Abstract
Aspergillus fumigatus is the most common pathogenic mold infecting humans and a significant cause of morbidity and mortality in immunocompromised patients. In invasive pulmonary aspergillosis, A. fumigatus spores are inhaled into the lungs, undergoing germination and invasive hyphal growth. The fungus occludes and disrupts the blood vessels, leading to hypoxia and eventual tissue necrosis. The ability of this mold to adapt to hypoxia is regulated in part by the sterol regulatory element binding protein (SREBP) SrbA and the DscA to DscD Golgi E3 ligase complex critical for SREBP activation by proteolytic cleavage. Loss of the genes encoding these proteins results in avirulence. To identify novel regulators of hypoxia sensing, we screened the Neurospora crassa gene deletion library under hypoxia and identified a novel rhomboid family protease essential for hypoxic growth. Deletion of the A. fumigatus rhomboid homolog rbdA resulted in an inability to grow under hypoxia, hypersensitivity to CoCl2, nikkomycin Z, fluconazole, and ferrozine, abnormal swollen tip morphology, and transcriptional dysregulation—accurately phenocopying deletion of srbA. In vivo, rbdA deletion resulted in increased sensitivity to phagocytic killing, a reduced inflammatory Th1 and Th17 response, and strongly attenuated virulence. Phenotypic rescue of the ΔrbdA mutant was achieved by expression and nuclear localization of the N terminus of SrbA, including its HLH domain, further indicating that RbdA and SrbA act in the same signaling pathway. In summary, we have identified RbdA, a novel putative rhomboid family protease in A. fumigatus that mediates hypoxia adaptation and fungal virulence and that is likely linked to SrbA cleavage and activation.
INTRODUCTION
The filamentous fungus Aspergillus fumigatus grows as an environmental saprophyte but can infect humans and animals through its dispersed asexual conidia. Upon inhalation by an immunocompromised host, conidia can germinate to form hyphae which infiltrate the surrounding lung tissue, causing often fatal invasive aspergillosis (IA). It is estimated that worldwide, more than 200,000 individuals suffer from life-threatening IA, with mortality rates ranging between 30 to 95% (1). These alarming statistics are a result of the generally poor health status of the patients, limited diagnostic and therapeutic options, and the arsenal of virulence-promoting properties of this ubiquitous mold (2, 3). Besides the abilities of the fungus to produce potent mycotoxins and to escape from the host innate immune cells, thermophilic growth and versatile nutrient acquisition are further general physiological attributes which favor growth on decaying matter as well as in the human host (4, 5). Adaptation to O2 concentrations below normal atmospheric levels of 21% is an additional challenge that A. fumigatus must cope with during colonization. O2 levels can drop to 2 to 4% on the surface of healthy alveolar tissue and be further diminished to less than 1% upon inflammation (6). Such levels of hypoxia are normally no threat for many aspergilli, as trace oxygen levels are often sufficient to support their growth. To sustain electron flow during such conditions, A. fumigatus fine-tunes components of its respiratory machinery (7–10). Enzymatic steps requiring molecular O2 in heme or ergosterol biosynthesis are highly sensitive to hypoxia and are under the control of the sterol regulatory element binding proteins (SREBPs) SrbA and SrbB. In the absence of both of these transcription factors, fungal growth is fully eliminated even at mildly hypoxic conditions of 1% O2, and its ability to cause disease in different mouse models for IA is abolished, suggesting a direct connection between virulence and hypoxia adaptation (11, 12). Further support is provided through the principal target genes of these transcription factors, which encode enzymes of the ergosterol biosynthetic pathway (13, 14). Azole-based antifungals inhibit these enzymes, and hence, the srbA-null mutant is highly sensitive to such treatments. Sterol biosynthesis is highly dependent on molecular O2, and the level of sterols can therefore be exploited as an indirect measure of O2 availability by SREBPs. This connection was first described for the fission yeast Schizosaccharomyces pombe (15) and later for Cryptococcus neoformans (16). The N-terminal helix-loop-helix (HLH) transcription factor domain is anchored to the membrane of the endoplasmic reticulum (ER) by two consecutive transmembrane segments and transported to the Golgi apparatus upon hypoxia-induced reduction of membrane sterols. Unlike mammals, fungi depend on the multisubunit machinery Dsc (defective for SREBP cleavage) consisting of a least six different membrane proteins (Dsc1 to -6 [Dsc1-6]) for sterol-mediated control at the Golgi apparatus (17, 18). The essential role of the Dsc complex in SrbA-dependent regulation is also conserved in A. fumigatus (19) and marks an important difference between higher mammals and the pathogenic fungus. The Dsc proteins are presumed to enable the proteolytic cleavage of SrbA that releases the N-terminal transcription factor domain to activate the hypoxic response. The identity of the SrbA-cleaving protease is not known. Here, we have exploited the Neurospora crassa gene deletion library (20) to identify genes which function during hypoxic growth. Besides gene members of the dsc complex, we have identified RbdA, a conserved putative rhomboid type protease that is essential for SrbA-dependent hypoxic growth in A. fumigatus and that is likely linked to SrbA cleavage and activation.
MATERIALS AND METHODS
Screening of the N. crassa genome deletion library.
The library, containing 121 96-well plates and 11,512 deletion strains (Fungal Genetics Stock Center, Kansas State University), was replicated twice with a 96-pin stainless steel replicator on 96-well plates containing 150 μl YAG broth medium/well. The plates were screened under normoxic or hypoxic (5% CO2, 1% O2, 94% N2) conditions for 24 h at 37°C. Results were recorded as growth or no growth when viewed under ×10 magnification. An srbA-null strain of A. fumigatus was used as a positive control under hypoxic conditions. Suspected hypoxia mutants were streaked on YAG-Triton 0.1% agar plates. Isolated colonies were inoculated on YAG agar plates and retested under normoxic and hypoxic conditions.
Strains and media.
The strains used in this study are detailed in Table S1 in the supplemental material. A. fumigatus conidia were harvested in 0.2% (vol/vol) Tween 20, resuspended in double-distilled water (DDW), and counted with a hemocytometer. Growth was performed in either (i) minimal medium (MM) containing 70 mM NaNO3, 1% (wt/vol) glucose, 12 mM potassium phosphate (pH 6.8), and 4 mM MgSO4, supplemented with trace elements and vitamins (1.5% [wt/vol] agar was added for MM agar plates); (ii) RPMI-MOPS containing RPMI 1640 medium (Biological Industries, Beit Haemek, Israel) plus 0.165 M MOPS buffer at pH 7.0; or (iii) rich YAG medium, which consists of 0.5% yeast extract, 1% glucose, and 10 mM MgCl2, supplemented with trace elements and vitamins (1.5% agar was added for both MM and YAG agar plates).
Generation of A. fumigatus mutant strains.
Generation of gene deletion/reconstituted strains and GFP-tagged strains was performed as described in Fig. S1 and S2 in the supplemental material. Transformation was performed as previously described (21).
MIC determination.
The MIC (the lowest drug concentration to completely inhibit fungal growth) was determined with the 2-fold microdilution method in a 96-well plate. A total of 2.5 × 103 A. fumigatus conidia per well were added in a total volume of 200 μl MM or RPMI-MOPS liquid medium per well. The plates were incubated at 37°C for 48 h unless otherwise specified. MIC values were determined by light microscopy.
Droplet growth inhibition assay.
Freshly harvested A. fumigatus conidia were diluted to 107, 106, 105, 104, and 103 spores/ml. A 10-μl droplet of spore suspension was placed on YAG plates containing 0, 1, 3, or 6 mg/ml CoCl2 (Sigma). Plates were incubated for 48 h at 37°C.
A. fumigatus organelle staining.
A total of 1.5 × 105 spores/ml were grown on rich YAG medium in a 6-well plate, containing round coverslips, for 8 h at 37°C. The coverslips were then washed twice with DDW and treated accordingly. Untreated coverslips were used for light microscopy. Fluorescence microscopy was used to determine differences in organelle morphology. Nuclear staining was performed for 20 min at room temperature using a DAPI (4′,6-diamidino-2-phenylindole) solution (50 mM KPO4 [pH 6.8], 0.2% [vol/vol] Triton X-100, 5% [vol/vol] glutaraldehyde, 0.05 μg/ml DAPI). Membrane staining was performed for 30 min at room temperature using FM4-64 solution (Invitrogen; final concentration, 4 μg/ml in rich YAG medium). Mitochondrial staining was performed for 30 min at 37°C using MitoTracker green FM solution (Invitrogen; final concentration, 1 μM). Cell wall staining was performed for 5 min at room temperature using calcoflour white (CFW) solution (Sigma; final concentration, 0.1 mg/ml). Dye staining was performed in the dark and was followed by two wash cycles in DDW, before imaging.
Biofilm formation.
Biofilm formation was assessed as previously described by Gravelat et al. (22). A. fumigatus spores/105 ml were inoculated in 50 ml RPMI-MOPS liquid medium in 250-ml Erlenmeyer flasks and incubated at 37°C with shaking at 250 rpm for 24 h. Biofilm formation on the glass rim was estimated visually.
Isolation of RNA and Northern hybridization.
For isolation of total RNA, mycelia were harvested by filtration through Miracloth tissue (Merck Chemicals GmbH, Darmstadt, Germany) and flash-frozen in liquid N2. Mycelia were disintegrated using a FastPrep 24 system (MP Biomedicals, Illkirch, France) in the presence of TRIsure (Bioline GmbH, Luckenwalde, Germany). After the addition of chloroform and phase separation centrifugation, RNA was precipitated and solubilized in H2O. RNA preparations were used for nonradioactive Northern hybridizations with DIGoxigenin (Jena Bioscience GmbH, Jena, Germany)-labeled PCR products as probes. Primers used for DNA amplification and DIG (digoxigenin) labeling of gene-specific probes are listed in Table S2 in the supplemental material. Semidry blotting, hybridization, and fluorescence-based detection were all performed as described previously (9).
Fluorescence microscopy.
For microscopic imaging of green fluorescent protein (GFP)-expressing A. fumigatus, hyphae were grown at 37°C on coverslips for 16 h in MM containing 1% (wt/vol) glucose and 20 mM glutamine. Xylose was added at the concentrations indicated in the figures, and hyphae were either left in air or transferred to a hypoxic workstation (model H35; Don Whitley Scientific Limited, West Yorkshire, United Kingdom) for 3 h. Before imaging, cells were fixed for 10 min in 3.7% (vol/vol) formaldehyde and nuclei were stained with Hoechst 33342 solution at a final concentration of 0.1 mg/ml. All images were captured using an Axio Imager M2 microscope (Carl Zeiss, Jena, Germany) with a 63× oil immersion objective lens (numerical aperture, 1.40). Images were documented using an AxioCam MRm camera (Carl Zeiss, Jena, Germany) and processed using ZEN 2012 imaging software (Carl Zeiss).
Galleria mellonella larval model of infection.
Sixth-instar-stage G. mellonella larvae were injected through the last proleg with 5 ×106 A. fumigatus conidia, which had been suspended in 10 μl of phosphate-buffered saline (PBS) as previously described (23). The larvae were kept for 8 days at 37°C, and death was monitored by visual inspection and prodding with a toothpick.
Immunocompromised murine model of aspergillosis.
Six-week-old female ICR mice were immunocompromised by subcutaneous injection with cortisone acetate (300 mg/kg) 3 days prior to infection, on the day of infection, and 3, 7, and 11 days postinfection. The mice were infected intranasally with 5 ×105 dormant spores, which had been suspended in 20 μl of PBS plus 0.2% Tween 20 (10 μl in each nostril). Mortality was monitored for 12 days. Experiments were ethically approved by the Ministry of Health (MOH) Animal Welfare Committee, Israel.
Nonimmunocompromised murine model of aspergillosis.
Six- to 8-week-old C57BL/6 mice purchased from Charles River (Calco, Italy) and genetically engineered homozygote Cftr−/− (Cftrtm1Unc) mice in the C57BL/6 background (24) were bred under specific-pathogen-free conditions at the Animal Facility of San Raffaele Hospital, Milan Italy. Murine experiments were performed according to the Italian Approved Animal Welfare Authorization 360/2015-PR and Legislative Decree 26/2014 under an animal license obtained from the Italian Ministry of Health lasting for 5 years (2015 to 2020). Viable conidia (95%) of A. fumigatus wild-type (WT) and mutant strains were obtained by growth on Sabouraud dextrose agar (Difco Laboratories, Detroit, MI, USA) supplemented with chloramphenicol for 4 days at room temperature. Mice were anesthetized by intraperitoneal (i.p.) injection of 2.5% 2,2,2-tribromoethanol (Avertin; Sigma Chemical Co, St. Louis, MO) before intranasal instillation of a suspension of 2 × 107 resting conidia/20 μl saline/mouse. Mice were monitored for fungal growth and histology. Fungal growth was expressed as log10 CFU per organ (CFU; mean ± standard deviation [SD]). For histology, paraffin-embedded sections (3 to 4 μm) were stained with periodic acid-Schiff (PAS). Histology sections and cytospin preparations were observed using a BX51 microscope (Olympus, Milan, Italy), and images were captured using a high-resolution DP71 camera (Olympus).
Conidiocidal assay.
Peritoneal polymorphonuclear leukocytes (PMNs) were incubated with unopsonized resting conidia at a 1:1 ratio at 37°C for determinations of conidiocidal activity (percentage of CFU inhibition, mean ± standard error [SE]) at 60 min.
Reverse transcriptase PCR (RT-PCR).
Total RNA extraction (TRIzol; Invitrogen Srl Life Technologies, Milan, Italy) and synthesis and PCR of cDNA were done as described previously (24). The PCR primers were as listed in Table S2 in the supplemental material. The mRNA-normalized data were expressed as the gene mRNA in mice infected with the WT strain relative to that in mice infected with the mutant strains.
RESULTS
Identification of novel hypoxia-sensitive mutants in N. crassa.
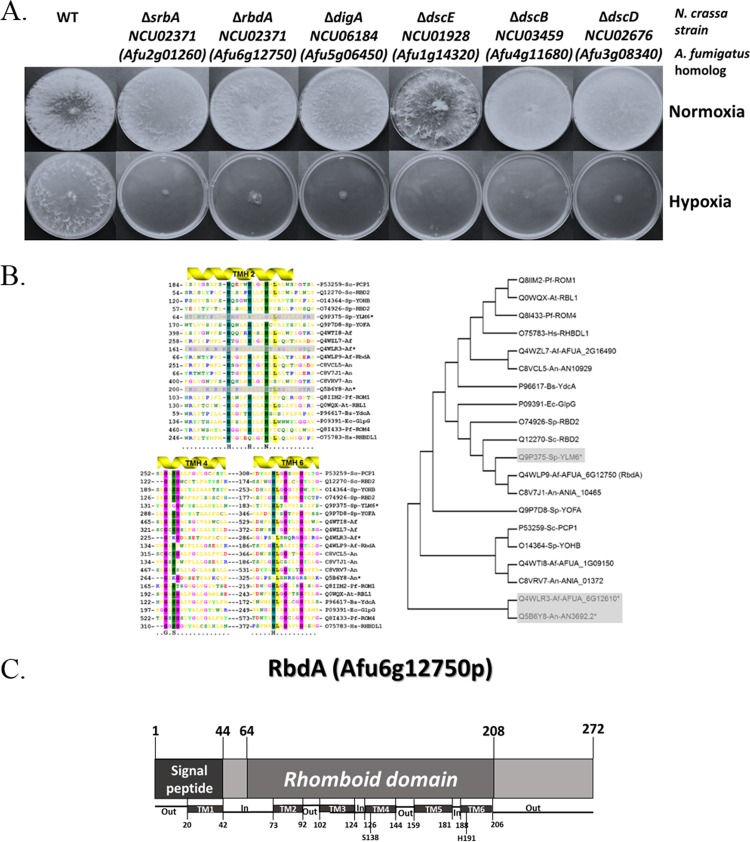
To identify novel genes involved in hypoxia adaptation in a filamentous fungus, we took advantage of the unique N. crassa gene knockout library. All 11,512 N. crassa library strains were pin-replicated into 96-well YAG liquid plates and grown in parallel under normoxic or hypoxic (1% O2) conditions for 24 h at 37°C. Six strains that grew under normoxia and did not grow under hypoxia were identified (Fig. 1A). Of these, four strains contained deletions in homologs of the previously discovered srbA, dscB, dscD, and dscE genes involved in hypoxia adaptation. dscE was selected for preliminary analysis in A. fumigatus since it has not been previously studied in the filamentous fungi. One strain contained a deletion in a gene encoding a homolog of Aspergillus nidulans DigA, which is involved in nuclear migration, mitochondrial morphology, and polarized growth (25). However, it is an essential gene in N. crassa that can survive only as a heterokaryon, and we were unable to generate digA deletion strains in A. fumigatus (data not shown). Another strain contained a deletion in a gene encoding a rhomboid family protease (NCU02371). This gene was selected for further detailed study in A. fumigatus, and we named it rbdA. Rhomboid family proteases are present in nearly all of the sequenced genomes of archaea, bacteria, and eukaryotes and function in diverse processes, including quorum sensing in bacteria and mitochondrial membrane fusion, apoptosis, and stem cell differentiation in eukaryotes (26). There are four putative rhomboid family members in A. fumigatus (Afu6g12750 [rbdA], Afu6g12610, Afu2g16490, and Afu1g09150), none previously studied. rbdA is closely related to Saccharomyces cerevisiae RBD2 (YPL246C) (27) and Schizosaccharomyces pombe RBD2 (SPCC790.03) (28) (Fig. 1B). Like all rhomboid family members, it is comprised of a conserved core structure of six transmembrane segments that contain the catalytic serine-histidine proteolytic pair (Fig. 1C).
FIG 1.
Identification of novel hypoxia-sensitive mutants of N. crassa. (A) Screening of the N. crassa gene knockout library identified six mutant strains with wild-type (WT) growth under normoxia (upper panel, 21% O2) and impaired growth under hypoxia (lower panel, <1% O2). Strains were point inoculated on YAG agar plates and incubated for 48 h at 37°C. (B) Identification of rhomboid proteases in fungal genomes. (Left) Proteins containing the peptidase S54 domain rhomboid (IPR002610) were identified from the sequenced genomes of S. cerevisiae (Sc), S. pombe (Sp), A. fumigatus (Af), and A. nidulans (An) using an InterPro domain scan, and the sequences were aligned with those of rhomboid-like proteins from human (Hs), Arabidopsis thaliana (At), Plasmodium falciparum (Pf), Bacillus subtilis (Bs), and Escherichia coli (Ec) using MUltiple Sequence Comparison by Log-Expectation (MUSCLE). Transmembrane helices (TMH) containing the conserved HXXXXHXXXN motif and the putative catalytic residues serine and histidine in THM 4 and 6 are indicated. The three potentially catalytically inactive, fungal rhomboid-like proteins are marked with an asterisk and shaded in gray. (Right) Phylogenetic tree of the aligned rhomboid-like proteins, generated using the tree explorer function within MEGA version 6. (C) A. fumigatus RbdA contains six transmembrane (TM) domains. The catalytically conserved serine protease residues S138 and H191 are contained within TM domains 4 and 6, respectively.
rbdA and dscE are required for hypoxia adaptation in A. fumigatus.
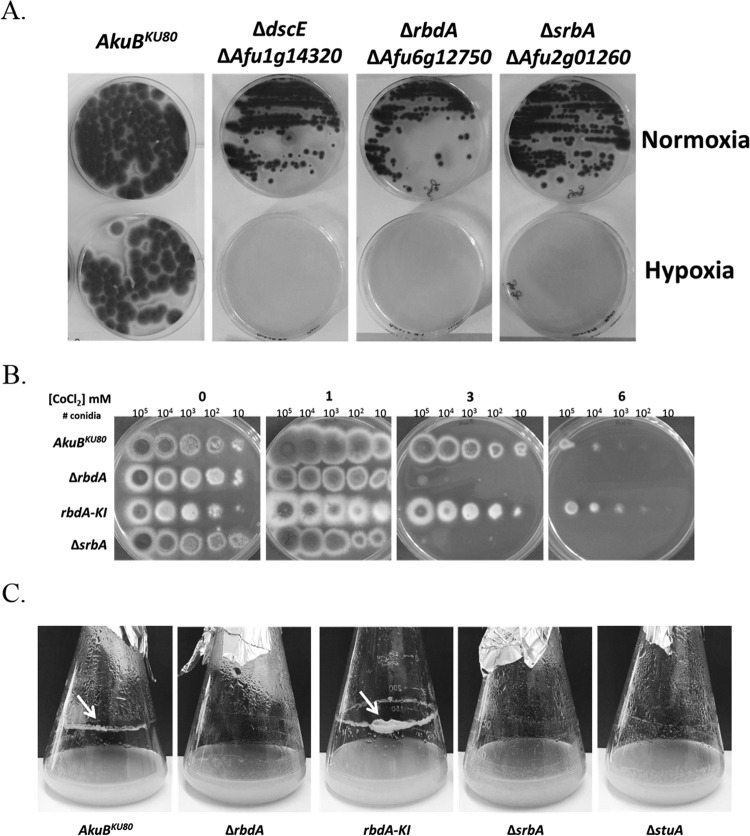
To identify the roles of rbdA and dscE in hypoxia adaptation in A. fumigatus, we generated null mutants of these genes and examined their growth under hypoxia in comparison to the well-characterized phenotype of the srbA deletion mutant (see the supplemental material for details on strain construction and verification). The ΔrbdA, ΔdscE, and ΔsrbA mutant strains grew normally under normoxic conditions but failed to grow under hypoxia (Fig. 2A). Furthermore, as previously shown for the ΔsrbA mutant strain, the ΔrbdA mutant strain was hypersensitive to the hypoxia-mimetic compound CoCl2 (Fig. 2B).
FIG 2.
Phenotypic in vitro characterization of the mutant strains. (A) rbdA and dscE are required for hypoxia adaptation in A. fumigatus. ΔrbdA, ΔdscE, and control AkuBKU80 and ΔsrbA strains were grown on YAG plates for 48 h at 37°C under normoxic (upper panel, 21% O2) and hypoxic (lower panel, <1% O2) conditions. (B) The ΔrbdA and ΔsrbA mutant strains are hypersensitive to CoCl2. The droplet dilution assay was used to evaluate strain sensitivity toward hypoxia. The strains were grown on YAG rich plates for 48 h at 37°C under increasing concentrations of CoCl2, as indicated above the images. An increase in sensitivity toward hypoxia was observed for the ΔrbdA and ΔsrbA mutant strains, whereas for the reconstituted strain, rbdA-KI, this sensitivity was restored and is similar to that of the AkuBKU80 background strain. (C) The ΔrbdA and ΔsrbA mutant strains do not form a biofilm rim. Strains were grown in shaking liquid culture (YAG) for 48 h at 37°C. AkuBKU80 control and reconstituted rbdA-KI strains form a biofilm rim (indicated by white arrows), whereas the ΔrbdA and ΔsrbA mutant strains do not. The ΔstuA mutant strain, which does not form a biofilm, was used as a negative control.
The ΔrbdA and ΔsrbA mutant strains do not form a biofilm.
A. fumigatus biofilms are formed under hypoxic conditions during lung infection, and the ability to form a biofilm is a key virulence determinant. We therefore examined the ability of the ΔrbdA and ΔsrbA mutant strains to form a biofilm rim while growing in shaking liquid culture (Fig. 2C). For a negative control, we included the A. fumigatus ΔstuA mutant, which does not form a biofilm (29). Results show that the AkuBKU80 parental strain and the rbdA knock-in (rbdA-KI) reconstituted strains form thick biofilm rims whereas the ΔrbdA and ΔsrbA mutant strains do not.
The ΔrbdA and ΔsrbA mutant strains are hypersensitive to CoCl2, nikkomycin Z, azoles, and ferrozine.
An extended phenotypic analysis was then performed on the ΔrbdA mutant strain, the focus of this report (Table 1). The reconstituted rbdA-KI control strain was prepared by ectopic integration of the rbdA gene under its endogenous promoter and terminator, as elaborated in the supplemental material. The AkuBKU80, ΔrbdA, rbdA-KI, and ΔsrbA strains were grown under increasing concentrations of various compounds in liquid MM or RPMI-MOPS, and MICs were determined (Table 1). Results show that the ΔrbdA and ΔsrbA mutant strains demonstrate identical hypersensitivities to CoCl2, azoles (ergosterol biosynthesis inhibitors), ferrozine (ferric iron chelator), and nikkomycin Z (chitin synthase inhibitor).
TABLE 1.
Hypersensitivity of ΔrbdA and ΔsrbA mutant strains to CoCl2, nikkomycin Z, fluconazole, voriconazole, and ferrozine
| Stressor compounda | MIC for: |
|||
|---|---|---|---|---|
| AkuBKU80 strain | ΔrbdA strain | rbdA-KI strain | ΔsrbA strain | |
| CoCl2 | 5 mM | 0.3 mM | 2.5 mM | 0.3 mM |
| Calcofluor white | 200 μg/ml | 200 μg/ml | 200 μg/ml | 200 μg/ml |
| Congo red | 12.5 μg/ml | 12.5 μg/ml | 12.5 μg/ml | 6.25 μg/ml |
| Caspofunginb | 0.5 μg/ml | 0.5 μg/ml | 0.5 μg/ml | 0.5 μg/ml |
| Nikkomycin Zb | >240 μg/ml | 120 μg/ml | >240 μg/ml | 120 μg/ml |
| Voriconazole | 0.3 μg/ml | 0.03 μg/ml | 0.6 μg/ml | 0.03 μg/ml |
| Fluconazole | >16 μg/ml | 8 μg/ml | >16 μg/ml | 8 μg/ml |
| Menadione | 0.6 μg/ml | 0.3 μg/ml | 0.3 μg/ml | 0.3 μg/ml |
| H2O2 | 3.125 mM | 6.25 mM | 6.25 mM | 6.25 mM |
| DTT | 10 mM | 10 mM | 10 mM | 10 mM |
| Ferrozine | >10 mM | 0.6 mM | >10 mM | 0.6 mM |
Ferrozine and nikkomycin Z were tested in RPMI-MOPS medium, and all other stressors were tested in liquid MM. DTT, dithiothreitol.
The minimal effective concentration (MEC), i.e., the lowest drug concentration to visibly inhibit fungal growth, was determined for caspofungin and nikkomycin Z.
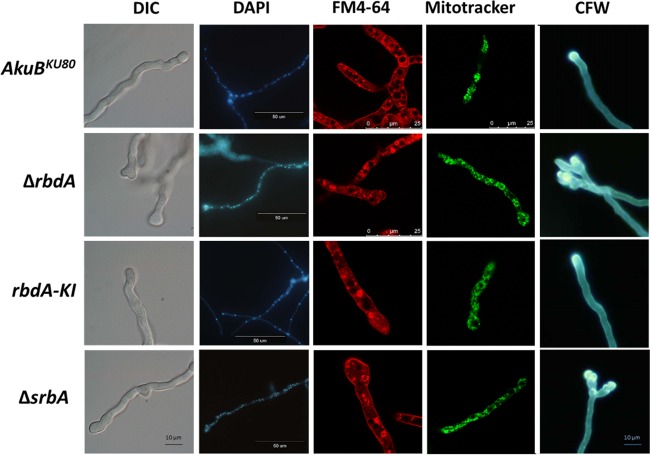
The ΔrbdA and ΔsrbA mutant strains exhibit abnormal tip branching and nuclear distribution.
We compared the microscopic structures of the ΔrbdA and ΔsrbA mutant strains under normoxic conditions. The ΔsrbA mutant strain was previously shown to display abnormal swollen-tip morphology (11). We found that the ΔrbdA mutant strain exhibited an identical swollen-tip phenotype (Fig. 3, differential interference contrast [DIC] images) and that in both ΔrbdA and ΔsrbA mutant strains, nuclei were abnormally clumped along the hyphae and in the swollen hyphal tips (Fig. 3, DAPI). Based on the hypersensitivity of the ΔrbdA and ΔsrbA mutant strains to nikkomycin Z and fluconazole, we also examined cell wall carbohydrate staining with calcofluor white and membrane staining with FM4-64. The swollen hyphal tips in both ΔrbdA and ΔsrbA mutant strains appeared to be stained to a greater extent by calcofluor white (Fig. 3, CFW). There were no differences in membrane staining with FM4-64 between the AkuBKU80 parental strain, the rbdA-KI reconstituted strain, and the ΔrbdA and ΔsrbA mutant strains (Fig. 3). We also examined mitochondrial morphology, because deletion of S. cerevisiae RBD1 results in the loss of mitochondrial integrity (27). However, the ΔrbdA and ΔsrbA mutant strains retained their normal wild-type mitochondrial morphology (Fig. 3, MitoTracker staining).
FIG 3.
The ΔrbdA and ΔsrbA mutant strains exhibit abnormal tip branching and nuclear distribution. Conidia were germinated on rich YAG medium under normoxic growth conditions for 16 h at 37°C. Swollen and split hyphal tips are evident in the ΔrbdA and ΔsrbA mutant strains in comparison to the hyphal tips of the AkuBKU80 background strain. This phenomenon is restored in the rbdA-KI reconstituted strain. The hyphae of the ΔrbdA and ΔsrbA strains contained unevenly distributed nuclei (DAPI). Moreover, the swollen hyphal tips of the ΔrbdA and ΔsrbA mutant strains contain large clusters of nuclei compared to the AkuBKU80 background strain or the rbdA-KI reconstituted strain. No other changes are evident in membranes stained with FM4-64, in mitochondria stained with MitoTracker green FM, or in cell wall carbohydrates stained with calcofluor white (CFW).
Hypoxic induction of SrbA target genes requires RbdA.
Since the ΔrbdA and ΔsrbA mutant strains displayed identical sensitivities under hypoxia and in the presence of sterol-targeting antifungals, we analyzed whether these phenotypic similarities were consequences of the same regulatory defects. Hypoxic expression of the genes encoding 14α sterol demethylase (erg11A) and C-4 sterol methyl oxidase (erg25A), two major monooxygenases of the ergosterol biosynthesis pathway, are under direct transcriptional control of SrbA (12). Northern hybridizations of erg25A and erg11A revealed that in wild-type cells, both genes are weakly expressed under normoxic conditions and are activated under hypoxia (Fig. 4). As expected, expression levels for both genes were just above detection levels in the ΔsrbA mutant and hypoxic induction was also fully abrogated in this strain (Fig. 4). Identical expression patterns were observed for erg11A and erg25A in the absence of RbdA, suggesting that the function of this protein is essential for SrbA-dependent gene regulation. Expression of the gene encoding hypoxia-inducible alcohol dehydrogenase, alcC, also depends at least partially on SrbA (12). Without SrbA, alcC mRNA expression was nearly silent, whereas some hypoxic induction of alcC mRNA expression was still functional in the ΔrbdA mutant strain, suggesting that the mode of regulation could be different than for the erg genes (Fig. 4). In contrast, regulation of the flavohemoprotein gene fhpA, whose transcription was found to be commonly responsive to NO and hypoxia (8, 30), did not depend on either SrbA or RbdA. These results suggest that RbdA is tightly connected to the transcriptional activator function of SrbA. SrbA is strongly upregulated in response to hypoxia in both the AkuBKU80 parental and ΔrbdA mutant strains, suggesting that srbA transcription is not affected by rbdA absence (see Fig. S3 in the supplemental material). Taken together, the identical chemical sensitivities, microscopic abnormalities, and transcriptional dysregulation of the ΔrbdA and ΔsrbA mutant strains suggest that the two genes are components of a shared hypoxia-sensing pathway.
FIG 4.
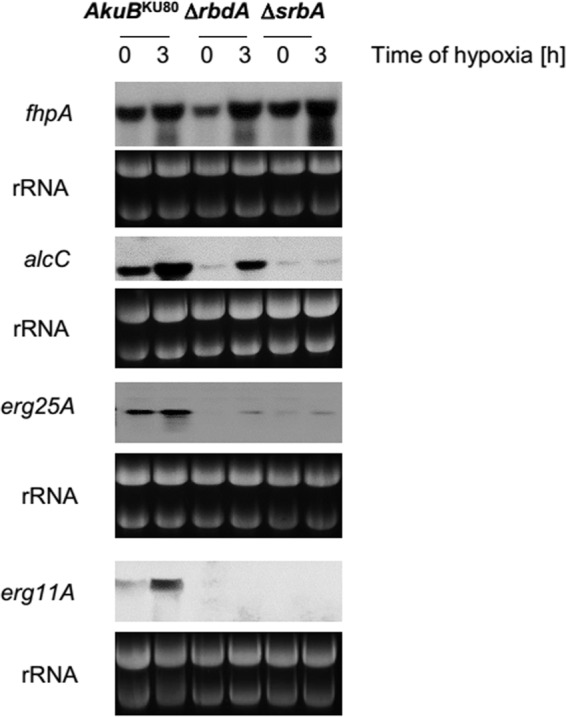
Expression of SrbA target genes in the AkuBKU80, ΔrbdA, and ΔsrbA strains. Northern blot analysis of the mRNA levels of SrbA target genes is shown for the AkuBKU80 wild-type and ΔrbdA and ΔsrbA mutant strains before (0 h) and after (3 h) a shift to a hypoxic atmosphere. All strains were initially cultivated for 16 h in MM (106 spores/ml inoculum) under normoxic conditions and then shifted for 3 h to hypoxic conditions (0.2% O2). Ten micrograms of RNA was loaded per lane, and rRNA bands are shown as a control for equal loading.
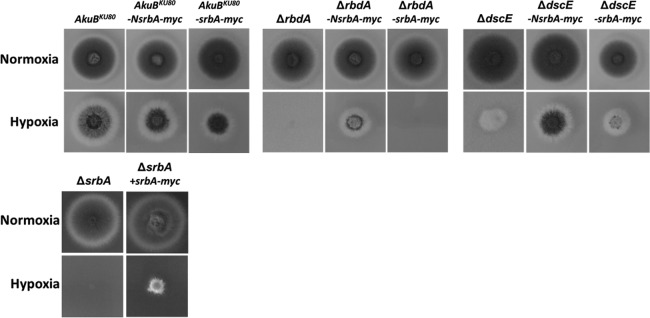
The HLH domain of SrbA rescues the hypoxic growth of the ΔrbdA mutant strain and localizes to the nucleus.
Based on the assumption that SrbA and RbdA are part of the same regulatory machinery, we hypothesized that correct processing and nuclear targeting of SrbA depends on a functional RbdA protein. If so, ectopic expression of the N terminus of SrbA, including its HLH domain, would complement the phenotype of the ΔrbdA mutant strain. To test this, we ectopically expressed myc-tagged N-terminal SrbA (NsrbA-myc, encoding the N-terminal 425 amino acids of SrbA [N-SrbA], under the endogenous srbA promoter and terminator) into the AkuBKU80, ΔrbdA, and ΔdscE strains to generate AkuBKU80-NsrbA-myc, ΔrbdA-NsrbA-myc, and ΔdscE-NsrbA-myc strains, respectively. For a control, we also ectopically expressed C-terminal myc-tagged full-length SrbA (srbA-myc, under the endogenous srbA promoter and terminator) to generate AkuBKU80-srbA-myc, ΔrbdA-srbA-myc, ΔdscE-srbA-myc, and ΔsrbA-srbA-myc strains (see the supplemental material for strain details). Both the ΔrbdA and ΔdscE mutant strains were functionally complemented for growth under hypoxia by transformation with the truncated N-terminally tagged NsrbA gene and not by the full-length srbA gene encoding SrbA-myc. The ΔsrbA strain was functionally complemented for growth under hypoxia by transformation with the full-length srbA gene encoding SrbA-myc, indicating that the SrbA-myc full-length protein is functional (Fig. 5). These results confirm that both the RbdA and DscE proteins are involved in cleavage and subsequent activation of SrbA. Despite repeated attempts, we could not visualize the full-length SrbA-myc protein by Western blotting and so cannot at this stage demonstrate that deletion of RbdA results in defective SrbA cleavage.
FIG 5.
The ΔrbdA and ΔdscE mutant strains are rescued for hypoxic growth by expression of cleaved N-SrbA but not full-length SrbA. AkuBKU80, ΔrbdA, and ΔdscE strains were transformed with plasmids expressing N-SrbA-myc (cleaved activated form) or full-length SrbA-myc under the endogenous srbA promoter. ΔrbdA-NsrbA-myc and ΔdscE-NsrbA-myc strains show partially restored growth under hypoxia, whereas ΔrbdA-srbA-myc and ΔdscE-srbA-myc strains do not, suggesting that RbdA and DscE are involved in srbAp activation by cleavage of its C-terminal domain.
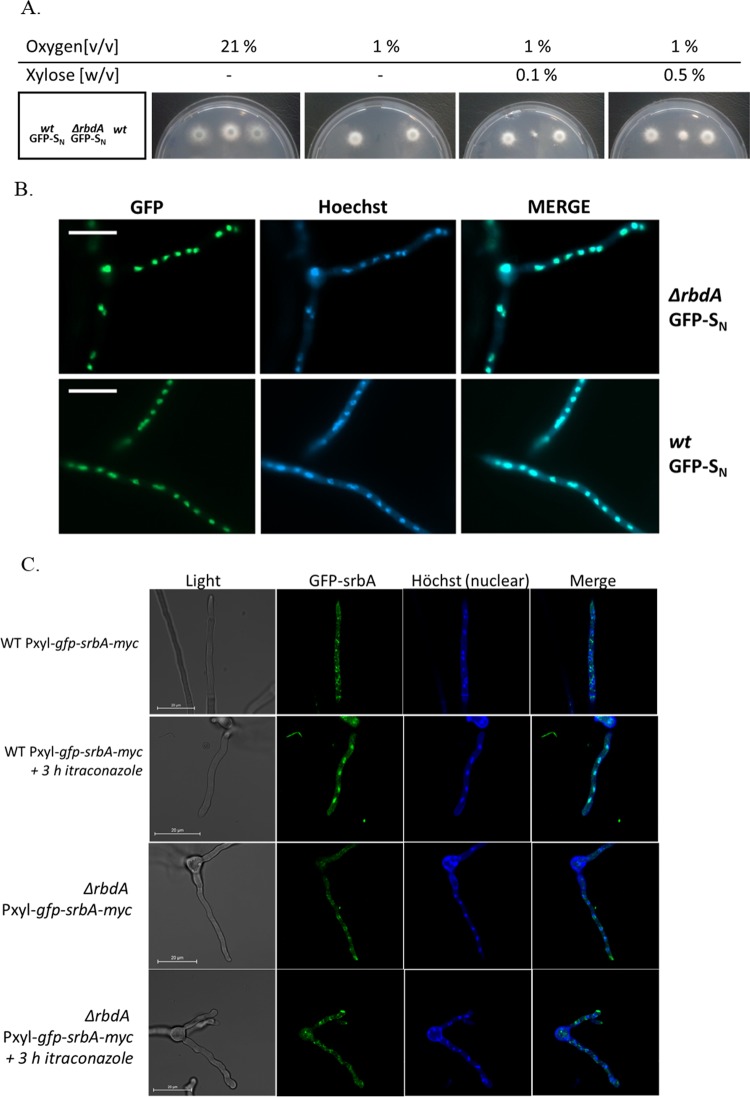
To follow the intracellular targeting of the N-SrbA domain, we fused it to GFP under the control of the endoxylanase promoter (xylp) from Penicillium chrysogenum to generate GFP-SrbANterm. The xylp promoter allows the xylose-inducible expression of target genes in A. fumigatus (31). Xylose-inducible expression of the GFP-SrbANterm domain in a wild-type background did not affect growth of A. fumigatus. The ΔrbdA mutant, in turn, failed to grow in a hypoxic atmosphere of 1% (vol/vol) O2 in the absence of the inducer, while xylose addition rescued the phenotype of the ΔrbdA mutant (Fig. 6A). At 0.5% xylose, hypoxic growth of the ΔrbdA mutant was nearly indistinguishable from that of the wild type. Phenotypic rescue of the ΔrbdA mutant coincided with the nuclear localization of GFP-SrbANterm (Fig. 6B) in the mutant, indicating that loss of the RbdA protein can be overcome by expression of the transcription factor SrbA in the nucleus.
FIG 6.
Hypoxic growth complementation and nuclear targeting of GFP-SrbANterm and GFP-SrbA. (A) A total of 105 conidia of parental A. fumigatus (AkuBKU80) or the ΔrbdA mutant strain expressing GFP-SrbANterm (GFP-SN) were point inoculated on MM with the indicated concentrations of xylose. After an initial incubation for 16 h in air, the plates were left in air (21% O2) or transferred to a hypoxic atmosphere (1% O2) for an additional 24 h. (B) Nuclear localization of GFP-SrbANterm was monitored by fluorescence micoscopy in the same strains using green and blue fluorescence channels for GFP- and Hoechst-stained nuclei, respectively. Bar, 50 μm. (C) Nuclear localization of GFP-SrbA is induced by azole treatment in the AkuBKU80 control strain and not in the ΔrbdA mutant strain. Bar, 20 μm.
SrbA-GFP does not undergo nuclear translocation in the ΔrbdA mutant.
To follow SrbA-GFP targeting to the nucleus in the AkuBKU80 control and ΔrbdA mutant strains, they were transformed with GFP-SrbA (full length; see the supplemental material), under the control of the endoxylanase promoter (xylp). Surprisingly, in both strains, GFP-SrbA showed a perinuclear localization under both normoxia and hypoxia, possibly as a result of the high level of expression induced by xylose (not shown). We therefore used itraconazole to reduce cellular ergosterol and strongly induce SrbA nuclear translocation. Results show that itraconazole (2.5 μM for 3 h) strongly induced SrbA nuclear translocation in the AkuBKU80 wild-type strain (Fig. 6C). In marked contrast, SrbA remained in a perinuclear location in the itraconazole-treated ΔrbdA strain, indicating that RbdA is essential for nuclear translocation of SrbA upon reduction of ergosterol levels.
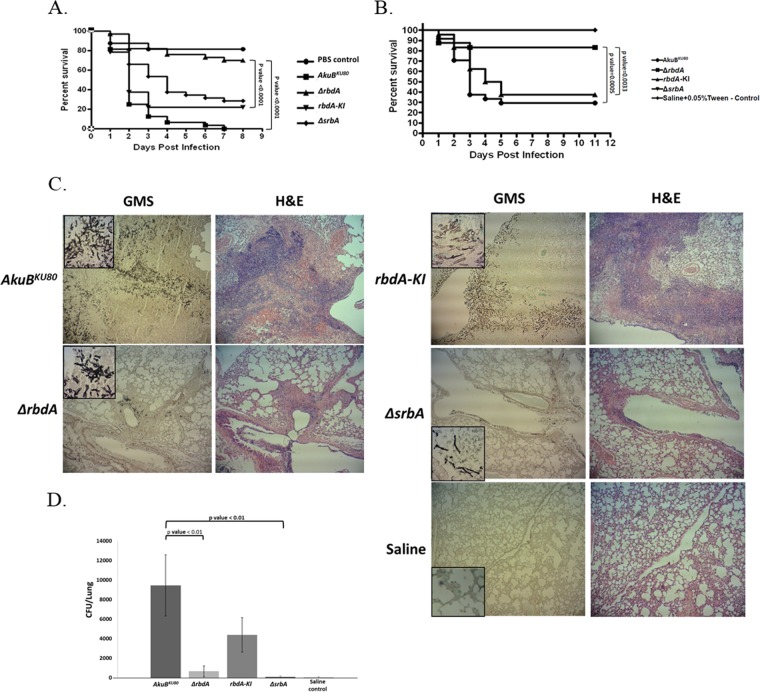
The ΔrbdA mutant strain shows reduced virulence in infected Galleria mellonella larvae and mice.
The ΔsrbA mutant has been previously shown to be avirulent in murine models of pulmonary aspergillosis (11). We therefore compared the levels of virulence of the AkuBKU80 parental and ΔsrbA, ΔrbdA, and rbdA-KI strains when injected into Galleria larvae or when administered intranasally into the lungs of cortisone acetate-immunocompromised mice, in which strong hypoxia is observed (32). Results show reduced virulence of the ΔrbdA strain in comparison to the AkuBKU80 parental strain in both Galleria mellonella (P < 0.0001) (Fig. 7A) and mice (P = 0.0005) (Fig. 7B). There was no statistically significant difference in the mortality rates of mice infected with either the ΔsrbA or ΔrbdA mutant strains. Histological examination of mouse lungs showed reduced hyphal growth, fungal colony size, host tissue damage, and inflammatory cell influx for mice infected with the ΔsrbA and ΔrbdA strains compared to those infected with the AkuBKU80 background strain and the reconstituted rbdA-KI strain (Fig. 7C). The lung fungal burdens of mice infected with the ΔsrbA and ΔrbdA mutant strains were significantly reduced in comparison to those of mice infected with the AkuBKU80 background strain (P < 0.01) (Fig. 7D). Taken together, these results indicate that the ΔsrbA and ΔrbdA mutant strains exhibit almost identical phenotypes of reduced virulence.
FIG 7.
The ΔrbdA mutant strain exhibits reduced virulence in the Galleria mellonella and murine models of infection. (A) G. mellonella larvae infected with the rbdA-null strain (ΔrbdA) showed a significant increase in their survival rates compared to those infected with the AkuBKU80 background strain or the rbdA-KI reconstituted strain (P < 0.0001, for both). (B) Survival curve of infected, cortisone acetate-immunocompromised mice, infected intranasally with A. fumigatus conidia. Mice infected with the ΔrbdA and ΔsrbA mutant strains showed a significant increase in their survival rates compared to the mice infected with the AkuBKU80 background strain (P = 0.0005) or the rbdA-KI reconstituted strain (P = 0.0033). (C) Mice infected with the ΔrbdA and ΔsrbA mutant strains showed reduced fungal colonization and lung pathology. Mice were infected as described in Materials and Methods. Three days after infection, the mice were sacrificed, and their lungs were removed and sent for histological staining with Grocott's methenamine silver stain (GMS; fungal staining) and hematoxylin and eosin (H&E; tissue and nuclear staining). The infecting ΔrbdA and ΔsrbA mutant strains showed reduced hyphal growth, colony size, tissue damage, and inflammatory cell influx compared to the AkuBKU80 background strain and the rbdA-KI reconstituted strain. (D) Mice infected with the ΔrbdA and ΔsrbA mutant strains showed reduced lung fungal loads. Infected mice were sacrificed on the third day postinfection, their lungs were removed and homogenized, and the homogenates were plated on YAG. The plates were incubated for 16 h, and the numbers of CFU were counted.
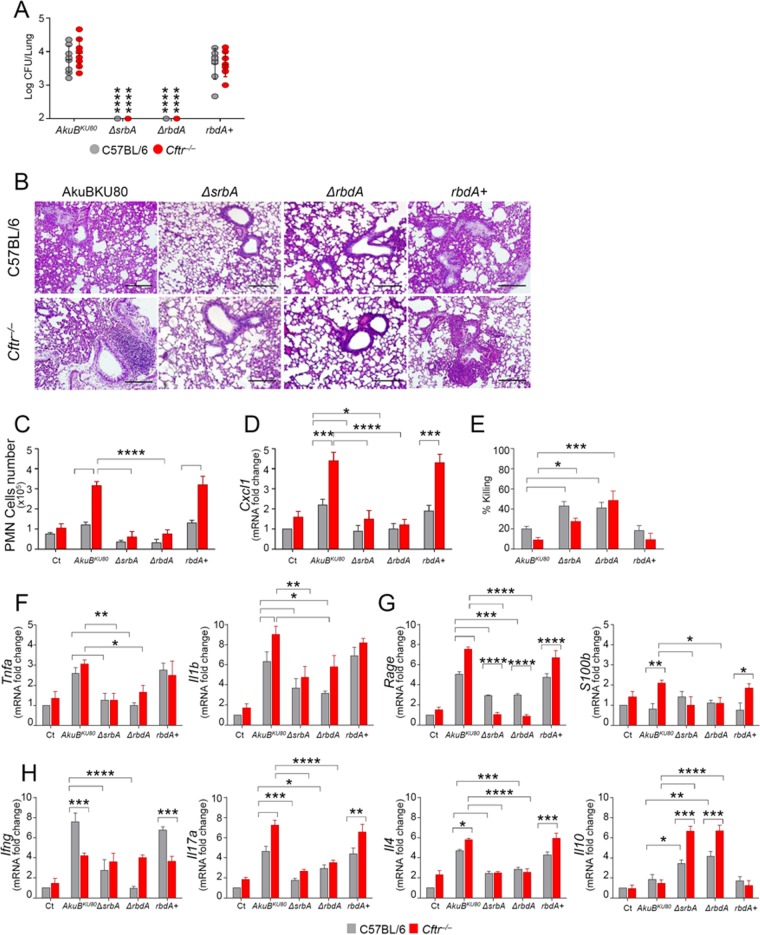
The ΔrbdA and ΔsrbA mutant strains induce a weakened inflammatory response in the lungs of infected mice and are more sensitive to phagocytic killing.
We evaluated the susceptibility of noncompromised C57BL/6 and Cftr−/− mice to infection with the Aspergillus mutant strains and evaluated the parameters of infection, lung inflammation, and cytokine production. We analyzed Cftr−/− mice because they are characterized by a marked hypoxic status in the lung compared to C57BL/6 parental strain mice (24). After intranasal infection of the mice, fungal growth in the lungs could not be detected with the hypoxic ΔsrbA and ΔrbdA mutant strains in either the Cftr−/− or control mice (Fig. 8A and B). A sustained inflammatory response, as revealed by the significantly heightened PMN influx, was observed in the lungs of C57BL/6 and Cftr−/− mice infected with the parental AkuBKU80 or the rbdA+ revertant strain (Fig. 8C). In contrast, A. fumigatus ΔsrbA and ΔrbdA mutant strains did not induce significant lung inflammation. The inflammatory response appeared to be higher in Cftr−/− mice than in C57BL/6 mice, as revealed by the significantly increased expression of the neutrophil chemoattractant CxCl1 (Fig. 8D) in the former. Effector phagocytes showed a greater ability to kill the ΔsrbA and ΔrbdA mutant strains than the WT strain (Fig. 8E). The reduced lung inflammatory response observed with the ΔsrbA or ΔrbdA mutant compared to that with the parental AkuBKU80 or rbdA+ revertant strain was associated with lower levels of Tnfa and Il1b (Fig. 8F) and RAGE/S100b gene expression (Fig. 8G), particularly in Cftr−/− mice, possibly reflecting the more hypoxic conditions in their lungs and the adverse effect of these conditions on ΔsrbA or ΔrbdA mutant growth.
FIG 8.
Susceptibility and cytokine response of Cftr−/− mice to A. fumigatus hypoxia mutant strains. C57BL/6 and Cftr−/− mice were infected intranasally with 2 ×107 live conidia. (A) Fungal growth (log10 CFU, mean ± SD) in the lungs. (B) Lung histology (periodic acid-Schiff staining). Photographs were taken with a high-resolution microscope (Olympus DP71) equipped with a 20× objective. Scale bar, 200 μm. (C) Cell count of PMNs from bronchoalveolar lavage (BAL) fluid determined by May-Grünwald Giemsa staining. (D) Cxcl1 gene expression (RT-PCR on total lung cells). (E) Conidiocidal activity (percentage of CFU inhibition) of PMNs from naive mice. (F to H) Cytokine (F and H) and Rage/S100b (G) gene expression (RT-PCR on total lung cells). Assays were done at 7 days postinfection. Data from three experiments were pooled and are presented as mean + SD for all bar graphs. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Two-way analysis of variance (ANOVA) with Bonferroni's posttest.
For the assessment of Th cell activation, we used quantitative PCR (qPCR) to measure the mRNA levels of Th cytokines in the lungs and the corresponding transcription factors in the draining thoracic lymph nodes of infected C57BL/6 and Cftr−/− mice. Gamma interferon (IFN-γ) (Th1), interleukin-17A (IL-17A) (Th17), and IL-4 (Th2) cytokine mRNA expression was significantly reduced in C57BL/6 mice infected with the ΔrbdA and ΔsrbA mutant strains (Fig. 8H). IL-17A (Th17) and IL-4 (Th2) cytokine mRNA expression was significantly reduced in Cftr−/− mice infected with the ΔrbdA and ΔsrbA mutant strains, whereas the IFN-γ (Th1) response to any of the fungal strains was low in these mice. Finally, Il10 mRNA expression (anti-inflammatory/Treg) was strongly increased in C57BL/6 mice infected with the ΔsrbA or ΔrbdA mutants and even more so in the Cftr−/− mice (Fig. 8H).
DISCUSSION
The SREBP/SrbA-mediated pathway in A. fumigatus is required for sensing and survival under hypoxia, adaptation to low iron levels, susceptibility to azoles, and virulence (11, 13, 33). A central unanswered question in our understanding of this pathway in ascomycete fungi is the identity of the proteases which cleave the DNA-binding N-terminal domain of SrbA, allowing it to translocate into the nucleus to activate selective gene expression. The SREBP pathway is best understood in mammals, where it acts via SREBP1 and SREBP2 to regulate fatty acid and cholesterol synthesis, respectively (34). SREBP1 and -2 are synthesized as inactive precursor proteins on the endoplasmic reticulum (ER) membrane, bound to the sterol-sensing protein Scap which anchors them to the ER-resident protein INSIG. A reduction in fatty acid or cholesterol levels disrupts the binding of Scap to INSIG, releasing the SREBP-Scap complex and allowing it to be transported to the Golgi apparatus via COPII vesicle-mediated transport. In the Golgi apparatus, SREBP is cleaved by a site-1 serine protease and a site-2 zinc metalloprotease, releasing the N-terminal transcription factor domain into the cytosol, where it is transported into the nucleus to drive gene expression leading to fatty acid (SREBP1) or cholesterol (SREBP2) synthesis.
In fungi, our understanding of the SREBP pathway and of Sre1/SrbA proteolytic cleavage is less complete. In the pathogenic basidiomycete fungus C. neoformans, release of the CnSre1 N-terminal transcription factor domain is apparently mediated by a single site-2 protease, Stp (16, 35). The model ascomycete yeast S. pombe lacks homologs of site-1 and site-2 proteases and instead utilizes the Dsc1-6 E3 ligase complex of proteins that are presumed to deliver Sre1 to its candidate protease, the proteasome (17, 18). However, it is unlikely that the proteasome is the target Sre1 protease. The proteasome usually completely degrades targeted proteins into short peptides, whereas Sre1 needs to be accurately and partially cleaved to generate the functional Sre1N transcription factor. In the filamentous ascomycete A. fumigatus, cleavage of the SREBP homolog SrbA to release the N-terminal transcription factor domain is mediated by DscA to DscD, but like S. pombe, it lacks homologs of site-1 and site-2 proteases and the identity of the protease responsible for SrbA cleavage has remained enigmatic (19).
In this report, we screened the N. crassa gene deletion library (20) to identify genes which function during hypoxic growth. We chose this approach because there is no full genome deletion library for A. fumigatus and N. crassa is the only filamentous ascomycete for which such a library exists. Six N. crassa mutant strains (ΔsrbA, ΔdscB, ΔdscD, ΔdscE, ΔdigA, and ΔrbdA mutants) that were unable to grow under hypoxia were identified from the 11,512 deletion strains screened. Gene deletions of the previously uncharacterized A. fumigatus dscE, digA, and rbdA homologs revealed that DscE (homologous to S. pombe Dsc5) and RbdA are essential for hypoxic growth. DscE contains a UBX domain found in ubiquitin regulatory proteins. In S. pombe, the Dsc5 UBX domain binds Cdc48 and recruits it to the Dsc complex (18). Cdc48 is an essential molecular chaperone with multiple cellular functions. In S. pombe, it is believed to promote conformational changes to Sre1 under hypoxia, allowing its dislocation for delivery to the proteasome.
Our work focused on the analysis of the RbdA rhomboid family protease, which has not been previously described. Rhomboid family proteases are one of four classes of unrelated intermembrane cleaving proteases (I-CLiPs) that also include the site-2 metalloproteases, presenilin aspartic proteases, and signal peptide peptidases (SPPs) (36). Uniquely, these diverse membrane-embedded proteases all carry out hydrolysis on the transmembrane region of their substrates. Rhomboid-like proteases are conserved throughout evolution from bacteria to humans. They exist in multiple copies in all examined genomes. They perform various functions, including metazoan growth factor signaling and inflammation, S. cerevisiae mitochondrial dynamics, apicomplexan parasite invasion, and the machinery of protein quality control (37).
Two functional rhomboid-like proteases have been identified in S. cerevisiae and were shown to control either membrane remodeling in mitochondria (PCP1/RBD1) or lipid organization and actin assembly (RBD2) (27, 38). Genome-based analysis revealed that RBD2 and PCP1 homologous proteins are conserved in S. pombe, A. fumigatus, and A. nidulans. Based on amino acid similarities, RbdA of A. fumigatus is a homologue of RBD2 and AFUA_1G09150 is likely to encode PCP1 (Fig. 1B). Unexpectedly, all three organisms encode two further rhomboid-like proteins, of which only one of each contains the essential catalytic residues and could therefore be functional. However, no putative function can be inferred from homology, and for both aspergilli, this third potential rhomboid protease clusters outside the PCP1 and RBD2 branches.
Here we demonstrate a novel function for the rhomboid family protease RbdA in the hypoxic response of A. fumigatus. A mutant rbdA deletion strain failed to grow under hypoxic conditions and was avirulent in murine models of infection. Several lines of evidence suggest that RbdA functions in the SrbA pathway. (i) The ΔrbdA mutant accurately phenocopies the ΔsrbA mutant in chemical sensitivity to iron, cell wall and ergosterol synthesis stresses, aberrant tip morphology, and avirulence in the infected host, including the induction of a reduced inflammatory Th1 and Th17 response in infected lungs and increased susceptibility to phagocytic killing. (ii) Hypoxic induction of the SrbA target genes erg11A and erg25A requires RbdA. This transcriptional dysregulation shared by the ΔrbdA and ΔsrbA mutant strains suggests that the two genes are components of a shared hypoxia-sensing pathway. (iii) The ΔrbdA strain is partially rescued for hypoxic growth by expression of cleaved active N-SrbA and is not rescued by expression of full-length SrbA. This bypass phenomenon was previously shown for the ΔdscA mutant and has been shown here for the ΔdscE strain. (iv) Inhibition of ergosterol biosynthesis by exposure to itraconazole induces SrbA nuclear translocation in wild-type A. fumigatus and not in the ΔrbdA strain. Additional work is necessary to confirm that full-length SrbA is not cleaved under hypoxia in the ΔrbdA mutant as well as in a point-mutated protease-dead RbdA-expressing strain.
During the writing of this paper, a genetic screen of the S. pombe nonessential haploid deletion collection identified the rhomboid protease Rbd2 as the candidate involved in proteolytic cleavage and release of the Sre1 N-terminal transcription factor domain (28), strengthening the validity of our results. Deletion of Rbd2 or mutation of the Rbd2 conserved serine-histidine catalytic residues resulted in a defect in Sre1 processing and an inability to grow under hypoxia. Of the four rhomboid family proteins in S. pombe (SPBC13E7.11 [Rbd1], SPCC790.03 [Rbd2], SPBP4H10.10 [mitochondrial Rbd3], and SPAC19B12.06c [mitochondrial Rbd4]), A. fumigatus RbdA is most closely related to Rbd2 (25% identity, 45% similarity), suggesting that this subclass of rhomboid proteases enable hypoxic growth by SrbA/Sre1 cleavage in both ascomycete yeast and molds. This is unlike the case of the basidiomycete yeast C. neoformans, in which a different class of CLiP, a site-2 metalloprotease, performs this action.
Furthermore, our work expands our understanding regarding the implications of RbdA/SrbA hypoxic signaling in the context of fungal virulence. Indeed, the more efficient elimination of the hypoxia-sensitive Aspergillus strains by effector phagocytes also affects the local inflammatory/anti-inflammatory cytokine balance. Thus, targeting hypoxia on the side of both the host and the fungus may help counteract pathogenic inflammation and microbial pathogenicity in cystic fibrosis.
Our work raises numerous interesting questions for future study. Where is the RbdA-dependent SrbA cleavage site? Are there additional proteases involved in cleaving SrbA? Does cleavage necessitate the proteasome? Do the Dsc complex and RbdA physically interact? Our findings also suggest a therapeutic avenue to treat invasive aspergillosis by inhibiting RbdA protease activity and thus blocking the ability of the fungus to proliferate at the hypoxic site of infection. Indeed, several potential selective rhomboid protease inhibitors have recently been identified, but their effect on fungal rhomboids has not been assessed (39).
Supplementary Material
ACKNOWLEDGMENTS
We thank Silke Steinbach for excellent technical assistance.
This study was supported by the German-Israeli Foundation for Scientific Research and Development (GIF no. 996-47.12/2008) to N.O. and O.K., the ERA-NET Pathogenomics project OXYstress (BMBF project no. 0315902B) to A.B., and the Hypoinfect Israel-Italy Cooperation Grant to N.O. and L.R.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00011-16.
REFERENCES
- 1.Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 2.Askew DS. 2008. Aspergillus fumigatus: virulence genes in a street-smart mold. Curr Opin Microbiol 11:331–337. doi: 10.1016/j.mib.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kontoyiannis DP, Lewis RE. 2015. Treatment principles for the management of mold infections. Cold Spring Harb Perspect Med 5:a019737. doi: 10.1101/cshperspect.a019737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Latge JP. 2001. The pathobiology of Aspergillus fumigatus. Trends Microbiol 9:382–389. doi: 10.1016/S0966-842X(01)02104-7. [DOI] [PubMed] [Google Scholar]
- 5.Brakhage AA. 2005. Systemic fungal infections caused by Aspergillus species: epidemiology, infection process and virulence determinants. Curr Drug Targets 6:875–886. doi: 10.2174/138945005774912717. [DOI] [PubMed] [Google Scholar]
- 6.Lewis JS, Lee JA, Underwood JC, Harris AL, Lewis CE. 1999. Macrophage responses to hypoxia: relevance to disease mechanisms. J Leukoc Biol 66:889–900. [DOI] [PubMed] [Google Scholar]
- 7.Grahl N, Dinamarco TM, Willger SD, Goldman GH, Cramer RA. 2012. Aspergillus fumigatus mitochondrial electron transport chain mediates oxidative stress homeostasis, hypoxia responses and fungal pathogenesis. Mol Microbiol 84:383–399. doi: 10.1111/j.1365-2958.2012.08034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kroll K, Pahtz V, Hillmann F, Vaknin Y, Schmidt-Heck W, Roth M, Jacobsen ID, Osherov N, Brakhage AA, Kniemeyer O. 2014. Identification of hypoxia-inducible target genes of Aspergillus fumigatus by transcriptome analysis reveals cellular respiration as an important contributor to hypoxic survival. Eukaryot Cell 13:1241–1253. doi: 10.1128/EC.00084-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hillmann F, Linde J, Beckmann N, Cyrulies M, Strassburger M, Heinekamp T, Haas H, Guthke R, Kniemeyer O, Brakhage AA. 2014. The novel globin protein fungoglobin is involved in low oxygen adaptation of Aspergillus fumigatus. Mol Microbiol 93:539–553. doi: 10.1111/mmi.12679. [DOI] [PubMed] [Google Scholar]
- 10.Hillmann F, Shekhova E, Kniemeyer O. 2015. Insights into the cellular responses to hypoxia in filamentous fungi. Curr Genet 61:441–455. doi: 10.1007/s00294-015-0487-9. [DOI] [PubMed] [Google Scholar]
- 11.Willger SD, Puttikamonkul S, Kim KH, Burritt JB, Grahl N, Metzler LJ, Barbuch R, Bard M, Lawrence CB, Cramer RA Jr. 2008. A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog 4:e1000200. doi: 10.1371/journal.ppat.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung D, Barker BM, Carey CC, Merriman B, Werner ER, Lechner BE, Dhingra S, Cheng C, Xu W, Blosser SJ, Morohashi K, Mazurie A, Mitchell TK, Haas H, Mitchell AP, Cramer RA. 2014. ChIP-seq and in vivo transcriptome analyses of the Aspergillus fumigatus SREBP SrbA reveals a new regulator of the fungal hypoxia response and virulence. PLoS Pathog 10:e1004487. doi: 10.1371/journal.ppat.1004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blosser SJ, Cramer RA. 2012. SREBP-dependent triazole susceptibility in Aspergillus fumigatus is mediated through direct transcriptional regulation of erg11A (cyp51A). Antimicrob Agents Chemother 56:248–257. doi: 10.1128/AAC.05027-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blosser SJ, Merriman B, Grahl N, Chung D, Cramer RA. 2014. Two C4-sterol methyl oxidases (Erg25) catalyse ergosterol intermediate demethylation and impact environmental stress adaptation in Aspergillus fumigatus. Microbiology 160:2492–2506. doi: 10.1099/mic.0.080440-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes AL, Todd BL, Espenshade PJ. 2005. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell 120:831–842. doi: 10.1016/j.cell.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 16.Chun CD, Liu OW, Madhani HD. 2007. A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS Pathog 3:e22. doi: 10.1371/journal.ppat.0030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stewart EV, Lloyd SJ, Burg JS, Nwosu CC, Lintner RE, Daza R, Russ C, Ponchner K, Nusbaum C, Espenshade PJ. 2012. Yeast sterol regulatory element-binding protein (SREBP) cleavage requires Cdc48 and Dsc5, a ubiquitin regulatory X domain-containing subunit of the Golgi Dsc E3 ligase. J Biol Chem 287:672–681. doi: 10.1074/jbc.M111.317370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart EV, Nwosu CC, Tong Z, Roguev A, Cummins TD, Kim DU, Hayles J, Park HO, Hoe KL, Powell DW, Krogan NJ, Espenshade PJ. 2011. Yeast SREBP cleavage activation requires the Golgi Dsc E3 ligase complex. Mol Cell 42:160–171. doi: 10.1016/j.molcel.2011.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willger SD, Cornish EJ, Chung D, Fleming BA, Lehmann MM, Puttikamonkul S, Cramer RA. 2012. Dsc orthologs are required for hypoxia adaptation, triazole drug responses, and fungal virulence in Aspergillus fumigatus. Eukaryot Cell 11:1557–1567. doi: 10.1128/EC.00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collopy PD, Colot HV, Park G, Ringelberg C, Crew CM, Borkovich KA, Dunlap JC. 2010. High-throughput construction of gene deletion cassettes for generation of Neurospora crassa knockout strains. Methods Mol Biol 638:33–40. doi: 10.1007/978-1-60761-611-5_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levdansky E, Kashi O, Sharon H, Shadkchan Y, Osherov N. 2010. The Aspergillus fumigatus cspA gene encoding a repeat-rich cell wall protein is important for normal conidial cell wall architecture and interaction with host cells. Eukaryot Cell 9:1403–1415. doi: 10.1128/EC.00126-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gravelat FN, Ejzykowicz DE, Chiang LY, Chabot JC, Urb M, Macdonald KD, al-Bader N, Filler SG, Sheppard DC. 2010. Aspergillus fumigatus MedA governs adherence, host cell interactions and virulence. Cell Microbiol 12:473–488. doi: 10.1111/j.1462-5822.2009.01408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fallon J, Kelly J, Kavanagh K. 2012. Galleria mellonella as a model for fungal pathogenicity testing. Methods Mol Biol 845:469–485. doi: 10.1007/978-1-61779-539-8_33. [DOI] [PubMed] [Google Scholar]
- 24.Iannitti RG, Casagrande A, De Luca A, Cunha C, Sorci G, Riuzzi F, Borghi M, Galosi C, Massi-Benedetti C, Oury TD, Cariani L, Russo M, Porcaro L, Colombo C, Majo F, Lucidi V, Fiscarelli E, Ricciotti G, Lass-Florl C, Ratclif L, Esposito A, De Benedictis FM, Donato R, Carvalho A, Romani L. 2013. Hypoxia promotes danger-mediated inflammation via receptor for advanced glycation end products in cystic fibrosis. Am J Respir Crit Care Med 188:1338–1350. doi: 10.1164/rccm.201305-0986OC. [DOI] [PubMed] [Google Scholar]
- 25.Geissenhoner A, Sievers N, Brock M, Fischer R. 2001. Aspergillus nidulans DigA, a potential homolog of Saccharomyces cerevisiae Pep3 (Vps18), is required for nuclear migration, mitochondrial morphology and polarized growth. Mol Genet Genomics 266:672–685. doi: 10.1007/s00438-001-0589-6. [DOI] [PubMed] [Google Scholar]
- 26.Urban S. 2009. Making the cut: central roles of intramembrane proteolysis in pathogenic microorganisms. Nat Rev Microbiol 7:411–423. doi: 10.1038/nrmicro2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McQuibban GA, Saurya S, Freeman M. 2003. Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature 423:537–541. doi: 10.1038/nature01633. [DOI] [PubMed] [Google Scholar]
- 28.Kim J, Ha HJ, Kim S, Choi AR, Lee SJ, Hoe KL, Kim DU. 2015. Identification of Rbd2 as a candidate protease for sterol regulatory element binding protein (SREBP) cleavage in fission yeast. Biochem Biophys Res Commun 468:606–610. doi: 10.1016/j.bbrc.2015.10.165. [DOI] [PubMed] [Google Scholar]
- 29.Gravelat FN, Beauvais A, Liu H, Lee MJ, Snarr BD, Chen D, Xu W, Kravtsov I, Hoareau CM, Vanier G, Urb M, Campoli P, Al Abdallah Q, Lehoux M, Chabot JC, Ouimet MC, Baptista SD, Fritz JH, Nierman WC, Latge JP, Mitchell AP, Filler SG, Fontaine T, Sheppard DC. 2013. Aspergillus galactosaminogalactan mediates adherence to host constituents and conceals hyphal beta-glucan from the immune system. PLoS Pathog 9:e1003575. doi: 10.1371/journal.ppat.1003575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lapp K, Vodisch M, Kroll K, Strassburger M, Kniemeyer O, Heinekamp T, Brakhage AA. 2014. Characterization of the Aspergillus fumigatus detoxification systems for reactive nitrogen intermediates and their impact on virulence. Front Microbiol 5:469. doi: 10.3389/fmicb.2014.00469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zadra I, Abt B, Parson W, Haas H. 2000. xylP promoter-based expression system and its use for antisense downregulation of the Penicillium chrysogenum nitrogen regulator NRE. Appl Environ Microbiol 66:4810–4816. doi: 10.1128/AEM.66.11.4810-4816.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grahl N, Puttikamonkul S, Macdonald JM, Gamcsik MP, Ngo LY, Hohl TM, Cramer RA. 2011. In vivo hypoxia and a fungal alcohol dehydrogenase influence the pathogenesis of invasive pulmonary aspergillosis. PLoS Pathog 7:e1002145. doi: 10.1371/journal.ppat.1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blatzer M, Barker BM, Willger SD, Beckmann N, Blosser SJ, Cornish EJ, Mazurie A, Grahl N, Haas H, Cramer RA. 2011. SREBP coordinates iron and ergosterol homeostasis to mediate triazole drug and hypoxia responses in the human fungal pathogen Aspergillus fumigatus. PLoS Genet 7:e1002374. doi: 10.1371/journal.pgen.1002374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bien CM, Espenshade PJ. 2010. Sterol regulatory element binding proteins in fungi: hypoxic transcription factors linked to pathogenesis. Eukaryot Cell 9:352–359. doi: 10.1128/EC.00358-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bien CM, Chang YC, Nes WD, Kwon-Chung KJ, Espenshade PJ. 2009. Cryptococcus neoformans site-2 protease is required for virulence and survival in the presence of azole drugs. Mol Microbiol 74:672–690. doi: 10.1111/j.1365-2958.2009.06895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolfe MS. 2009. Intramembrane-cleaving proteases. J Biol Chem 284:13969–13973. doi: 10.1074/jbc.R800039200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freeman M. 2014. The rhomboid-like superfamily: molecular mechanisms and biological roles. Annu Rev Cell Dev Biol 30:235–254. doi: 10.1146/annurev-cellbio-100913-012944. [DOI] [PubMed] [Google Scholar]
- 38.Cortesio CL, Lewellyn EB, Drubin DG. 2015. Control of lipid organization and actin assembly during clathrin-mediated endocytosis by the cytoplasmic tail of the rhomboid protein Rbd2. Mol Biol Cell 26:1509–1522. doi: 10.1091/mbc.E14-11-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolf EV, Zeissler A, Vosyka O, Zeiler E, Sieber S, Verhelst SH. 2013. A new class of rhomboid protease inhibitors discovered by activity-based fluorescence polarization. PLoS One 8:e72307. doi: 10.1371/journal.pone.0072307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.