased from Sigma-Aldrich, Hamburg, Germany, unless otherwise stated. C. burnetii NMI (NMI strain RSA493) from the strain collection of the Institute for Hygiene and Infectious Diseases of Animals (JLU [28]; NMI was propagated in mice, reisolated, and subsequently passaged 9 times in cell culture before being used in this study), and NMII (strain 439, clone 4; generously supplied by Anja Lührmann, Institute for Clinical Microbiology, Immunology and Hygiene, University Hospital Erlangen, Erlangen, Germany) were propagated in African green monkey kidney (BGM) cells and purified as described previously (29). Bacterial concentrations were determined by counting C. burnetii-like particles in Gimenez-stained smears using an Ortholux II microscope with a counting tube (Leitz-Leica, Mannheim, Germany). Briefly, suspensions were diluted 1:100 with sterile saline, and 10-μl aliquots were air dried on a 1-cm2 section of a slide and fixed with 100% methanol for 1 h. Slides were incubated for 6 min in 0.5 ml carbol fuchsine working solution (1.5 mg/ml Neofuchsine [Merck, Hellerup, Denmark], 3 mg/ml phenol [Merck] in phosphate buffer). Slides were rinsed twice with water and counterstained with 0.5 ml malachite green working solution (8 g malachite green [Merck] in 1,000 ml distilled water) for 1 min. Bacterial concentration was calculated on basis of C. burnetii particles counted in 100 small ocular squares at 787.5-fold magnification (30). The bacterial concentration was used for multiplicity of infection (MOI) determination. C. burnetii suspensions in NaCl solution were aliquoted (1 × 109 bacterial cells per ml) and stored at −80°C until further use. Immediately prior to inoculation, aliquots were thawed at ambient temperature and resuspended thoroughly for at least 1 min to separate bacterial agglutination. Relative abundances of full-length and truncated LPS molecules forming the cell envelope of the C. burnetii strains were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (4 to 20% Tris-glycine gel) and subsequent silver staining of the gels (31).
Chlamydia psittaci strain DC15, propagated and purified in BGM cells as described before (32), was used for control experiments.
Preparation of bovine MDM and alveolar macrophages.
Bovine MDM were isolated following an established protocol (33–35). Briefly, a citrated whole-blood sample (5:1 diluted with 3.8% sodium citrate solution) was centrifuged (2,380 × g, 20 min), and the buffy coat was collected. After several washing steps with phosphate-buffered saline (PBS)-EDTA buffer (PBS supplemented with 5.4 mM EDTA; 800 × g, 10 min), the remaining erythrocytes were lysed by incubation of the resuspended pellet in lysis buffer (8.26 g NH4Cl, 1.09 g NaHCO3, 0.037 g Na3EDTA, and 1,000 ml distilled water) for 10 min. Remaining cells were washed three more times (PBS-EDTA buffer; 300 × g, 4°C, 10 min) and layered onto Pancoll (PAN-Biotech, Aidenbach, Germany) for density centrifugation (800 × g, 45 min at ambient temperature). Mononuclear cells were collected from the interphase, centrifuged at 800 × g for 10 min, and washed twice with 0.89% NaCl solution (300 × g, 10 min). Cells were adjusted to 4 × 106/ml in Iscove's modified Dulbecco's medium (IMDM) culture medium 1 (IMDM without phenol red, supplemented with 20% heat-inactivated fetal calf serum [FCS], 1% penicillin-streptomycin, 1% amphotericin B, 0.05% 100 mM β-mercaptoethanol). Twenty-five milliliters of this cell suspension was transferred to Teflon bags (VueLife Bags, American Fluoroseal Corp., Gaithersburg, MD, USA) and incubated for 7 days (37°C, 5% CO2). At the end of the incubation period, the cells were harvested and cultured in IMDM culture medium 2 (IMDM without phenol red, 2% FCS, 1% penicillin-streptomycin, 1% amphotericin B, 0.05% 100 mM β-mercaptoethanol). Cells were seeded into uncoated cell culture plates (Cellstar for suspension cultures; Greiner, Frickenhausen, Germany), unless otherwise stated. After 18 h of incubation, nonadherent cells, i.e., mostly lymphocytes, were carefully washed away with 0.89% NaCl solution, and adherent macrophages were left. Cell culture medium 3 (IMDM culture medium 2 devoid of antibiotics) was added 1 to 2 further days before inoculation with C. burnetii. For each preparation, the cell composition of the culture was determined by fluorescence-activated cell sorter (FACS) analysis (see below). Cultures consisted of 81% + 2.8% (mean + standard deviation [SD] for 8 cultures) CD14+ cells with light scatter characteristics (size and granulation) of macrophages.
For the preparation of alveolar macrophages, bronchoalveolar lavage (BAL) fluid was extracted from the lungs of clinically healthy calves with a protocol described before (36). After a washing step, cells were harvested and cultured in IMDM culture medium 2. Cells were seeded into cell culture plates and incubated overnight for adherence. Cells were washed again, and the medium was changed to IMDM culture medium 3 1 to 2 days before inoculation with C. burnetii. Cultures consisted of 98% cells with morphological characteristics (size and granulation) of macrophages. All animal experiments were conducted according to the rules laid down in the German Animal Protection Act and approved by the competent authority (Thuringian State Office for Consumer Protection, reg. no. 22-2684-04-04-102/13 and reg. no. 04-004/1).
Cultivation of macrophages for studying C. burnetii invasion and replication kinetics.
Invasion and replication of C. burnetii strains NMI and NMII were studied with macrophages (5 ×105 cells/well) cultivated in polystyrene tubes (Greiner). MDM cultures were inoculated at an MOI of 100 by addition of bacteria to IMDM culture medium 3 for 1 h. Preceding experiments had revealed that increasing the MOI to 200 and/or the inoculation time to 2 h did not result in significantly higher bacterial numbers 7 days after inoculation. After 1 h, macrophages were washed 3 times with 0.89% NaCl solution and replenished with IMDM culture medium 3. Triplicate cell cultures were harvested at different time points to monitor C. burnetii invasion and replication efficacy. Cells were lysed by three freeze/thaw cycles and extensive vortexing (5 min). For quantification by PCR, cell suspension was additionally inactivated by boiling (95°C, 20 min). Replication rates were calculated by quantitation of the icd gene by absolute quantitative real-time PCR (37). CT values (where CT is threshold cycle) of technical duplicates varied by less than 0.51 and were used to calculate genome equivalents (GE) considering values obtained with an entrained icd-harboring plasmid standard. To enumerate viable bacteria, infected macrophages were lysed by a 3-fold thaw-freeze cycle. From the resulting suspension, the median tissue culture infective doses (TCID50) were defined by endpoint titration on BGM cells (38). In a 4-fold approach, confluent BGM cells in a microtiter plate (Nunc F96 MicroWell; Nunc, Wiesbaden, Germany) were inoculated with a log4-dilution series of the sample (dilution with cell culture medium, consisting of 500 ml Eagle's minimum essential medium with Earle's salts supplemented with NaHCO3 [0.85 g/liter], 5 ml l-glutamine, 5 ml Vitamin 100X solution, and 50 ml FCS [all from Biochrom, Berlin, Germany]). After sealing the wells with adhesive film (Nunc) and centrifugation at 2,400 × g for 60 min at 30°C, the plate was incubated at 37°C. On day 8, wells containing C. burnetii-infected BGM cells were identified by phase-contrast microscopy (Nikon Eclipse TS100-F). C. burnetii-specific vacuoles were considered, and each well having at least one C. burnetii vacuole was counted as positive. The TCID50 was calculated according to the method of Spearman and Kärber (39).
Cultivation of macrophages for RNA isolation.
Macrophages (2 × 106 cells/well) were cultured in 6-well culture plates and inoculated with C. burnetii strains NMI or NMII as described above. As controls, cells were challenged with either a heat-killed suspension of NMI (95°C, 10 min) with a theoretical MOI of 100 for 1 h or with LPS of Escherichia coli O111:B4 (6 μg/ml) or C. psittaci (MOI, 2) for the duration of the experiment. Cells were lysed with RLT buffer (RNeasy minikit [Qiagen, Hilden, German]) for RNA isolation at different time points. Total RNA was isolated with the RNeasy minikit (Qiagen) according to the instructions of the manufacturer. To avoid DNA contamination, RNA was purified with the RNase-free DNase set (Qiagen).
RT and cytokine-specific real-time PCR.
Equal amounts of RNA from each sample were reverse transcribed into cDNA. First, each sample was denatured for 5 min at 65°C and directly cooled on ice. After that, samples were incubated for 60 min at 37°C using 4 U reverse transcriptase (Qiagen) and the appropriate buffer containing 0.5 mM deoxynucleoside triphosphates (dNTPs; Qiagen), 1 μM random primer, and 20 U RNase inhibitor (Promega, Mannheim, Germany). The reaction was terminated by heating to 93°C for 5 min. For excluding the presence of DNA, reverse transcription (RT) controls that were composed of sample and master-mix without reverse transcriptase were included and gave yield to negative results throughout. Thereafter, the levels of relative gene expression of different host-specific cytokines in comparison to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a housekeeping gene were determined by quantitative real-time SYBR green-based (Applied Biosystems, Waltham, MA, USA) PCR using an ABI Prism7500 (Applied Biosystems). All primers (Table 1) were exon-intron spanning and run at an annealing temperature of 60°C. The PCR profile was as follows: denaturation (15 s, 95°C), annealing (1 min, 60°C; 39 cycles), and melting step (15 s, 60°C). PCR products quantified over cycle 38 are not valid. Relative gene expression levels were calculated by using the relative expression software REST (40).
TABLE 1.
Sequences of primers used in this study
| Primer | Sequence (5′–3′)a |
|---|---|
| GAPDH | F, GCG ATA CTC ACT CTT CTA CCT TCG A |
| R, TCG TAC CAG GAA ATG AGC TTG AC | |
| IL-1-β | F, ACC TGA ACC CAT CAA CGA AAT G |
| R, TAG GGT CAT CAG CCT CAA ATA ACA | |
| IL-10 | F, GTG ATG CCA CAG GCT GAG AA |
| R, TGC TCT TGT TTT CGC AGG GCA | |
| IL-12 | F, GCA GCT TCT TCA TCA GGG ACA T |
| R, CCT CCA CCT GCC GAG AAT T | |
| INF-γ | F, TTC TTG AAC GGC AGC TCT GAG |
| R, TGG CGA CAG GTC ATT CAT CA | |
| TGF-β | F, GGC CCT GCC CTT ACA TCT G |
| R, CGG GTT GTG CTG GTT GTA CA | |
| TNF-α | F, TCT TCT CAA GCC TCA AGT AAC AAG T |
| R, CCA TGA GGG CAT TGG CAT AC |
F, forward; R, reverse.
Cultivation of macrophages for flow cytometry analysis.
Macrophages (2 × 106 cells/well, 6-well plates) were inoculated with C. burnetii (MOI, 100) for 1 h. After 24 h of incubation, cells were detached by incubation with Accutase (PAA Laboratories, Germany), transferred to microtiter plates (V-shape; Greiner), and pelleted by centrifugation (400 × g, 4 min and 4°C). For detection of surface proteins (CD40, CD80, CD86, major histocompatibility complex class I [MHC-I] and MHC-II, CD11b [CR3], CD61 [αVβ3]), cells were incubated with 50 μl diluted primary antibody for 20 min (Table 2). After washing in PBS–0.5% FCS, cells were fixed with paraformaldehyde (4% in PBS, 4°C, 24 h). Thereafter, cells were washed and incubated with secondary antibody (anti-mouse IgG1-APC, anti-mouse IgG2a-APC, anti-mouse IgG2b-PE; Southern Biotech, USA) diluted 1:1,000 in PBS for 20 min. Finally, cells were washed again and analyzed with BD FACSCantoII (Becton-Dickinson, Heidelberg, Germany). Data were analyzed with BD FACSDIVA software (version 6).
TABLE 2.
Murine antibodies against bovine cell surface antigens used in this study
| Antigen | Antibody clone (source) | Isotype | Dilution |
|---|---|---|---|
| CD11b | MM12A (VMRD, Pullman, WA, USA) | IgG1 | 1:250 |
| CD14 | CAM36A (VMRD, Pullman, WA, USA) | IgG1 | 1:500 |
| CD40 | IL-A156 (Dirk Werling; Royal Veterinary College, Hatfield, UK) | IgG1 | 1:10 |
| CD61 | MCA2263GA (AbD Serotec, Düsseldorf, Germany) | IgG1 | 1:100 |
| CD80 | IL-A159 (Werling, UK) | IgG1 | 1:10 |
| CD86 | IL-A190 (Werling, UK) | IgG1 | 1:10 |
| MHC-I | PT85A (VMRD, Pullman, WA, USA) | IgG2a | 1:500 |
| MHC-II | H34A (VMRD, Pullman, WA, USA) | IgG2b | 1:500 |
Interleukin-1β ELISA.
For interleukin-1β enzyme-linked immunosorbent assay (ELISA), infected macrophages (2 × 106 cells/well; MOI, 100) were incubated for 24 h in 6-well plates. Supernatant was collected and irradiated for 2 h with UV light. Pretests had shown that this procedure inactivated C. burnetii (no vacuole formation suggesting C. burnetii replication after four passages on BGM cells). Samples were concentrated by using Amicon Ultra 4 columns (Millipore, Darmstadt, Germany). Microtiter plates (MaxiSorb; Nunc-Thermo Fisher Scientific, Braunschweig, Germany) were coated with capture antibody (MCA1658; AbD Serotec, Puchheim, Germany; 100 ng/well in coating buffer [15 mM Na2CO3, 34 mM NaHCO3, and 500 ml distilled water ]) overnight at 4°C. After washing the wells twice with washing buffer (PBS, 0.05% Tween 20), samples and a dilution series (400 ng/ml through 1:10) of recombinant interleukin-1β standard (PBP008; AbD Serotec) were added for 1 h. After another washing step with blocking buffer (PBS, 0.05% bovine serum albumin [BSA]), primary antibody (AHP423, 1:1,000 in blocking buffer; AbD Serotec) was added and incubated for 1 h. Following one washing step, wells were incubated with horseradish peroxidase-coupled secondary antibody (STAR124P; AbD Serotec; 1:50,000 in reagent buffer [washing buffer supplemented with 0.5% BSA]) for 1 h. After a final washing step, substrate (3,3′,5,5′ tetramethylbenzidin; Thermo Fisher Scientific, Waltham, MA, USA) was added and incubated for 10 min in the dark. Absorbance at 450 nm was measured (Sunrise reader; Tecan, Mannedorf, Switzerland) after the reaction was stopped by addition of 100 μl of 1 N H2SO4.
Western blot analysis.
MDM (2 × 106 cells/well, 6-well plates) were inoculated with C. burnetii (MOI, 100) for 24 h. Total cellular protein was sampled by lysing cells with 500 μl Laemmli buffer (0.5 M Tris-HCl, 87% glycerol, 10% SDS, 4% β-mercaptoethanol, 0.05% bromophenol blue) and boiling at 100°C for 10 min. Protein concentration was measured by the Bradford method (41). Equivalent protein amounts from each sample and a positive control (recombinant bovine IL-1β protein) were separated by SDS-PAGE (4 to 12%) and blotted onto a polyvinylidene difluoride (PVDF) membrane (Thermo Fisher Scientific). Nonspecific binding sites were blocked by incubation with skimmed milk. Tris-buffered saline with Tween 20 (100 mM NaCl, 100 mM Tris, 5 mM MgCl2 · 6H2O, 0.05% Tween 20) was used for washing steps. The membrane was incubated overnight with primary antibody (AHP 423, 1:1,000 in PBS-Tween). After washing, an alkaline phosphatase-conjugated secondary antibody (anti-mouse-IgG1-AP, 1:2,000 in PBS-Tween) was applied for 1.5 h. Protein was visualized by using a 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium chloride detection system.
Statistical analysis.
Statistical comparisons were conducted using the t test or U test with statistic software XLSTAT. Real-time PCR data were analyzed by a randomization test with pairwise reallocation (software REST [40]).
RESULTS
C. burnetii internalization and replication in bovine macrophages.
Cultured MDM expressed the C. burnetii receptors CR3 and αVβ3 integrin (42) on their surface (Fig. 1A). The same was true for alveolar macrophages, which surface expressed C. burnetii receptors when freshly isolated from BAL fluid.
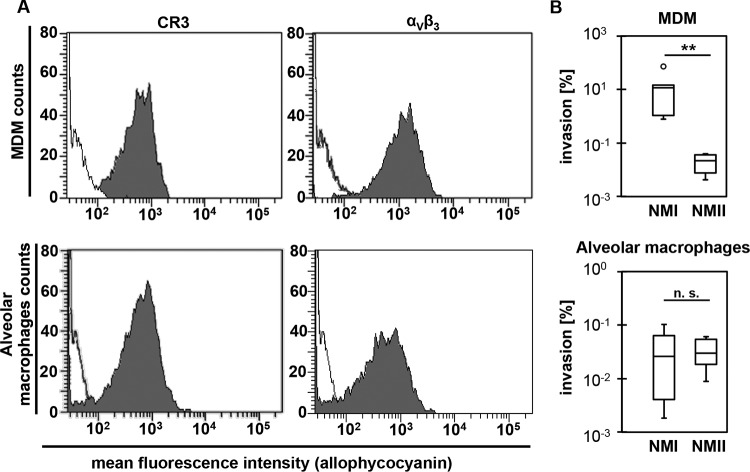
FIG 1.
Expression of Coxiella burnetii-specific receptors and internalization of C. burnetii into bovine macrophages. (A) Uninfected bovine MDM (top) and alveolar macrophages (bottom) were analyzed by FACS for expression of CR3 and αVβ3 on their surface. Gray-shaded curves depict detection of the respective antigens, and black lines represent the secondary antibody control (representative results of two technical replicates in 2 independent experiments each). (B) The numbers of cell-associated C. burnetii bacteria at 1 dpi were calculated based on the number of genome equivalents (GE) quantified by icd-specific real-time PCR and expressed as percentage of GE in the inoculum (box plot of twice-performed technical determinations in 3 independent experiments; **, P ≤ 0.01; n. s., not significantly different).
Macrophages from both sources internalized C. burnetii strains NMI and NMII in general but with different efficiencies. The numbers of C. burnetii genome equivalents (GE) determined varied considerably between biological replicates, i.e., different cultures of primary cells. However, the invasion efficacy of NMI into bovine MDM was significantly higher, with approximately 10% of the C. burnetii bacteria being cell bound 1 day postinoculation (dpi), while only a small portion of the NMII inoculum was cell associated at that point in time (Fig. 1B). Different from what was seen with MDM, similar low mean percentages of the inoculated NMI and NMII bacteria were associated with alveolar macrophage cells within 1 dpi.
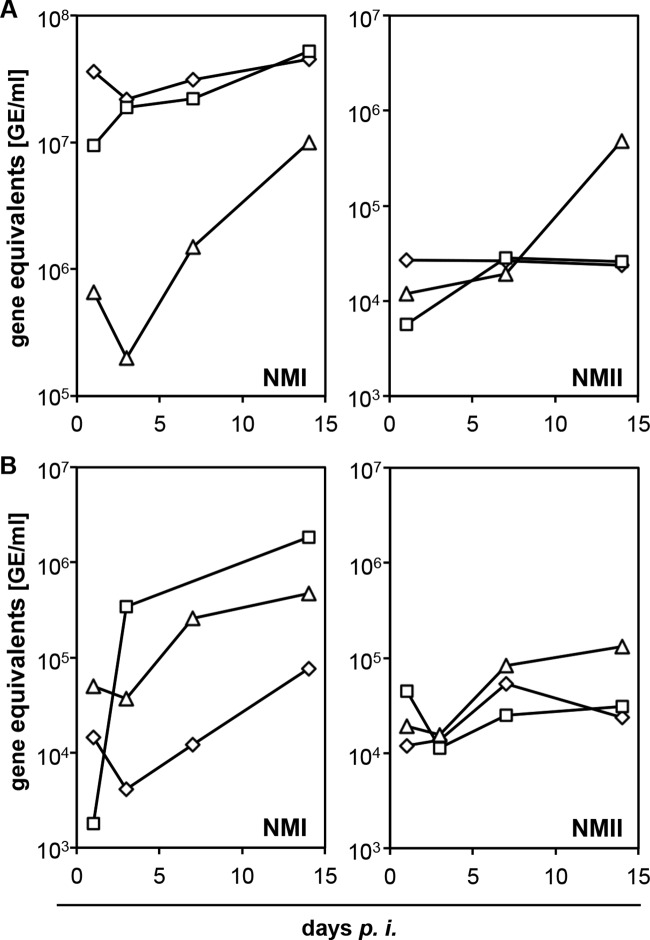
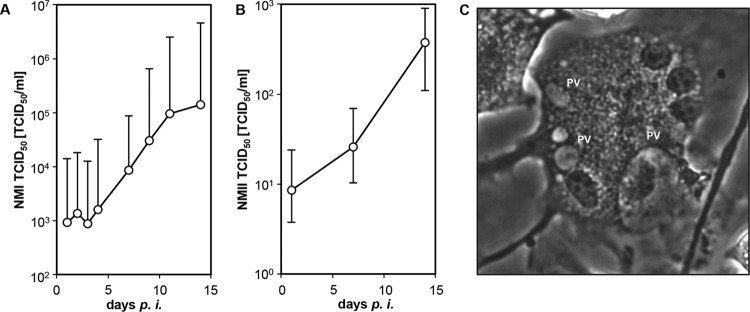
Despite marked biological variation between cultures, bovine MDM and alveolar macrophages generally permitted C. burnetii replication. After an initial decline in absolute GE numbers from day 1 to day 3 of inoculation in MDM (NMI) and alveolar macrophages (NMI, NMII), bacterial replication started mostly at 3 dpi and did not cease until day 14. However, no explicit replication was evident in NMII-infected bovine MDM (Fig. 2). When applying a retitration method on BGM cells to more closely monitor the number of viable C. burnetii cells in bovine MDM cultures, NMI and NMII both showed an increase in numbers by about 2 orders of magnitude within 14 dpi (Fig. 3A and B). While the numbers of NMI had increased 243- ± 219-fold (mean values for three independent experiments and respective SD), the numbers of NMII increased 56- ± 35-fold, but the values were not statistically significantly different (P = 0.2). The ability of C. burnetii to establish an intracellular replicative niche in bovine MDM was further corroborated by the formation of parasitophorous vacuoles (Fig. 3C), which were indistinguishable in shape and size between the two strains (data not shown). Small PVs were located mostly in groups close to the cellular membrane and fused in later stages of the replication cycle to larger PVs. Coxiella-infected macrophages retained their viability and integrity over the entire time course of the experiments as demonstrated by light microscopy and trypan blue exclusion tests at days 1, 7, and 14 after inoculation.
FIG 2.
Replication of C. burnetii in bovine MDM (A) and in alveolar macrophages (B). Cells were inoculated with C. burnetii strains NMI or NMII (MOI of 100 for 1 h), and C. burnetii-specific genome equivalents (GE) were quantified in duplicate at different time points by icd-specific real-time PCR and calculated by use of a standard curve. Kinetic of C. burnetii replication was monitored in 3 independent experiments depicted by different symbols within the curves.
FIG 3.
Replication of C. burnetii in bovine MDM. Cells were inoculated with C. burnetii strain NMI (A) or NMII (B) (MOI of 100 for 1 h), and viable cell-associated C. burnetii cells were quantified at different time points by TCID50 assay (geomean and geometric deviation values for three independent experiments. (C) Mature parasitophorous vacuole (PV) in NMI-infected bovine MDM (7 dpi).
Bovine macrophages rapidly responded to C. burnetii by differential transcription of cytokine genes.
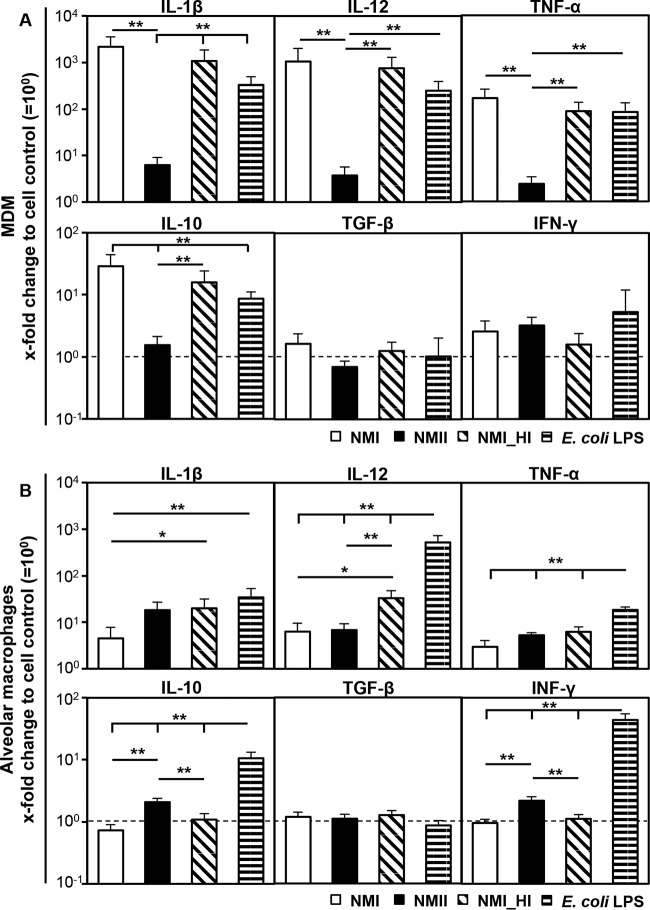
MDM responded to C. burnetii by transcription of different cytokine genes as early as 3 hpi (Fig. 4A). NMI significantly increased the amounts of mRNA specific for some T helper cell 1 (Th1)-related cytokines, such as IL-1β, IL-12, and tumor necrosis factor alpha (TNF-α), but not for gamma interferon (IFN-γ). Induction of Th2-related cytokines was less prominent (IL-10), or the amounts of specific mRNA were not different from those of uninoculated MDM cultures (transforming growth factor beta [TGF-β]). Inoculation with E. coli LPS (6 μg/ml) and a heat-inactivated NMI suspension were used as stimulation controls. Both induced a significant upregulation of proinflammatory cytokine transcription 3 hpi (Fig. 4A), whereas inoculation with a heat-killed suspension of NMI led to MDM responses indistinguishable from those obtained with inoculation with viable NMI. Induction of cytokine expression correlated with the LPS phase, as the MDM response to NMII inoculation was not different from the medium control or significantly lower than the response to NMI. In contrast to MDM, alveolar macrophages responded to C. burnetii in a more restrained manner (Fig. 4B). Remarkably, viable NMII induced a 4.5-fold-more-pronounced increase in the IL-1β mRNA level than that induced by viable NMI, while a heat-killed NMI suspension led to marked transcription of IL-1β and IL-12. NMII only slightly induced transcription of IL-10 and IFN-γ, whereas C. burnetii did not affect TGF-β transcription in alveolar macrophages.
FIG 4.
Influence of C. burnetii on cytokine expression 3 h after inoculation of bovine MDM (A) and alveolar macrophage (B) cultures. Cells were inoculated with C. burnetii strain NMI or NMII (MOI of 100 for 1 h). Addition of a heat-killed suspension of NMI (NMI_HI) at an equivalent bacterial cell-to-macrophage ratio or E. coli LPS (6 μg/ml) served as a control. Amounts of cytokine mRNA were normalized to GAPDH mRNA and determined relative to the cell control (set to 10°). Results are depicted as mean values with their respective SD. A randomization test with a pairwise reallocation was used to compare ΔΔCT values from 3 independent experiments (*, P ≤ 0.05; **, P ≤ 0.01; significantly different from cell control results).
The cytokine response of bovine MDM to prolonged exposure to C. burnetii was transient.
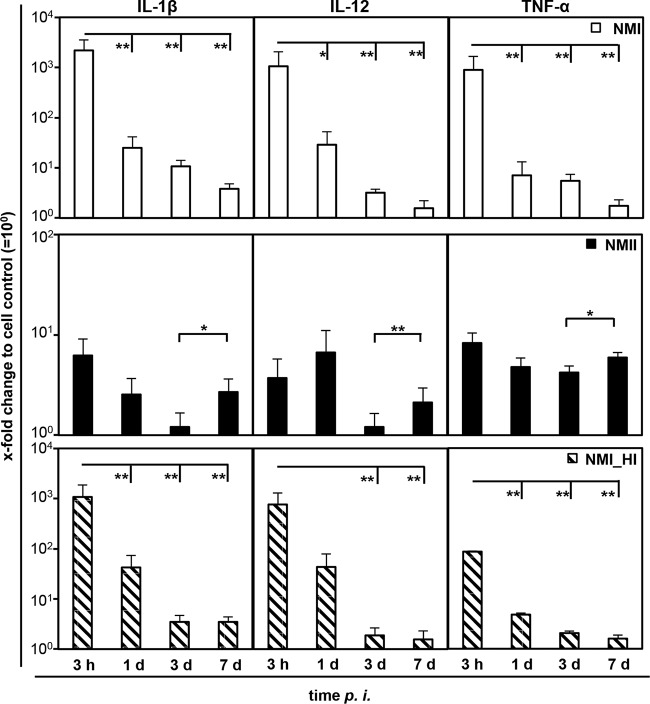
Increased transcription of cytokines rapidly waned during C. burnetii infection of MDM. While NMI induced large amounts of mRNA specific for IL-1β, IL-12, and TNF-α at 3 hpi, the amounts decreased to near-background values (medium control) with increasing intracellular C. burnetii numbers following bacterial growth (Fig. 5). The transient character of the cytokine response was independent of bacterial replication, as inoculation with a heat-killed NMI suspension yielded similar results. These findings contrast with results from two control experiments showing that E. coli LPS as well as C. psittaci inoculation led to a further increase of at least IL-12 transcription from 3 or 4 h to 1 day after addition of the stimulus (Fig. 6).
FIG 5.
Influence of C. burnetii on cytokine expression by bovine MDM over time. Cells were inoculated with C. burnetii strain NMI or NMII (MOI of 100 for 1 h) or with a heat-killed suspension of NMI (NMI_HI) at an equivalent bacterial cell-to-macrophage ratio. Amounts of mRNA of different cytokines were normalized to GAPDH mRNA and determined relative to the cell control (set to 10°) at different time points. A randomization test with a pairwise reallocation was used to compare ΔΔCT values from 3 independent experiments. Results are depicted as mean values with their respective SD (*, P ≤ 0.05; **, P ≤ 0.01).
FIG 6.
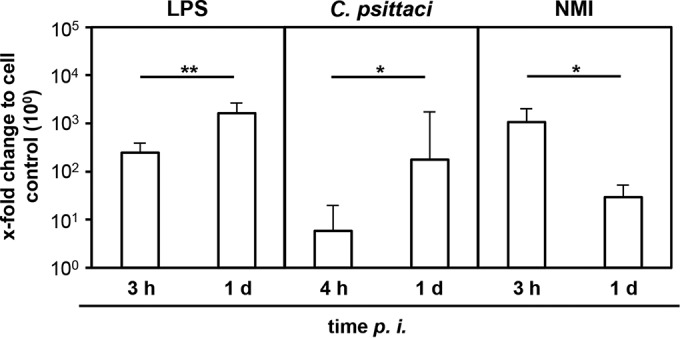
IL-12 expression by bovine MDM upon exposure to E. coli LPS or infection with Chlamydia psittaci for 3 or 4 and 24 h. Amounts of IL-12-specific mRNA were normalized to GAPDH mRNA and determined relative to cell control (set to 10°) at different time points. Results are depicted as mean values + SD for 3 independent experiments. Statistical differences are indicated (*, P ≤ 0.05; **, P ≤ 0.01).
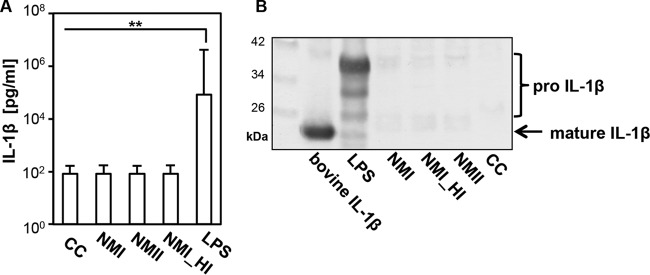
C. burnetii prevented IL-1β translation and release in bovine MDM culture.
In contrast to other proinflammatory cytokines, IL-1β is expressed as a precursor protein and requires an enzyme-controlled cleavage process for formation of the bioactive cytokine. Quantitative analysis by ELISA revealed that after 24 h of incubation, IL-1β protein was only detectable in supernatants of LPS-stimulated MDM cultures (Fig. 7A). Inoculation of MDM cultures with NM variants did not result in detectable release of IL-1β beyond that of an uninfected control culture. These findings were corroborated by Western blotting showing that stimulation with E. coli LPS led to expression of precursor protein and cleavage products of different sizes while inoculation with viable and heat-killed C. burnetii suspensions did not (Fig. 7B).
FIG 7.
C. burnetii induced IL-1β activation in bovine MDM. (A) Cells were inoculated with C. burnetii strain NMI or NMII (MOI of 100 for 1 h), with a heat-killed suspension of NMI (NMI_HI) at an equivalent bacterial cell-to-macrophage ratio or stimulated with E. coli LPS or mock inoculated with NaCl solution (CC). Supernatants were collected 24 h later. The amount of mature IL-1β was analyzed by ELISA. Results from 3 independent experiments are depicted as mean values + SD, and statistical differences are indicated (**, P ≤ 0.01). (B) Total protein was isolated from infected or stimulated MDM at 24 hpi. Equal protein amounts from each sample were separated on a 4 to 12% SDS-polyacrylamide gel and immunolabeled for IL-1β. Bovine recombinant IL-1β served as a control for the mature protein.
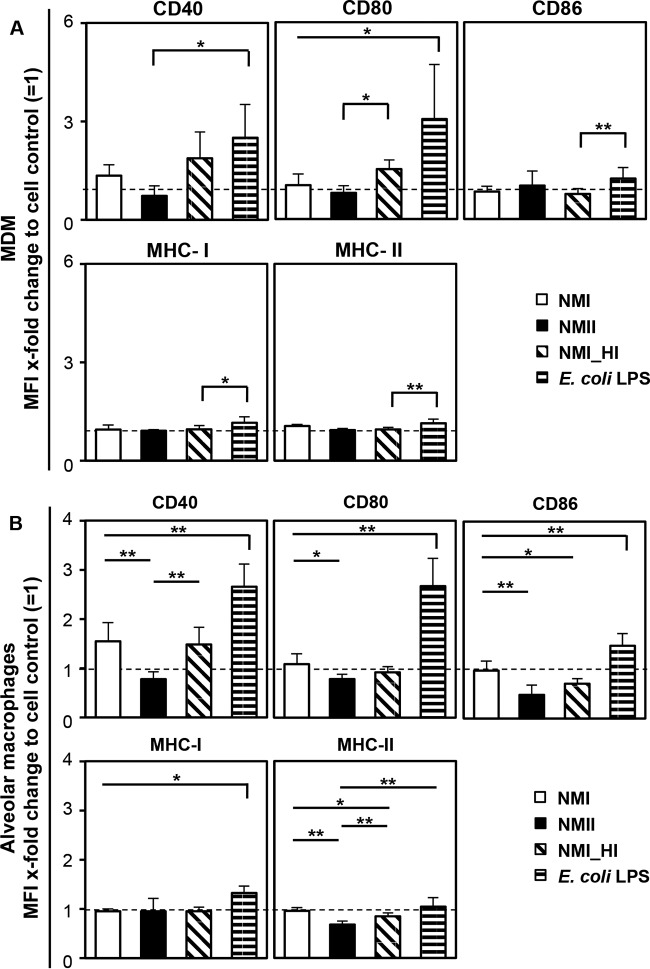
C. burnetii only marginally affected macrophage maturation.
E. coli LPS and the heat-killed NMI suspension intensified MDM maturation triggered by plastic adherence as deduced from an increase in CD40 and CD80 expression, while CD86 and MHC expression were only slightly affected by E. coli LPS stimulation (Fig. 8A). In contrast, cell maturation was barely affected by viable NMII. Only viable NMI induced a weak expression of CD40 (P = 0.0001) and CD80 compared to the untreated control. Although alveolar macrophages responded less vigorously to stimulation, the overall reaction pattern to C. burnetii was similar to that of MDM (Fig. 8B).
FIG 8.
Surface marker expression by bovine MDM (A) and alveolar macrophages (B) in response to C. burnetii infection. Cells were inoculated with C. burnetii strain NMI or NMII (MOI of 100 for 1 h) or with a heat-killed suspension of NMI (NMI_HI) at an equivalent bacterial cell to macrophage ratio or stimulated with E. coli LPS. Twenty-four hours later, cells were immunolabeled for the surface markers CD40, CD80, CD86, and MHC-I and -II. Results of 3 to 8 independent experiments are shown as mean values + SD, and statistical differences are indicated (*, P ≤ 0.05; **, P ≤ 0.01; significantly different from cell control results).
DISCUSSION
The currently limited understanding of the infection route of C. burnetii in ruminants has direct implications for designing strategies to control the epizootic pathogen and to protect humans from a zoonotic disease. Macrophages are accepted as the major host cell for C. burnetii infection, and rodent studies reported high numbers of C. burnetii in alveolar macrophages (43). Infection models for studying host-pathogen interactions have already been established with human, mice, and monkey macrophage cultures (18, 44, 45). Here, we present the first in vitro study demonstrating that primary macrophages from cattle can be utilized in an infection model for investigating the lifestyle of C. burnetii in the ruminant host.
Internalization of Coxiella into macrophages occurs by phagocytosis and is mediated by αVβ3 integrin for virulent phase I particles, whereas avirulent phase II particles additionally require the CR3 receptor (42). We found that bovine MDM and alveolar macrophages express high numbers of both receptors on their surface. Internalization of virulent NMI was more efficient than that of the avirulent strain NMII. Interestingly, this prominent difference between the strains was observed only in bovine MDM cultures but not when alveolar macrophage cultures were inoculated in which equally low numbers of C. burnetii bacteria expressing either LPS phase became cell bound. As opposed to what is seen with bovine macrophages, phase I particles are only poorly internalized by human monocytes, whereas phase II particles are better ingested (46). It remains to be determined whether these apparent species differences can be traced back to quantitative differences in the expression of αVβ3, CR3, or other surface molecules like, e.g., Toll-like receptor 4 (TLR4) by phagocyte subsets.
Despite quantitative differences in internalization, phase I and phase II strains replicated with similar kinetics in bovine MDM. NMI and NMII replication became particularly apparent when viable bacteria were retitrated in an independent cell system commonly deployed for C. burnetii isolation for diagnostic purposes. Irrespective of LPS phase, C. burnetii initiated proliferation once bacteria were intracellular and apparently had escaped from the antibacterial activities of the phagocytes as in human MDM (47). C. burnetii was reported to productively proliferate in human alveolar macrophages as well (18). Due to the limited number of bovine alveolar macrophages available for this study and the laborious replating assay, only quantitation of genome equivalents (GE) was applied to assess C. burnetii replication in this cell type (37). While NMI-inoculated alveolar macrophages showed a clear increase in GE numbers from day 1 to day 14 of culture, GE numbers in NMII-inoculated cultures increased slightly between days 3 and 7 (P = 0.041). Counting the infected cells in a replating assay has the big advantage of allowing the quantification of viable cells, whereas RT-PCR may also quantify C. burnetii particles that are deficient in infectivity or replication. In order to permit massive parallel testing of a large number of samples, the assay read-out was by phase-contrast microscopy, which may have impaired the sensitivity of this approach. The characteristics of the methods likely explain the discrepancies in absolute numbers of genome equivalents and 50% tissue culture infective doses observed in this study. It remains to be determined, by titration on susceptible cells, whether bovine alveolar macrophages are capable of effectively limiting growth of phase II organisms or whether the abundance of GE from nonviable bacteria had masked the detection of replication.
Macrophages are one of the first barriers of the innate immune system against pathogens. During the early phase of infection, C. burnetii induced a rapid expression of Th1-related cytokines like IL-1β, IL-12, and TGF-β in bovine macrophages. The same response pattern was observed in murine macrophages with strong expression of IL-1α, IL-12, and TNF-α at 3 hpi (17). Cytokine expression in bovine macrophages differed after inoculation with strains possessing different LPS phase types, with NMI inducing more-pronounced responses than NMII under the experimental conditions applied, i.e., exposure to C. burnetii particles at an MOI of 100. Phase-dependent induction of inflammatory host cell responses also occurs in human dendritic cells, but full-length LPS of virulent C. burnetii appears to mask TLR ligands from innate immune recognition in this setting (48). If the target cell does become activated by C. burnetii, TLR2 recognition of the lipid A moiety of LPS (49) is implicated in the induction of proinflammatory responses (50). LPS phases in Coxiella vary in the carbohydrate composition of the O chain. Lack of certain sugars results in a shorter chain length and loss of the TLR ligand masking function. In contrast, differences between the two C. burnetii phase types in their lipid A structure have not been reported (51). Other microbe-associated molecular patterns of C. burnetii, the binding of which to bovine macrophages is not efficiently prevented by phase I LPS, may therefore account for the differences observed.
C. burnetii and other intracellular bacteria induce proinflammatory responses by host cell attachment independent of the internalization process or its efficiency (52–54). Particularly, TNF expression was shown to be induced by binding of C. burnetii to the cell (52). A heat-killed suspension of NMI induced a cytokine expression pattern in bovine macrophages similar to that induced by viable NMI. Heat-killed NMI was detected intracellularly by staining inoculated bovine MDM with anti-NMI antibodies (data not shown), proving that the cells recognize nonviable NMI particles as to-be-phagocytosed material. C. burnetii uptake is not solely an actively steered process, and surface-located factors, such as outer membrane protein (Omp) A or heat shock proteins (Hsp), could be implicated in the regulation process. OmpA is a highly conserved outer membrane protein among Gram-negative bacteria that has an important role in pathogenesis, including adherence, invasion, and activation of host responses (55) through binding to TLR2 (56, 57). Coxiella-expressed OmpA triggers an adhesion-mediated internalization of bacteria by nonphagocytic cells, e.g., epithelial cell, but not by macrophages (58). Heat-stable Hsps govern the early phase of Legionella infection in macrophages and induce a rapid increase of IL-1β mRNA by a protein kinase C (PKC)-dependent signaling pathway (59). Hsp70 is located on the surface of C. burnetii and homologous to Hsps of Legionella (60). C. burnetii of the two LPS phase types bound to and invaded bovine alveolar macrophages equally well, followed by minor differences in the accompanying host cell response. In bovine MDM, strikingly different invasion rates of NMI and NMII mirrored quantitative variations in proinflammatory cytokine expression. The early host cell activation in bovine macrophages thus seems to result from bacterial cell adhesion and/or internalization and occurs independently of bacterial metabolic activity.
While C. burnetii productively replicated in bovine MDM, enhanced expression of certain cytokines rapidly declined during later stages of infection to the levels of uninfected cell controls (7 dpi). In order to determine if the observed effect is specific for infection with C. burnetii, we exemplarily compared the temporal response pattern of IL-12 transcription in bovine MDM cultures to that in E. coli LPS and another intracellular pathogen, C. psittaci. C. psittaci has a broad host range, including cattle (61), and replicated efficiently in the bovine MDM cultures (K. Hillarius and A. Berndt, data not shown). Contrary to NMI infection, both treatments induced increasing IL-12 expression during the first 24 h, implying that C. burnetii specifically controls the host response in bovine macrophages to allow replication without significant inflammatory cytokine production. Probably by different molecular mechanisms but similar to that of human DCs (48), this immune evasion strategy may allow C. burnetii to productively infect and replicate in an immunocompetent host.
IL-1β is one of the most potent endogenous pyrogens and instrumental for the host defense against pathogens (62). The synthesis of this protein is strictly regulated. A precursor of IL-1β is cleaved by caspase 1 to mature and highly active IL-1β, which then is secreted into the extracellular space (63, 64). Bovine MDM responded to C. burnetii infection (3 hpi) with profound transcription of the IL-1β gene. However, in contrast to E. coli LPS stimulation, we found neither mature IL-1β in culture supernatants of these macrophages nor the precursor in their whole-cell lysates after 24 h. A heat-killed suspension of NMI also prevented IL-1β translation, implying that the conversion of gene transcription activation into protein-encoded information was blocked by processes not requiring active bacterial metabolism. Different from monocytes, caspase 1 is not constitutively activated in macrophages, which require free ATP in addition to, e.g., stimulation by LPS, to activate the enzyme (65, 66). Splicing forms of IL-1β were detectable in LPS-stimulated bovine MDM, indicating that ATP was available in the system and probably released from cells that underwent cell death during the isolation or cultivation process of these primary cell cultures. Intracellular bacteria, such as Chlamydia or Mycobacterium, inhibit the influx of ATP to avoid the maturation and secretion of the IL-1β protein (67, 68). It needs to be elucidated whether the response of bovine macrophages is restricted by C. burnetii at the level of posttranscriptional modification and whether this also applies to other proinflammatory cytokines.
The generation of an adaptive immune response emanates from T cell activation by antigen-presenting cells and involves specific surface proteins (MHC molecules) as well as costimulatory molecules (CD40, CD80, CD86) (69). Tissue-resident macrophages normally express only few such costimulatory molecules on their surfaces. Only after ingestion of foreign material may cells change their surface phenotype, downregulate adhesion molecules, migrate to draining lymph nodes, and convert into effective antigen-presenting cells by increasing the surface expression of molecules essential for the interaction with naive T cells. Interestingly, neither infection with the virulent strain NMI nor infection with the avirulent strain NMII led to a substantial maturation of bovine macrophages in our study. While formalin-killed NMI failed to induce cell maturation in human DCs (48), a heat-killed NMI suspension induced an increase of CD40 and CD80 expression in bovine MDM. Further studies are needed to determine whether the maturation of bovine antigen-presenting cells induced by preformed Coxiella factors is counteracted by components secreted by actively replicating bacteria.
The depression of macrophage maturation and the short duration of proinflammatory cytokine expression (IL-1β, IL-12, TNF-α) during the first 3 hpi demonstrates that C. burnetii induced a combination of M1 and M2 polarization of bovine macrophages, i.e., an atypical M2 polarization resulting in M2-typical properties dominating over M1-typical properties as reported for human macrophages (70). It is well established that intracellular bacteria resist antimicrobial effectors to survive in the host cell (71). During acute Q fever infection in humans, an atypical M2 polarization of macrophages promotes long-term survival of C. burnetii with a slow replication rate, whereas M1-polarized monocytes control the infection. During chronic Q fever, monocytes and macrophages exhibit an M2 polarization with an intense replication of C. burnetii (72).
In conclusion, bovine macrophages may serve (i) as a vehicle for C. burnetii dissemination through the organism to the intended host cells for replication (e.g., epithelial cells in placenta, udder, or gut) and (ii) as a niche for bacterial long-term survival, given the long life span of macrophages (73) and the comparably low replication efficiency of C. burnetii in bovine macrophages. Although cattle are not the main source of human infection, the bovine macrophage model established and characterized here can be used to study host-pathogen interactions in a reservoir host at the cellular level, allowing to characterize replication and the pathogen-induced host cell response to different C. burnetii strains, e.g., the species-specific adaptations of the highly prevalent C. burnetii genotype ST 20 recently identified in bovine milk in the United States (74).
ACKNOWLEDGMENTS
We thank Bettina Hopf (Institute of Hygiene and Infectious Diseases of Animals) and Anke Hinsching (Friedrich-Loeffler-Institut, Institute for Molecular Pathogenesis) for their excellent technical assistance. We also thank Gernot Schmoock and his technical assistant Nadine Lemser (both at Friedrich-Loeffler-Institut, Institute of Bacterial Infections and Zoonoses) for their help in establishing the icd PCR. We appreciate the valuable suggestions of Hermann Willems (Clinic for Ruminants and Swine/Internal Medicine & Surgery, Justus-Liebig-University, Giessen, Germany). Andreas Henke (Institute for Virology and Antiviral Therapy, University Hospital Jena, Jena, Germany) and Christian Berens (Friedrich-Loeffler-Institut, Institute for Molecular Pathogenesis) are acknowledged for helpful discussions and critical reading of the manuscript.
K.S. and M.M. were financially supported by the German Federal Ministry of Education and Research (BMBF) as part of the “Network Q-Fever: Investigation of molecular pathogenesis of Q-fever and its application in diagnostics and epidemiology in Germany” (grant number 01KI1001G).
We declare that we have no conflict of interest.
REFERENCES
- 1.Maurin M, Raoult D. 1999. Q fever. Clin Microbiol Rev 12:518–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carcopino X, Raoult D, Bretelle F, Boubli L, Stein A. 2009. Q fever during pregnancy: a cause of poor fetal and maternal outcome. Ann N Y Acad Sci 1166:79–89. doi: 10.1111/j.1749-6632.2009.04519.x. [DOI] [PubMed] [Google Scholar]
- 3.Langley JM. 1990. Perinatal Q fever: is Coxiella burnetii a human perinatal pathogen?, p 201–212. In Marrie TJ. (ed), Q fever, the disease. CRC Press, Inc., Boca Raton, FL. [Google Scholar]
- 4.Hellenbrand W, Breuer T, Petersen L. 2001. Changing epidemiology of Q fever in Germany, 1947–1999. Emerg Infect Dis 7:789–796. doi: 10.3201/eid0705.010504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodolakis A, Berri M, Hechard C, Caudron C, Souriau A, Bodier CC, Blanchard B, Camuset P, Devillechaise P, Natorp JC, Vadet JP, Arricau-Bouvery N. 2007. Comparison of Coxiella burnetii shedding in milk of dairy bovine, caprine, and ovine herds. J Dairy Sci 90:5352–5360. doi: 10.3168/jds.2006-815. [DOI] [PubMed] [Google Scholar]
- 6.Benson WW, Brock DW, Mather J. 1963. Serologic analysis of a penitentiary group using raw milk from a Q fever infected herd. Public Health Rep 78:707–710. doi: 10.2307/4591908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fishbein DB, Raoult D. 1992. A cluster of Coxiella burnetii infections associated with exposure to vaccinated goats and their unpasteurized dairy products. Am J Trop Med Hyg 47:35–40. [DOI] [PubMed] [Google Scholar]
- 8.Kazar J. 2005. Coxiella burnetii infection. Ann N Y Acad Sci 1063:105–114. doi: 10.1196/annals.1355.018. [DOI] [PubMed] [Google Scholar]
- 9.Roest HJ, van Gelderen B, Dinkla A, Frangoulidis D, van Zijderveld F, Rebel J, van Keulen L. 2012. Q fever in pregnant goats: pathogenesis and excretion of Coxiella burnetii. PLoS One 7:e48949. doi: 10.1371/journal.pone.0048949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roest HI, Post J, van Gelderen B, van Zijderveld FG, Rebel JM. 2013. Q fever in pregnant goats: humoral and cellular immune responses. Vet Res 44:67. doi: 10.1186/1297-9716-44-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ammerdorffer A, Roest HI, Dinkla A, Post J, Schoffelen T, van Deuren M, Sprong T, Rebel JM. 2014. The effect of C. burnetii infection on the cytokine response of PBMCs from pregnant goats. PLoS One 9:e109283. doi: 10.1371/journal.pone.0109283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khavkin T, Tabibzadeh SS. 1988. Histologic, immunofluorescence, and electron microscopic study of infectious process in mouse lung after intranasal challenge with Coxiella burnetii. Infect Immun 56:1792–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baumgartner W, Bachmann S. 1992. Histological and immunocytochemical characterization of Coxiella burnetii-associated lesions in the murine uterus and placenta. Infect Immun 60:5232–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emikpe B, Yussouf SM, Ezeasor CK, Tanko PN. 2013. Immunohistochemical detection of Brucella mellitensis and Coxiella burnetii antigens in formalin-fixed tissues of West African Dwarf goats. Arch Clin Microbiol 4(2). [Google Scholar]
- 15.Marriott HM, Dockrell DH. 2007. The role of the macrophage in lung disease mediated by bacteria. Exp Lung Res 33:493–505. doi: 10.1080/01902140701756562. [DOI] [PubMed] [Google Scholar]
- 16.Voth DE, Heinzen RA. 2007. Lounging in a lysosome: the intracellular lifestyle of Coxiella burnetii. Cell Microbiol 9:829–840. doi: 10.1111/j.1462-5822.2007.00901.x. [DOI] [PubMed] [Google Scholar]
- 17.Tujulin E, Lilliehook B, Macellaro A, Sjostedt A, Norlander L. 1999. Early cytokine induction in mouse P388D1 macrophages infected by Coxiella burnetii. Vet Immunol Immunopathol 68:159–168. doi: 10.1016/S0165-2427(99)00023-9. [DOI] [PubMed] [Google Scholar]
- 18.Graham JG, MacDonald LJ, Hussain SK, Sharma UM, Kurten RC, Voth DE. 2013. Virulent Coxiella burnetii pathotypes productively infect primary human alveolar macrophages. Cell Microbiol 15:1012–1025. doi: 10.1111/cmi.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andoh M, Naganawa T, Hotta A, Yamaguchi T, Fukushi H, Masegi T, Hirai K. 2003. SCID mouse model for lethal Q fever. Infect Immun 71:4717–4723. doi: 10.1128/IAI.71.8.4717-4723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willems H, Jager C, Baljer G. 1998. Physical and genetic map of the obligate intracellular bacterium Coxiella burnetii. J Bacteriol 180:3816–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang G, To H, Russell KE, Hendrix LR, Yamaguchi T, Fukushi H, Hirai K, Samuel JE. 2005. Identification and characterization of an immunodominant 28-kilodalton Coxiella burnetii outer membrane protein specific to isolates associated with acute disease. Infect Immun 73:1561–1567. doi: 10.1128/IAI.73.3.1561-1567.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eckart RA, Bisle S, Schulze-Luehrmann J, Wittmann I, Jantsch J, Schmid B, Berens C, Luhrmann A. 2014. Antiapoptotic activity of Coxiella burnetii effector protein AnkG is controlled by p32-dependent trafficking. Infect Immun 82:2763–2771. doi: 10.1128/IAI.01204-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baca OG, Roman MJ, Glew RH, Christner RF, Buhler JE, Aragon AS. 1993. Acid phosphatase activity in Coxiella burnetii: a possible virulence factor. Infect Immun 61:4232–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mertens K, Lantsheer L, Samuel JE. 2005. A minimal set of DNA repair genes is sufficient for survival of Coxiella burnetii under oxidative stress. Ann N Y Acad Sci 1063:73–75. doi: 10.1196/annals.1355.009. [DOI] [PubMed] [Google Scholar]
- 25.Confer AW, Ayalew S. 2013. The OmpA family of proteins: roles in bacterial pathogenesis and immunity. Vet Microbiol 163:207–222. doi: 10.1016/j.vetmic.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 26.Moos A, Hackstadt T. 1987. Comparative virulence of intra- and interstrain lipopolysaccharide variants of Coxiella burnetii in the guinea pig model. Infect Immun 55:1144–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hotta A, Kawamura M, To H, Andoh M, Yamaguchi T, Fukushi H, Hirai K. 2002. Phase variation analysis of Coxiella burnetii during serial passage in cell culture by use of monoclonal antibodies. Infect Immun 70:4747–4749. doi: 10.1128/IAI.70.8.4747-4749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thiele D, Willems H. 1994. Is plasmid based differentiation of Coxiella burnetii in ‘acute’ and ‘chronic’ isolates still valid? Eur J Epidemiol 10:427–434. doi: 10.1007/BF01719667. [DOI] [PubMed] [Google Scholar]
- 29.Jager C, Willems H, Thiele D, Baljer G. 1998. Molecular characterization of Coxiella burnetii isolates. Epidemiol Infect 120:157–164. doi: 10.1017/S0950268897008510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gimenez DF. 1964. Staining rickettsiae in yolk-sac cultures. Stain Technol 39:135–140. doi: 10.3109/10520296409061219. [DOI] [PubMed] [Google Scholar]
- 31.Hackstadt T. 1986. Antigenic variation in the phase I lipopolysaccharide of Coxiella burnetii isolates. Infect Immun 52:337–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goellner S, Schubert E, Liebler-Tenorio E, Hotzel H, Saluz HP, Sachse K. 2006. Transcriptional response patterns of Chlamydophila psittaci in different in vitro models of persistent infection. Infect Immun 74:4801–4808. doi: 10.1128/IAI.01487-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adler H, Peterhans E, Jungi TW. 1994. Generation and functional characterization of bovine bone marrow-derived macrophages. Vet Immunol Immunopathol 41:211–227. doi: 10.1016/0165-2427(94)90098-1. [DOI] [PubMed] [Google Scholar]
- 34.Werling D, Howard CJ, Niederer E, Straub OC, Saalmüller A, Langhans W. 1998. Analysis of the phenotype and phagocytic activity of monocytes/macrophages from cattle infected with the bovine leukaemia virus. Vet Immunol Immunopathol 62:185–195. doi: 10.1016/S0165-2427(98)00074-9. [DOI] [PubMed] [Google Scholar]
- 35.Menge C, Loos D, Bridger PS, Barth S, Werling D, Baljer G. 2015. Bovine macrophages sense Escherichia coli Shiga toxin 1. Innate Immun 21:655–664. doi: 10.1177/1753425915581215. [DOI] [PubMed] [Google Scholar]
- 36.Prohl A, Ostermann C, Lohr M, Reinhold P. 2014. The bovine lung in biomedical research: visually guided bronchoscopy, intrabronchial inoculation and in vivo sampling techniques. J Vis Exp 2014(89):51557 doi: 10.3791/51557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klee SR, Tyczka J, Ellerbrok H, Franz T, Linke S, Baljer G, Appel B. 2006. Highly sensitive real-time PCR for specific detection and quantification of Coxiella burnetii. BMC Microbiol 6:2. doi: 10.1186/1471-2180-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dörner J. 2011. Wirksamkeitsprüfung chemischer Verfahren zur Desinfektion von Coxiella burnetii in kontaminierten Bodenmatrizes. VVB Laufersweiler Verlag, Gießen, Germany. [Google Scholar]
- 39.Spearman C, Kärber G. 1974. Virologische Arbeitsmethoden, vol 1. Gustav-Fischer-Verlag, Stuttgart, Germany. [Google Scholar]
- 40.Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 42.Capo C, Lindberg FP, Meconi S, Zaffran Y, Tardei G, Brown EJ, Raoult D, Mege JL. 1999. Subversion of monocyte functions by Coxiella burnetii: impairment of the cross-talk between alphavbeta3 integrin and CR3. J Immunol 163:6078–6085. [PubMed] [Google Scholar]
- 43.Stein A, Louveau C, Lepidi H, Ricci F, Baylac P, Davoust B, Raoult D. 2005. Q fever pneumonia: virulence of Coxiella burnetii pathovars in a murine model of aerosol infection. Infect Immun 73:2469–2477. doi: 10.1128/IAI.73.4.2469-2477.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voth DE, Howe D, Heinzen RA. 2007. Coxiella burnetii inhibits apoptosis in human THP-1 cells and monkey primary alveolar macrophages. Infect Immun 75:4263–4271. doi: 10.1128/IAI.00594-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brennan RE, Russell K, Zhang G, Samuel JE. 2004. Both inducible nitric oxide synthase and NADPH oxidase contribute to the control of virulent phase I Coxiella burnetii infections. Infect Immun 72:6666–6675. doi: 10.1128/IAI.72.11.6666-6675.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghigo E, Pretat L, Desnues B, Capo C, Raoult D, Mege JL. 2009. Intracellular life of Coxiella burnetii in macrophages. Ann N Y Acad Sci 1166:55–66. doi: 10.1111/j.1749-6632.2009.04515.x. [DOI] [PubMed] [Google Scholar]
- 47.Howe D, Shannon JG, Winfree S, Dorward DW, Heinzen RA. 2010. Coxiella burnetii phase I and II variants replicate with similar kinetics in degradative phagolysosome-like compartments of human macrophages. Infect Immun 78:3465–3474. doi: 10.1128/IAI.00406-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shannon JG, Howe D, Heinzen RA. 2005. Virulent Coxiella burnetii does not activate human dendritic cells: role of lipopolysaccharide as a shielding molecule. Proc Natl Acad Sci U S A 102:8722–8727. doi: 10.1073/pnas.0501863102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borrello S, Nicolo C, Delogu G, Pandolfi F, Ria F. 2011. TLR2: a crossroads between infections and autoimmunity? Int J Immunopathol Pharmacol 24:549–556. [DOI] [PubMed] [Google Scholar]
- 50.Meghari S, Honstettre A, Lepidi H, Ryffel B, Raoult D, Mege JL. 2005. TLR2 is necessary to inflammatory response in Coxiella burnetii infection. Ann N Y Acad Sci 1063:161–166. doi: 10.1196/annals.1355.025. [DOI] [PubMed] [Google Scholar]
- 51.Zamboni DS, Campos MA, Torrecilhas AC, Kiss K, Samuel JE, Golenbock DT, Lauw FN, Roy CR, Almeida IC, Gazzinelli RT. 2004. Stimulation of toll-like receptor 2 by Coxiella burnetii is required for macrophage production of pro-inflammatory cytokines and resistance to infection. J Biol Chem 279:54405–54415. doi: 10.1074/jbc.M410340200. [DOI] [PubMed] [Google Scholar]
- 52.Dellacasagrande J, Ghigo E, Hammami SM, Toman R, Raoult D, Capo C, Mege JL. 2000. alpha(v)beta(3) integrin and bacterial lipopolysaccharide are involved in Coxiella burnetii-stimulated production of tumor necrosis factor by human monocytes. Infect Immun 68:5673–5678. doi: 10.1128/IAI.68.10.5673-5678.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Netea MG, Kullberg BJ, Galama JM, Stalenhoef AF, Dinarello CA, Van der Meer JW. 2002. Non-LPS components of Chlamydia pneumoniae stimulate cytokine production through Toll-like receptor 2-dependent pathways. Eur J Immunol 32:1188–1195. [DOI] [PubMed] [Google Scholar]
- 54.Yamamoto Y, Okubo S, Klein TW, Onozaki K, Saito T, Friedman H. 1994. Binding of Legionella pneumophila to macrophages increases cellular cytokine mRNA. Infect Immun 62:3947–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koebnik R, Locher KP, Van Gelder P. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol Microbiol 37:239–253. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- 56.Jeannin P, Magistrelli G, Goetsch L, Haeuw JF, Thieblemont N, Bonnefoy JY, Delneste Y. 2002. Outer membrane protein A (OmpA): a new pathogen-associated molecular pattern that interacts with antigen presenting cells-impact on vaccine strategies. Vaccine 20(Suppl 4):A23–A27. doi: 10.1016/S0264-410X(02)00383-3. [DOI] [PubMed] [Google Scholar]
- 57.Bhowmick R, Pore D, Chakrabarti MK. 2014. Outer membrane protein A (OmpA) of Shigella flexneri 2a induces TLR2-mediated activation of B cells: involvement of protein tyrosine kinase, ERK and NF-kappaB. PLoS One 9:e109107. doi: 10.1371/journal.pone.0109107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Martinez E, Cantet F, Fava L, Norville I, Bonazzi M. 2014. Identification of OmpA, a Coxiella burnetii protein involved in host cell invasion, by multi-phenotypic high-content screening. PLoS Pathog 10:e1004013. doi: 10.1371/journal.ppat.1004013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Retzlaff C, Yamamoto Y, Okubo S, Hoffman PS, Friedman H, Klein TW. 1996. Legionella pneumophila heat-shock protein-induced increase of interleukin-1 beta mRNA involves protein kinase C signalling in macrophages. Immunology 89:281–288. doi: 10.1046/j.1365-2567.1996.d01-735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Macellaro A, Tujulin E, Hjalmarsson K, Norlander L. 1998. Identification of a 71-kilodalton surface-associated Hsp70 homologue in Coxiella burnetii. Infect Immun 66:5882–5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Knittler MR, Berndt A, Bocker S, Dutow P, Hanel F, Heuer D, Kagebein D, Klos A, Koch S, Liebler-Tenorio E, Ostermann C, Reinhold P, Saluz HP, Schofl G, Sehnert P, Sachse K. 2014. Chlamydia psittaci: new insights into genomic diversity, clinical pathology, host-pathogen interaction and anti-bacterial immunity. Int J Med Microbiol 304:877–893. doi: 10.1016/j.ijmm.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 62.Dinarello CA. 1997. Interleukin-1. Cytokine Growth Factor Rev 8:253–265. doi: 10.1016/S1359-6101(97)00023-3. [DOI] [PubMed] [Google Scholar]
- 63.Dinarello CA. 1998. Interleukin-1 beta, interleukin-18, and the interleukin-1 beta converting enzyme. Ann N Y Acad Sci 856:1–11. doi: 10.1111/j.1749-6632.1998.tb08307.x. [DOI] [PubMed] [Google Scholar]
- 64.van de Veerdonk FL, Netea MG, Dinarello CA, Joosten LA. 2011. Inflammasome activation and IL-1beta and IL-18 processing during infection. Trends Immunol 32:110–116. doi: 10.1016/j.it.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 65.Netea MG, Nold-Petry CA, Nold MF, Joosten LA, Opitz B, van der Meer JH, van de Veerdonk FL, Ferwerda G, Heinhuis B, Devesa I, Funk CJ, Mason RJ, Kullberg BJ, Rubartelli A, van der Meer JW, Dinarello CA. 2009. Differential requirement for the activation of the inflammasome for processing and release of IL-1beta in monocytes and macrophages. Blood 113:2324–2335. doi: 10.1182/blood-2008-03-146720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vitiello L, Gorini S, Rosano G, la Sala A. 2012. Immunoregulation through extracellular nucleotides. Blood 120:511–518. doi: 10.1182/blood-2012-01-406496. [DOI] [PubMed] [Google Scholar]
- 67.Zaborina O, Li X, Cheng G, Kapatral V, Chakrabarty AM. 1999. Secretion of ATP-utilizing enzymes, nucleoside diphosphate kinase and ATPase, by Mycobacterium bovis BCG: sequestration of ATP from macrophage P2Z receptors? Mol Microbiol 31:1333–1343. doi: 10.1046/j.1365-2958.1999.01240.x. [DOI] [PubMed] [Google Scholar]
- 68.Coutinho-Silva R, Perfettini JL, Persechini PM, Dautry-Varsat A, Ojcius DM. 2001. Modulation of P2Z/P2X(7) receptor activity in macrophages infected with Chlamydia psittaci. Am J Physiol Cell Physiol 280:C81–C89. [DOI] [PubMed] [Google Scholar]
- 69.Aimanianda V, Haensler J, Lacroix-Desmazes S, Kaveri SV, Bayry J. 2009. Novel cellular and molecular mechanisms of induction of immune responses by aluminum adjuvants. Trends Pharmacol Sci 30:287–295. doi: 10.1016/j.tips.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 70.Benoit M, Barbarat B, Bernard A, Olive D, Mege JL. 2008. Coxiella burnetii, the agent of Q fever, stimulates an atypical M2 activation program in human macrophages. Eur J Immunol 38:1065–1070. doi: 10.1002/eji.200738067. [DOI] [PubMed] [Google Scholar]
- 71.Monack DM, Mueller A, Falkow S. 2004. Persistent bacterial infections: the interface of the pathogen and the host immune system. Nat Rev Microbiol 2:747–765. doi: 10.1038/nrmicro955. [DOI] [PubMed] [Google Scholar]
- 72.Benoit M, Desnues B, Mege JL. 2008. Macrophage polarization in bacterial infections. J Immunol 181:3733–3739. doi: 10.4049/jimmunol.181.6.3733. [DOI] [PubMed] [Google Scholar]
- 73.van Furth R, Cohn ZA. 1968. The origin and kinetics of mononuclear phagocytes. J Exp Med 128:415–435. doi: 10.1084/jem.128.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pearson T, Hornstra HM, Hilsabeck R, Gates LT, Olivas SM, Birdsell DM, Hall CM, German S, Cook JM, Seymour ML, Priestley RA, Kondas AV, Clark Friedman CL, Price EP, Schupp JM, Liu CM, Price LB, Massung RF, Kersh GJ, Keim P. 2014. High prevalence and two dominant host-specific genotypes of Coxiella burnetii in U.S. milk. BMC Microbiol 14:41. doi: 10.1186/1471-2180-14-41. [DOI] [PMC free article] [PubMed] [Google Scholar]