ABSTRACT
Escherichia coli O26 is the second most important enterohemorrhagic E. coli (EHEC) serogroup worldwide. Serogroup O26 strains are categorized mainly into two groups: enteropathogenic (EPEC) O26, carrying a locus of enterocyte effacement (LEE) and mostly causing mild diarrhea, and Shiga-toxigenic (STEC) O26, which carries the Shiga toxin (STX) gene (stx), responsible for more severe outcomes. stx-negative O26 strains can be further split into two groups. One O26 group differs significantly from O26 EHEC, while the other O26 EHEC-like group shows all the characteristics of EHEC O26 except production of STX. In order to determine the different populations of O26 E. coli present in U.S. cattle, we sequenced 42 O26:H11 strains isolated from feedlot cattle and compared them to 37 O26:H11 genomes available in GenBank. Phylogenetic analysis by whole-genome multilocus sequence typing (wgMLST) showed that O26:H11/H− strains in U.S. cattle were highly diverse. Most strains were sequence type 29 (ST29). By wgMLST, two clear lineages could be distinguished among cattle strains. Lineage 1 consisted of O26:H11 EHEC-like strains (ST29) (4 strains) and O26:H11 EHEC strains (ST21) (2 strains), and lineage 2 (36 strains) consisted of O26:H11 EPEC strains (ST29). Overall, our analysis showed U.S. cattle carried pathogenic (ST21; stx1+ ehxA+ toxB+) and also potentially pathogenic (ST29; ehxA+ toxB+) O26:H11 E. coli strains. Furthermore, in silico analysis showed that 70% of the cattle strains carried at least one antimicrobial resistance gene. Our results showed that whole-genome sequence analysis is a robust and valid approach to identify and genetically characterize E. coli O26:H11, which is of importance for food safety and public health.
IMPORTANCE Escherichia coli O26 is the second most important type of enterohemorrhagic E. coli (EHEC) worldwide. Serogroup O26 strains are categorized into two groups: enteropathogenic (EPEC) carrying LEE, causing mild diarrhea, and Shiga toxigenic (STEC) carrying the stx gene, responsible for more severe outcomes. However, there are currently problems in distinguishing one group from the other. Furthermore, several O26 stx-negative strains are consistently misidentified as either EHEC-like or EPEC. The use of whole-genome sequence (WGS) analysis of O26 strains from cattle in the United States (i) allowed identification of O26 strains present in U.S. cattle, (ii) determined O26 strain diversity, (iii) solved the misidentification problem, and (iv) screened for the presence of antimicrobial resistance and virulence genes in the strains. This study provided a framework showing how to easily and rapidly use WGS information to identify and genetically characterize E. coli O26:H11, which is important for food safety and public health.
INTRODUCTION
In the United States, enterohemorrhagic Escherichia coli (EHEC) O157:H7 has caused the majority of food-borne outbreaks in the last 2 decades (1). However, among the non-O157:H7 EHEC strains, Shiga toxin-producing E. coli O26:H11/H− (STEC) is the second most common cause of EHEC food-borne infections in the United States (2, 3) and worldwide (4–7). STEC O26 can cause a wide range of infections in humans, from mild diarrhea to hemorrhagic colitis (HC), in some cases leading to hemolytic-uremic syndrome (HUS) (6). Outbreaks of O26 infection have been linked to beef, contact with cattle (4, 8), produce (http://www.cdc.gov/ecoli/2012/o26-02-12/index.html), and person-to-person transmission (9). Therefore, in 2011, the U.S. Department of Agriculture Food Safety and Inspection Services (USDA FSIS) declared O26 and five other non-O157 STEC serogroups, O45, O113, O111, O121, and O145, adulterants in ground beef and nonintact beef products and began testing for these pathogens in June 2012 in domestic and imported beef trimmings (10).
Pathogenic O26 strains are divided into two main groups: enteropathogenic E. coli (EPEC), characterized by carrying the locus of enterocyte effacement (LEE) and causing mild diarrhea, and EHEC, which in addition to the LEE carries the stx1 and/or stx2 gene and is responsible for more severe outcomes, such as HC and HUS (7, 11). EPEC O26 has been further divided into two subgroups. One subgroup carries most virulence markers (including an EHEC virulence plasmid carrying a gene, ehxA, encoding an enterohemolysin) for O26 EHEC except the stx gene(s). This subgroup has been called EHEC-like O26 (or atypical EPEC O26). They also have the same sequence type (ST), as determined by multilocus sequence typing (MLST), and the same multilocus variable-number tandem-repeat analysis (MLVA) profiles as some O26 EHEC strains (12, 13). It is believed that atypical EPEC O26 evolved into a highly pathogenic clone (EHEC O26) through acquisition of a Shiga toxin (STX)-producing phage (6, 14, 15). The second EPEC group is differentiated from EHEC or EHEC-like O26 based on its fermentative profiles, by carrying a different virulence plasmid (encoding an alpha-hemolysin), and by having the arcA allele type 1 gene (13). Serotype O26:H11/H− can be divided, based on MLST, into several STs, with ST21 and ST29 associated with disease in humans (16). Although present worldwide (6), O26:H11/H− has been predominantly described in Europe, with little data on its prevalence on other continents.
Recently, the diversity of O26:H11 strains of cattle origin in the United States based on a combination of molecular markers and clustered regularly interspaced short palindromic repeat (CRISPR) typing has been described (17). It was suggested that some of the strains may have the potential to become pathogenic to humans by acquisition of a stx-carrying phage, as they contained a CRISPR PCR target (SP_O26-E) previously identified only in stx2-positive O26:H11 clinical strains (17). Further analysis of the O26 cattle strains from the United States using 48 informative single nucleotide polymorphisms (SNPs) found that EHEC-like O26 strains (stx negative) displayed SNP genotypes synonymous with two publicly available clinical strains (ST29; O26:H11 EHEC strains). This result suggested that the EHEC-like O26 strains (stx negative) were genetically closer to the clinical O26 EHEC (stx+) strains (18). However, the actual phylogeny of EHEC, EHEC-like, and EPEC O26 strains has not been determined.
In order to further characterize E. coli O26 in U.S. cattle and to confirm previously suggested evolutionary and phylogenetic relationships among EHEC, EHEC-like, and EPEC O26 strains, we conducted a whole-genome sequence (WGS) analysis of selected O26:H11/H− strains isolated from commercial feedlot cattle (19) that had been characterized by various molecular techniques (17, 18) and compared them to other clinical and environmental O26:H11 genomes available at GenBank (37 genomes). The 42 O26:H11/H− strains were analyzed for virulence genes, in silico MLST, and antibiotic resistance genes. Finally, their phylogenetic relationships and diversity were determined by whole-genome phylogeny analysis using two methods: (i) allele-based whole-genome MLST (wgMLST) and (ii) targeted SNP analysis.
MATERIALS AND METHODS
Bacterial strains and media.
The E. coli O26:H11 strains (n = 42) used in this study are listed in Table 1. Each strain was assigned a U.S. FDA Center for Food Safety and Applied Nutrition (CFSAN) number for future tracking. The strains were isolated from cattle feces in a commercial feedlot located in the Midwest (19). The isolation procedure included enrichment of the feces in E. coli broth, immunomagnetic separation (IMS) with O26-specific IMS beads, and plating on selective medium, followed by confirmation of the presumptive isolate by a multiplex PCR targeting the O26 wzx gene (wzxO26) and the three major virulence genes, stx1, stx2, and eae (20). The strains were stored on tryptone soy broth containing glycerol at −70°C until further use. The strains were plated onto blood agar (Remel, Lenexa, KS) and grown overnight at 37°C for further analysis.
TABLE 1.
Characteristics of the E. coli O26:H11/H− strains of cattle origin analyzed in this study
| Straina | CFSAN no. | Serotype | Accession no. (WGS) | STb | stx1b | stx2b | ehxAb |
|---|---|---|---|---|---|---|---|
| 1895-A | CFSAN025090 | O26:H11 | LPTU00000000 | 21 | + | − | + |
| 1914-A | CFSAN025091 | O26:H11 | LPTV00000000 | 21 | + | − | + |
| 1357-A | CFSAN025092 | O26:H11 | LPTW00000000 | 29 | − | − | − |
| 1270-A | CFSAN025093 | O26:H11 | LPXY00000000 | 29 | − | − | − |
| 2228-A | CFSAN025094 | O26:H11 | LPXZ00000000 | 29 | − | − | − |
| 2152-B | CFSAN025095 | O26:H11 | LPYA00000000 | 29 | − | − | − |
| 1341-A | CFSAN025096 | O26:H11 | LPYB00000000 | 29 | − | − | + |
| 1676-A | CFSAN025098 | O26:H11 | LPYC00000000 | 29 | − | − | − |
| 1692-A | CFSAN025099 | O26:H11 | LPYD00000000 | 29 | − | − | − |
| 1740-A-A | CFSAN025100 | O26:H11 | LPYE00000000 | 29 | − | − | − |
| 1740-A-B | CFSAN025101 | O26:H11 | LPYF00000000 | 29 | − | − | − |
| 1668-A-A | CFSAN025102 | O26:H11 | LPYG00000000 | 29 | − | − | + |
| 1668-A-B | CFSAN025103 | O26:H11 | LPYH00000000 | 29 | − | − | + |
| 1958-A-B | CFSAN025104 | O26:H11 | LPYI00000000 | 29 | − | − | − |
| 2194-B | CFSAN025106 | O26:H11 | LPYJ00000000 | 29 | − | − | − |
| 2105-G | CFSAN025107 | O26:H11 | LPYK00000000 | 29 | − | − | − |
| 2223-B | CFSAN025108 | O26:H11 | LPYL00000000 | 29 | − | − | − |
| 2139-A | CFSAN025109 | O26:H11 | LPYM00000000 | 29 | − | − | − |
| 2176-A | CFSAN025110 | O26:H11 | LPYN00000000 | 29 | − | − | − |
| 4131 | CFSAN025114 | O26:H11 | LPYO00000000 | 29 | − | − | − |
| 4170 | CFSAN025115 | O26:H11 | LPYP00000000 | 29 | − | − | − |
| 4196 | CFSAN025116 | O26:H11 | LPYQ00000000 | 29 | − | − | − |
| 4277-H-A | CFSAN025117 | O26:H11 | LPYR00000000 | 29 | − | − | − |
| 4368-A | CFSAN025118 | O26:H11 | LPYS00000000 | 29 | − | − | − |
| 4435-B | CFSAN025119 | O26:H11 | LPYT00000000 | 29 | − | − | − |
| 4271-C | CFSAN025121 | O26, H?c | LPYU00000000 | 29 | − | − | − |
| 4468-A-B | CFSAN025122 | O26:H11 | LPYV00000000 | 29 | − | − | − |
| 4848-A-B | CFSAN025123 | O26, H?c | LPYW00000000 | 29 | − | − | − |
| 4730-C | CFSAN025124 | O26:H11 | LPYX00000000 | 29 | − | − | − |
| 4513 | CFSAN025126 | O26:H11 | LPYY00000000 | 29 | − | − | − |
| 4592-A | CFSAN025127 | O26:H11 | LPYZ00000000 | 29 | − | − | − |
| 5583-H | CFSAN025128 | O26:H11 | LPZA00000000 | 29 | − | − | − |
| 5687-B | CFSAN025129 | O26:H11 | LPZB00000000 | 29 | − | − | − |
| 5206-E-B | CFSAN025130 | O26:H11 | LPZC00000000 | 29 | − | − | − |
| 1802-A | CFSAN025105 | O26:H11 | LPZD00000000 | 29 | − | − | − |
| 4822-A | CFSAN025125 | O26:H?c | LPZE00000000 | 29 | − | − | − |
| 4468-A-A | CFSAN025133 | O26:H11 | LPZF00000000 | 29 | − | − | − |
| 4860-A | CFSAN025135 | O26:H11 | LPZG00000000 | 29 | − | − | − |
| 4863-A | CFSAN025136 | O26:H11 | LPZH00000000 | 29 | − | − | + |
| 5196-B | CFSAN025137 | O26:H11 | LPZI00000000 | 29 | − | − | − |
| 5019-A | CFSAN025138 | O26:H11 | LPZJ00000000 | 29 | − | − | − |
| 3674-A | CFSAN025132 | O26:H11 | LPZK00000000 | 29 | − | − | − |
All strains were isolated in Nebraska, USA.
Determined by in silico analysis of the WGS assemblies. +, present; −, absent.
Insertion sequence (probably IS2) disrupting fliC gene integrity.
DNA preparation.
Genomic DNA from each strain was isolated from overnight cultures using the DNeasy blood and tissue kit (Qiagen, Valencia, CA), following the manufacturer's instructions. The resultant DNA extract was stored at −20°C until it was used as a template for whole-genome sequencing. The concentration was determined using a Qubit double-stranded DNA HS assay kit and a Qubit 2.0 fluorometer (Thermo Scientific), according to the manufacturer's instructions.
Whole-genome sequencing, contig assembly, and annotation.
The genomes of the strains were sequenced using an Illumina MiSeq sequencer (Illumina, San Diego, CA), with 250-bp paired-end chemistry, according to the manufacturer's instructions, at approximately 80 times the average coverage. The genome libraries were constructed using the Nextera XT DNA sample preparation kit (Illumina). Genomic sequence contigs were de novo assembled using default settings within CLC Genomics Workbench v7.6.1 (Qiagen) with a minimum contig size threshold of 500 bp. The draft genomes were annotated using the NCBI Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP) (http://www.ncbi.nlm.nih.gov/genomes/static/Pipeline.html; 21).
In silico serotyping.
The serotype of each strain analyzed in this study was confirmed using the genes deposited in the Center for Genomic Epidemiology (http://www.genomicepidemiology.org) for E. coli as part of their Web-based serotyping tool (SerotypeFinder 1.1 [https://cge.cbs.dtu.dk/services/SerotypeFinder]) (22, 23). We used Ridom to perform batch screening of the genomes analyzed. Briefly, all the genes were divided into O-type (wzx and wzy) and H-type (fliC) genes in FASTA format (e.g., all the wzx alleles were in a single FASTA file) and used as the task template. For virulence screening, a project was created using all three task templates, and each whole-genome sequence was screened for the presence of each gene type (O-type or H-type gene). The results were similar to those with SerotypeFinder, and as with the virulence genes previously, the data were now in a database and new alleles (if found) could be added to the task templates.
In silico MLST phylogenetic analysis.
The initial analysis and identification of the strains were performed using an in silico E. coli MLST approach, based on the information available at the E. coli MLST website (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli) and using Ridom SeqSphere+ software v2.4.0 (Ridom, Münster, Germany). Seven housekeeping genes (dnaE, gyrB, recA, dtdS, pntA, pyrC, and tnaA), described previously for E. coli (24), were used for MLST analysis. The same E. coli MLST database was also used to assign numbers to alleles and STs.
In silico determination of virulence genes.
Virulence genes were determined using the genes deposited in the Center for Genomic Epidemiology (http://www.genomicepidemiology.org) for E. coli as part of their VirulenceFinder 1.5 Web-based tool (https://cge.cbs.dtu.dk/services/VirulenceFinder; 23), but we used Ridom to perform batch screening of the genomes analyzed. Briefly, all the genes were divided into classes or groups by homology in FASTA format (e.g., all the astA alleles were in a single FASTA file) and used as a task template. Afterward, a project was created using all the task templates, and each WGS was screened for the presence of each gene class (virulence gene). Table S1 in the supplemental material shows the 95 virulence genes analyzed by this method. The stx gene variants analyzed are available at https://cge.cbs.dtu.dk/services/VirulenceFinder. The result was very similar to the one displayed at VirulenceFinder, except that the data were now in a database and new alleles (if found) could be added to the task templates.
In silico antimicrobial resistance gene identification.
Antimicrobial resistance genes present in the sequenced genomes, as well as in those retrieved from GenBank (Table 2), were identified by using the genes deposited in the Center for Genomic Epidemiology (http://www.genomicepidemiology.org) as part of their Resfinder 2.1 Web-based tool (https://cge.cbs.dtu.dk/services/ResFinder) (25), but we used Ridom to perform batch screening of the genomes analyzed. Briefly, all the genes were divided into classes or groups by homology in FASTA format (e.g., all the blaTEM alleles were located in a single FASTA file) and used as the task template. Later, a project was created using all the task templates, and each WGS was screened for the presence of each gene class (antimicrobial resistance gene). The result was very similar to that of ResFinder, except that the data were now in a database and new alleles (if found) could be added to the task templates.
TABLE 2.
E. coli O26:H11 genomes available at GenBank used for phylogenetic and in silico virulence analyses
| Strain | GenBank accession no. | Serotype | Yr isolated | Country | STb | stx1c | stx2c | ehxAc |
|---|---|---|---|---|---|---|---|---|
| 11368a,e | NC_013369 | O26:H11 | 2001 | Japan | 21 | + | − | + |
| 21765e | CDLB00000000 | O26:H11 | 2005 | France | 29 | − | + | − |
| 05-3646e | JHOE01000000 | O26:H11 | 2005 | USA | 21 | + | − | + |
| 2011C-3506e | JHLS01000000 | O26:H11 | 2011 | USA | 21 | + | − | + |
| 2011C-3387e | JHLV00000000 | O26:H11 | 2011 | USA | 21 | + | − | − |
| 2011C-3655e | JHLN00000000 | O26:H11 | 2011 | USA | 21 | + | − | + |
| 06-3464e | JHNO00000000 | O26:H11 | 2006 | USA | 21 | + | − | + |
| 2009C-3612e | JHGZ00000000 | O26:H11 | 2009 | USA | 29 | − | + | + |
| 2009C-4760e | JHGK00000000 | O26:H11 | 2009 | USA | 21 | + | − | + |
| 2010C-4430e | JHND00000000 | O26:H11 | 2010 | USA | 21 | + | − | + |
| 2010C-4819e | JHMP00000000 | O26:H11 | 2010 | USA | 21 | + | + | + |
| 2011C-3282e | JHLX00000000 | O26:H11 | 2011 | USA | 21 | + | − | + |
| 2011C-3270e | JHLY00000000 | O26:H11 | 2011 | USA | 21 | + | − | + |
| 2010EL-1699e | JHMF00000000 | O26:H11 | 2010 | USA | 21 | + | − | + |
| 2010C-5028e | JHMI00000000 | O26:H11 | 2010 | USA | 21 | + | − | + |
| 2010C-4834e | JHMN00000000 | O26:H11 | 2010 | USA | 21 | + | − | + |
| 2010C-4788e | JHMS00000000 | O26:H11 | 2010 | USA | 21 | + | − | + |
| 2010C-4347e | JHFB00000000 | O26:H11 | 2010 | USA | 21 | + | − | + |
| 2010C-4244e | JHFD00000000 | O26:H11 | 2010 | USA | 21 | + | − | + |
| 2010C-3902e | JHFH00000000 | O26:H11 | 2010 | USA | 21 | + | + | + |
| 2010C-3871e | JHFJ00000000 | O26:H11 | 2010 | USA | 21 | + | + | + |
| 2010C-3472e | JHFX00000000 | O26:H11 | 2010 | USA | 21 | + | − | + |
| 2010C-3051e | JHGA00000000 | O26:H11 | 2010 | USA | 21 | + | − | + |
| 2009C-4826e | JHGI00000000 | O26:H11 | 2009 | USA | 21 | + | − | + |
| 2009C-4747e | JHGM00000000 | O26:H1 | 2009 | USA | 21 | + | − | + |
| 2009C-3996e | JHGV00000000 | O26:H11 | 2009 | USA | 21 | + | − | + |
| 2009C-3689e | JHGX00000000 | O26:H11 | 2009 | USA | 29 | − | + | + |
| 03-3500e | JHNT00000000 | O26:H11 | 2001 | USA | 21 | + | − | + |
| CVM10021d | AKAZ00000000 | O26:H11 | 1995 | Unknown | 21 | + | − | − |
| CVM9952f | AKBC00000000 | O26:H11 | 1985 | Unknown | 21 | + | − | − |
| CVM9942d | AJVW00000000 | O26:H11 | 1983 | Unknown | 21 | + | − | − |
| CVM10026d | AJVX00000000 | O26:H11 | 1995 | Unknown | 21 | + | − | + |
| CVM10030d | AKBA00000000 | O26:H11 | 1995 | Unknown | 21 | + | − | + |
| CVM10224e | AKBB00000000 | O26:H11 | 1997 | Unknown | 21 | + | − | − |
| CFSAN001629e | AMXO00000000 | O26:H11 | 1997 | Unknown | 21 | + | − | − |
| 2011C-3274e | JAST00000000 | O26:H11 | 2011 | USA | 21 | + | − | + |
| ATCC BAA-2196e | AYOF00000000 | O26:H11 | 2003 | USA | 21 | + | + | + |
Closed genome. The rest are all draft genomes.
In silico ST.
Determined by in silico analysis. +, present; −, absent.
Cattle origin.
Clinical origin.
Pig origin.
Phylogenetic relationship of strains by wgMLST analysis.
The phylogenetic relationship of the strains was assessed by a wgMLST analysis using Ridom SeqSphere+ v2.4.0 software. The genome of O26:H11 strain 11368 (NC_013361.1) was used as a reference. After eliminating loci that were missing from the genome of any strain used in our analyses, we performed a wgMLST analysis. These remaining loci were considered the core genome shared by the analyzed strains. We used the DNA distance method of Nei et al. (26) to calculate the matrix of genetic distance, taking only the numbers of same/different alleles in the core genes into consideration. A neighbor-joining (NJ) tree using the appropriate genetic distances was built after the wgMLST analysis. The discriminatory index was calculated with the Ridom software using Simpson's discriminatory index as described previously (27). wgMLST uses the allele number of each locus to determine the genetic distance and to build the phylogenetic tree. The use of allele numbers reduces the influence of recombination in the data set studied and allows fast clustering determination of genomes.
Phylogenetic relationship of strains by targeted SNP analysis.
The SNP analysis was conducted in parallel with the wgMLST analysis in order to validate the results obtained by wgMLST and to conduct phylogenetic analysis. After the identification of core genes among the strains, SNPs present in each locus were extracted using Ridom SeqSphere+ software. This core SNP matrix was used for phylogenetic analysis. The maximum-likelihood (ML) phylogeny was reconstructed with Mega v6 software (28), using the Kimura 2-parameter model to estimate genetic distances. The statistical support of the nodes in the ML tree was assessed by 1,000 bootstrap resamplings. The use of SNPs allowed the determination of the true phylogeny and the discovery of informative SNPs in the data set.
Nucleotide sequence accession numbers.
The draft genome sequences of all 42 E. coli O26 strains used in our study are available in GenBank under the accession numbers listed in Table 1.
RESULTS
In silico serotyping, MLST, and virulence gene profiles of cattle O26:H11/H− strains.
The serotypes of cattle O26:H11/H− strains were determined previously by PCR assay for the wzxO26 and fliCH11 genes; all the strains were O26, although several strains possessed an undetermined H antigen (20). In silico serotyping confirmed the lack of integrity of the H11 marker (fliCH11) in the strains with the unknown H antigen (4271-C, 4822-A, and 4848-A-B). This lack of integrity of the fliCH11 gene explained the observed failure to detect the H11 antigen with the fliCH11 PCR employed previously (Table 1). Most cattle O26:H11/H− strains analyzed (n = 40/42) were ST29, while the remaining two (1895-A and 1914-A) were ST21.
The results of the WGS-based virulence typing are shown in Tables 1 and 3. The genomes were screened for 95 known E. coli virulence genes and two arcA allele type genes (see Table S1 in the supplemental material). The 42 strains were negative for 74 of the 95 virulence genes tested and described for E. coli (23) (see Table S1 in the supplemental material). A majority of the strains were positive for the eae-β1, tir, cif, pssA, iss, lpfA, nleA, nleB, prfB, espA, espB, espJ, and espF genes (Table 3). The exceptions were strain 5019-A, which was negative for espF, and strains 5583-H and 4592-A, which were negative for nleA. The stx1 gene was present only in the two ST21 strains. Interestingly, the two ST21 strains also carried the microcin H47 system (mchB, mchC, mchF, and mcmA genes).
TABLE 3.
Virulence genes present in the O26:H11/H− E. coli strains sequenced in this study by in silico analysisa
| Strain | astA | cba | cma | efa1 | espI | espK | espP | f17A | f17G | gad | iha | nleC | tccP | toxB | α-hlyAc | arcA allele 2 | arcA allele 1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1270-A | − | − | − | + | − | − | − | − | − | + | − | + | − | − | + | − | + |
| 1357-A | + | − | − | − | − | − | − | − | − | − | − | + | + | − | + | − | + |
| 1676-A | − | − | − | + | − | − | − | + | + | − | − | − | − | − | + | − | + |
| 1692-A | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + |
| 1740-A-A | − | − | − | + | − | − | − | + | + | − | − | − | − | − | + | − | + |
| 1740-A-B | − | − | − | + | − | − | − | + | + | + | − | − | − | − | + | − | + |
| 1802-A | − | − | − | + | − | − | − | + | + | − | − | − | + | − | + | − | + |
| 1958-A-B | + | − | − | − | − | − | − | − | − | − | − | + | + | − | + | − | + |
| 2105-G | + | − | − | − | − | − | − | + | + | − | − | − | − | − | + | − | + |
| 2139-A | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + |
| 2152-B | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + |
| 2176-A | + | + | + | + | − | − | − | + | + | − | − | − | − | − | + | − | + |
| 2194-B | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + |
| 2223-B | + | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | + |
| 2228-A | + | − | − | − | − | − | − | − | − | − | − | + | − | − | + | − | + |
| 3674-A | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + |
| 4131 | − | − | − | + | − | − | − | − | − | − | − | + | − | − | + | − | + |
| 4170 | + | − | − | − | − | − | − | − | − | − | − | − | + | − | + | − | + |
| 4196 | − | − | − | + | − | − | − | − | − | − | − | + | − | − | + | − | + |
| 4271-C | − | − | + | + | − | − | − | − | − | − | − | − | − | − | + | − | + |
| 4277-H-A | + | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | + |
| 4368-A | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + |
| 4435-B | + | − | − | − | − | − | − | − | − | + | − | − | − | − | + | − | + |
| 4468-A-A | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + |
| 4468-A-B | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + |
| 4513 | − | − | − | − | − | − | − | − | − | + | − | − | − | − | + | − | + |
| 4592-A | − | − | − | + | − | − | − | − | − | − | − | + | − | − | + | − | + |
| 4730-C | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + |
| 4822-A | − | − | + | + | − | − | − | − | − | + | − | − | − | − | + | − | + |
| 4848-A-B | − | − | + | − | − | − | − | − | − | − | − | − | − | − | + | − | + |
| 4860-A | + | − | − | − | − | − | − | − | − | + | + | − | − | − | − | − | + |
| 5019-A | + | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | + |
| 5196-B | + | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | + |
| 5206-E-B | + | − | − | − | − | − | − | + | + | − | − | − | − | − | + | − | + |
| 5583-H | − | − | − | + | − | − | − | − | − | − | − | + | − | − | + | − | + |
| 5687-B | + | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + |
| 1341-A | − | − | − | − | + | + | + | − | − | − | + | + | − | + | − | + | − |
| 1668-A-A | − | − | − | − | + | + | + | − | − | − | + | + | − | + | − | + | − |
| 1668-A-B | − | − | − | − | + | + | + | − | − | − | + | + | − | + | − | + | − |
| 1895-Ab | + | + | + | + | − | + | + | − | − | − | − | + | − | + | − | + | − |
| 1914-Ab | + | + | + | + | − | + | + | − | − | − | − | + | − | + | − | + | − |
| 4863-A | − | − | − | − | + | + | + | − | − | + | + | + | − | + | − | + | − |
+, present; −, absent. The functions of the genes can be found in Table S1 in the supplemental material. All the samples carried the following virulence genes: cif, eae-β1, pssA, tir, espA, espB, espF (except 5019-A), nleA (5583-H and 4592-A), nleB, prfB, espJ, iss, and lpfA. The exhA gene content of each strain is shown in Table 1.
1895-A and 1914-A were the only ST21 strains among the O26:H11 isolates from cattle. These were positive for Shiga toxin gene type 1 (stx type 1, a 100% match to GenBank accession number AF461168) and carried plasmid genes mchB, mchC, mchF, and mcmA.
Alpha-hemolysin-pEO5 (α-hlyA; NG_036728).
According to the classification proposed by Bugarel et al. (6), most O26 cattle strains (n = 36/42) analyzed in this study were classified as EPEC (eae-β+, alpha-hemolysin gene positive, and arcA allele type 1 gene). The remaining six cattle strains were classified as EHEC (1895-A and 1914-A) (stx+, eae-β+, espK+, and arcA allele type 2 gene), and EHEC-like (1341-A, 1668-A-A, 1668-A-B, and 4863-A) (eae-β+, espK+, and arcA allele type 2 gene). These six strains also carried the ehxA and toxB genes (two ST21 and four ST29 strains), and hence, also carried an EHEC virulence plasmid.
In silico serotyping, MLST, and virulence of O26 E. coli genomes available in GenBank.
Thirty-seven O26:H11 STEC genomes (32 clinical, 4 from cattle, and 1 from pig) were downloaded from GenBank, and in silico MLST showed that the majority (92%) were ST21 (Table 2). All the strains were confirmed as O26:H11 by in silico serotyping. The results of the WGS-based virulence typing are shown in Table 4. Thirty-one of the 37 strains were positive for stx1 variant a (stx1a) only and 3 were positive for both stx1a and stx2a, while the remaining 3 were positive only for stx2a (Table 4). The strains positive only for stx2a were ST29. Most strains were positive for the presence of cif, eae-β1, pssA (except CVM9952), tir (except 2009C-4826), espA, espB, espF (except 2009C-4826 and 2009C-3689), nleA (except CVM10021, CVM9952, CVM9942, 2009C-3612, 2010C-4819, and 2010C-4788), nleB, prfB, espJ (except 2010EL-1699, 2009C-4826, and 2009C-4747), iss (except CVM9942), and lpfA. All the strains were considered EHEC (stx+, eae-β+, espK+, and arcA allele type 2 gene), except for strain 21765, which was an EPEC carrying a stx2 gene (eae-β+, alpha-hemolysin gene positive, and arcA allele type 1 gene). The virulence genes found in plasmids showed high diversity among the strains, with most carrying ehxA (n = 30), while only 27 also carried toxB, indicating the presence of different virulence plasmids. As observed for cattle O26 strains, three of the GenBank genomes (CVM10021, 2011C-3506, and 2010C-3871) showed the presence of the microcin H47 system (mchB, mchC, mchF, and mcmA genes).
TABLE 4.
Virulence genes present in the O26:H11/H− E. coli strains available at GenBank by in silico analysisa
| Strain | astA | cba | celb | cma | efa1 | espI | espK | espP | gad | iha | nleC | tccP | toxB | katP | arcA allele 2 | arcA allele 1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CVM9952 | − | − | − | − | + | − | + | − | − | − | − | − | − | − | + | − |
| CVM10030 | − | − | − | − | + | − | + | − | − | − | + | − | − | − | + | − |
| CFSAN001629 | − | + | − | + | + | − | + | − | − | − | + | − | − | − | + | − |
| CVM9942 | − | − | + | − | + | − | + | − | − | − | + | − | − | − | + | − |
| 2011C-3387 | − | − | − | − | + | − | + | − | + | − | − | − | − | + | + | − |
| 05-3646 | − | − | − | − | + | − | − | − | − | − | + | − | − | + | + | − |
| CVM10026 | − | − | + | − | − | − | + | − | − | − | + | − | − | − | + | − |
| CVM10224 | − | − | − | − | + | − | + | − | − | − | + | − | − | − | + | − |
| CVM10021b | + | − | + | − | + | − | − | − | − | − | − | − | − | + | + | − |
| 21765c | − | − | − | − | + | − | − | − | + | − | − | + | − | − | − | + |
| 2010C-3051 | − | − | − | − | + | − | + | + | − | − | + | − | + | + | + | − |
| 2009C-3689 | − | − | − | − | − | + | + | + | − | + | + | − | + | − | + | − |
| ATCC BAA-2196 | − | − | − | − | + | − | − | + | − | − | + | − | + | + | + | − |
| 2010C-4834 | − | − | − | − | + | − | + | + | − | − | + | − | + | + | + | − |
| 2010C-5028 | − | − | − | − | + | − | + | + | − | − | + | − | + | + | + | − |
| 2010C-4430 | − | − | − | − | + | − | + | + | − | − | + | − | + | + | + | − |
| 11368 | − | − | − | − | + | − | + | − | − | − | + | + | + | + | + | − |
| 2011C-3270 | − | − | − | − | + | − | + | + | − | − | + | − | + | + | + | − |
| 2010C-4788 | − | − | − | − | + | − | + | + | − | − | + | − | + | + | + | − |
| 2011C-3282 | − | − | − | − | + | − | + | + | − | − | + | − | + | + | + | − |
| 2011C-3274 | − | + | − | + | + | − | + | + | − | − | + | + | + | + | + | − |
| 2011C-3655 | − | − | − | − | + | − | + | + | − | − | + | − | + | + | + | − |
| 2011C-3506c | − | − | − | − | + | − | + | + | + | + | + | + | + | + | + | − |
| 03-3500 | − | − | + | − | + | − | + | + | − | − | + | − | + | + | + | − |
| 2010C-3902 | − | − | + | − | + | − | + | + | − | − | + | − | + | + | + | − |
| 2010C-4819 | − | − | − | − | + | − | + | + | − | − | + | − | + | + | + | − |
| 2010EL-1699 | − | − | − | − | + | − | + | + | − | − | + | − | + | + | + | − |
| 2010C-4347 | − | − | − | − | + | − | + | + | − | − | + | − | + | + | + | − |
| 2009C-4760 | − | − | − | − | + | − | + | + | − | − | + | − | + | + | + | − |
| 2009C-4747 | − | − | − | − | + | − | + | + | − | − | + | − | + | + | + | − |
| 2010C-4244 | − | − | − | − | + | − | + | + | − | − | + | − | + | + | + | − |
| 2010C-3472 | − | − | − | − | + | − | + | + | − | − | + | − | + | + | + | − |
| 2009C-4826 | − | − | + | − | + | − | + | + | − | + | + | − | + | + | + | − |
| 06-3464 | − | − | − | − | + | − | + | + | − | − | + | − | + | + | + | − |
| 2009C-3996 | − | − | − | − | + | − | + | + | − | − | + | − | + | + | + | − |
| 2010C-3871b | + | − | − | − | + | − | − | + | − | − | + | − | + | + | + | − |
| 2009C-3612 | − | − | − | − | − | + | + | + | − | + | + | − | + | − | + | − |
+, present; −, absent. A majority of the strains carried the following virulence genes: cif, eae-β1, pssA (except CVM9952), tir (except 2009C-4826), espA, espB, espF (except 2009C-4826 and 2009C-3689), nleA (except CVM10021, CVM9952, CVM9942, 2009C-3612, 2010C-4819, and 2010C-4788), nleB, prfB, espJ (except 2010EL-1699, 2009C-4826, and 2009C-4747), iss (except CVM9942), and lpfA. The Shiga toxin and exhA gene contents of each strain are shown in Table 2.
Isolate positive for alpha-hemolysin–pEO5 (α-hlyA; NG_036728).
Isolate positive for mchB, mchC, mchF, and mcmA.
Presence of antimicrobial resistance genes.
Twenty-nine of the 42 cattle strains (70%) carried at least one antibiotic resistance gene, while only 6 of the 37 genomes available at GenBank carried at least one antibiotic resistance gene (16%) (Table 5). Of the cattle strains positive for antibiotic resistance, 16 carried only a tetracycline resistance gene (tetC), while the remaining strains carried multiple antibiotic resistance genes (aminoglycoside and tetracycline; beta-lactamase and tetracycline; aminoglycoside and tetracycline; or aminoglycoside, phenicol, and sulfonamide) (Table 5). Among the six genomes from GenBank positive for antibiotic resistance, three carried only a single resistance gene (tetA, tetB, or blaTEM-1B), while the remaining three carried multiple antibiotic resistance genes.
TABLE 5.
Presence of antimicrobial resistance genes identified by in silico analysis in the genomes of the strains analyzed in this studya
| Strainb | aadAc | aph3c | aph6c | strAc | strBc | blaTEMd | floRe | sul2f | tetAg | tetBg | tetCg | tetMg |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1357-A | − | − | − | − | − | − | − | − | − | − | + | − |
| 1676-A | − | − | − | − | − | − | − | − | − | − | + | − |
| 1692-A | − | + | + | − | − | − | − | − | − | + | − | − |
| 1740-A-A | − | − | − | − | − | − | − | − | − | − | + | − |
| 1740-A-B | − | − | − | − | − | − | − | − | − | − | + | − |
| 1802-A | − | − | − | − | − | − | − | − | − | − | + | − |
| 1895-A | − | − | − | + | + | − | − | − | + | − | − | − |
| 1914-A | − | − | − | + | + | − | − | − | + | − | − | − |
| 1958-A-B | − | − | − | − | − | − | − | − | − | − | + | − |
| 2139-A | − | − | − | + | + | − | − | − | − | − | − | − |
| 2152-B | − | − | − | + | + | − | + | + | − | − | − | − |
| 2194-B | − | − | − | + | + | − | − | + | − | + | − | − |
| 2228-A | − | − | − | − | − | − | − | − | − | − | + | − |
| 3674-A | − | − | − | + | + | − | + | + | − | − | − | − |
| 4170 | − | − | − | − | − | − | − | − | − | − | + | − |
| 4196 | − | − | − | − | − | − | − | − | − | − | + | − |
| 4271-C | − | − | − | − | − | + | − | − | + | − | − | + |
| 4277-H-A | − | − | − | − | − | − | − | − | − | − | + | − |
| 4368-A | − | − | − | + | + | − | + | + | − | − | − | − |
| 4435-B | − | − | − | − | − | − | − | − | − | − | + | − |
| 4592-A | − | − | − | − | − | − | − | − | − | − | + | − |
| 4730-C | − | − | − | + | + | − | − | + | − | + | − | − |
| 4822-A | − | − | − | − | − | + | − | − | − | − | − | + |
| 4848-A-B | − | − | − | − | − | + | − | − | + | − | − | + |
| 4860-A | − | − | − | − | − | − | − | − | − | − | + | − |
| 5019-A | − | − | − | − | − | − | − | − | − | − | + | − |
| 5196-B | − | − | − | − | − | − | − | − | − | − | + | − |
| 5583-H | − | − | − | + | + | − | − | − | − | + | − | − |
| 5687-B | − | − | − | − | − | − | − | − | − | − | + | − |
| ATCC BAA-2196 | − | − | − | + | + | − | − | + | − | − | − | − |
| CVM10026 | − | − | − | + | + | + | − | + | − | + | − | − |
| CVM9942 | + | − | − | − | − | − | − | − | − | + | − | − |
| CVM9952 | − | − | − | − | − | − | − | − | − | + | − | − |
| 2010C-4788 | − | − | − | − | − | + | − | − | − | − | − | − |
| 2011C-3274 | − | − | − | − | − | − | − | − | + | − | − | − |
+, present; −, absent.
The strains sequenced in this study are shown in boldface. Only strains that carried antimicrobial resistance genes are shown.
Aminoglycoside resistance.
Beta-lactamase resistance.
Phenicol resistance.
Sulfonamide resistance.
Tetracycline resistance.
Phylogenetic relationship of strains by wgMLST analysis.
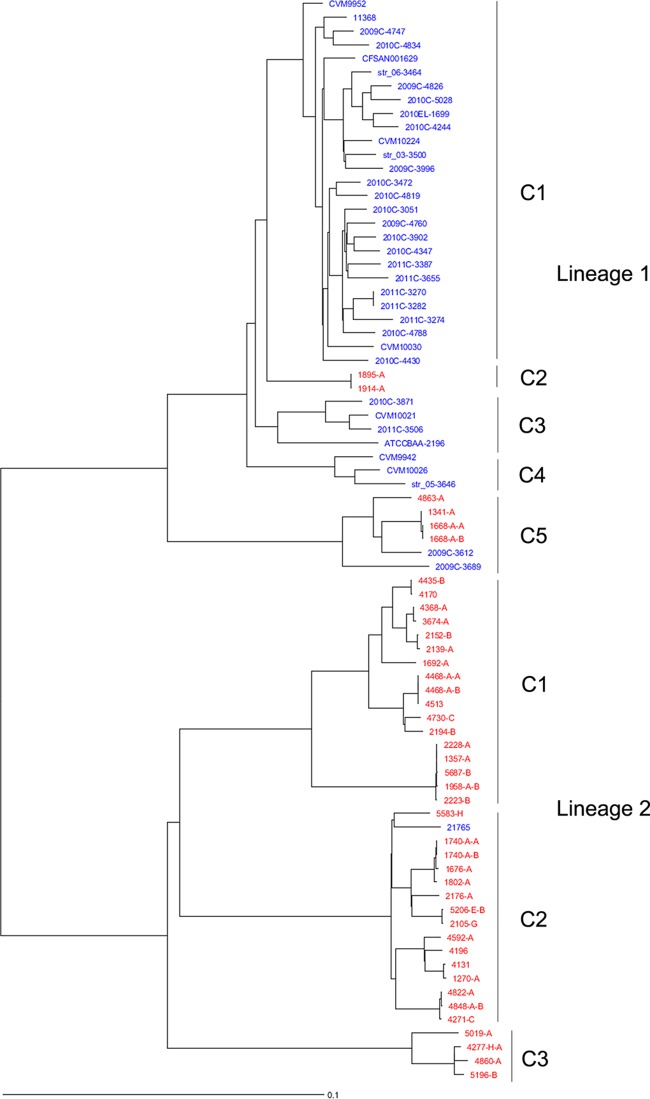
The phylogenetic relationships among the E. coli O26:H11/H− strains sequenced in this study and the genomes available at GenBank as determined by wgMLST analysis are shown in Fig. 1. The genome of strain 11368 (NC_013361.1) was used as a reference. This E. coli strain has 4,554 genes, 2,965 of which (core genes) were present in all the analyzed strains, with 1,048 genes identical among all the strains (see Table S2 in the supplemental material). The resultant NJ tree showed that the O26 genomes analyzed were highly diverse and polyphyletic. The genomes were divided into two main lineages (1 and 2), each with multiple clades. This wgMLST analysis using the core genes was highly discriminatory, with a discriminatory index of 0.997 (72 types/79 samples).
FIG 1.
Phylogenetic analysis of the O26:H11/H− E. coli strains sequenced in this study and the genomes available at GenBank by wgMLST. Ridom SeqSphere+ identified 2,965 core genes. The evolutionary history was inferred by using an NJ tree built using the genetic distance and showing the existence of two clear lineages within O26 strains, with lineage 1 composed of EHEC and EHEC-like O26:H11/H− strains, while lineage 2 was composed of EPEC O26:H11/H strains. The E. coli strains sequenced in this study are in red, and strains from GenBank are in blue. C1 to C5, clades.
Lineage 1 was defined as the EHEC O26 lineage that comprised O26:H11 EHEC-like (ST29; stx negative) and O26:H11 EHEC (ST21) cattle strains, as well as most clinical O26:H11 E. coli GenBank genomes. Lineage 1 was composed of five different clades, with cattle O26:H11 strains belonging to clades C2 (ST21) and C5 (ST29). The ST29 EHEC-like cattle strains (lineage 1) clustered with the two O26:H11 stx+ strains isolated from clinical samples in 2009, showing nearly identical virulence profiles except for the stx gene, indicating the pathogenic potential of these strains. The other ST21 lineage 1 cattle strains clustered separately as a single clade and can be considered potentially pathogenic, as well, because they carried all the virulence genes known in an O26:H11 EHEC strain (6).
Lineage 2 was composed of EPEC O26 cattle strains (ST29) (Fig. 1) and was divided into three diverse clades, with different virulence gene contents (Table 3). Interestingly, strain 21765, which was isolated from humans during the first raw milk cheese outbreak described in France in 2005 (5), belonged to lineage 2 (EPEC) but was described in the original genome publication as an EHEC strain. Most O26 EPEC strains from U.S. cattle (4/5) were negative for the alpha-hemolysin gene (indicative of the presence of the virulence plasmid), clustered in clade C3 of lineage 2, and might be considered less pathogenic than the EPEC strains from the other two clusters due to the absence of the virulence plasmid and other virulence genes present in the other two clusters (Fig. 1).
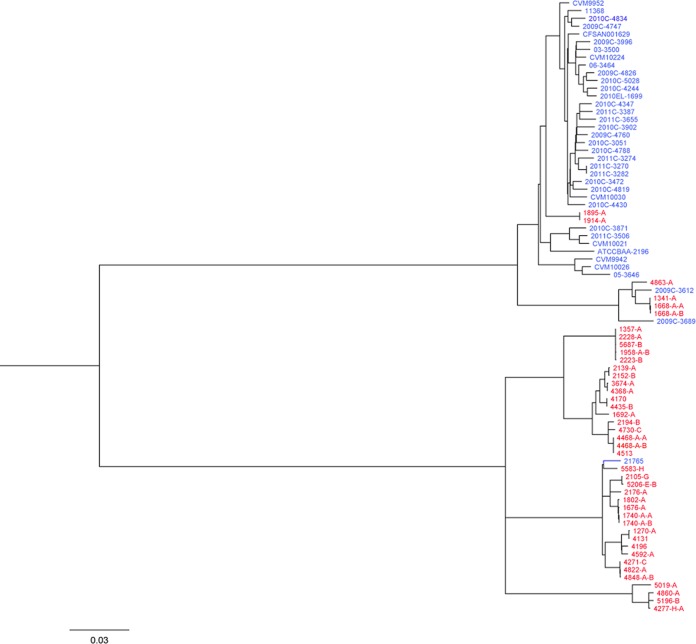
Phylogenetic relationship of strains by targeted SNP analysis.
To evaluate the results obtained by wgMLST and to determine the actual evolutionary phylogenetic relationships among the E. coli O26:H11/H− strains sequenced in this study and the genomes available at GenBank, a targeted SNP analysis was performed (Fig. 2). The SNP matrix consisted of 5,050 SNP positions in 2,965 genes (identified previously by wgMLST) (see Table S3 in the supplemental material). A maximum-likelihood tree was generated, which agreed overall with the NJ tree generated with the wgMLST data with strong bootstrap support on each node (Fig. 2). Consistent with the wgMLST NJ tree, two main lineages with individual clades were observed in the SNP ML tree. This targeted SNP analysis demonstrated the robustness of the wgMLST analysis for determining lineages and clades within E. coli O26:H11/H−.
FIG 2.
Molecular phylogenetic analysis of the O26:H11/H− E. coli strains sequenced in this study and the genomes available at GenBank by the maximum-likelihood method. The evolutionary history was inferred by using the maximum-likelihood method based on the Kimura 2-parameter model (55). The tree is drawn to scale, with branch lengths measured in substitutions per site. All positions containing gaps and missing data were eliminated. There were a total of 4,916 positions in the final data set. The bootstrap values are not shown for visualization purposes. The lineages and clades are the same as in Fig. 1. The E. coli strains sequenced in this study are in red, and strains from GenBank are in blue.
DISCUSSION
E. coli O26:H11/H− is the second most frequent EHEC serotype, causing diarrhea and HUS in the United States (3, 29) and worldwide (4, 6, 9, 30). In recent years there has been an increase in the occurrence of this serotype and other non-O157 serotypes in humans associated with consumption of contaminated food, mainly beef and dairy products. A new O26:H11/H− clone (ST29; stxa+, eae+, plasmid gene profile ehxA+, and etpD+) has been recently reported to be distributed all over Europe (30) and has been found in cattle (31). Few studies have been conducted on O26:H11/H− in the United States (17, 18), and none has investigated if this new emerging clone exists in the United States. Our preliminary analysis of O26:H11/H− strains from the United States showed the presence of different virulence plasmids and suggested that these strains could be divided further into different lineages (unpublished results). In the present study, we performed an in-depth analysis of 42 O26:H11/H− strains isolated from cattle in 2011 using whole-genome sequence phylogenetic analysis by wgMLST and targeted SNPs. We compared them to the genomes of other O26:H11 strains available at GenBank (37 genomes).
We showed that these 42 O26:H11/H− strains were highly diverse, as previously described (17), with most of them belonging to ST29 (n = 40), either EHEC-like (n = 4) or EPEC (n = 36). The remaining two strains were ST21 and formed an independent, unique clade within lineage 1. In contrast to the cattle strains, the majority of the O26 genomes retrieved from GenBank were ST21 (92%). By wgMLST and targeted SNP phylogeny, we were able to distinguish two clearly distinct lineages among the E. coli O26 strains analyzed, with EHEC-like strains clustering together and differing by 76 (1668-A-A, 1668-A-B, and 1341-A) and 160 (4863-A) SNPs (out of 5,050 SNPs) from two clinical EHEC strains, 2009C-3612 and 2009C-3689, respectively. Therefore, EHEC-like O26:H11 (stx-negative) strains seem to be EHEC strains that have lost or could acquire the Shiga toxin prophage, confirming a previously suggested hypothesis (6, 18). A search for possible known insertion sites for stx2 phages (wrbA, sbcB, yehV, yecE, and Z2577) (32) found that all the sites were intact in the four stx-negative ST29 EHEC strains and that the two ST21 O26 E. coli cattle strains have wrbA and yehV sites occupied by a phage. These phylogenetic analyses confirmed that hypothesis and that these stx-negative O26:H11/H− “EHEC-like” strains are not EPEC but can be grouped in O26 lineage 1 and therefore can be called EHEC O26. These EHEC O26 (stx-negative) strains can be distinguished from pathogenic EPEC O26 by more than 689 alleles and more than 1,900 SNPs, plus the presence of the EHEC virulence plasmid. Furthermore, within the two lineages, at least five and three different clades could be distinguished, respectively, showing the high genomic diversity among the genomes analyzed.
One interesting finding of this study was that the French O26:H11 strain 21765, isolated in 2005 during a milk cheese outbreak (5), belonged to one of the three clades within O26 lineage 2 (EPEC) and not to the EHEC lineage, as originally described (5). This strain carried all the virulence genes described in O26 EPECs but differed from the other members of that clade by also carrying a stx2a phage. The strain was isolated prior to the E. coli O104:H4 strain that caused a large outbreak in Germany in 2011 (33). The O104:H4 strain was characterized as a new pathotype or atypical E. coli, being phylogenetically close to enteroaggregative E. coli (EAEC) and carrying virulence genes often observed in those strains, but carrying a stx2a phage, as well (34, 35). These O104:H4 strains caused a large number of disease cases, with 22.3% of the patients developing HUS (36). Several authors called for a new pathotype definition for this strain as enteroaggregative-hemorrhagic E. coli (EAHEC) (37), with numerous researchers attempting to explain its origin (38). Our phylogenetic analysis showed that other E. coli strains from a different pathogenic background (EPEC) can also acquire a stx phage and cause disease in humans, which may be a more common trait in E. coli strains than previously thought. Our research clearly showed that WGS reveals more about strains that were previously incorrectly or not entirely characterized. For example, by wgMLST analysis, we were also able to show that some O26:H11 strains isolated at the Gastrointestinal Bacteria Reference Unit in the United Kingdom from 2009 to 2013, whose genomes were reported (9) and recently available at GenBank (only Sequence Read Archive [SRA]), were in fact EPEC strains: 670/13, 2290-502/12, and 680/13 (results not shown). These three strains were stx negative and clustered separately from the remaining EHEC O26 strains on the phylogenetic tree (9). However, in that publication, the actual nature of these strains (EHEC, EHEC-like, or EPEC) was not stated.
Some of the U.S. cattle strains (n = 3) were untypeable for their H antigen, resulting in O26:H− serotypes using conventional serotyping techniques. After in silico serotyping analysis, we found that the H antigen gene was fragmented; further analysis of the fliC genes of these three strains showed that the gene was disrupted by an undetermined insertion sequence (IS). However, analysis of the short sequences at either end of the disruption showed that the IS present might be IS2. The lack of H antigen production was described previously for O157:H− German strains (39), though in that case, there was a deletion of 12 bp in the fliC H7 gene. The evolution of pathogens is largely influenced by IS elements, which have contributed to the diversification of EHEC strains, specifically O157 (40–43). Recently, an IS excision enhancer (IEE) element was discovered in EHEC O157 promoting the excision and deletion of IS3 family members, resulting in genomic rearrangements and strain diversification (44, 45). In the strains analyzed, the IEE element (IEE cluster I) was present solely in O26 lineage 1, suggesting that these strains have a higher potential to increase their diversity and virulence than those of lineage 2 (46). IEE cluster I has been found in different pathogenic strains from diverse serotypes (O157:H7, O26:H11, O111:H11, O118:NM, O145:H28, and O139:NM/H38) that are frequently involved in serious outbreaks (2, 47).
Analysis of known E. coli virulence genes (see Table S1 in the supplemental material) in O26 strains sequenced in this study and GenBank O26 strains showed they were very similar, with some exceptions. The main difference was in the presence of stx, efa1, espK, katP, espP, alpha-hemolysin (α-hlyA), and ehxA genes (Tables 3 and 4). The lack of observation of some of these genes may be due to the technique implemented to generate the genomes (e.g., 454, which generates insertions/deletions in homopolymer tracks), which did not allow optimal assembly. The stx genes were found exclusively in strains belonging to O26 lineage 1 (the EHEC lineage), regardless of their origins, except for strain 21765 (stx+, O26 lineage 2 [EPEC], clade C2 [this study]), a clinical strain isolated during the French outbreak (5). The efa1 gene, encoding an E. coli factor for adherence, was present in most EHEC lineage 1 strains, except ST29 strains. Some of the virulence genes carried by plasmids in EHEC strains are espK, katP, espP, ehxA, etp, and toxB. espK was present in most O26 EHEC lineage 1 strains (90%), except four strains, while the O26 lineage 2 strains were all negative for espK. Strain 21765 was negative for espK, consistent with its being an EPEC O26 strain. Therefore, the espK gene is still a good marker for differentiating O26 EHEC strains from O26 EPEC strains, as proposed previously (6). The katP (catalase/peroxidase) gene was not present in any of the U.S. cattle strains analyzed and in only one of the GenBank cattle genomes (CVM10021), while it was present in 27 of the remaining 36 O26 E. coli GenBank genomes. The katP gene is a plasmid-borne gene and protects E. coli against peroxide damage (48). The gene is absent in the new ST29 clone that recently emerged in Europe (30, 49), showing that the majority of the clinical U.S. strains for which genomes are available in GenBank do not belong to this newly described clone. The espP (serine protease) gene was absent in all cattle strains from O26 lineage 2 (EPEC) and present in the majority (76%) of O26 lineage 1 (EHEC) strains. espP is one of the virulence genes located on the plasmid that participate actively in the colonization of the gut by EHEC strains by downregulating complement (50). The other two important virulence plasmid genes, ehxA and toxB, were present only in O26 lineage 1 (EHEC) strains but were not found in 6 and 9 of them, respectively. This indicates diverse plasmid contents in these strains. toxB was missing in the new European O26:H11/H− clone (30, 49). The etp genes were missing in all EHEC O26 genomes analyzed and have been reported to be present in the newly emerged O26:H11/H− clone in Europe (30). On the other hand, the α-hlyA gene, another plasmid-borne virulence gene described for EPEC strains, was mostly present in O26 lineage 2 (EPEC), except in five strains, with four of them forming an independent clade within lineage 2 and appearing to lack a virulence plasmid; therefore, they may be considered less pathogenic.
The prevalence of antimicrobial resistance genes in the U.S. cattle O26 strains was higher (70%) than in the clinical O26 E. coli GenBank genomes (16%). Of the EHEC strains from U.S. cattle, only ST21 (stx1+) carried antibiotic resistance genes. Some cattle strains carried multiple antimicrobial resistance genes (31%), while only 9% of the clinical GenBank strains carried more than one antibiotic resistance gene. Resistance to antibiotics has been described in other O26 strains from cattle (Scotland; 2004), although the percent resistance to one or several antibiotics was below 2% (51). The mechanism for resistance to multiple antibiotics in O26:H11 strains is not new, and antibiotic resistance has been described previously in a stx+ O26:H11 strain (O6877) isolated from a clinical case in Australia as part of an integron element (52). The integron was located in the 111-kb virulence plasmid (pO26-CRL) (52). The plasmid is similar to another virulence plasmid in O26:H11 strain H30 (165 kb) (53) based on its virulence gene content (espP, exhA, katP, and toxB). These differences in virulence plasmid contents between the two EHEC O26 strains reaffirm the high diversity inside EHEC lineage O26:H11. A BLAST search for integron class 1, 2, and 3 integrase genes (intI1, intI2, and intI3) showed that they were absent among the cattle strains analyzed, and only one strain was positive for the intI1 gene (CVM9942, a strain isolated from cattle that carried aad and tetB genes) among the GenBank strains. Interestingly, some of the O26 genomes analyzed carried the microcin H47 system (mchB, mchC, mchF, and mcmA genes). This system has been reported to produce microcin H47, which acts as a bactericidal antibiotic and is produced by naturally occurring E. coli (54). The presence of this system in some of the O26 strains analyzed highlights the plasticity of O26 genomes in the environment.
Overall, the results presented here showed that cattle in the United States can be a reservoir of two different O26:H11 phylogenetic lineages: O26 lineage 1—EHEC O26:H11/H− (either stx+ or stx negative, but with the potential to acquire a stx prophage)—and O26 lineage 2—EPEC O26:H11/H−. The wgMLST analysis allowed the differentiation of O26 strains into two different lineages, with O26 EHEC-like strains grouping with EHEC and therefore being EHEC strains that have either lost or not gained the stx prophage. The wgMLST also allowed us to correctly identify strains that were misidentified as EHEC (strain 21765) or not categorized at all (strains 670/13, 2290-502/12, and 680/13). We have also provided a description of their virulence and antimicrobial resistance gene profiles. The strains from these two observed lineages are potentially pathogenic and produce different outcomes of the illnesses (EPEC produces watery diarrhea, while EHEC produces bloody diarrhea), with lineage 1 infection potentially progressing to HUS. Most U.S. cattle strains also carried multiple antibiotic resistance genes. Finally, this study showed that most O26:H11 clinical strains from the United States (2009 to 2011) belonged to an O26:H11 EHEC clone (ST21 or ST29; stx1+ ehxA+ toxB+) different from the newly described O26:H11 EHEC European clone (ST29; stx1+ and/or stx2+ ehxA+ etpD+).
Supplementary Material
ACKNOWLEDGMENTS
We thank George Kastanis and Elizabeth Reed for their helpful comments on the manuscript.
The findings and conclusions in this report are ours and do not necessarily represent the official position of the Food and Drug Administration.
Funding Statement
This work was supported by the FDA Food Science and Research Intramural Program and was based, in part, upon previous work supported by Agriculture and Food Research Initiative competitive grant 2012-68003-30155 from the USDA National Institute of Food and Agriculture.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00498-16.
REFERENCES
- 1.Rangel JM, Sparling PH, Crowe C, Griffin PM, Swerdlow DL. 2005. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982-2002. Emerg Infect Dis 11:603–609. doi: 10.3201/eid1104.040739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brooks JT, Sowers EG, Wells JG, Greene KD, Griffin PM, Hoekstra RM, Strockbine NA. 2005. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983-2002. J Infect Dis 192:1422–1429. doi: 10.1086/466536. [DOI] [PubMed] [Google Scholar]
- 3.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chase-Topping ME, Rosser T, Allison LJ, Courcier E, Evans J, McKendrick IJ, Pearce MC, Handel I, Caprioli A, Karch H, Hanson MF, Pollock KG, Locking ME, Woolhouse ME, Matthews L, Low JC, Gally DL. 2012. Pathogenic potential to humans of bovine Escherichia coli O26, Scotland. Emerg Infect Dis 18:439–448. doi: 10.3201/eid1803.111236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galia W, Mariani-Kurkdjian P, Loukiadis E, Blanquet-Diot S, Leriche F, Brugere H, Shima A, Oswald E, Cournoyer B, Thevenot-Sergentet D. 2015. Genome sequence and annotation of a human infection isolate of Escherichia coli O26:H11 involved in a raw milk cheese outbreak. Genome Announc 3:e01568-14. doi: 10.1128/genomeA.01568-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bugarel M, Beutin L, Scheutz F, Loukiadis E, Fach P. 2011. Identification of genetic markers for differentiation of Shiga toxin-producing, enteropathogenic, and avirulent strains of Escherichia coli O26. Appl Environ Microbiol 77:2275–2281. doi: 10.1128/AEM.02832-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenkins C, Evans J, Chart H, Willshaw GA, Frankel G. 2008. Escherichia coli serogroup O26—a new look at an old adversary. J Appl Microbiol 104:14–25. doi: 10.1111/j.1365-2672.2007.03465.x. [DOI] [PubMed] [Google Scholar]
- 8.Ethelberg S, Smith B, Torpdahl M, Lisby M, Boel J, Jensen T, Nielsen EM, Molbak K. 2009. Outbreak of non-O157 Shiga toxin-producing Escherichia coli infection from consumption of beef sausage. Clin Infect Dis 48:e78–e81. doi: 10.1086/597502. [DOI] [PubMed] [Google Scholar]
- 9.Dallman TJ, Byrne L, Launders N, Glen K, Grant KA, Jenkins C. 2015. The utility and public health implications of PCR and whole genome sequencing for the detection and investigation of an outbreak of Shiga toxin-producing Escherichia coli serogroup O26:H11. Epidemiol Infect 143:1672–1680. doi: 10.1017/S0950268814002696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.U.S. Department of Agriculture, Food Safety and Inspection Service. 2012. Shiga toxin-producing Escherichia coli in certain raw beef products. Fed Regist 77:31975–31981. [Google Scholar]
- 11.Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin Microbiol Rev 11:142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leomil L, Pestana de Castro AF, Krause G, Schmidt H, Beutin L. 2005. Characterization of two major groups of diarrheagenic Escherichia coli O26 strains which are globally spread in human patients and domestic animals of different species. FEMS Microbiol Lett 249:335–342. doi: 10.1016/j.femsle.2005.06.030. [DOI] [PubMed] [Google Scholar]
- 13.Miko A, Lindstedt BA, Brandal LT, Lobersli I, Beutin L. 2010. Evaluation of multiple-locus variable number of tandem-repeats analysis (MLVA) as a method for identification of clonal groups among enteropathogenic, enterohaemorrhagic and avirulent Escherichia coli O26 strains. FEMS Microbiol Lett 303:137–146. doi: 10.1111/j.1574-6968.2009.01874.x. [DOI] [PubMed] [Google Scholar]
- 14.Bielaszewska M, Middendorf B, Kock R, Friedrich AW, Fruth A, Karch H, Schmidt MA, Mellmann A. 2008. Shiga toxin-negative attaching and effacing Escherichia coli: distinct clinical associations with bacterial phylogeny and virulence traits and inferred in-host pathogen evolution. Clin Infect Dis 47:208–217. doi: 10.1086/589245. [DOI] [PubMed] [Google Scholar]
- 15.Bielaszewska M, Prager R, Kock R, Mellmann A, Zhang W, Tschape H, Tarr PI, Karch H. 2007. Shiga toxin gene loss and transfer in vitro and in vivo during enterohemorrhagic Escherichia coli O26 infection in humans. Appl Environ Microbiol 73:3144–3150. doi: 10.1128/AEM.02937-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bletz S, Bielaszewska M, Leopold SR, Kock R, Witten A, Schuldes J, Zhang W, Karch H, Mellmann A. 2013. Evolution of enterohemorrhagic Escherichia coli O26 based on single-nucleotide polymorphisms. Genome Biol Evol 5:1807–1816. doi: 10.1093/gbe/evt136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ison SA, Delannoy S, Bugarel M, Nightingale KK, Webb HE, Renter DG, Nagaraja TG, Loneragan GH, Fach P. 2015. Genetic diversity and pathogenic potential of attaching and effacing Escherichia coli O26:H11 strains recovered from bovine feces in the United States. Appl Environ Microbiol 81:3671–3678. doi: 10.1128/AEM.00397-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ison SA, Delannoy S, Bugarel M, Nagaraja TG, Renter DG, den Bakker HC, Nightingale KK, Fach P, Loneragan GH. 2015. Targeted amplicon sequencing for SNP genotyping of attaching and effacing Escherichia coli O26:H11 cattle strains using a high-throughput library preparation technique. Appl Environ Microbiol 82:640–649. doi: 10.1128/AEM.03182-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cull CA, Paddock ZD, Nagaraja TG, Bello NM, Babcock AH, Renter DG. 2012. Efficacy of a vaccine and a direct-fed microbial against fecal shedding of Escherichia coli O157:H7 in a randomized pen-level field trial of commercial feedlot cattle. Vaccine 30:6210–6215. doi: 10.1016/j.vaccine.2012.05.080. [DOI] [PubMed] [Google Scholar]
- 20.Paddock ZD, Renter DG, Cull CA, Shi X, Bai J, Nagaraja TG. 2014. Escherichia coli O26 in feedlot cattle: fecal prevalence, isolation, characterization, and effects of an E. coli O157 vaccine and a direct-fed microbial. Foodborne Pathog Dis 11:186–193. doi: 10.1089/fpd.2013.1659. [DOI] [PubMed] [Google Scholar]
- 21.Klimke W, Agarwala R, Badretdin A, Chetvernin S, Ciufo S, Fedorov B, Kiryutin B, O'Neill K, Resch W, Resenchuk S, Schafer S, Tolstoy I, Tatusova T. 2009. The National Center for Biotechnology Information's Protein Clusters Database. Nucleic Acids Res 37:D216–D223. doi: 10.1093/nar/gkn734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joensen KG, Tetzschner AM, Iguchi A, Aarestrup FM, Scheutz F. 2015. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J Clin Microbiol 53:2410–2426. doi: 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM. 2014. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nei M, Tajima F, Tateno Y. 1983. Accuracy of estimated phylogenetic trees from molecular data. II. Gene frequency data. J Mol Evol 19:153–170. [DOI] [PubMed] [Google Scholar]
- 27.Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol 26:2465–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown JA, Hite DS, Gillim-Ross LA, Maguire HF, Bennett JK, Patterson JJ, Comstock NA, Watkins AK, Ghosh TS, Vogt RL. 2012. Outbreak of Shiga toxin-producing Escherichia coli serotype O26:H11 infection at a child care center in Colorado. Pediatr Infect Dis J 31:379–383. doi: 10.1097/INF.0b013e3182457122. [DOI] [PubMed] [Google Scholar]
- 30.Bielaszewska M, Mellmann A, Bletz S, Zhang W, Kock R, Kossow A, Prager R, Fruth A, Orth-Holler D, Marejkova M, Morabito S, Caprioli A, Pierard D, Smith G, Jenkins C, Curova K, Karch H. 2013. Enterohemorrhagic Escherichia coli O26:H11/H−: a new virulent clone emerges in Europe. Clin Infect Dis 56:1373–1381. doi: 10.1093/cid/cit055. [DOI] [PubMed] [Google Scholar]
- 31.Zweifel C, Cernela N, Stephan R. 2013. Detection of the emerging Shiga toxin-producing Escherichia coli O26:H11/H- sequence type 29 (ST29) clone in human patients and healthy cattle in Switzerland. Appl Environ Microbiol 79:5411–5413. doi: 10.1128/AEM.01728-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serra-Moreno R, Jofre J, Muniesa M. 2007. Insertion site occupancy by stx2 bacteriophages depends on the locus availability of the host strain chromosome. J Bacteriol 189:6645–6654. doi: 10.1128/JB.00466-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frank C, Werber D, Cramer JP, Askar M, Faber M, an der Heiden M, Bernard H, Fruth A, Prager R, Spode A, Wadl M, Zoufaly A, Jordan S, Kemper MJ, Follin P, Muller L, King LA, Rosner B, Buchholz U, Stark K, Krause G. 2011. Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N Engl J Med 365:1771–1780. doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- 34.Beutin L, Hammerl JA, Reetz J, Strauch E. 2014. Shiga toxin 2A-encoding bacteriophages in enteroaggregative Escherichia coli O104:H4 strains. Emerg Infect Dis 20:1567–1568. doi: 10.3201/eid2009.131373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beutin L, Hammerl JA, Strauch E, Reetz J, Dieckmann R, Kelner-Burgos Y, Martin A, Miko A, Strockbine NA, Lindstedt BA, Horn D, Monse H, Huettel B, Muller I, Stuber K, Reinhardt R. 2012. Spread of a distinct Stx2-encoding phage prototype among Escherichia coli O104:H4 strains from outbreaks in Germany, Norway, and Georgia. J Virol 86:10444–10455. doi: 10.1128/JVI.00986-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beutin L, Martin A. 2012. Outbreak of Shiga toxin-producing Escherichia coli (STEC) O104:H4 infection in Germany causes a paradigm shift with regard to human pathogenicity of STEC strains. J Food Prot 75:408–418. doi: 10.4315/0362-028X.JFP-11-452. [DOI] [PubMed] [Google Scholar]
- 37.Brzuszkiewicz E, Thurmer A, Schuldes J, Leimbach A, Liesegang H, Meyer FD, Boelter J, Petersen H, Gottschalk G, Daniel R. 2011. Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: entero-aggregative-haemorrhagic Escherichia coli (EAHEC). Arch Microbiol 193:883–891. doi: 10.1007/s00203-011-0725-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beutin L, Hammerl JA, Reetz J, Strauch E. 2013. Shiga toxin-producing Escherichia coli strains from cattle as a source of the Stx2a bacteriophages present in enteroaggregative Escherichia coli O104:H4 strains. Int J Med Microbiol 303:595–602. doi: 10.1016/j.ijmm.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 39.Monday SR, Minnich SA, Feng PC. 2004. A 12-base-pair deletion in the flagellar master control gene flhC causes nonmotility of the pathogenic German sorbitol-fermenting Escherichia coli O157:H- strains. J Bacteriol 186:2319–2327. doi: 10.1128/JB.186.8.2319-2327.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kusumoto M, Fukamizu D, Ogura Y, Yoshida E, Yamamoto F, Iwata T, Ooka T, Akiba M, Hayashi T. 2014. Lineage-specific distribution of insertion sequence excision enhancer in enterotoxigenic Escherichia coli isolated from swine. Appl Environ Microbiol 80:1394–1402. doi: 10.1128/AEM.03696-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kusumoto M, Ooka T, Nishiya Y, Ogura Y, Saito T, Sekine Y, Iwata T, Akiba M, Hayashi T. 2011. Insertion sequence-excision enhancer removes transposable elements from bacterial genomes and induces various genomic deletions. Nat Commun 2:152. doi: 10.1038/ncomms1152. [DOI] [PubMed] [Google Scholar]
- 42.Rump LV, Fischer M, Gonzalez-Escalona N. 2011. Prevalence, distribution and evolutionary significance of the IS629 insertion element in the stepwise emergence of Escherichia coli O157:H7. BMC Microbiol 11:133. doi: 10.1186/1471-2180-11-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rump LV, Fischer M, Gonzalez-Escalona N. 2011. Different IS629 transposition frequencies exhibited by Escherichia coli O157:H7 strains in the stepwise evolutionary model. Appl Environ Microbiol 77:5030–5033. doi: 10.1128/AEM.00249-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol Mol Biol Rev 62:725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ooka T, Ogura Y, Asadulghani M, Ohnishi M, Nakayama K, Terajima J, Watanabe H, Hayashi T. 2009. Inference of the impact of insertion sequence (IS) elements on bacterial genome diversification through analysis of small-size structural polymorphisms in Escherichia coli O157 genomes. Genome Res 19:1809–1816. doi: 10.1101/gr.089615.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toro M, Rump LV, Cao G, Meng J, Brown EW, Gonzalez-Escalona N. 2015. Simultaneous presence of insertion sequence excision enhancer and insertion sequence IS629 correlates with increased diversity and virulence in Shiga toxin-producing Escherichia coli. J Clin Microbiol 53:3466–3473. doi: 10.1128/JCM.01349-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malloy CD, Marr JS. 2001. Evolution of the Control of Communicable Diseases Manual: 1917 to 2000. J Public Health Manag Pract 7:97–104. doi: 10.1097/00124784-200107050-00013. [DOI] [PubMed] [Google Scholar]
- 48.Uhlich GA, Chen CY, Cottrell BJ, Irwin PL, Phillips JG. 2012. Peroxide resistance in Escherichia coli serotype O157:H7 biofilms is regulated by both RpoS-dependent and -independent mechanisms. Microbiology 158:2225–2234. doi: 10.1099/mic.0.059535-0. [DOI] [PubMed] [Google Scholar]
- 49.Januszkiewicz A, Wolkowicz T, Chrost A, Szych J. 2015. Characterization of the Shiga toxin-producing Escherichia coli O26 isolated from human in Poland between 1996 and 2014. Lett Appl Microbiol 60:605–608. doi: 10.1111/lam.12413. [DOI] [PubMed] [Google Scholar]
- 50.Orth D, Ehrlenbach S, Brockmeyer J, Khan AB, Huber G, Karch H, Sarg B, Lindner H, Wurzner R. 2010. EspP, a serine protease of enterohemorrhagic Escherichia coli, impairs complement activation by cleaving complement factors C3/C3b and C5. Infect Immun 78:4294–4301. doi: 10.1128/IAI.00488-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vali L, Hamouda A, Hoyle DV, Pearce MC, Whitaker LH, Jenkins C, Knight HI, Smith AW, Amyes SG. 2007. Antibiotic resistance and molecular epidemiology of Escherichia coli O26, O103 and O145 shed by two cohorts of Scottish beef cattle. J Antimicrob Chemother 59:403–410. doi: 10.1093/jac/dkl491. [DOI] [PubMed] [Google Scholar]
- 52.Venturini C, Beatson SA, Djordjevic SP, Walker MJ. 2010. Multiple antibiotic resistance gene recruitment onto the enterohemorrhagic Escherichia coli virulence plasmid. FASEB J 24:1160–1166. doi: 10.1096/fj.09-144972. [DOI] [PubMed] [Google Scholar]
- 53.Fratamico PM, Yan X, Caprioli A, Esposito G, Needleman DS, Pepe T, Tozzoli R, Cortesi ML, Morabito S. 2011. The complete DNA sequence and analysis of the virulence plasmid and of five additional plasmids carried by Shiga toxin-producing Escherichia coli O26:H11 strain H30. Int J Med Microbiol 301:192–203. doi: 10.1016/j.ijmm.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 54.Lavina M, Gaggero C, Moreno F. 1990. Microcin H47, a chromosome-encoded microcin antibiotic of Escherichia coli. J Bacteriol 172:6585–6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.