ABSTRACT
The oxidation of soluble Mn(II) to insoluble Mn(IV) is a widespread bacterial activity found in a diverse array of microbes. In the Mn(II)-oxidizing bacterium Pseudomonas putida GB-1, two Mn(II) oxidase genes, named mnxG and mcoA, were previously identified; each encodes a multicopper oxidase (MCO)-type enzyme. Expression of these two genes is positively regulated by the response regulator MnxR. Preliminary investigation into putative additional regulatory pathways suggested that the flagellar regulators FleN and FleQ also regulate Mn(II) oxidase activity; however, it also revealed the presence of a third, previously uncharacterized Mn(II) oxidase activity in P. putida GB-1. A strain from which both of the Mn(II) oxidase genes and fleQ were deleted exhibited low levels of Mn(II) oxidase activity. The enzyme responsible was genetically and biochemically identified as an animal heme peroxidase (AHP) with domain and sequence similarity to the previously identified Mn(II) oxidase MopA. In the ΔfleQ strain, P. putida GB-1 MopA is overexpressed and secreted from the cell, where it actively oxidizes Mn. Thus, deletion of fleQ unmasked a third Mn(II) oxidase activity in this strain. These results provide an example of an Mn(II)-oxidizing bacterium utilizing both MCO and AHP enzymes.
IMPORTANCE The identity of the Mn(II) oxidase enzyme in Pseudomonas putida GB-1 has been a long-standing question in the field of bacterial Mn(II) oxidation. In the current work, we demonstrate that P. putida GB-1 employs both the multicopper oxidase- and animal heme peroxidase-mediated pathways for the oxidation of Mn(II), rendering this model organism relevant to the study of both types of Mn(II) oxidase enzymes. The presence of three oxidase enzymes in P. putida GB-1 deepens the mystery of why microorganisms oxidize Mn(II) while providing the field with the tools necessary to address this question. The initial identification of MopA as a Mn(II) oxidase in this strain required the deletion of FleQ, a regulator involved in both flagellum synthesis and biofilm synthesis in Pseudomonas aeruginosa. Therefore, these results are also an important step toward understanding the regulation of Mn(II) oxidation.
INTRODUCTION
Biomineralization, the formation of minerals by living things, is best known as the process by which hardened body structures, like bones or shells, are formed. However, this process is also thought to contribute to the formation of geological features, like the ferromanganese crusts and nodules found at the sediment-water column interface (1–3). While the transition metal manganese (Mn) can cycle abiotically between a soluble, reduced Mn(II) form and an insoluble, oxidized Mn(IV) form in the environment (4, 5), both the oxidation and reduction of Mn can be driven by microbial activity. For example, the dissimilatory metal-reducing bacteria Shewanella sp. and Geobacter sp. utilize Mn(III, IV) oxide minerals as terminal electron acceptors during anaerobic respiration, resulting in their reduction to soluble Mn(II) compounds (6, 7).
The ability to form Mn(III, IV) minerals through the oxidation of Mn(II) is found in a diverse array of bacteria, including Gram-positive and Gram-negative species, and can be found in many environments, including deep-sea vents, freshwater lakes, rivers, and soil (8, 9). There are, however, common themes among Mn(II)-oxidizing bacteria. In most cases, Mn(II) oxidation begins in stationary phase. The Mn(II) oxidase activity is localized to the outer surface of the cell, for example, the exosporium of Bacillus sp. SG-1 or the sheath of Leptothrix discophora SS-1 (8). As a consequence, the cells become covered in Mn minerals. The oxidation reaction is thermodynamically favorable, raising the possibility that bacteria could derive energy from Mn(II) oxidation (10). Recent studies have also shown that Mn(II)-oxidizing bacteria exhibit increased resistance to the reactive oxygen species hydrogen peroxide (11). Nonetheless, the physiological function of bacterial Mn(II) oxidation remains unclear.
The Mn(II) oxidase complex from Bacillus sp. PL-12 is made up of multiple subunits, including a multicopper oxidase (MCO) named MnxG. Because the active Mnx complex can be heterologously expressed in Escherichia coli (12), this Mn(II) oxidase is the best studied to date. The putative Mn(II) oxidase enzymes of Leptothrix discophora SS-1 and Pedomicrobium sp. ACM 3067 have been identified as MCO enzymes called MofA and MoxA, respectively (13, 14). However, the Mn(II) oxidases in Aurantimonas manganoxydans SI85-9A1, Erythrobacter sp. SD21 (15, 16), and Roseobacter sp. AzwK-3b (17) are animal heme peroxidases (AHPs) which, in A. manganoxydans SI85-9A1 and Erythrobacter sp. SD-21, have been named MopA [Mn(II)-oxidizing peroxidase]. Similarly, species of white rot fungus employ MCO proteins called laccases and a variety of peroxidase family enzymes (Mn peroxidases and lignin peroxidases) in order to degrade lignin via an Mn(III) intermediate (18, 19). Thus, while the organisms that oxidize Mn(II) and the environments they inhabit are quite diverse, the variety of enzymes responsible appears to be limited to MCOs and peroxidases.
The gammaproteobacterium Pseudomonas putida GB-1 is a prominent model system for the investigation of bacterial Mn(II) oxidation (20). Recent work revealed the presence of two Mn(II) oxidase genes in this strain: mnxG, encoding an MCO with a low level of homology to the Bacillus sp. PL-12 Mn(II) oxidase subunit MnxG, and mcoA, encoding a second MCO distinct from MnxG (21). A strain from which both genes had been deleted failed to oxidize Mn(II) under all conditions tested. Quantitative PCR (qPCR) revealed that both mnxG and mcoA are upregulated in the presence of the σ54-dependent response regulator MnxR (21). MnxR is part of a two-component regulatory pathway (the Mnx TCR) that is required for Mn(II) oxidation (22). Thus, a simple pathway could be proposed whereby the Mnx TCR, in response to an as-yet-unknown environmental signal, phosphorylates MnxR, stimulating its function and resulting in upregulation of mnxG and mcoA and, ultimately, in Mn(II) oxidase activity.
The physiological role of Mn(II) oxidation in bacteria remains unclear; however, elucidating the pathways that regulate expression of this activity may provide clues about this role. Therefore, we chose to further investigate the Mnx regulatory pathway using a genetic approach. Specifically, we screened for second site mutations that could suppress the oxidation defect of the ΔmnxR strain. Our results support a connection between Mn(II) oxidation and biofilm formation while revealing the surprising presence of a third Mn(II) oxidase enzyme in P. putida GB-1.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains and plasmids used in this study are shown in Table 1. P. putida GB-1 was grown at room temperature (RT) in LB or Lept medium (23). Escherichia coli strains were grown in LB medium at 37°C. Solid media contained 1.5% (wt/vol) agar. Antibiotics were used at the following concentrations: kanamycin (Km) 30 μg/ml, ampicillin (Amp) 100 μg/ml, and gentamicin (Gm) 50 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype, characteristic, or construction | Antibiotic resistancea | Reference or source |
|---|---|---|---|
| Strain | |||
| Pseudomonas putida | |||
| GB-1 | Manganese oxidizer, wild type | Ampr | 45 |
| ΔfleN | GB-1 with in-frame deletion of fleN | 24 | |
| ΔmnxR | GB-1 with in-frame deletion of mnxR | 22 | |
| ΔfleQ | In-frame deletion of fleQ generated by conjugation of pKG289 into GB-1 | This work | |
| MopA-OE | In-frame deletion of mnxG, mcoA, and fleQ generated by conjugation of pKG289 into ΔmnxG ΔmcoA | This work | |
| MopA-KO | In-frame deletion of mnxG, mcoA, fleQ, and mopA by conjugation of pKG168 into MopA-OE | This work | |
| ΔfleN ΔmnxR | In-frame deletion of fleN and mnxR by conjugation of pKG214 into ΔmnxR | This work | |
| ΔmnxR ΔfleQ | In-frame deletion of mnxR and fleQ by conjugation of pKG289 into ΔmnxR | This work | |
| ΔmcoA ΔmnxG | GB-1 with in-frame deletions of mcoA and mnxG | 21 | |
| Escherichia coli | |||
| TAM1 | mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Active Motif | |
| Plasmid | |||
| pEX18Gm | Gene replacement vector; oriT sacB | Gmr | 46 |
| pJET1.2/blunt | Commercial cloning vector | Ampr | Thermo Scientific |
| pKG168 | ∼500 bp upstream of mopA fused in frame with ∼500 bp downstream cloned into pEX18Gm for making in-frame deletion | Gmr | This work |
| pKG178 | mopA amplified with mop1-F and mop2-R, cloned into pJET1.2/blunt and subcloned into pUCP22 with XbaI and XmaI | Ampr Gmr | This work |
| pKG214 | ∼500 bp upstream of fleN fused in frame with ∼500 bp downstream cloned into pEX18Gm for making in-frame deletion | Gmr | 24 |
| pKG224 | pUCP22 carrying fleN under the control of PlacZ | Ampr Gmr | This work |
| pKG289 | ∼500 bp upstream of fleQ fused in frame with ∼500 bp downstream cloned into pEX18Gm for making in-frame deletion | Gmr | This work |
| pRK2013 | Helper plasmid for conjugation | Kmr | 47 |
| pRL27 | Tn5 | Kmr | 48 |
| pUCP22 | Broad-host-range plasmid, ColE1 replicon | Ampr Gmr | 49 |
Ampr, ampicillin resistance; Gmr, gentamicin resistance; Kmr, kanamycin resistance.
Transposon mutagenesis.
pRL27, the plasmid carrying transposon Tn5, was moved into the P. putida GB-1 ΔmnxR strain (Table 1) by triparental mating using helper plasmid pRK2013, as previously described (24). The oxidation phenotype of the transposon (Tn) mutants was assessed by replica plating transconjugants onto Lept plates with 100 μM MnCl2. The ΔmnxR strain failed to oxidize Mn(II) under these conditions; suppressors of the ΔmnxR oxidation defect were identified as colonies exhibiting visible Mn(IV) oxides after 4 days of incubation at RT. At least 5,000 colonies were screened in each of two independent conjugation experiments. The sites of transposon insertion were determined as previously described (24).
Generation of in-frame deletions.
The in-frame deletions of mopA, fleQ, and fleN were generated as described previously (22). Briefly, DNA upstream and downstream of the gene of interest was amplified (primers, Table 2) (24) and fused by PCR splicing by overhang extension (SOE) (25). The fusion construct was cloned into the gene replacement vector pEX18Gm (Table 1), which could then be moved via conjugation into various P. putida GB-1 backgrounds. Genes were deleted through double recombination with the fusion construct and verified by PCR amplification across the site of deletion.
TABLE 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| Per-upstream-F | 5′CCTCCCTTTATCGCTAAGCGGG3′ |
| Per-junction-F | 5′TCTCCCGAAAGATCGAGGGCAGACTTCATCCTGGCGGGTTGA3′ |
| Per-junction-R | 5′TCAACCCGCCAGGATGAAGTCTGCCCTCGATCTTTCGGGAGA3′ |
| Per-downstream-R | 5′AGAAGAACCGCCTGGTGGC3′ |
| fleQ_1-F | 5′CTCAGCCTGCCAGCTGAAGG3′ |
| fleQ_2-R | 5′CGCTGAGTTCGTCAATGCGCTG3′ |
| fleQ_3-F | 5′TGCTATTGCATGTGGCGTGAAACCCAGGCAGAAGATTGACCTTGTTGGG3′ |
| fleQ_4-R | 5′CCCAACAAGGTCAATCTTCTGCCTGGGTTTCACGCCACATGCAATAGCA3′ |
| mop1-F | 5′CCCGGGACCCATGACCAGATACTGCGC3′, adds an XmaI site |
| mop2-R | 5′TCTAGACCCCCGGCCTTTGACACC3′, adds an XbaI site |
Complementation.
The mopA gene was PCR amplified with primers designed to incorporate an XmaI site at the 5′ end and an XbaI site at the 3′ end of the gene (Table 2). The PCR product was ligated into pJET1.2/blunt (Fermentas, Glen Burnie, MD) and then subcloned into pUCP22. The mopA gene is expressed from PlacZ in this construct. The plasmid was moved into P. putida GB-1 strains via heat shock transformation (22), and complementation was assessed by Mn(II) oxidation assays and SDS-PAGE, with pUCP22 as the empty vector (EV) control.
Complementation by fleN was performed using the plasmid pKG224 (Table 1). This plasmid expresses fleN from a PlacZ promoter in the pUCP22 backbone. The plasmids were transformed into the suppressor strains as described above, and Mn(II) oxidation was assessed on Lept plates after growth at RT for 5 days.
Motility assay.
Selected strains were grown overnight in LB media and then diluted 25-fold into Lept media containing 100 μM MnCl2. After approximately 2.5 h growth with shaking at RT, 10-μl aliquots were placed on a microscope slide and examined using a Leica DM750 compound microscope at ×100 magnification.
Quantification of Mn(II) oxidation.
Selected strains were grown overnight at RT in LB (MopA-KO + empty vector and MopA-KO + pmopA were grown in LB with 50 μg/ml amp). The following morning, triplicate cultures were set up as follows: 40 μl was subcultured into 2 ml Lept with 100 μM MnCl2 in 14-ml polystyrene snap-cap tubes. Cultures were incubated with shaking at RT for 3 days. To measure oxide production, 500 μl 0.016% leucoberbelin blue (LBB) (23, 26) in 1% acetic acid was added to each culture, and tubes were shaken at RT for 2 h. Then, 300 μl of this mixture was transferred to a 96-well plate, and absorbance was scanned from 360 to 750 nm with 4-nm steps in a SpectraMax M2e plate reader (Molecular Devices). The LBB absorbance at 624 nm was normalized to a baseline determined from the slope of the absorbance at 480 nm and 700 nm.
Generation of concentrated culture supernatants.
Single colonies were inoculated into 5 ml LB and grown at RT overnight. The following morning, cultures were diluted 50-fold into Lept medium. These cultures were grown at RT with continuous shaking. After 48 h, cells were pelleted by centrifugation at 6,000 rpm for 15 min at 4°C. After transfer of the supernatants to clean 50-ml tubes, any remaining cells were removed by filtering through 0.2-μm-pore-size Acrodisc syringe filters (Pall Corporation). The ∼34 ml of supernatant was concentrated using Amicon Ultra-15 50-kDa MWCO filters (Amicon) to a volume of 1 to 2 ml. Then, 500 μl of this was further concentrated using Amicon Ultra-0.5 30-kDa centrifugal filter devices to a final volume of ∼60 μl. Concentrated culture supernatants were stored at 4°C.
Analysis of protein.
Concentrated culture supernatants were loaded in duplicate and separated on a Mini-Protean TGX precast 4% to 15% PAGE gel (Bio-Rad) using 25 mM Tris, 192 mM glycine buffer (pH 8.3). After electrophoresis, the gel was rinsed three times in double-distilled water (dH2O) and cut in half. One half of the gel was incubated in 10 mM HEPES (pH 7.5) and 100 μM MnCl2 overnight at RT (23). Brown bands formed at the location of Mn(II)-oxidizing protein complexes and were recorded photographically. The gel was then rinsed in dH2O, briefly submerged in 0.0032% LBB to stain oxides blue, and rephotographed. The other half of the gel was stained using imperial protein stain (Thermo Scientific), destained with H2O, and photographed. Separation of proteins in the concentrated culture supernatants by denaturing gel electrophoresis was performed as described above, except the running buffer contained 0.1% SDS. Proteins were visualized by staining with imperial protein stain.
Liquid chromatography-tandem mass spectrometry.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis was performed by the Oregon Health & Science University proteomics shared resource (http://www.ohsu.edu/xd/research/research-cores/proteomics/). The sample was reduced with dithiothreitol, alkylated with iodoacetamide, and digested overnight with trypsin (27). The digested sample was then separated using liquid chromatography with a NanoAcquity ultraperformance liquid chromatography system (Waters) and delivered to an LTQ Velos dual pressure linear ion trap mass spectrometer (Thermo Fisher). Samples were applied at 15 μl/min to a Symmetry C18 trap cartridge (Waters) for 10 min and then switched onto a 75-μm to 250-mm NanoAcquity BEH 130 C18 column with 1.7-μm particles (Waters) using mobile phases water and acetonitrile containing 0.1% formic acid, 7% to 30% acetonitrile gradient over 60 min, and a 300-nl/min flow rate. Comet (version 2015.01, revision 1) was used to search MS2 Spectra against a July 2015 version of the Uniprot Pseudomonas putida (strain GB-1) FASTA protein database, with concatenated sequence-reversed entries to estimate error thresholds and with 179 common contaminant sequences and their reversed forms. The database processing was performed with python scripts available at http://www.proteomicanalysisworkbench.com. Comet searches for all samples were performed with trypsin enzyme specificity (28, 29). Peptide-to-protein mapping and protein filtering were performed using PAW_results_7.py (version 7.0). The in-house Python scripts have been described previously (29).
RESULTS
Suppression of the oxidation defect of the ΔmnxR deletion strain.
To further investigate the pathways that regulate Mn(II) oxidation, we screened for mutations that could suppress the oxidation defect of the mnxR deletion (ΔmnxR) strain. This was done by randomly mutagenizing the ΔmnxR strain with the transposon Tn5 and screening for colonies with restored Mn(II) oxidation (Fig. 1A and B). In two separate screens, we identified colonies with Mn(II) oxidase activity, for a total of 29 ΔmnxR,Tn5 suppressor strains (Table 3). We sequenced DNA flanking the transposon for a subset of the strains in order to determine the site of transposon insertion. Four transposons mapped to the gene fleN, and the remaining eight mapped to flhF (Table 3), a gene located immediately upstream of fleN on the P. putida GB-1 chromosome (30).
FIG 1.
Suppression of ΔmnxR. (A) The nonoxidizing ΔmnxR strain is a white oval. (B) Random mutagenesis with the Tn5 transposon resulted in ΔmnxR strains with Mn oxidase activity, i.e., suppressor strains. Mn(II)-oxidizing strains are represented by an orange oval. (C) Complementation, restoring to the strain a wild-type copy of the gene disrupted by the suppressor mutation, should result in the suppressor strain reverting to the nonoxidizing phenotype.
TABLE 3.
ΔmnxR suppressors
| Suppressor | Transposon insertion site | Mapping method |
|---|---|---|
| Sup3 | fleN | Sequenced/complemented by pFleN |
| Sup4 | flhF | Sequenced/complemented by pFleN |
| Sup5 | flhF | Sequenced |
| Sup6 | fleN or flhF | Complemented by pFleN |
| Sup7 | fleN or flhF | Complemented by pFleN |
| Sup8 | flhF | Sequenced |
| Sup9 | fleN or flhF | Complemented by pFleN |
| Sup10 | fleN or flhF | Complemented by pFleN |
| Sup11 | fleN or flhF | Complemented by pFleN |
| Sup13 | fleN or flhF | Complemented by pFleN |
| Sup14 | fleN or flhF | Complemented by pFleN |
| Sup15 | fleN or flhF | Complemented by pFleN |
| Sup17 | fleN or flhF | Complemented by pFleN |
| Sup18 | fleN or flhF | Complemented by pFleN |
| Sup19 | fleN or flhF | Complemented by pFleN |
| Sup20 | fleN or flhF | Complemented by pFleN |
| Sup21 | fleN or flhF | Complemented by pFleN |
| Sup22 | fleN or flhF | Complemented by pFleN |
| Sup23 | fleN or flhF | Complemented by pFleN |
| Sup26 | fleN or flhF | Complemented by pFleN |
| Sup27 | fleN or flhF | Complemented by pFleN |
| KGsup1 | fleN | Sequenced |
| KGsup2 | fleN | Sequenced |
| KGsup3 | flhF | Sequenced |
| KGsup4 | flhF | Sequenced |
| KGsup5 | flhF | Sequenced |
| KGsup6 | flhF | Sequenced |
| KGsup7 | fleN | Sequenced |
| KGsup8 | flhF | Sequenced |
Loss of the flagellar regulator FleN suppresses ΔmnxR.
It is possible that the flhF::Tn5 insertions affect expression of the downstream fleN gene through polar effects. Also, previous work showed that a strain with an in-frame deletion of fleN exhibited increased Mn(II) oxidation (24). Therefore, we hypothesized that disruption of FleN function was responsible for the restoration of Mn(II) oxidation observed in the ΔmnxR,Tn5 suppressor strains. If so, it should be possible to complement the oxidation phenotype of the ΔmnxR,Tn5 strains (Fig. 1C) with a plasmid expressing FleN (pKG224) (Table 1). In each case, pKG224 caused the ΔmnxR,Tn5 strains to behave like the ΔmnxR strain, while the empty vector control did not; in other words, the ΔmnxR,Tn5 strains carrying the FleN-expressing plasmid failed to oxidize Mn(II), while the ΔmnxR,Tn5 strains carrying the empty vector continued to oxidize Mn(II). For controls, we also transformed pKG224 and the empty vector into suppressor strains for which the site of Tn5 insertion had been mapped by sequencing (Sup3, fleN::Tn5 and Sup4, flhF::Tn5) (Table 3). Both strains were complemented by the FleN-expressing plasmid. Because multicopy expression of FleN restored the ΔmnxR phenotype to each of the ΔmnxR::Tn5 strains, we concluded that the suppression of the ΔmnxR oxidation defect in each of the suppressor strains was due to disruption of FleN activity.
To verify that the loss of FleN suppressed the oxidation defect of the ΔmnxR strain, we generated a double-mutant strain in which both fleN and mnxR had been deleted. As previously published (22, 24), the ΔmnxR strain failed to oxidize Mn(II) and the ΔfleN strain exhibited increased Mn(II) oxidation on solid media relative to wild type (Fig. 2). The ΔmnxR ΔfleN mutant strain exhibited an oxidation phenotype on plates similar to that of the ΔfleN strain (Fig. 2). Therefore, consistent with the results of the transposon mutagenesis, deletion of fleN suppressed the oxidation defect of the ΔmnxR strain.
FIG 2.
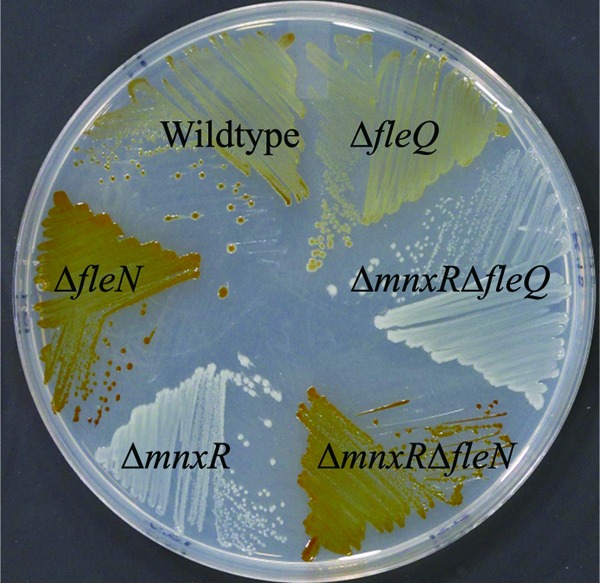
Suppression of ΔmnxR. The indicated strains were streaked onto Lept media and grown at RT for 3 days.
In Pseudomonas aeruginosa PAO1, FleN has been shown to regulate flagellar number through inhibition of the transcription factor FleQ (31, 32). The P. putida GB-1 genome encodes a gene with extensive similarity to P. aeruginosa PAO1 FleQ (PputGB1_3934, 84% identical). In-frame deletion of this gene resulted in cells that were nonmotile when viewed under a microscope during the early exponential phase, supporting the identification of PputGB1_3934 as the master flagellar regulatory gene, fleQ. The ΔfleQ strain, when grown on solid media, appeared to oxidize Mn(II) to a slightly lesser extent than wild-type bacteria (Fig. 2). This slight decrease was also present in cultures grown in liquid media (Fig. 3). Thus, FleN may regulate Mn(II) oxidation at least in part through its inhibition of the positive regulator FleQ, in a manner similar to the regulation of flagella synthesis.
FIG 3.
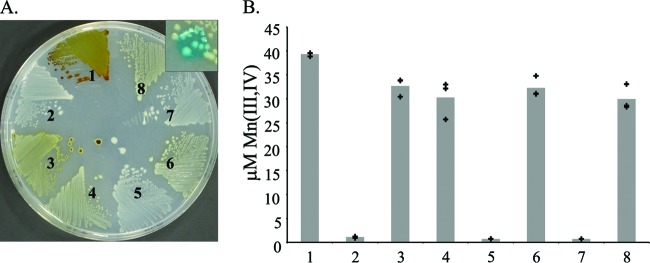
Deletion of fleQ reveals the presence of a third Mn(II) oxidase, MopA. (A) Strains were streaked onto Lept medium and grown at RT for 7 days. Inset shows colonies after 10 μl leucoberbelin blue was dropped on top. (B) Quantification of Mn(III, IV) oxides in liquid cultures after shaking at RT for 3 days. Bars represent the average of three replicates; crosses indicate individual values. Strains shown: 1, wild type; 2, ΔmnxR; 3, ΔfleQ; 4, ΔmnxG ΔmcoA ΔfleQ; 5, ΔmnxG ΔmcoA ΔfleQ ΔmopA; 6, ΔmnxR ΔfleQ; 7, ΔmnxG ΔmcoA ΔfleQ ΔmopA + pUCP22; 8, ΔmnxG ΔmcoA ΔfleQ ΔmopA + pKG178.
ΔfleQ reveals a third Mn(II) oxidase activity.
If both FleQ and MnxR are positive regulators of Mn(II) oxidation, we predict that deletion of both genes—ΔmnxR ΔfleQ—would result in a nonoxidizing phenotype. This is initially what was observed (Fig. 2). However, after prolonged incubation on solid media (7 days), the ΔmnxR ΔfleQ mutant strain produced a diffuse brown color that resembled Mn oxides (Fig. 3A). We verified that the faint brown color was the result of Mn(III, IV) oxide formation by dropping a solution of leucoberbelin blue (LBB) (23, 26) onto the colonies; the reaction of LBB with Mn(III, IV) oxides produces a blue color (Fig. 3A, inset). Quantification of Mn(III, IV) oxide formation in liquid culture gave a similar result: the ΔmnxR ΔfleQ mutant strain produced nearly 30-fold higher concentration of Mn(III, IV) oxides than the ΔmnxR strain (Fig. 3B).
We identified two possible explanations for the Mn(II) oxidation seen in the ΔfleQ ΔmnxR strain: either one or both of the two oxidase genes are expressed in the ΔfleQ ΔmnxR mutant through an MnxR-independent regulatory pathway, or the ΔfleQ ΔmnxR mutant had unmasked a third Mn(II) oxidase activity. To test these possibilities, we generated a triple mutant in which the two known Mn(II) oxidase genes and fleQ had each been deleted (ΔmnxG ΔmcoA ΔfleQ) (Table 1). In previous work, the ΔmnxG ΔmcoA strain failed to oxidize Mn(II) (21). However, the ΔmnxG ΔmcoA ΔfleQ strain retained a level of Mn(II) oxidase activity similar to that of the ΔfleQ ΔmnxR double mutant (Fig. 3A and B) despite each of the known Mn(II) oxidase genes having been deleted from the chromosome. Therefore, the ΔfleQ deletion unmasked a third Mn(II) oxidase activity. While the role of FleN, FleQ, and MnxR in regulating Mn(II) oxidation is an important subject of ongoing research in our group, we here address our efforts to identify and characterize this third Mn(II) oxidase enzyme.
The third Mn(II) oxidase in P. putida GB-1 is MopA.
Previous work (21) had shown that the P. putida GB-1 genome encodes an AHP similar to the AHP proteins implicated in Mn(II) oxidation in A. manganoxydans SI85-9A1, Erythrobacter sp. SD-21, and Roseobacter sp. AzwK-3b (15–17). However, deletion of this gene (mopA) from the P. putida GB-1 genome had little reproducible effect on Mn(II) oxidation. Furthermore, genes homologous to mopA appeared to be present in the genomes of both oxidizing and nonoxidizing pseudomonads (21). For these reasons, we had previously discounted a role for mopA in Mn(II) oxidation by P. putida GB-1 (21). To determine if MopA is responsible for the cryptic Mn(II) oxidation seen in the ΔfleQ ΔmnxR and ΔmnxG ΔmcoA ΔfleQ strains, we deleted mopA from the ΔmnxG ΔmcoA ΔfleQ triple mutant. This resulted in complete loss of Mn(II) oxidation (Fig. 3A and B). Oxidation activity could be restored to the quadruple mutant by a plasmid carrying mopA (pKG178) (Table 1). Therefore, MopA is a third Mn(II) oxidase enzyme in P. putida GB-1.
MopA is present in the culture supernatant.
When wild-type cells oxidize Mn(II) on plates, the oxides remain associated with the cells, turning the colonies dark brown. In the fleQ mutants (especially ΔmnxG ΔmcoA ΔfleQ and ΔmnxR ΔfleQ), the oxides are light brown and diffuse into the agar of the plate (Fig. 3A, inset). This diffusion suggests that the oxidase is secreted away from the cells, into the culture medium. To test this, we isolated cell-free culture supernatants from wild type, ΔmnxG ΔmcoA ΔfleQ, and ΔmnxG ΔmcoA ΔfleQ ΔmopA strains and separated the protein by SDS-PAGE (Fig. 4). A large band that migrated above the 200-kDa marker was clearly visible in the supernatant prepared from the ΔmnxG ΔmcoA ΔfleQ strain but not from the wild-type or ΔmnxG ΔmcoA ΔfleQ ΔmopA + empty vector strains. This band was restored to the ΔmnxG ΔmcoA ΔfleQ ΔmopA strain by the pMopA plasmid (Fig. 4). MopA is 3,608 amino acids long, or roughly 373 kDa. Thus, this >200-kDa protein was likely MopA. The MopA band was also visible in whole-cell lysates prepared from the ΔmnxG ΔmcoA ΔfleQ and the ΔfleQ strains but not in wild-type whole-cell lysates (data not shown). Thus, the fleQ deletion resulted in substantially increased levels of MopA protein. In the remainder of the text, we refer to the ΔmnxG ΔmcoA ΔfleQ strain as the MopA overexpression (MopA-OE) strain and the ΔmnxG ΔmcoA ΔfleQ ΔmopA strain as the MopA knockout (MopA-KO). To confirm that the protein in the >200-kDa band was MopA, we excised it from the MopA-OE lane (Fig. 4, box) and submitted it for mass spectrometry. The major component of the mass spectra was MopA, with 1,345 peptides mapping to this protein. Protein sequence coverage was 40.7% (Table 4).
FIG 4.
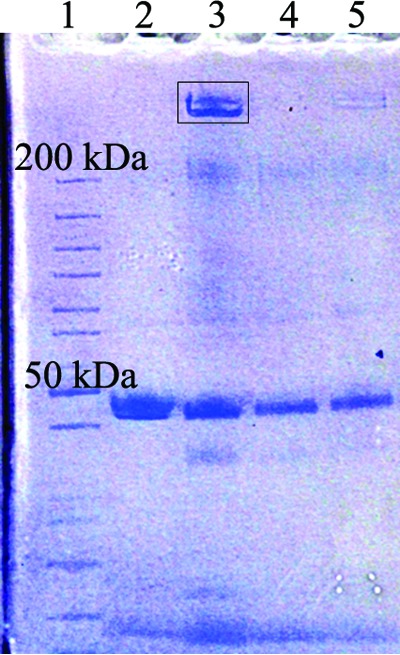
Deletion of fleQ results in overexpression and secretion of MopA. Lane 1, PAGERuler unstained protein ladder (Thermo Scientific); lane 2, wild type; lane 3, MopA-OE; lane 4, MopA-KO + EV; lane 5, MopA-KO + pmopA. The top protein in the marker lane is 200 kDa. The box indicates the portion of the gel submitted for LC-MS/MS.
TABLE 4.
LC-MS/MS peptide identification of proteins present in SDS-PAGE band
| Locus tag | Protein description | Estimated molecular mass (kDa) | Total spectral counts | % coverage |
|---|---|---|---|---|
| PputGB1_3353 | MopA | 373 | 1,345 | 40.7 |
| PputGB1_3075 | Outer membrane adhesion-like protein | 359 | 19 | 4.2 |
| PputGB1_1837 | Leucine-rich repeat protein | 168 | 2 | 1.8 |
| PputGB1_3392 | Uncharacterized | 248 | 2 | 1.9 |
| PputGB1_0792 | Probable malate:quinone oxidoreductase | 54 | 2 | 6 |
| PputGB1_0945 | Uncharacterized | 138 | 2 | 2.8 |
| PputGB1_3939 | Flagellin | 49 | 4 | 10.4 |
| PputGB1_1040 | Uncharacterized | 74 | 2 | 4.5 |
MopA actively oxidizes Mn(II).
It is possible to detect Mn(II) oxidase activity in proteins separated by nondenaturing gel electrophoresis (17, 23, 33). Therefore, if the secreted MopA is an Mn(II) oxidase, this activity should be apparent after separation of culture supernatant on a native gel. After separating concentrated cell-free culture supernatant by native PAGE, we soaked the gel overnight in a buffer solution containing MnCl2 (see Materials and Methods). A smear of brown Mn(IV) oxides was visible in the MopA-OE lane, and a sharper brown band was present in the MopA-KO + pmopA lane (Fig. 5B). After soaking the gel briefly in LBB, these brown bands turned blue, supporting their identification as Mn(III, IV) oxides (Fig. 5C). This result supported the conclusion that MopA is capable of oxidizing Mn(II). In addition, a faint blue spot was visible in the wild-type lane, migrating higher in the gel than those in the MopA-OE and MopA-KO + pmopA lanes (Fig. 5C). MnxG is predicted to weigh 209 kDa; McoA, 123 kDa. Therefore, this faint blue band at the top of the gel may represent a complex of two or three of the Mn(II) oxidase enzymes.
FIG 5.
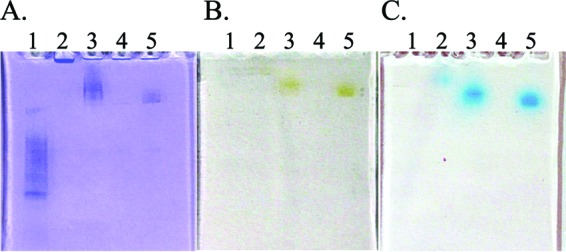
Native PAGE analysis results for protein stain (A); activity assay using gel incubated in 10 mM HEPES (pH 7.5) + 100 μM MnCl2 overnight at RT (B); activity assay using gel described for panel B after soaking in 0.0032% LBB (C). Lane 1, PAGERuler unstained protein ladder; lane 2, wild type; lane 3, MopA-OE; lane 4, MopA-KO + EV; lane 5, MopA-KO + pmopA.
DISCUSSION
This work began as an effort to understand the regulation of Mn(II) oxidation in Pseudomonas putida GB-1 but ultimately revealed the presence of a third Mn(II) oxidase enzyme in this strain. Deletion of fleQ resulted in the overexpression of a large, secreted protein capable of oxidizing Mn(II), as shown by in-gel oxidase assays. Mass spectrometry and directed mutagenesis identified this protein as MopA, an animal heme peroxidase with sequence similarity to the MopA Mn(II) oxidase proteins of A. manganoxydans SI85-9A1 and other Mn(II)-oxidizing bacteria. Therefore, P. putida GB-1 employs three separate enzymes for the oxidation of Mn: the two MCOs, MnxG and McoA, and the AHP, MopA.
FleQ is a σ54-dependent transcription factor that initiates the tightly controlled regulatory cascade that results in flagellum synthesis (34). In P. aeruginosa PAO1, FleQ also regulates extracellular polysaccharide synthesis and biofilm formation (35), and, in P. putida KT2440, FleQ has been shown to play a role in expression of the adhesion protein LapA (36). Thus, the fleQ deletion may have pleiotropic effects on the physiology of P. putida GB-1. In addition, the overexpression of MopA seen in the fleQ mutants could cause the enzyme to exhibit nonnative activity. This raises the question of whether Mn(II) oxidation is a normal function of MopA in P. putida GB-1. Deletion of mopA from a fleQ-positive strain does not reproducibly result in decreased Mn(II) oxidation. However, under normal laboratory growth conditions, the bulk of Mn(II) oxidation may be performed by either MnxG or McoA, making the contribution of MopA difficult to detect. In agreement with this, the oxidation defect of a mnxG-mutant strain is exacerbated by the further deletion of mopA (data not shown). Thus, removing one of the MCO Mn(II) oxidases renders it possible to detect a role for MopA in Mn(II) oxidation in a fleQ-positive strain background.
A connection between Mn(II) oxidation and flagellum synthesis has been previously identified (24). In a genetic screen for transposon insertions that resulted in increased Mn(II) oxidation, several insertions were identified within flagellar regulatory and structural genes. Curiously, these flagellar mutants exhibited very different oxidation phenotypes depending on the growth conditions, with some of the mutants failing to oxidize Mn(II) in liquid media while exhibiting increased Mn(II) oxidation on solid media. The results reported here suggest that deletion of fleQ results in increased expression of one Mn(II) oxidase gene (mopA) (Fig. 4), while the activity of the other two oxidases may be decreased (suggested by the decreased oxidation of the ΔfleQ strain) (Fig. 3A and B). In addition, the precise amount of Mn(II) oxidation exhibited by strains with single oxidase gene deletions varies with growth conditions (21) and from experiment to experiment (data not shown). Therefore, the three oxidase genes are likely not expressed at the same times and under the same conditions. Future studies will need to focus on the regulation of individual genes, rather than the regulation of Mn(II) oxidation. Nonetheless, the fact that FleQ regulates not just flagellum synthesis but biofilm formation and surface adhesion suggests that the different Mn(II) oxidases play different roles in planktonic versus sessile lifestyles.
The AHP Mn(II) oxidase enzymes are characterized by the presence of one or two conserved heme-binding domains and multiple repeat-in-toxin family calcium-binding domains located carboxy-terminal to the heme domains (17). In Roseobacter sp. AzwK-3b, it has been proposed that one or more of its four secreted AHP enzymes indirectly oxidize Mn(II) through the production of superoxide and the removal of hydrogen peroxide (17). However, work done with the heterologously expressed heme-binding domain from Erythrobacter sp. SD-21 MopA showed that Mn(II) oxidation by this enzyme was insensitive to superoxide dismutase (16), suggesting that superoxide was not involved in oxidation by Erythrobacter sp.SD-21 MopA. Mn(II) oxidation by Roseobacter sp. AzwK-3b cultures and culture supernatants is stimulated by light (37) and by the reductant NADH (17, 38). Preliminary experiments examining Mn(II) oxidation by concentrated culture supernatants prepared from the MopA-OE strain indicated that the oxidase activity of GB-1 MopA is also stimulated by light (data not shown) but is completely inhibited by NADH (data not shown). Therefore, the mechanism(s) by which AHP enzymes contribute to Mn(II) oxidation remains unclear. However, the identification of a Mn(II) oxidation-associated AHP in the genetically tractable P. putida GB-1 strain may simplify investigation of these mechanisms.
The results reported here describe a Mn(II)-oxidizing bacterium utilizing both MCO and AHP Mn(II) oxidase enzymes. As previously reported (21), Leptothrix cholodnii SP-6 encodes in its genome two different putative MCO-type Mn(II) oxidases. In addition, L. cholodnii SP-6 has a potential mopA gene, while the A. manganoxydans SI85-9A1 genome has not just mopA but also a gene with similarity to an mcoA homolog (30) and two genes with sequence similarity to the Pedomicrobium sp. ACM 3067 Mn(II) oxidase MoxA (39). Therefore, P. putida GB-1 may not be unique in harboring Mn(II) oxidase enzymes of both the MCO and AHP families. In the basidiomycete fungus Stropharia rugosoannulata, degradation of lignin is accomplished through the cooperation of a laccase (an MCO-type enzyme) and a Mn peroxidase, with the peroxidase utilizing H2O2 generated during the oxidation of Mn(II) to Mn(III) by the laccase (19). Therefore, it is possible that MopA works together with one or both of the MCO-type Mn(II) oxidases during oxidation.
Mn(II) oxidation occurs on the outside surface of the cell, coating the cell with Mn oxides (8). The Mn(II) oxidase enzymes that have been identified so far are found loosely bound to the exosporium (40) or the outer membrane (15) or are secreted from the cell into the culture milieu (17). P. putida GB-1 MopA is secreted into the culture supernatant (Fig. 4); the cellular localization of MnxG and McoA is unknown, although the proteins are likely exported (41). Furthermore, the results shown here suggest that Mn(II) oxidation is regulated as part of the planktonic to biofilm lifestyle switch. Thus, Mn(II) oxidase enzymes can be considered a public good or a resource available to the bacterial biofilm community at large, not just the cell actively secreting the protein (42). What function does Mn(II) oxidation play in the bacterial community such that the cost of secreting large oxidase enzymes is evolutionarily affordable? Mn(II) oxidation has been shown to protect P. putida GB-1 from hydrogen peroxide (11). In the closely related plant-beneficial strain P. putida KT2440, deletion of the mopA homolog PP2561 resulted in increased sensitivity to compounds like tert butyl hydroperoxide (43), supporting a role for Mn(II) oxidation in the oxidative stress response. This mutant strain also had a competitive defect in colonization of plant hosts that may be linked to its increased sensitivity to reactive oxygen species (43, 44). Understanding how and why bacteria oxidize Mn(II) has been a long-standing goal in this field; the identification of the oxidase enzymes in P. putida GB-1 makes it possible to further address the question of the physiological role of Mn(II) oxidation.
ACKNOWLEDGMENTS
The OHSU proteomics shared resource received partial support from NIH core grants P30EY010572 and P30CA069533. The Velos2 instrument was funded by grant R01DC002368-15S1. Portions of this work were supported by a National Science Foundation grant (CHE-1410688). L.S. was supported by the School of Chemical, Biological, and Environmental Engineering (CBEE) Johnson internship program at Oregon State University and the Center for Coastal Margin Observation and Prediction undergraduate internship program at Oregon Health & Science University.
Funding Statement
The OHSU proteomics shared resource received partial support from NIH core grants P30EY010572 and P30CA069533. The Velos2 instrument was funded by grant R01DC002368-15S1. L.S. was supported by the School of Chemical, Biological, and Environmental Engineering (CBEE) Johnson internship program at Oregon State University and the Center for Coastal Margin Observation and Prediction undergraduate internship program at Oregon Health & Science University.
REFERENCES
- 1.Wang X, Müller WEG. 2009. Marine biominerals: perspectives and challenges for polymetallic nodules and crusts. Trends Biotechnol 27:375–383. doi: 10.1016/j.tibtech.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Blöthe M, Wegorzewski A, Müller C, Simon F, Kuhn T, Schippers A. 2015. Manganese-cycling microbial communities inside deep-sea manganese nodules. Environ Sci Technol 49:7692–7700. doi: 10.1021/es504930v. [DOI] [PubMed] [Google Scholar]
- 3.Tully BJ, Heidelberg JF. 2013. Microbial communities associated with ferromanganese nodules and the surrounding sediments. Front Microbiol 4:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spiro TG, Bargar JR, Sposito G, Tebo BM. 2010. Bacteriogenic manganese oxides. Acc Chem Res 43:2–9. doi: 10.1021/ar800232a. [DOI] [PubMed] [Google Scholar]
- 5.Geszvain K, Butterfield C, Davis RE, Madison AS, Lee SW, Parker DL, Soldatova A, Spiro TG, Luther GW, Tebo BM. 2012. The molecular biogeochemistry of manganese(II) oxidation. Biochem Soc Trans 40:1244–1248. doi: 10.1042/BST20120229. [DOI] [PubMed] [Google Scholar]
- 6.Kouzuma A, Hashimoto K, Watanabe K. 2012. Roles of siderophore in manganese-oxide reduction by Shewanella oneidensis MR-1. FEMS Microbiol Lett 326:91–98. doi: 10.1111/j.1574-6968.2011.02444.x. [DOI] [PubMed] [Google Scholar]
- 7.Richter K, Schicklberger M, Gescher J. 2012. Dissimilatory reduction of extracellular electron acceptors in anaerobic respiration. Appl Environ Microbiol 78:913–921. doi: 10.1128/AEM.06803-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tebo BM, Geszvain K, Lee S-W. 2010. The molecular geomicrobiology of bacterial manganese(II) oxidation, p 285–308. In Barton LL, Mandl M, Loy A (ed), Geomicrobiology: molecular and environmental perspective. Springer, Dordrecht, Netherlands. [Google Scholar]
- 9.Stein LY, La Duc MT, Grundl TJ, Nealson KH. 2001. Bacterial and archaeal populations associated with freshwater ferromanganous micronodules and sediments. Environ Microbiol 3:10–18. doi: 10.1046/j.1462-2920.2001.00154.x. [DOI] [PubMed] [Google Scholar]
- 10.Tebo BM, Johnson HA, McCarthy JK, Templeton AS. 2005. Geomicrobiology of manganese(II) oxidation. Trends Microbiol 13:421–428. doi: 10.1016/j.tim.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 11.Banh A, Chavez V, Doi J, Nguyen A, Hernandez S, Ha V, Jimenez P, Espinoza F, Johnson HA. 2013. Manganese (Mn) oxidation increases intracellular Mn in Pseudomonas putida GB-1. PLoS One 8:e77835. doi: 10.1371/journal.pone.0077835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butterfield CN, Soldatova AV, Lee S-W, Spiro TG, Tebo BM. 2013. Mn(II,III) oxidation and MnO2 mineralization by an expressed bacterial multicopper oxidase. Proc Natl Acad Sci U S A 110:11731–11735. doi: 10.1073/pnas.1303677110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corstjens PLAM, de Vrind JPM, Goosen T, de Vrind-de Jong EW. 1997. Identification and molecular analysis of the Leptothrix discophora SS-1 mofA gene, a gene putatively encoding a manganese-oxidizing protein with copper domains. Geomicrobiol J 14:91–108. doi: 10.1080/01490459709378037. [DOI] [Google Scholar]
- 14.Ridge JP, Lin M, Larsen EI, Fegan M, McEwan AG, Sly LI. 2007. A multicopper oxidase is essential for manganese oxidation and laccase-like activity in Pedomicrobium sp. ACM 3067. Environ Microbiol 9:944–953. [DOI] [PubMed] [Google Scholar]
- 15.Anderson CR, Johnson HA, Caputo N, Davis RE, Torpey JW, Tebo BM. 2009. Mn(II) oxidation is catalyzed by heme peroxidases in “Aurantimonas manganoxydans” strain SI85-9A1 and Erythrobacter sp. strain SD-21. Appl Environ Microbiol 75:4130–4138. doi: 10.1128/AEM.02890-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakama K, Medina M, Lien A, Ruggieri J, Collins K, Johnson HA. 2014. Heterologous expression and characterization of the manganese-oxidizing protein from Erythrobacter sp. strain SD21. Appl Environ Microbiol 80:6837–6842. doi: 10.1128/AEM.01873-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andeer PF, Learman DR, McIlvin M, Dunn JA, Hansel CM. 2015. Extracellular heme peroxidases mediate Mn(II) oxidation in a marine Roseobacter bacterium via superoxide production. Environ Microbiol 17:3925–3936. doi: 10.1111/1462-2920.12893. [DOI] [PubMed] [Google Scholar]
- 18.Höfer C, Schlosser D. 1999. Novel enzymatic oxidation of Mn2+ to Mn3+ catalyzed by a fungal laccase. FEBS Lett 451:186–190. doi: 10.1016/S0014-5793(99)00566-9. [DOI] [PubMed] [Google Scholar]
- 19.Schlosser D, Hofer C. 2002. Laccase-catalyzed oxidation of Mn2+ in the presence of natural Mn3+ chelators as a novel source of extracellular H2O2 production and its impact on manganese peroxidase. Appl Environ Microbiol 68:3514–3521. doi: 10.1128/AEM.68.7.3514-3521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brouwers GJ, de Vrind JP, Corstjens PL, Cornelis P, Baysse C, de Vrind-de Jong EW. 1999. cumA, a gene encoding a multicopper oxidase, is involved in Mn2+ oxidation in Pseudomonas putida GB-1. Appl Environ Microbiol 65:1762–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geszvain K, McCarthy JK, Tebo BM. 2013. Elimination of manganese(II, III) oxidation in Pseudomonas putida GB-1 by a double knockout of two putative multicopper oxidase genes. Appl Environ Microbiol 79:357–366. doi: 10.1128/AEM.01850-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geszvain K, Tebo BM. 2010. Identification of a two-component regulatory pathway essential for Mn(II) oxidation in Pseudomonas putida GB-1. Appl Environ Microbiol 76:1224–1231. doi: 10.1128/AEM.02473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boogerd FC, de Vrind JP. 1987. Manganese oxidation by Leptothrix discophora. J Bacteriol 169:489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geszvain K, Yamaguchi A, Maybee J, Tebo BM. 2011. Mn(II) oxidation in Pseudomonas putida GB-1 is influenced by flagella synthesis and surface substrate. Arch Microbiol 193:605–614. doi: 10.1007/s00203-011-0702-0. [DOI] [PubMed] [Google Scholar]
- 25.Horton RM. 1997. In vitro recombination and mutagenesis of DNA: SOEing together tailor-made genes. Methods Mol Biol 67:141–149. [DOI] [PubMed] [Google Scholar]
- 26.Tebo B, Clement BG, Dick GJ. 2007. Biotransformations of manganese, p 1316 In Hurst CJ, Crawford RL, Garland JL, Lipson DA, Mills AL, Stetzenbach LD (ed), Manual of environmental microbiology, 3rd ed ASM Press, Washington, DC. [Google Scholar]
- 27.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1:2856–2860. [DOI] [PubMed] [Google Scholar]
- 28.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 29.Wilmarth PA, Riviere MA, David LL. 2009. Techniques for accurate protein identification in shotgun proteomic studies of human, mouse, bovine, and chicken lenses. J Ocul Biol Dis Infor 2:223–234. doi: 10.1007/s12177-009-9042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. 2012. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res 40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dasgupta N, Arora SK, Ramphal R. 2000. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol 182:357–364. doi: 10.1128/JB.182.2.357-364.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dasgupta N, Ramphal R. 2001. Interaction of the antiactivator FleN with the transcriptional activator FleQ regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol 183:6636–6644. doi: 10.1128/JB.183.22.6636-6644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okazaki M, Sugita T, Shimizu M, Ohode Y, Iwamoto K, de Vrind-de Jong EW, de Vrind JP, Corstjens PL. 1997. Partial purification and characterization of manganese-oxidizing factors of Pseudomonas fluorescens GB-1. Appl Environ Microbiol 63:4793–4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, Ramphal R. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol 50:809–824. doi: 10.1046/j.1365-2958.2003.03740.x. [DOI] [PubMed] [Google Scholar]
- 35.Baraquet C, Murakami K, Parsek MR, Harwood CS. 2012. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res 40:7207–7218. doi: 10.1093/nar/gks384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martínez-Gil M, Ramos-González MI, Espinosa-Urgel M. 2014. Roles of cyclic di-GMP and the gac system in transcriptional control of the genes coding for the Pseudomonas putida adhesins LapA and LapF. J Bacteriol 196:1484–1495. doi: 10.1128/JB.01287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansel CM, Francis CA. 2006. Coupled photochemical and enzymatic Mn(II) oxidation pathways of a planktonic Roseobacter-like bacterium. Appl Environ Microbiol 72:3543–3549. doi: 10.1128/AEM.72.5.3543-3549.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diaz JM, Hansel CM, Voelker BM, Mendes CM, Andeer PF, Zhang T. 2013. Widespread production of extracellular superoxide by heterotrophic bacteria. Science 340:1223–1226. doi: 10.1126/science.1237331. [DOI] [PubMed] [Google Scholar]
- 39.Dick GJ, Podell S, Johnson HA, Rivera-Espinoza Y, Bernier-Latmani R, McCarthy JK, Torpey JW, Clement BG, Gaasterland T, Tebo BM. 2008. Genomic insights into Mn(II) oxidation by the marine alphaproteobacterium Aurantimonas sp. strain SI85-9A1. Appl Environ Microbiol 74:2646–2658. doi: 10.1128/AEM.01656-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Francis CA, Casciotti KL, Tebo BM. 2002. Localization of Mn(II)-oxidizing activity and the putative multicopper oxidase, MnxG, to the exosporium of the marine Bacillus sp. strain SG-1. Arch Microbiol 178:450–456. doi: 10.1007/s00203-002-0472-9. [DOI] [PubMed] [Google Scholar]
- 41.De Vrind J, De Groot A, Brouwers GJ, Tommassen J, De Vrind-De Jong E. 2003. Identification of a novel Gsp-related pathway required for secretion of the manganese-oxidizing factor of Pseudomonas putida strain GB-1. Mol Microbiol 47:993–1006. doi: 10.1046/j.1365-2958.2003.03339.x. [DOI] [PubMed] [Google Scholar]
- 42.West SA, Griffin AS, Gardner A, Diggle SP. 2006. Social evolution theory for microorganisms. Nat Rev Microbiol 4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 43.Matilla MA, Ramos JL, Bakker PAHM, Doornbos R, Badri DV, Vivanco JM, Ramos-González MI. 2010. Pseudomonas putida KT2440 causes induced systemic resistance and changes in Arabidopsis root exudation. Environ Microbiol Rep 2:381–388. [DOI] [PubMed] [Google Scholar]
- 44.Matilla MA, Espinosa-Urgel M, Rodríguez-Herva JJ, Ramos JL, Ramos-González MI. 2007. Genomic analysis reveals the major driving forces of bacterial life in the rhizosphere. Genome Biol 8:R179. doi: 10.1186/gb-2007-8-9-r179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corstjens PL, de Vrind JP, Westbroek P, de Vrind-de Jong EW. 1992. Enzymatic iron oxidation by Leptothrix discophora: identification of an iron-oxidizing protein. Appl Environ Microbiol 58:450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 47.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Larsen R, Wilson M, Guss A, Metcalf W. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol 178:193–201. doi: 10.1007/s00203-002-0442-2. [DOI] [PubMed] [Google Scholar]
- 49.West SE, Schweizer HP, Dall C, Sample AK, Runyen-Janecky LJ. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]