Abstract
Coxiella burnetii is an intracellular pathogen and the cause of Q fever. Gamma interferon (IFN-γ) is critical for host protection from infection, but a role for type I IFN in C. burnetii infection has not been determined. Type I IFN supports host protection from a related pathogen, Legionella pneumophila, and we hypothesized that it would be similarly protective in C. burnetii infection. In contrast to our prediction, IFN-α receptor-deficient (IFNAR−/−) mice were protected from C. burnetii-induced infection. Therefore, the role of type I IFN in C. burnetii infection was distinct from that in L. pneumophila. Mice treated with a double-stranded-RNA mimetic were protected from C. burnetii-induced weight loss through an IFNAR-independent pathway. We next treated mice with recombinant IFN-α (rIFN-α). When rIFN-α was injected by the intraperitoneal route during infection, disease-induced weight loss was exacerbated. Mice that received rIFN-α by this route had dampened interleukin 1β (IL-1β) expression in bronchoalveolar lavage fluids. However, when rIFN-α was delivered to the lung, bacterial replication was decreased in all tissues. Thus, the presence of type I IFN in the lung protected from infection, but when delivered to the periphery, type I IFN enhanced disease, potentially by dampening inflammatory cytokines. To better characterize the capacity for type I IFN induction by C. burnetii, we assessed expression of IFN-β transcripts by human macrophages following stimulation with lipopolysaccharide (LPS) from C. burnetii. Understanding innate responses in C. burnetii infection will support the discovery of novel therapies that may be alternative or complementary to the current antibiotic treatment.
INTRODUCTION
There is growing recognition of both beneficial and detrimental outcomes of type I interferon (IFN) signaling in bacterial infections. Type I IFNs clearly promote infection with Listeria monocytogenes (1), Mycobacterium tuberculosis (2), and Staphylococcus aureus (3) and are disadvantageous to the host. In contrast, recent evidence suggests that type I IFNs are critical for host protection from Streptococcus pneumoniae and Pseudomonas aeruginosa infection (4). Coxiella burnetii is an intracellular bacterial pathogen of the lung and the cause of Q fever. Acute Q fever is associated with flu-like symptoms that can lead to a complicated chronic infection potentially involving endocarditis, hepatitis, or chronic fatigue (5). Because of these common symptoms, Q fever has been misdiagnosed and underreported in the United States (6). Initially classified as a member of the genus Rickettsia, C. burnetii has been reclassified as a member of the Gammaproteobacteria and is closely related to Legionella pneumophila (7).
A role for type I IFN in L. pneumophila has been characterized (8–10). Treatment of cultured macrophages with type I IFN protects the cells from in vitro infection by L. pneumophila (10), and this protection was independent of gamma interferon (IFN-γ) signaling (8). Furthermore, type I IFN-α receptor-deficient (IFNAR−/−) macrophages were more permissive to L. pneumophila infection in vitro than were wild-type cells (11). There was no difference in bacterial detection in the lungs when IFNAR2−/− and wild-type mice were compared 48 h after L. pneumophila infection (12), nor was there a difference in lung bacterial burden between wild-type and IFNAR−/− mice (13). Despite this, most reports suggest that type I IFN supports cellular resistance to L. pneumophila infection.
Proinflammatory cytokines are expressed in Q fever in human patients (14, 15) and following injection of inactivated bacteria in mice (14, 15). Expression of tumor necrosis factor alpha (TNF-α) occurs soon after bacterial sensing and is associated with a reduction in C. burnetii replication in human macrophages. Production of the anti-inflammatory mediator interleukin 10 (IL-10), which downmodulates TNF-α, reverses this effect (16). Whereas TNF-α and IL-6 are innate cytokines produced early, expression of IL-10 appears to occur later in infection and has largely been associated with development of chronic Q fever in human patients (14, 17, 18). IFN-γ, produced by T cells and natural killer cells, is critical for protection of mice from C. burnetii infection and eventual clearance of the bacteria (18–21). A primary downstream effect of type I IFN signaling production is increased expression of IL-10, resulting in dampened responses and cytokine expression (22). Type I IFN can also dampen inflammasome signaling (23), and this is a primary mechanism by which type I IFN promotes infection with M. tuberculosis (2, 24). Thus, in some situations, type I IFN can be distinctly anti-inflammatory, resulting in a state that can promote infection. The pro- or anti-inflammatory role of type I IFNs during Q fever has not been addressed.
Type I IFNs are likely to be produced during C. burnetii infection, as would be expected with an intracellular pathogen. For example, injection of inactivated phase I C. burnetii cells protected mice from infection with a variety of type I IFN-sensitive viruses, but not a type I IFN-insensitive virus (15). Also, a fraction extracted from C. burnetii under investigation as a potential vaccine was proposed as a broad-spectrum antiviral prophylactic therapy because it strongly induced type I IFN and protected mice from viral infection (25). In this case, it was determined that both type I IFNs and IFN-γ were induced by the fraction. Thus, it appears that phase I C. burnetii induces expression of type I IFNs, but a role for type I IFNs in innate protection against C. burnetii has not been characterized. While infection of human cells with C. burnetii in vitro does not induce overt expression of inflammatory cytokines (26), here, we demonstrated that the lipopolysaccharide (LPS) from the Nine Mile phase I (NMI) strain of C. burnetii can induce expression of IFN-β transcripts in human cells. The cytokine is also induced during intracellular infection by M. tuberculosis, primarily through induction of intracellular pathways following cell damage (27). Similarly, as C. burnetii NMI modifies the phagolysosome and replicates in infected macrophages, bacterial components, such as type IV effector proteins, are eventually released into the cytoplasm (28). We hypothesized that type I IFN production during C. burnetii infection is beneficial and promotes bacterial clearance. The most compelling support for this hypothesis is the fact that infection with L. pneumophila, a close relative of C. burnetii (7), induces type I IFNs, and these cytokines provide innate immune protection from infection (13).
To address the role of type I IFNs during C. burnetii infection, we utilized a mouse model of infection with the virulent NMI strain. Contrary to our original hypothesis, our results suggested that type I IFN signaling promoted C. burnetii replication and pathogenesis. IFNAR-deficient mice were significantly protected from weight loss following NMI infection and harbored fewer bacteria in their spleens. When treated with poly(I·C), a potent innate agonist that primarily induces type I IFN and other innate cytokines, wild-type and IFNAR−/− mice were protected from infection. Thus, poly(I·C) protected through a mechanism that was independent of type I IFN. To directly address the role of a specific type I IFN, we then treated mice with recombinant type I IFN-α (rIFN-α). In this case, the outcome of the infection depended on the route of rIFN-α delivery. When delivered intraperitoneally (i.p.), rIFN-α enhanced C. burnetii disease, whereas intratracheal (i.t.) rIFN-α delivery to the lung protected mice from infection. Unlike the case with L. pneumophila, our results suggest that type I IFN does not always protect against C. burnetii replication in vivo, and rather, may promote infection outside the lung. Interventions in the type I IFN signaling pathway that enhance proinflammatory cytokine expression represent novel therapeutic approaches to counter C. burnetii infection.
MATERIALS AND METHODS
Bacterial strain, mice, and in vivo infection.
C. burnetii strain NMI (RSA493) was kindly donated by Robert Heinzen at Rocky Mountain Laboratories, Hamilton, MT, USA. All work with the NMI strain was done in a CDC-certified biosafety level 3 (BSL-3) facility approved for category B select agent use. All animal procedures performed were approved by the Institutional Animal Care and Use Committee of Montana State University, Bozeman, MT. The mice (at least 5 mice per group) were 6- to 12-week-old C57BL/6 (wild-type) or IFNAR−/− (on a C57BL/6 background) mice. For all experiments, the mice were lightly anesthetized with isoflurane and inoculated i.t. with 1 × 102 to 1 × 105 genome equivalents (GE) of NMI suspended in 100 μl sterile phosphate-buffered saline (PBS). The mice were weighed daily and sacrificed between 1 and 30 days postinfection. The data shown are the average percentages of the starting weights of all the mice in the group. In some experiments, mice were treated with poly(I·C) i.p. (50 μg) or i.t. (10 μg) or recombinant universal type I IFN i.p. (rIFN-α; 20,000 to 200,000 U/kg of body weight; R&D Systems) or i.t. (200,000 U/kg). Treatments were on day 1 postinfection and subsequently during infection on alternating days.
Purification of C. burnetii DNA.
One-half of the spleen and all but the left lobe of the lung were weighed, homogenized in Hanks balanced salt solution (HBSS), and centrifuged at 500 × g for 5 min. The tissue cell pellets were suspended in 1 ml water, vortexed for 1 min, and then centrifuged again. The supernatant fluids were centrifuged at 14.2 × g for 30 min to pellet the bacteria. The supernatant fluids were decanted, and the pelleted bacteria were suspended in 1 ml PBS, vortexed for 1 min, and centrifuged at 500 × g for 5 min to pellet the cellular debris. The supernatant fluids were again centrifuged at 14.2 × g for 30 min to wash and pellet the bacteria. The supernatant fluids were decanted, and the bacterial pellet was stored at −20°C until DNA extraction.
At these time points and infectious doses, there are enough bacteria in lungs and spleens to detect using PCR and too many to easily count using histological staining. In contrast, in livers and hearts, there are insufficient bacteria to detect with PCR, but infectious foci can be enumerated on histological sections. To assess and compare bacterial burdens in spleen and lung tissue under various experimental conditions, quantitative PCR (qPCR) was performed as previously reported (29, 30). Briefly, whole sample DNA was extracted using an UltraClean Microbial DNA isolation kit (MoBio Laboratories, Inc.) following the manufacturer's protocol. Genome copies of bacterial DNA were determined by amplification of rpoS gene copies, as previously reported (29, 30). This gene was amplified in a quantitative real-time PCR with SYBR green PCR master mixture from Applied Biosystems (in vivo; BSL-3) and the Applied Biosystems 7500 real-time PCR system.
Immunohistochemistry.
Unless otherwise indicated, PCR results were confirmed with immunofluorescent bacterial staining of histological sections from lung, liver, heart, and spleen. The left pulmonary lobe, the right lateral liver lobe, and half the spleen from each infected mouse were harvested for histology. The tissues were fixed in 10% formalin overnight, followed by 70% ethyl alcohol (EtOH) overnight, and then processed and embedded in paraffin. Serial 5-μm sections were cut, deparaffinized with xylene, and rehydrated. Immunostaining was performed at room temperature. Cells in paraffin-embedded tissue sections were incubated with 100 mM glycine for 20 min and then rinsed with PBS. Proteinase K (Dako, Carpinteria, CA) was then applied for 6 min. Samples were washed with Tris-buffered saline (TBS) and blocked with Rodent Block M (Biocare Medical, Concord, CA, USA) for 30 min. They were then washed with TBS with 0.05% Tween 20 (TBST) before being incubated with polyclonal rabbit anti-C. burnetii at 1:8,000 in 0.5% TBST for 1 h at room temperature. After being washed again with TBST, a Rabbit on Rodent AP-Polymer (Biocare Medical, Concord, CA, USA) antibody was added for 30 min. Immunodetection was performed with Permanent Red (Dako, Carpinteria, CA) for 2 min and stopped with water. Tissues were counterstained with hematoxylin, and coverslips were mounted with aqueous Vectamount AQ mounting medium (Vector Laboratories, Burlingame, CA). Images were acquired using a Nikon DS-Ri-1 camera mounted on a Nikon Eclipse 80i microscope, and the bacterial burden was determined by counting the vacuoles containing bacteria in 20 random fields at ×20 or ×40 magnification and verified to be within a cell wall by bright-field microscopy. Hematoxylin and eosin (H&E) staining was done following the manufacturer's protocol. Pictures were taken and analyzed by a blinded histotechnician. Counts of fluorescently stained vacuoles containing bacteria in the lung and spleen sections closely confirmed the DNA PCR results, unless otherwise stated, and there were no substantial day 9 pathological differences in these in vivo infections (data not shown). Of note, most stained day 9 lung sections contained too many infected vacuoles to enumerate.
Cytometric bead array.
Cytokines in bronchoalveolar lavage (BAL) fluid and serum were quantified using the BD Cytometric Bead Array (BD Biosciences). Initial experiments assessed the cytokines in a 7-plex: IL-12, IFN-γ, IL-10, IL-17, granulocyte-macrophage colony-stimulating factor (GM-CSF), granulocyte colony-stimulating factor (G-CSF), and IL-1β. Subsequent experiments were expanded to include in an 11-plex: IL-12, IFN-γ, IL-10, IL-17, G-CSF, IL-1β, IL-2, KC, IL-6, TNF-α, and CD62L. BAL fluids and sera were minimally diluted, and the assay was performed according to the manufacturer's instructions. Data were analyzed using BD FCAP Array software, Microsoft Excel, and GraphPad (Prism).
Stimulation of human cells.
Studies involving blood from human subjects were carried out in compliance with guidelines of the Montana State University Institutional Review Board, and as such, each subject signed informed consent documentation. Leukocytes were separated from whole blood using Histopaque 1077 (Sigma-Aldrich) according to the manufacturer's instructions. Blood was collected with ACD solution B anti-coagulant tubes (BD Biosciences). Whole blood was diluted 1:2 with HBSS and underlaid with Histopaque 1077. The tubes were centrifuged, and the resulting buffy layer containing peripheral blood mononuclear cells (PBMCs) was aspirated. The cells were washed once with HBSS, and then red blood cells (RBCs) were removed by water lysis and the cells were cultured in human macrophage colony-stimulating factor (M-CSF) (60 ng/ml) at starting concentrations of 5 × 106 cells/ml for up to 1 week in X-vivo 15 medium (Lonza) before stimulation. C. burnetii NMI LPS (kindly provided by Michael Minnick, University of Montana) was extracted using the method of Westphal and Jann (31). Using this method with no further purification steps, there is a potential for contamination of the LPS with other bacterial components. The cells were stimulated with C. burnetii NMI LPS (6 × 10−4 endotoxin units [EU]/ml) and E. coli LPS (Sigma; used at a standard dose of 10 ng/ml, or approximately 100 EU/ml).
Cells were stimulated for 6 to 18 h and lysed for RNA extraction and transcript analyses. RNA from human cells was extracted and purified using an RNeasy minikit (Qiagen), including elimination of genomic DNA, according to the manufacturer's instructions. Real-time reverse transcription (RT)-PCR was performed using Superscript II (Invitrogen Life Technologies) and ∼700 ng of RNA according to the manufacturer's protocol. The relative specific mRNA in the cells was quantified by measuring SYBR green incorporation during real-time quantitative RT-PCR using the relative standard curve method. Each RT reaction mixture (1 μl) was used in the 25-μl real-time PCRs performed in triplicate. Primers specific for the human genes encoding IFN-β, IL-1β, and β-actin were designed using NCBI Primer-BLAST. The PCR was set up and cycled, data were collected on the MyiQ Single-Color Real-Time PCR detection system, and calculations were performed using standard curves for each primer pair and normalized to β-actin detection as described in the manufacturer's protocol (Bio-Rad). Data are presented as fold induction of transcripts over those from control cells treated with medium only.
Statistical methods.
Statistical significance is based on comparisons made using two-way analysis of variance (ANOVA) or the unpaired Student t test. Two-way ANOVA comparisons take into account differences between mouse strains or treatments and the change that occurs over time postinfection.
RESULTS
IFNAR-deficient mice are protected from C. burnetii infection.
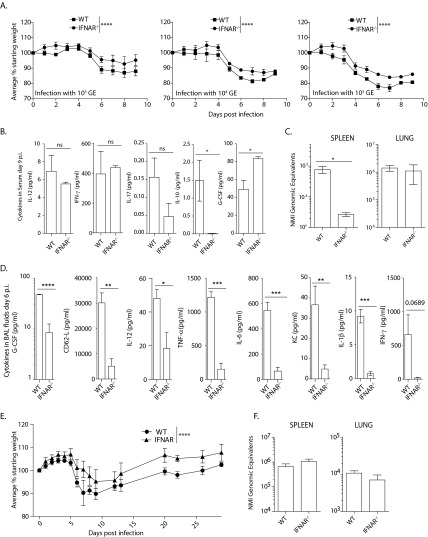
We utilized virulent NMI infection in wild-type and IFNAR−/− mice to determine the role of type I IFN signaling during in vivo infection with C. burnetii. Mice were infected with C. burnetii NMI at a range of doses from 103 to 105 bacteria per mouse delivered intratracheally, the most relevant infection route (32). Weight loss during a 9-day disease course was assessed as the primary indicator of disease progression. At each dose, wild-type mice lost significantly more weight than did IFNAR−/− mice (Fig. 1A). At day 9 postinfection, tissues were harvested and assessed for damage and bacterial burden. Despite the more severe weight loss seen in wild-type mice, there were no differences in tissue weights, tissue damage, or bacterial counts between the two mouse strains at day 9 after infection (data not shown). Serum was collected at day 9 postinfection, and a 7-plex panel of cytokines was assessed. The only notable differences detected included IL-10, where wild-type mice expressed more of the cytokine than did IFNAR-deficient mice but the overall quantities of the cytokine were very low (Fig. 1B). Also, there was a significant increase in G-CSF in sera from IFNAR−/− mice compared to sera from wild-type mice. There were few differences in expression of other cytokines assessed in the sera of wild-type and IFNAR−/− mice (Fig. 1B). While the 9-day experiments were important for determining differences in weight loss between the strains as a measure of disease severity, the interval and dose were clearly not optimal for assessing differences in bacterial counts or cytokine expression.
FIG 1.
IFNAR-deficient mice have less severe disease and diminished bacterial replication, despite decreased inflammatory cytokines. (A) Infection of wild-type (WT) mice was compared to that of IFNAR−/− mice at three difference infectious doses, and in each case, the IFNAR−/− mice were protected from C. burnetii-induced weight loss compared to wild-type mice. Weight loss differences between the groups over the entire course of infection were compared by two-way ANOVA. (B) Following infection with 104 bacteria, cytokines in serum were assessed at day 9 postinfection. The only significant differences in a total of 7 assessed cytokines in the sera are shown in the two rightmost graphs. (C) Mice were infected with 102 bacteria, and tissues were collected at day 6 postinfection. Spleens and lungs were assessed for bacterial burden. (D) Cytokines in BAL supernatant fluids, collected 6 days postinfection, were quantified. IFNAR−/− mice had decreased inflammatory cytokines in the lungs. (E) Differences in weight loss that were observed 9 days postinfection continued until day 30 postinfection. IFNAR−/− mice recovered faster from C. burnetii-induced weight loss after infection with 103 GE. (F) There were no differences in C. burnetii GE as detected by PCR 30 days postinfection. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant. The error bars indicate standard deviations.
In an attempt to better resolve differences in bacterial replication, we next infected the two mouse strains with a lower dose of bacteria (102 bacteria per mouse) and assessed tissues for bacterial burden at 6 days postinfection. Day 6 was chosen because it was the earliest time at which disease symptoms (weight loss) were apparent (Fig. 1A, left). Tissues, BAL fluids, and sera were collected for assessment of bacterial GE and cytokine quantification. In this case, there was no difference in bacterial GE in the lungs, but there were significantly fewer bacteria detected in the spleens of IFNAR−/− mice (Fig. 1C). Thus, as suggested by weight loss differences, mice lacking IFN signaling appear to be relatively protected from C. burnetii replication. Although differences between the two strains in serum cytokines were not detected, there were substantial differences detected in the BAL fluids. As shown in Fig. 1D, IFNAR−/− mice had substantially reduced expression of 7 of 11 cytokines tested. Finally, we also assessed weight loss trends over a longer period. There was little to no detection of IL-2, IL-17, or IL-10 in these samples. During a 30-day monitoring period after infection with 103 GE of C. burnetii, similar weight trends were maintained; IFNAR−/− mice remained healthier than wild-type mice (Fig. 1E). Bacteria were measured at day 30 postinfection in lungs and spleens, but there were no differences between the two strains (data not shown), similar to day 9 postinfection. Thus, it appears that while C. burnetii replicates in the lungs of IFNAR-deficient mice, the mice also clearly mount a diminished innate immune cytokine response. These studies were consistent with IFN signaling promoting C. burnetii replication outside the lung, in particular.
Treatment of mice with poly(I·C) during C. burnetii infection.
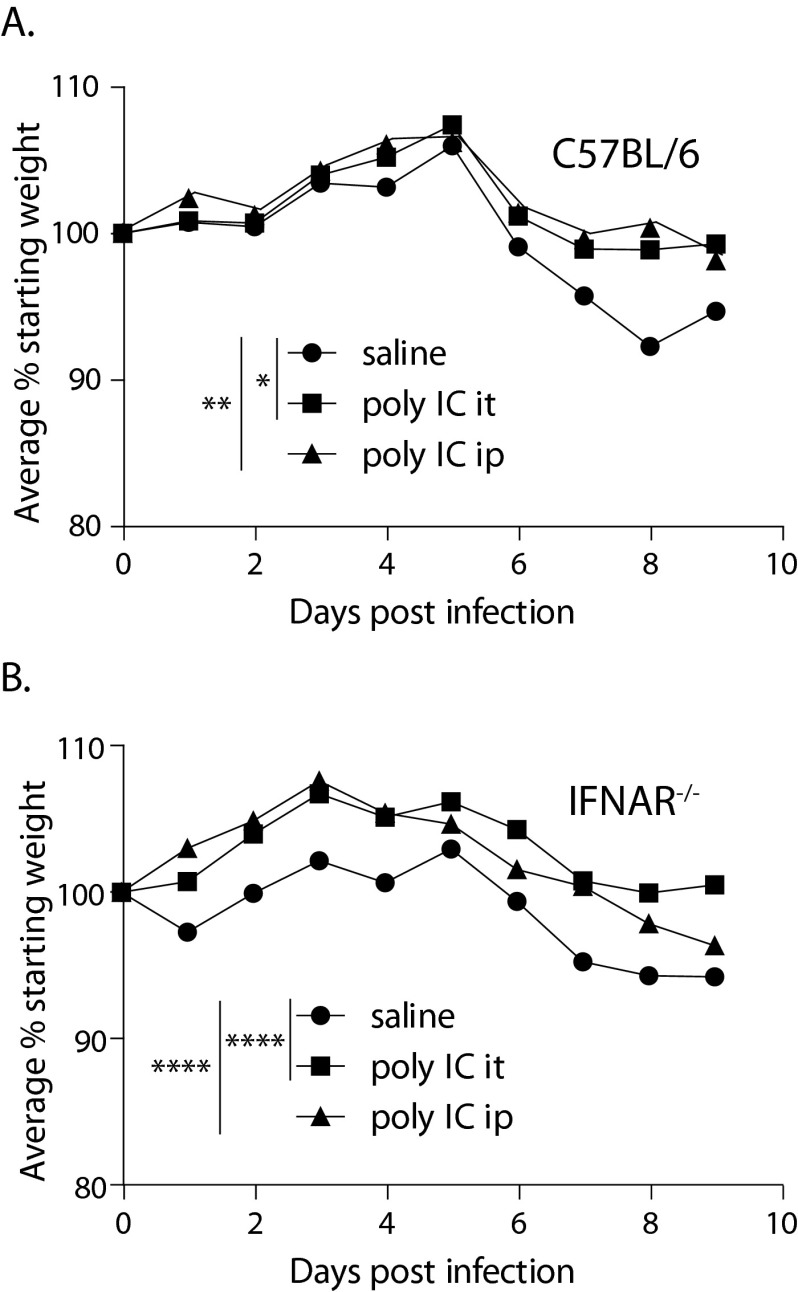
Developmental differences between wild-type and IFNAR mice may be a reason for differences in innate responses; thus, to demonstrate that type I IFN enhances infection in vivo, we next utilized poly(I·C) treatment, which is commonly used to induce expression of type I IFN. Mice were treated at day 0 and on alternate days during the 9-day course of infection (with 102 GE C. burnetii) with poly(I·C) delivered either intratracheally or intraperitoneally. Surprisingly, poly(I·C) delivered by either route significantly alleviated disease by diminishing weight loss in wild-type mice (Fig. 2A) and thus provided a benefit to the treated mice. Poly(I·C) was also protective in NMI-infected IFNAR-deficient mice (Fig. 2B). A similar outcome resulted following infection of mice with 104 bacteria (data not shown). Poly(I·C) treatment did not result in differences in bacterial-load detection at day 9 postinfection (data not shown). Poly(I·C) is a potent innate agonist, and although it is well known for inducing type I IFN, it has been shown to have immune-enhancing effects other than type I IFN signaling (33). Our data suggest that immune pathways induced by poly(I·C) that are independent of IFNAR signaling can protect against C. burnetii-induced weight loss.
FIG 2.
Poly(I·C) protects against C. burnetii morbidity through an IFNAR-independent mechanism. (A) Wild-type mice infected with 102 GE of C. burnetii and treated by the i.t. or i.p. route with poly(I·C) had diminished disease symptoms compared to saline-treated mice. (B) In IFNAR-deficient mice, poly(I·C) also provided protection from C. burnetii-induced disease, suggesting that poly(I·C) protects through a mechanism that is independent of IFNAR. The data are representative of the results of at least 2 replicate experiments. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Treatment of mice with intraperitoneal rIFN-α during C. burnetii infection.
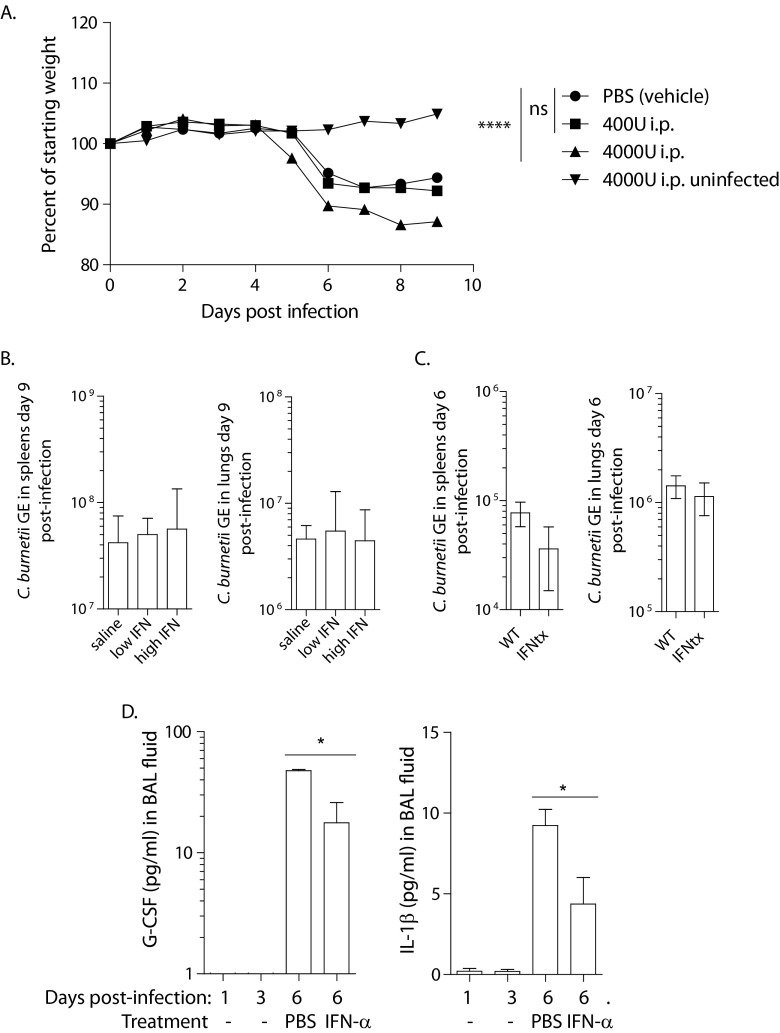
Since poly(I·C) treatment reduced weight loss independently of type I IFN following infection, we utilized a more specific approach to IFN signaling by measuring the effect of rIFN-α delivery on weight loss and bacterial replication following in vivo infection with C. burnetii. rIFN-α was injected i.p. on the day of infection and every other day thereafter for the 9-day infection course. For this study, two different rIFN-α doses were tested. The mice that received the lower dose (20,000 U/kg, approximately 400 U per mouse) by i.p. injection did not differ in weight loss from those injected with saline (Fig. 3A). Mice injected with 4,000 U experienced significantly greater weight loss than saline-injected mice. As shown in Fig. 3A, mice infected with 103 bacteria lost weight on day 6, but those treated with rIFN-α began to lose weight 1 day earlier. This dose of IFN-α did not induce weight loss in uninfected mice (Fig. 3A). These observations concerning weight loss were not accompanied by significant differences in bacterial counts in the spleens or lungs at day 9 following an infectious dose of 103 bacteria or at day 6 following infection with 102 bacteria (Fig. 3B and C). Different counts of macrophages, lymphocytes, neutrophils, and eosinophils in BAL fluids also indicated no differences in these cell subsets in the BAL fluids at day 6 postinfection with 102 bacteria (data not shown). Cytokines were measured in the sera and BAL fluids of mice treated i.p. with IFN-α at day 6 postinfection with 102 bacteria. The only significant differences were in the expression of G-CSF and IL-1β in BAL fluids (Fig. 3D). While G-CSF and IL-1β were not detected at days 1 and 3 postinfection, they were induced by NMI infection at day 6 postinfection. Peripheral treatment with rIFN-α caused a reduction in G-CSF and IL-1β in BAL fluids (Fig. 3D). Thus, peripheral injection of rIFN-α resulted in enhanced weight loss and a subtle decrease in two potentially protective cytokines in the lung.
FIG 3.
Intraperitoneal injection of rIFN-α enhanced C. burnetii-induced weight loss. (A) When wild-type mice were treated with rIFN-α by i.p. injection and infected with 103 GE C. burnetii, the higher dose of rIFN-α promoted C. burnetii pathogenesis. When this high dose was delivered to uninfected mice, there was no change in weight. The lower dose of rIFN-α did not affect weight loss compared to saline-treated mice. (B and C) There were no differences induced by IFN-α treatment (IFNtx) in bacterial quantities in lungs or spleens 9 (B) or 6 (C) days postinfection. (D) Following infection with 102 bacteria and high-dose rIFN-α treatment, cytokines in BAL fluids and serum were assessed. The only two cytokines that changed significantly were G-CSF and IL-1β in BAL fluid. These cytokines were not detected in day 1 and day 3 untreated infected mice. They were present in PBS-treated mice at day 6 postinfection and decreased in response to i.p. rIFN-α treatment. The data are representative of the results of at least 2 replicate experiments. *, P < 0.05; ****, P < 0.0001; ns, not significant.
Treatment of mice with intratracheal rIFN-α during C. burnetii infection.
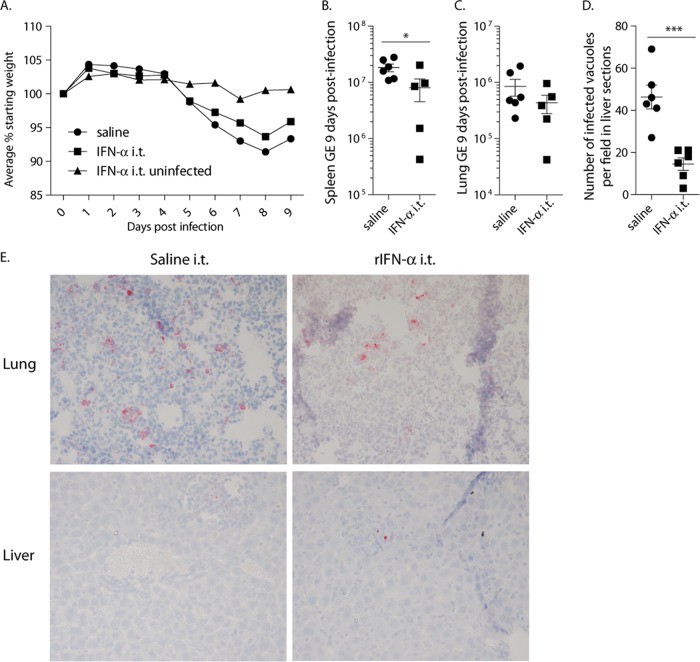
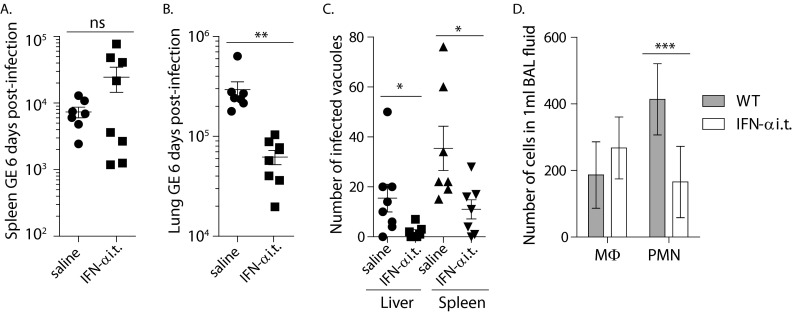
Because the lung is the route for a majority of C. burnetii infections, we also treated mice with rIFN-α by the i.t. route to study a tissue-specific response. Surprisingly, and in contrast to peritoneal treatment, delivery of rIFN-α to the lung protected against bacterial replication. Mice were infected with 103 bacteria and treated intratracheally with 4,000 U rIFN-α on the day of infection and every other day thereafter. The difference in weight loss between saline- and cytokine-treated groups was not significant, and treatment with rIFN-α alone, in the absence of infection, did not result in weight loss (Fig. 4A). At day 9 postinfection, fewer bacterial GE were detected by PCR in the spleens of treated mice, whereas there was not a significant difference in the lungs (Fig. 4B and C). Histological staining of liver sections indicated that there were also significantly fewer bacteria in the livers of mice treated i.t. with IFN-α (Fig. 4D). To refine this experiment, mice were similarly treated with either saline or rIFN-α and infected with 102 bacteria for 6 days. There was no weight loss with this dose of bacteria in this short time frame. PCR analyses suggested that rIFN-α-treated mice had similar amounts of bacteria in the spleens but significantly fewer bacteria in the lungs (Fig. 5A and B). Histological staining of livers and spleen sections, however, indicated that there were significantly fewer bacteria in both tissues in rIFN-α-treated mice (Fig. 5C). The PCR results from the spleen shown in Fig. 5A suggest that the type I IFN-treated mice segregated into two distinct groups. This may suggest differences in how mice respond to an injection of a bolus of recombinant type I IFN. However, such segregation was not seen in our histological stains of the same tissues. As such, at this time, we do not know the reason for the segregation seen in Fig. 5A or if this is a consistent observation. These data from the spleens are a rare case in which the results from PCR and histological quantification are not the same. However, the data from all the tissues taken together clearly support the idea that treatment with type I IFN in the lung protects against bacterial replication both in the lung and in other tissues. The morphologies of cell populations contained in BAL fluids were also analyzed. Type I IFN treatment did not appear to change the number of macrophages recruited to or replicating in the lung but resulted in significantly fewer neutrophils. This suggests that the lungs of type I IFN-treated mice were less inflamed than those of mice treated with saline. Whether this is a cause of decreased numbers of bacteria or an effect of less bacterial replication in the lung is not known.
FIG 4.
Intratracheal injection of rIFN-α protected mice from C. burnetii replication 9 days after infection with 103 C. burnetii bacteria. (A) Mice treated with saline and IFN-α by the i.t. route and infected with 103 bacteria did not differ significantly in weight loss during the course of the infection. Treatment of mice with IFN-α by the i.t. route in the absence of infection did not cause weight loss. (B and C) There was a significant difference in bacterial replication in the spleen (B) but not in the lung (C) by qPCR analyses of C. burnetii GE. (D) Histological assessment of mouse livers indicated fewer bacteria in the livers of i.t. rIFN-α-treated mice than in those of mice treated with saline. (E) Partial fields of representative histological sections in which bacterial foci can be seen. *, P < 0.05; ***, P < 0.001. The error bars indicate standard deviations.
FIG 5.
Intratracheal injection of rIFN-α protected mice from C. burnetii replication 6 days after infection with 102 C. burnetii bacteria. (A and B) Mice treated with rIFN-α by the i.t. route had no significant difference in the spleen (A) but significantly fewer bacteria in the lung (B) as measured by qPCR. (C) Histological assessment of spleens and livers indicated fewer bacteria in both organs following treatment with IFN-α than following treatment with saline. (D) Mice treated with rIFN-α had numbers of cells with macrophage (MΦ) morphology in BAL fluids similar to those of saline-treated mice but significantly fewer cells with polymorphonuclear cell (PMN) morphology. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant. The error bars indicate standard deviations.
Induction of IFN-β transcripts in human cells by NMI LPS stimulation.
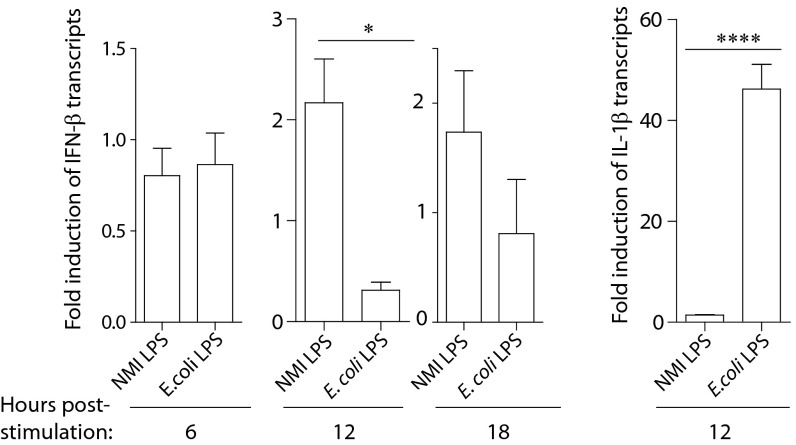
Our experiments comparing wild-type mice to IFNAR−/− mice suggest that type I IFN is induced and has a function during C. burnetii infection. However, the virulent form of C. burnetii is known to have very subtle and noninflammatory effects on human cells because of the unique branched terminal sugar-containing O-polysaccharide chain on the LPS molecule (26, 34). To understand the potential for induction of type I IFN during C. burnetii infection in human disease, we utilized human macrophages to assess transcript changes induced by C. burnetii NMI LPS stimulation compared to E. coli LPS stimulation. Macrophages were differentiated using M-CSF, and then the cells were treated with medium or LPS from either C. burnetii NMI or E. coli. The fold induction of transcript expression over untreated cells was calculated for IFN-β and IL-1β, with normalization to β-actin transcripts. Consistent with findings described in the literature with extracts from phase I organisms (15, 25), LPS from the virulent strain of C. burnetii increased expression of IFN-β transcripts by about 2-fold compared to human cells treated with control medium after stimulation for 12 h. Cells were also stimulated for 6 or 18 h, and this increased expression of IFN-β was not apparent at these intervals (Fig. 6). At the same 12-h interval, E. coli LPS at a greater but commonly used EU concentration did not affect IFN-β transcripts but caused a dramatic increase in IL-1β transcript expression that was not affected by NMI LPS. Similarly, using differentiated mouse macrophages, NMI LPS did not result in expression of TNF-α, IL-6, or CXCL10, whereas E. coli LPS robustly induced expression of these proteins (data not shown). Because of the greatly diminished capacity of LPS from C. burnetii NMI to activate cells (34), the detection of even a small but consistent increase in IFN-β transcripts in human cells in response to NMI LPS stimulation is notable. Our data support induction of type I IFN by C. burnetii and the idea that the cytokine can affect infection differently depending on the location in the body.
FIG 6.
Human inflammatory transcripts changed in response to NMI LPS treatment in vitro. Human macrophages (n = 4 donors) were treated with medium only, NMI LPS, or E. coli LPS for 6, 12, or 18 h. RNA was extracted, and transcripts encoding IFN-β, IL-1β, and β-actin were assessed by qPCR. After normalization to β-actin expression, the fold increase over controls treated with medium only was calculated. Transcripts expressing IFN-β were increased by an average of 2-fold over the medium-only control following 12 h of NMI LPS stimulation, but not after 6 h of stimulation, and diminished from 2-fold after 18 h of stimulation. The transcripts were unaltered by E. coli LPS stimulation at these time points. In contrast, IL-1β transcripts were strongly increased by E. coli LPS after 12 h of stimulation and were not changed using NMI LPS. *, P < 0.05; ****, P < 0.0001. The error bars indicate standard deviations.
DISCUSSION
Type I IFN is induced by L. pneumophila and suppresses its replication in macrophages (10, 13). The mechanism was demonstrated to be independent of IL-1β, which protects from L. pneumophila infection. Here, we have demonstrated that type I IFN promotes C. burnetii replication in the periphery but may counter infection in the lung. This was a surprising finding, since these bacteria are genetically closely related (7), and both cause primary infections in the lung. Wild-type mice had more severe weight loss due to infection than did IFNAR-deficient mice, as well as increased bacteria in spleens, but not the lung, at day 6 postinfection. This suggests that when the bacteria exit the lung, type I IFN signaling may promote their dissemination and/or replication. Cytokine expression in BAL fluids was generally dampened in the absence of type I IFN signaling (in IFNAR−/− mice), yet this state did not promote bacterial replication in the lung. Thus, these cytokines, more highly expressed in wild-type mice, are apparently not effective in the defense against C. burnetii infection and may be a side effect that promotes weight loss. Diminished inflammatory cytokines may be the reason for better outcomes of C. burnetii infection in terms of weight loss in IFNAR-deficient mice. Interestingly, despite an increased innate cytokine response in the lung in wild-type mice, it appears that IFN signaling in the periphery promoted bacterial replication so that more bacteria were measured in wild-type spleens. Developmental or microbiome differences may affect disease outcomes in these two strains. For example, we consistently observe that healthy IFNAR−/− mice have smaller spleens than do wild-type mice (unpublished observation). Since type I IFN affects hematopoiesis (35, 36), developmental differences between IFNAR-deficient and wild-type mice that affect cell activation likely exist. However, IFNAR deficiency did not appear to affect the pool of susceptible macrophages in the lung and their capacity to be infected by C. burnetii. Because of these questions and uncertainties, we utilized alternative methods to investigate a role for type I IFN in C. burnetii infection: induction by poly(I·C) and injection of rIFN-α.
We expected to detect strong expression of IL-10, and changes to the expression of the cytokine in response to changes in type I IFN, in our studies. Expression of IL-10 is a well-characterized anti-inflammatory consequence of type I IFN signaling (37). Also, IL-10 is increased in patients with Q fever and serves to suppress TNF-α expression (16). Furthermore, it has been demonstrated that mice that express excess IL-10 are more susceptible to C. burnetii infection than are wild-type mice (18). There was a slight decrease in IL-10 in IFNAR-deficient mice compared to wild-type mice, but the expression levels of the cytokine were very low in general. Possibly, the reagents for detecting IL-10 in these in vivo studies are less efficient than those designed to detect other cytokines or IL-10 expression and function are localized tissue microenvironment-specific effects, and thus, its detection in BAL fluids and serum is difficult. Considering that antibodies to IL-10 are used in patients to alter immunity (38), experiments involving in vivo blockade of IL-10 in C. burnetii infection may warrant investigation.
In most, if not all, cases reported in the literature, treatment with poly(I·C) results in outcomes similar to those of treatment with recombinant type I IFN, presumably due to the fact that poly(I·C) primarily stimulates type I IFN expression. However, in virulent C. burnetii infection, poly(I·C) treatment protected from C. burnetii pathogenesis in an IFN-independent manner, but bacterial replication did not appear to be altered. Poly(I·C) can also stimulate other pathways, including the NF-κB pathway (33), and the downstream effects of this stimulation can vary, depending on the cytokine and the costimulatory milieu (39). Even a slight increase in IFN-γ would be highly protective against C. burnetii infection (20, 40). The results with poly(I·C) introduce the possibility that stimulation with other innate agonists may be effective for use against C. burnetii infection. Considering the length of time currently required for antibiotic treatment of chronic C. burnetii infection, it is worth considering alternative or complementary approaches that might diminish the duration or dose of antibiotic treatment or increase the effectiveness of the current dose.
When rIFN-α was delivered to the periphery, infection with C. burnetii was enhanced, consistent with findings using IFNAR-deficient mice. The only detectible differences in cytokine expression were decreases in G-CSF and IL-1β, and these cytokines were not detected at days 1 or 3 postinfection. Considering that IFNAR-deficient mice were protected from C. burnetii-induced weight loss, we expected exacerbated weight loss following treatment with rIFN-α. Usually, however, increased weight loss is due to increased, not decreased, inflammatory cytokines. The decreased inflammatory cytokine expression detected in the lung following peripheral type I IFN treatment may be localized to and specific to that tissue. Although chronic Q fever patients have increased expression of IL-1β (21) and C. burnetii likely subverts the inflammasome and downstream effects to facilitate intracellular replication, little is known about the interactions between this pathway and C. burnetii (41). These results are the first to suggest the induction of IL-1β during acute C. burnetii infection in mice and that type I IFN treatment can dampen its expression. IL-1β protects macrophages from intracellular infection in part by enhancing TNF signaling (42). The observation that type I IFN promotes C. burnetii replication and decreases expression of IL-1β is similar to that described for M. tuberculosis infection, in which bacterial replication is promoted by type I IFN and a critical balance between type I IFN and IL-1β exists (2). This inverse relationship between type I IFN and IL-1β has also been demonstrated in the absence of infection (23). IL-1β is likely to have a role in innate protection from C. burnetii infection because C. burnetii is an obligate intracellular pathogen that targets macrophages, although IL-1β appears to be less effective later when the infection is chronic (21).
When delivered to the lung, type I IFN in the lung was protective from C. burnetii infection in all tissues assessed, suggesting a tissue-specific response for the cytokine. Similarly, type I IFN promotes inflammatory responses in the respiratory tract following respiratory syncytial virus (RSV) infection (43). In contrast, during influenza virus infection, type I IFN decreased proinflammatory cytokines, which was directly linked to robust IL-10 expression. The expression of IL-10 likely depends on several other factors in addition to type I IFNs. In our experiments, IL-10 expression was minimal, and the presence of intact type I IFN signaling seems to have promoted the expression of inflammatory cytokines in wild-type mice. In contrast, when type I IFN was delivered to the lung, significantly decreased neutrophils indicated decreased inflammatory cytokine presence, yet the treatment protected against infection. Similarly, although i.p. rIFN-α treatment promoted peripheral infection, it also had specific anti-inflammatory effects in the lung, resulting in decreases in G-CSF and IL-1β. Thus, the mechanism for protection in the lung may be subtle activation of specific or infected cells by the combination of bacterial infection and type I IFN that occurs in the absence of overt inflammation.
The literature suggests C. burnetii has the capacity to trigger production of type I IFN, but the precise mechanism has not been determined (15, 25). One way to induce the cytokine is LPS signaling through Toll-like receptor 4 (TLR4). Signaling downstream of TLR4 through a MyD88-independent pathway known as Toll–IL-1 receptor (TIR) domain-containing adaptor-inducing IFN-β (TRIF) induces expression of type I IFN (44). The LPS molecule of C. burnetii is distinct from other LPS molecules because it is hypoacylated (45). Whereas normally acylated LPS structures stimulate mouse and human TLR4s similarly, hypoacylated LPS has been shown to have a greatly diminished inflammatory capacity when interacting with human TLR4 compared to mouse TLR4 (46). As such, hypoacylated LPS from Yersinia pestis induced blunted inflammatory responses through human TLR4 compared to mouse TLR4 (46). Thus, it may be that the inflammatory state induced by C. burnetii infection in mice is distinct from that induced in humans. We utilized human cells to determine if type I IFN could be expressed in response to C. burnetii LPS in comparison to E. coli LPS. We confirmed that, unlike E. coli LPS, C. burnetii NMI LPS induced few inflammatory products, but after 12 h of stimulation, expression of IFN-β transcripts increased. Other researchers have determined that a 10-fold-higher concentration of LPS (100 ng/ml) induces IFN-β transcripts at 1 h posttreatment in vitro, which is diminished at later time points (47, 48). Similarly, we identified a specific interval (12 h of stimulation) in which C. burnetii LPS induces IFN-β. During infection in vivo, this response is likely less uniform and more robust. Thus, in human cells, NMI LPS could potentially stimulate the TRIF pathway to induce type I IFN.
Q fever is relatively rare compared to many other infectious diseases but is likely underreported and misdiagnosed (6, 49). Not only are people who engage in agricultural work at risk for infection, but Q fever is increasingly prevalent in military populations. Furthermore, domestic infections are also increasingly unassociated with contact with animals (49). Our data suggest that strong systemic induction of the type I IFN pathway, as would occur during concurrent viral infection, would enhance C. burnetii pathogenesis and replication in humans. This is the opposite of what would be expected with the closely related lung pathogen L. pneumophila and underscores the need to consider the interaction between type I IFN signaling and bacterial pathogens on a case-by-case basis. Understanding innate responses during C. burnetii infection will likely lead to novel complementary treatment approaches to augment the current antibiotic regimen.
ACKNOWLEDGMENTS
This project was primarily funded through NIH COBRE (P20 RR020185), with partial funding through NIH-NCCAM (AT0004986-01), an NIH grant (1R21AI094261), the M. J. Murdock Charitable Trust, and The Montana State University Agricultural Experimental Station.
REFERENCES
- 1.O'Connell RM, Saha SK, Vaidya SA, Bruhn KW, Miranda GA, Zarnegar B, Perry AK, Nguyen BO, Lane TF, Taniguchi T, Miller JF, Cheng G. 2004. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J Exp Med 200:437–445. doi: 10.1084/jem.20040712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mayer-Barber KD, Andrade BB, Oland SD, Amaral EP, Barber DL, Gonzales J, Derrick SC, Shi R, Kumar NP, Wei W, Yuan X, Zhang G, Cai Y, Babu S, Catalfamo M, Salazar AM, Via LE, Barry CE III, Sher A. 2014. Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk. Nature 511:99–103. doi: 10.1038/nature13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin FJ, Gomez MI, Wetzel DM, Memmi G, O'Seaghdha M, Soong G, Schindler C, Prince A. 2009. Staphylococcus aureus activates type I IFN signaling in mice and humans through the Xr repeated sequences of protein A. J Clin Invest 119:1931–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parker D, Prince A. 2011. Type I interferon response to extracellular bacteria in the airway epithelium. Trends Immunol 32:582–588. doi: 10.1016/j.it.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helbig KJ, Heatley SL, Harris RJ, Mullighan CG, Bardy PG, Marmion BP. 2003. Variation in immune response genes and chronic Q fever. Concepts: preliminary test with post-Q fever fatigue syndrome. Genes Immun 4:82–85. [DOI] [PubMed] [Google Scholar]
- 6.Dahlgren FS, Haberling DL, McQuiston JH. 2015. Q fever is underestimated in the United States: a comparison of fatal Q fever cases from two national reporting systems. Am J Trop Med Hyg 92:244–246. doi: 10.4269/ajtmh.14-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams KP, Gillespie JJ, Sobral BWS, Nordberg EK, Snyder EE, Shallom JM, Dickerman AW. 2010. Phylogeny of Gammaproteobacteria. J Bacteriol 192:2305–2314. doi: 10.1128/JB.01480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schiavoni G, Mauri C, Carlei D, Belardelli F, Castellani Pastoris M, Proietti E. 2004. Type I IFN protects permissive macrophages from Legionella pneumophila infection through an IFN-γ-independent pathway. J Immunol 173:1266–1275. doi: 10.4049/jimmunol.173.2.1266. [DOI] [PubMed] [Google Scholar]
- 9.Opitz B, Vinzing M, van Laak V, Schmeck B, Heine G, Günther S, Preissner R, Slevogt H, N′Guessan PD, Eitel J, Goldmann T, Flieger A, Suttorp N, Hippenstiel S. 2006. Legionella pneumophila induces IFNβ in lung epithelial cells via IPS-1 and IRF3, which also control bacterial replication. J Biol Chem 281:36173–36179. doi: 10.1074/jbc.M604638200. [DOI] [PubMed] [Google Scholar]
- 10.Plumlee CR, Lee C, Beg AA, Decker T, Shuman HA, Schindler C. 2009. Interferons direct an effective innate response to Legionella pneumophila infection. J Biol Chem 284:30058–30066. doi: 10.1074/jbc.M109.018283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coers J, Vance RE, Fontana MF, Dietrich WF. 2007. Restriction of Legionella pneumophila growth in macrophages requires the concerted action of cytokine and Naip5/Ipaf signalling pathways. Cell Microbiol 9:2344–2357. doi: 10.1111/j.1462-5822.2007.00963.x. [DOI] [PubMed] [Google Scholar]
- 12.Ang DKY, Oates CVL, Schuelein R, Kelly M, Sansom FM, Bourges De, Boon L, Hertzog PJ, Hartland EL, van Driel IR. 2010. Cutting edge: pulmonary Legionella pneumophila is controlled by plasmacytoid dendritic cells but not type I IFN. J Immunol 184:5429–5433. doi: 10.4049/jimmunol.1000128. [DOI] [PubMed] [Google Scholar]
- 13.Lippmann J, Müller HC, Naujoks J, Tabeling C, Shin S, Witzenrath M, Hellwig K, Kirschning CJ, Taylor GA, Barchet W, Bauer S, Suttorp N, Roy CR, Opitz B. 2011. Dissection of a type I interferon pathway in controlling bacterial intracellular infection in mice. Cell Microbiol 13:1668–1682. doi: 10.1111/j.1462-5822.2011.01646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honstettre A, Imbert G, Ghigo E, Gouriet F, Capo C, Raoult D, Mege JL. 2003. Dysregulation of cytokines in acute Q fever: role of interleukin-10 and tumor necrosis factor in chronic evolution of Q fever. J Infect Dis 187:956–962. doi: 10.1086/368129. [DOI] [PubMed] [Google Scholar]
- 15.Waag DM, Kende M, Damrow TA, Wood OL, Williams JC. 1990. Injection of inactivated phase I Coxiella burnetii increases non-specific resistance to infection and stimulates lymphokine production in mice. Ann N Y Acad Sci 590:203–214. doi: 10.1111/j.1749-6632.1990.tb42221.x. [DOI] [PubMed] [Google Scholar]
- 16.Ghigo E, Capo C, Raoult D, Mege JL. 2001. Interleukin-10 stimulates Coxiella burnetii replication in human monocytes through tumor necrosis factor down-modulation: role in microbicidal defect of Q fever. Infect Immun 69:2345–2352. doi: 10.1128/IAI.69.4.2345-2352.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghigo E, Honstettre Al, Capo C, Gorvel JP, Raoult D, Mege JL. 2004. Link between impaired maturation of phagosomes and defective Coxiella burnetii killing in patients with chronic Q fever. J Infect Dis 190:1767–1772. doi: 10.1086/425041. [DOI] [PubMed] [Google Scholar]
- 18.Meghari S, Bechah Y, Capo C, Lepidi H, Raoult D, Murray PJ, Mege JL. 2008. Persistent Coxiella burnetii infection in mice overexpressing IL-10: an efficient model for chronic Q fever pathogenesis. PLoS Pathog 4:e23. doi: 10.1371/journal.ppat.0040023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andoh M, Zhang G, Russell-Lodrigue KE, Shive HR, Weeks BR, Samuel JE. 2007. T cells are essential for bacterial clearance, and gamma interferon, tumor necrosis factor alpha, and B cells are crucial for disease development in Coxiella burnetii infection in mice. Infect Immun 75:3245–3255. doi: 10.1128/IAI.01767-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dellacasagrande J, Capo C, Raoult D, Mege JL. 1999. IFN-γ mediated control of Coxiella burnetii survival in monocytes: the role of cell apoptosis and TNF. J Immunol 162:2259–2265. [PubMed] [Google Scholar]
- 21.Capo C, Zugun F, Stein A, Tardei G, Lepidi H, Raoult D, Mege JL. 1996. Upregulation of tumor necrosis factor alpha and interleukin-1 beta in Q fever endocarditis. Infect Immun 64:1638–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kole A, He J, Rivollier A, Silveira DD, Kitamura K, Maloy KJ, Kelsall BL. 2013. Type I IFNs regulate effector and regulatory T cell accumulation and anti-inflammatory cytokine production during T cell-mediated colitis. J. Immunol 191:2771–2779. doi: 10.4049/jimmunol.1301093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guarda G, Braun M, Staehli F, Tardivel A, Mattmann C, Förster I, Farlik M, Decker T, Du Pasquier RA, Romero P, Tschopp J. 2011. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity 34:213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Novikov A, Cardone M, Thompson R, Shenderov K, Kirschman KD, Mayer-Barber KD, Myers TG, Rabin RL, Trinchieri G, Sher A, Feng CG. 2011. Mycobacterium tuberculosis triggers host type I IFN signaling to regulate IL-1β production in human macrophages. J Immunol 187:2540–2547. doi: 10.4049/jimmunol.1100926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zvilich M, Williams JC, Waag D, Rill WR, Malli RJ, Bell P, Kende M. 1995. Characterization of the non-specific humoral and cellular antiviral immunity stimulated by the chloroform-methanol residue (CMR) fraction of Coxiella burnetii. Antiviral Res 27:389–404. doi: 10.1016/0166-3542(95)00022-E. [DOI] [PubMed] [Google Scholar]
- 26.Graham JG, MacDonald LJ, Hussain SK, Sharma UM, Kurten RC, Voth DE. 2013. Virulent Coxiella burnetii pathotypes productively infect primary human alveolar macrophages. Cell Microbiol 15:1012–1025. doi: 10.1111/cmi.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pandey AK, Yang Y, Jiang Z, Fortune SM, Coulombe F, Behr MA, Fitzgerald KA, Sassetti CM, Kelliher MA. 2009. NOD2, RIP2 and IRF5 play a critical role in the type I interferon response to Mycobacterium tuberculosis. PLoS Pathog 5:e1000500. doi: 10.1371/journal.ppat.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larson CL, Beare PA, Voth DE, Howe D, Cockrell DC, Bastidas RJ, Valdivia RH, Heinzen RA. 2015. Coxiella burnetii effector proteins that localize to the parasitophorous vacuole membrane promote intracellular replication. Infect Immun 83:661–670. doi: 10.1128/IAI.02763-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Read AJ, Erickson S, Harmsen AG. 2010. Role of CD4+ and CD8+ T cells in clearance of primary pulmonary infection with Coxiella burnetii. Infect Immun 78:3019–3026. doi: 10.1128/IAI.00101-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coleman SA, Fischer ER, Howe D, Mead DJ, Heinzen RA. 2004. Temporal analysis of Coxiella burnetii morphological differentiation. J Bacteriol 186:7344–7352. doi: 10.1128/JB.186.21.7344-7352.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Westphal O, Jann K. 1965. Bacterial lipopolysaccharides. Extraction with phenol-water and further applications of the procedure, p 83–91. In Whistler RL, Wolfan ML (ed), Methods in carbohydrate chemistry. Academic Press, New York, NY. [Google Scholar]
- 32.Raoult D, Marrie TJ, Mege JL. 2005. Natural history and pathophysiology of Q fever. Lancet Infect Dis 5:219–226. doi: 10.1016/S1473-3099(05)70052-9. [DOI] [PubMed] [Google Scholar]
- 33.Zou J, Kawai T, Tsuchida T, Kozaki T, Tanaka H, Shin KS, Kumar H, Akira S. 2013. Poly IC triggers a cathepsin D- and IPS-1-dependent pathway to enhance cytokine production and mediate dendritic cell necroptosis. Immunity 38:717–728. doi: 10.1016/j.immuni.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 34.Shannon JG, Howe D, Heinzen RA. 2005. Virulent Coxiella burnetii does not activate human dendritic cells: role of lipopolysaccharide as a shielding molecule. Proc Natl Acad Sci U S A 102:8722–8727. doi: 10.1073/pnas.0501863102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seo SU, Kwon HJ, Ko HJ, Byun YH, Seong BL, Uematsu S, Akira S, Kweon MN. 2011. Type I interferon signaling regulates Ly6Chi monocytes and neutrophils during acute viral pneumonia in mice. PLoS Pathog 7:e1001304. doi: 10.1371/journal.ppat.1001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor D, Wilkison M, Voyich J, Meissner N. 2011. Prevention of bone marrow cell apoptosis and regulation of hematopoiesis by type I IFNs during systemic responses to pneumocystis lung infection. J. Immunol 186:5956–5967. doi: 10.4049/jimmunol.1003558. [DOI] [PubMed] [Google Scholar]
- 37.Zhang L, Yuan S, Cheng G, Guo B. 2011. Type I IFN promotes IL-10 production from t cells to suppress Th17 cells and Th17-associated autoimmune inflammation. PLoS One 6:e28432. doi: 10.1371/journal.pone.0028432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Llorente L, Richaud-Patin Y, Garcia-Padilla C, Claret E, Jakez-Ocampo J, Cardiel MH, Alcocer-Varela J, Grangeot-Keros L, Alarcin-Segovia D, Wijdenes J, Galanaud P, Emilie D. 2000. Clinical and biologic effects of anti-interleukin-10 monoclonal antibody administration in systemic lupus erythematosus. Arthritis Rheum 43:1790–1800. doi:. [DOI] [PubMed] [Google Scholar]
- 39.Jensen S, Gad M. 2010. Differential induction of inflammatory cytokines by dendritic cells treated with novel TLR-agonist and cytokine based cocktails: targeting dendritic cells in autoimmunity. J Inflamm 7:37. doi: 10.1186/1476-9255-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghigo E, Capo C, Tung CH, Raoult D, Gorvel JP, Mege JL. 2002. Coxiella burnetii survival in THP-1 monocytes involves the impairment of phagosome maturation: IFNγ mediates its restoration and bacterial killing. J Immunol 169:4488–4495. doi: 10.4049/jimmunol.169.8.4488. [DOI] [PubMed] [Google Scholar]
- 41.Cunha LD, Zamboni DS. 2013. Subversion of inflammasome activation and pyroptosis by pathogenic bacteria. Front Cell Infect Microbiol 3:76. doi: 10.3389/fcimb.2013.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jayaraman P, Sada-Ovalle I, Nishimura T, Anderson AC, Kuchroo VK, Remold HG, Behar SM. 2013. IL-1β promotes antimicrobial immunity in macrophages by regulating TNFR signaling and caspase-3 activation. J Immunol 190:4196–4204. doi: 10.4049/jimmunol.1202688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goritzka M, Durant LR, Pereira C, Salek-Ardakani S, Openshaw PJM, Johansson C. 2014. Alpha/beta interferon receptor signaling amplifies early proinflammatory cytokine production in the lung during respiratory syncytial virus infection. J Virol 88:6128–6136. doi: 10.1128/JVI.00333-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kolb JP, Casella CR, SenGupta S, Chilton PM, Mitchell TC. 2014. Type I interferon signaling contributes to the bias that Toll-like receptor 4 exhibits for signaling mediated by the adaptor protein TRIF. Sci Signal 7:ra108. doi: 10.1126/scisignal.2005442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toman R, Garidel P, Andra J, Slaba K, Hussein A, Koch MH, Brandenburg K. 2004. Physicochemical characterization of the endotoxins from Coxiella burnetii strain Priscilla in relation to their bioactivities. BMC Biochem 5:1. doi: 10.1186/1471-2091-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hajjar AM, Ernst RK, Fortuno ES, Brasfield AS, Yam CS, Newlon LA, Kollmann TR, Miller SI, Wilson CB. 2012. Humanized TLR4/MD-2 mice reveal LPS recognition differentially impacts susceptibility to Yersinia pestis and Salmonella enterica. PLoS Pathog 8:e1002963. doi: 10.1371/journal.ppat.1002963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheikh F, Dickensheets H, Gamero AM, Vogel SN, Donnelly RP. 2014. An essential role for IFN-β in the induction of IFN-stimulated gene expression by LPS in macrophages. J Leukoc Biol 96:591–600. doi: 10.1189/jlb.2A0414-191R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacobs AT, Ignarro LJ. 2001. LPS-induced expression of interferon-beta mediates the timing of nitric-oxide synthase induction in RAW 264.7 macrophages. J Biol Chem 276:47950–47957. [DOI] [PubMed] [Google Scholar]
- 49.Dahlgren FS, McQuiston JH, Massung RF, Anderson AD. 2015. Q fever in the United States: summary of case reports from two national surveillance systems, 2000-2012. Am J Trop Med Hyg 92:247–255. doi: 10.4269/ajtmh.14-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]