ABSTRACT
The microbial ecology of cheese involves a rich and complex interaction between starter lactic acid bacteria and nonstarter lactic acid bacteria (NSLAB), mainly originating from raw milk and/or from the environment, that can contribute to the final characteristics of cheese. The aim of the present research was the exploration of the active microbiota by RNA-based approaches during the manufacturing and ripening of a Grana-like cheese. Reverse transcriptase PCR (RT-PCR)-denaturing gradient gel electrophoresis (DGGE) and RNA-based high-throughput sequencing were applied to profile microbial populations, while the enumeration of active bacteria was carried out by using quantitative PCR (qPCR). Three different cheese productions (named D, E, and F) collected in the same month from the same dairy plant were analyzed. The application of the qPCR protocol revealed the presence of 7 log CFU/ml of bacterial load in raw milk, while, during ripening, active bacterial populations ranged from <4 to 8 log CFU/ml. The natural whey starters used in the three productions showed the same microbiota composition, characterized by the presence of Lactobacillus helveticus and Lactobacillus delbrueckii. Nevertheless, beta-diversity analysis of the 16S rRNA sequencing data and RT-PCR-DGGE showed a clear clustering of the samples according to the three productions, probably driven by the different milks used. Milk samples were found to be characterized by the presence of several contaminants, such as Propionibacterium acnes, Acidovorax, Acinetobacter, Pseudomonas, and NSLAB. The core genera of the starter tended to limit the development of the spoilage bacteria only in two of the three batches. This study underlines the influence of different factors that can affect the final microbiota composition of the artisanal cheese.
IMPORTANCE This study highlights the importance of the quality of the raw milk in the production of a hard cheese. Independent from the use of a starter culture, raw milk with low microbiological quality can negatively affect the populations of lactic acid bacteria and, as a consequence, impact the quality of the final product due to metabolic processes associated with spoilage bacteria.
INTRODUCTION
Grana-like hard cheese is a typical raw cow's milk cheese produced in the Piedmont region in northwest Italy. Curd fermentation is carried out by natural thermophilic whey cultures obtained from the manufacturing of the previous day, after heating at 53 to 55°C, and the ripening is commonly carried out for 12 months.
Natural whey starters (NWS) are used for several protected designation of origin (PDO) cheeses. They are usually dominated by thermophilic lactic acid bacteria (LAB), but subdominant species, including mesophilic LAB, may also occur. NWS have a complex microbial association of various species as well as a large number of biotypes (1).
Temperature is the main factor associated with the variability of the microbiota composition of the NWS. Cooking treatment and whey cooling lead to the selection of a characteristic microbiota consisting of thermophilic, aciduric, and moderately heat-resistant LAB (2). Changes of these variable parameters can affect the development of the final cultures.
Previous studies have described in detail the microbial composition of this kind of cheese and showed the predominance of Lactobacillus helveticus, Lactobacillus delbrueckii subsp. lactis, Lactobacillus fermentum, and Streptococcus thermophilus as the main species isolated and identified (3, 4).
It is well reported that the production is characterized by microbial dynamic changes and that bacteria play a primary role in defining quality (5). Different microbial populations coexist and interact, contributing through their metabolism to the development of taste, aroma, texture, shelf-life, and safety (6). In particular, the microbial ecology of long-ripened cheeses, produced from raw milk and using whey starters, is based on the complex interaction among starter lactic acid bacteria (SLAB) and nonstarter lactic acid bacteria (NSLAB) (7). SLAB give their contributions in the early stages of the cheese making, while NSLAB, which are able to use other carbon sources apart from lactose, become the dominant microbiota of the ripened cheeses (8) and are responsible for the flavor and texture of the cheeses due to their proteolytic and lipolytic activities (7). NSLAB are mainly mesophilic microorganisms originating from raw milk and/or from the environment, and their presence may introduce variability into the ripening process (9). For this reason, the development of beneficial NSLAB coming from the natural whey starter, from the raw milk, or from the manufacturing environment is crucial to minimize microbial variability during the ripening process and obtain the desired organoleptic characteristics of the cheese. However, several contaminants with spoilage potential may also occur, in most cases with negative effects on the quality of the final product (10).
The use of 16S rRNA genes through high-throughput sequencing (HTS) has emerged as a new culture-independent tool and may allow a quantitative investigation of the structure of microbial communities, beside being much more sensitive to detect subdominant populations (11, 12). Several studies based on target amplicon sequencing showed that milk source, processing (raw or pasteurized), and addition of various ingredients affect the composition of the microbiota with an impact on the final attributes of the products (13). However, more information is needed regarding the function and the activity of the microbiota during the process. Few studies based on metatranscriptomic analysis are applied on cheese matrix aiming at exploring the function of the cheese-associated microbiota (14–16).
The aim of this work was to study the microbial dynamics of the active fraction of the microbiota during the manufacturing and ripening of a raw-milk, long-ripened, hard-cooked, Grana-type cheese, with particular emphasis on the contribution of milk and whey starter, by coupling reverse transcriptase PCR (RT-PCR)-denaturing gradient gel electrophoresis (DGGE), quantitative PCR (qPCR), and rRNA pyrosequencing. The molecular target used in this ecology study was rRNA, which has been described as an indicator for metabolically active microbiota, allowing for a greater understanding of microbial community structure and functionality (17, 18).
MATERIALS AND METHODS
Sampling.
Three different cheese productions (D, E, and F) from the same dairy plant, located in the Piedmont region (in northwest Italy), were studied until the 10th month of ripening. The cheese studied was a hard-pressed Grana-type cheese. All of the productions were carried out in the spring season, in three successive weeks. A full description of the samples is reported in Table 1. The samples were whey, raw milk (Frisona cow), milk after the addition of whey, and curd before and after cutting, after pressing, after storage room at 46°C, after salting, after thermostatic room at 25°C, and from the 1st to the 10th ripening month. During the first 4 months of ripening, the temperature ranged from 16 to 17°C with a relative humidity of 80%, while, in the following months, the temperature reached 20°C with a humidity of 82%. Samples were transported under refrigeration to the laboratory and were subjected to analysis within 2 h of collection. Sampling on cheese loafs was carried out using a sterile punch, which was inserted perpendicular to the center of the cheese and then rotated 360° for digging samples.
TABLE 1.
Results of the total active bacterial counts obtained by qPCR
| Sample | Sample description | Log CFU/ml or log CFU/g for: |
||
|---|---|---|---|---|
| Production D | Production E | Production F | ||
| 1 | Whey starter | 9.82 | 6.67 | 7.9 |
| 2 | Raw milk | 7.52 | 7.59 | *a |
| 3 | Milk + whey | 9.76 | 7.2 | * |
| 4 | Curd after cutting | 8.22 | 7.9 | * |
| 5 | Curd after heating | 8.33 | 7.47 | * |
| 6 | Curd after pressing | 8.43 | 6.91 | * |
| 7 | Curd after storage room | 7.91 | 8.16 | * |
| 8 | Cheese after salting | 8.05 | 7.8 | * |
| 9 | Cheese after thermostatic room | 8.35 | 7.21 | * |
| 10 | 1st ripening month | 7.77 | 7.94 | * |
| 11 | 2nd ripening month | 7.23 | 8.17 | * |
| 12 | 3rd ripening month | 8.15 | 7.71 | * |
| 13 | 4th ripening month | 7.58 | 7.67 | * |
| 14 | 5th ripening month | 8.02 | 6.23 | * |
| 15 | 6th ripening month | 7.99 | 7.98 | * |
| 16 | 7th ripening month | 7.01 | 6.3 | * |
| 17 | 8th ripening month | 7 | 6.73 | * |
| 18 | 9th ripening month | 6.5 | 6.97 | * |
| 19 | 10th ripening month | 5.5 | 6.5 | * |
Result below quantification limit indicated by *.
RT-PCR-DGGE.
Sample preparation and RNA extraction were performed according to the protocol reported by Rantsiou et al. (19) by using the Master-Pure complete DNA and RNA purification kit (Epicentre, Madison, WI, USA) following the manufacturer's instructions. Resuspended RNA was treated with Turbo DNase (Ambion, Italy) in order to eliminate the DNA. Complete DNA digestion was confirmed using 1 μl of extracted RNA in PCR with primers 518r and 338f (20); when a PCR product was obtained, the DNase treatment was repeated.
Reverse transcription reactions were performed using Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega, Milan, Italy). A total of 200 ng of RNA was mixed with 1 μl of 100 μM 518r primer, and the reaction was carried out at 42°C for 1 h. Then, 1 μl of the obtained cDNA was used as the template for the amplification of the bacterial V3 region of the 16S rRNA gene, and the PCR products were analyzed by means of DGGE, as described in reference 21.
A database of fingerprints was created using the Bionumerics software, version 4.6 (Applied Maths, Sint Marten Latem, Belgium). A combined data matrix that included all of the fingerprints from RNA was obtained, and dendrograms of similarity were retrieved using the Dice coefficient and the unweighted pair group method with the arithmetic average clustering algorithm (20). The similarity distance matrix generated through the Bionumerics software was used to build a partial least-squares discriminant analysis (PLS-DA) utilizing the mixOmics R package (www.r-project.org).
Selected DGGE bands were extracted from the gels using sterile pipette tips, transferred into 50 μl of sterile water, and incubated overnight at 4°C. Then, 2 μl of the eluted bands was reamplified using the conditions described above and checked by means of DGGE. The PCR products that gave a single band, comigrating with the RNA control, were then amplified with the same primers without a guanine-cytosine (GC) clamp and sequenced by MWG Biotech (Ebersberg, Germany). The Sanger sequences were aligned in GenBank using the BLAST search program (http://www.ncbi.nlm.nih.gov/blast/).
Construction of LAB standard curve for bacterial enumeration.
In order to obtain an enumeration of the active bacterial cells, different standard curves were constructed. Ten milliliters of milk, whey, and 10 g of curd and grated ripened cheese were contaminated with 10 ml of 10-fold serial dilutions of an overnight-pure-culture mix of strains of Lactococcus lactis subsp. lactis, Lactobacillus plantarum, Lactobacillus pentosus, Enterococcus faecium, and Lb. helveticus (from the culture collection of the Department of Agricultural, Forest and Food Science, University of Turin), previously identified by using molecular methods described by Bautista-Gallego et al. (22) and supplemented with 90 ml of sterile Ringer's solution (Oxoid, Milan, Italy). The samples were homogenized in a Stomacher (Interscience Rockland, MA, USA) for 1 min, and 1 ml of each mixture was subjected to RNA extraction as described above. Reverse transcription was performed using 9 μl of RNA, and the resulting cDNA sample was submitted to qPCR. Standard curves were constructed by plotting the threshold cycle (CT) values against log CFU/g or log CFU/ml on MRS agar. MRS agar plates were incubated at 37°C for 48 h in microaerobic conditions. Correlation coefficients (R2) and efficiency of amplification were calculated as previously described (23).
Detection of total bacteria by qPCR.
Quantitative PCR targeting the V3 region of the 16S rRNA gene was used for the active bacterium quantification. Amplifications were performed in a final volume of 25 μl in the Chromo 4 real-time PCR detection system (Bio-Rad, Milan, Italy) with the use of SSo advanced Sybr green supermix (Bio-Rad, Italy) and 1 μl of cDNA was amplified with 338f and 518r primers (21) at a final concentration of 400 nM. Samples were amplified in triplicate using the following conditions: initial denaturation at 95°C for 5 min and 40 cycles of 95°C for 15 s, then of 60°C for 30 s.
RNA analysis by pyrosequencing.
Samples were carefully selected from the three productions in order to describe the microbial diversity during the whole manufacturing process (see Table 3). cDNA obtained as explained above was used to study the microbial diversity of the active populations by pyrosequencing of the amplified V1 to V3 region of the 16S rRNA gene by using primers and the PCR condition previously reported (24). PCR products were purified twice by Agencourt AMPure kit (Beckman Coulter, Milan, Italy) and quantified using QuantiFluor (Promega, Milan, Italy), and an equimolar pool was obtained prior to further processing. The amplicon pool was used for pyrosequencing on a GS junior platform (454 Life Sciences, Roche, Monza, Italy) according to the manufacturer's instructions by using titanium chemistry.
TABLE 3.
Numbers of sequences analyzed, observed diversity, and estimated sample coverage for 16S rRNA amplicons from selected samples from the three productions
| Sample no. | Sample description | Batch |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D |
F |
E |
||||||||||||||
| Reads | OTUs | chao1 | Shannon | ESCa | Reads | OTUs | chao1 | Shannon | ESC | Reads | OTUs | chao1 | Shannon | ESC | ||
| 1 | Whey starter | 4,570 | 19 | 19.17 | 1.31 | 1.00 | 3,081 | 26 | 32.00 | 1.55 | 1.00 | 2,998 | 21 | 35.00 | 1.28 | 1.00 |
| 2 | Raw milk | 2,603 | 53 | 72.46 | 1.46 | 0.99 | 7,951 | 103 | 104.50 | 4.69 | 1.00 | 6,337 | 109 | 126.65 | 3.72 | 1.00 |
| 3 | Milk + whey | 3,375 | 23 | 35.00 | 1.33 | 1.00 | 4,286 | 97 | 115.33 | 4.20 | 1.00 | 1,843 | 19 | 20.50 | 1.40 | 1.00 |
| 4 | Curd after cutting | 3,272 | 21 | 49.00 | 1.60 | 1.00 | 5,761 | 106 | 109.60 | 3.63 | 1.00 | 3,061 | 85 | 114.00 | 3.77 | 0.99 |
| 5 | Curd after heating | 1,990 | 19 | 20.50 | 1.79 | 1.00 | 3,115 | 107 | 111.50 | 4.30 | 1.00 | 3,147 | 29 | 51.00 | 1.57 | 1.00 |
| 6 | Curd after pressing | 3,359 | 17 | 20.75 | 1.28 | 1.00 | 3,785 | 70 | 78.25 | 2.43 | 1.00 | 2,744 | 21 | 30.33 | 1.34 | 1.00 |
| 7 | Curd after storage room | 3,657 | 29 | 35.88 | 1.32 | 1.00 | 3,750 | 151 | 166.11 | 5.95 | 1.00 | 3,191 | 22 | 23.20 | 1.85 | 1.00 |
| 8 | Cheese after salting | 4,348 | 49 | 73.00 | 2.22 | 1.00 | 3,661 | 65 | 72.56 | 1.89 | 1.00 | 8,116 | 52 | 71.43 | 2.29 | 1.00 |
| 11 | 2nd ripening month | 3,111 | 137 | 151.50 | 4.13 | 0.99 | 9,489 | 102 | 108.50 | 2.54 | 1.00 | 2,197 | 35 | 62.20 | 1.92 | 0.99 |
| 13 | 4th ripening month | 4,602 | 108 | 129.12 | 3.64 | 0.99 | 4,722 | 99 | 106.50 | 3.91 | 1.00 | 2,828 | 49 | 119.00 | 3.22 | 0.99 |
| 15 | 6th ripening month | 3,883 | 48 | 58.11 | 2.39 | 1.00 | 3,858 | 79 | 85.11 | 3.76 | 1.00 | 2,639 | 36 | 40.00 | 2.48 | 1.00 |
| 17 | 8th ripening month | 3,906 | 188 | 201.20 | 4.52 | 0.99 | 5,334 | 41 | 87.00 | 0.26 | 1.00 | 2,495 | 34 | 34.43 | 2.76 | 1.00 |
| 19 | 10th ripening month | 3,723 | 140 | 159.03 | 4.03 | 0.99 | 4,891 | 147 | 156.00 | 4.05 | 1.00 | 2,582 | 62 | 72.00 | 2.95 | 0.99 |
ESC, estimated sample coverage.
Bioinformatics analysis.
16S rRNA data were analyzed by using QIIME 1.9.0 software (25) and a previously described pipeline (26). Operational taxonomic units (OTUs) were picked at 99% similarity by the UCLUST clustering methods (27), and representative sequences of each cluster were used to assign taxonomy using the Greengenes 16S rRNA gene database version 2013 by the RDP classifier (28). When the assigned taxonomy was only at the genus level, representative sequences belonging to Lactobacillaceae were checked using the BLAST (BLASTN) search program (http://www.ncbi.nlm.nih.gov/blast/) to get the species level as best hit. Statistics and plotting were carried out in the R environment (www.r-project.org). Alpha-diversity indices were calculated by using the diversity function of the vegan package (29). Weighted UniFrac distance matrices obtained through QIIME were imported in R to obtain principal coordinates analysis (PCoA) plots. OTU tables filtered at 0.5% abundance in at least two samples were used for cooccurrence/coexclusion analysis, carried out by using the psych package of R (www.r-project.org). The correlation matrix was plotted by using the corrplot package of R (30).
Nucleotide sequence accession number.
All of the sequencing data were deposited at the Sequence Read Archive of the National Center for Biotechnology Information (SRP044294).
RESULTS
Active bacteria during the production and ripening of the cheeses.
Different standard curves were built considering the evolution of the samples during the cheese manufacturing (milk, whey, curd, and ripened cheese) in order to obtain a direct quantification of viable bacteria in the samples. For all of the samples, the limit of quantification was 4 log CFU/g or log CFU/ml. The efficiencies were different based on the matrix, and the R2 values were always >0.922 (data not shown). The results of the quantification, expressed as log CFU/ml or log CFU/g, are reported in Table 1. Milk D and E showed microbial loads around 7 log CFU/ml, while 7 to 8 log CFU/g were detected in the curds. A slight decrease was observed during the ripening, reaching values that ranged from 6 to 7 log CFU/g, underlining a large amount of active bacteria also at the 10th month of ripening for batches D and E. The application of qPCR highlighted lower counts in most of the samples from production F, where it was not possible to reach the quantification, since the CT values obtained were out of the linearity range (value < 4 log CFU/ml or log CFU/g).
RT-PCR-DGGE analysis.
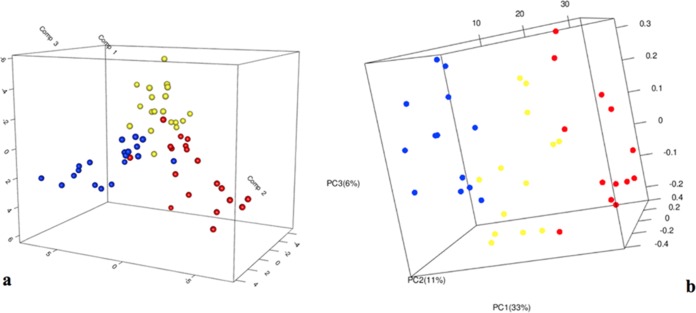
Nineteen samples, from raw milk to cheese ripened for 10 months, for each of the three different productions were analyzed by DGGE. The PLS-DA regression, built using the production batch as discriminatory class (Fig. 1A), showed a certain gradient of separation between the three series of samples. The results of the identification of selected bands are reported in Table 2. NWS used in the three productions showed a similar profile characterized by Lb. helveticus and Lactobacillus delbrueckii subsp. bulgaricus, while Streptococcus thermophilus was detected only in production F. Milks used for productions F and D showed the presence of bands identified as Lb. helveticus, Lb. delbrueckii subsp. bulgaricus, and Lactobacillus acidophilus, while milk from production E was characterized by Acinetobacter baumannii, Acinetobacter haemolyticus, Pseudomonas sp., and Streptococcus sanguinis. Regarding the common species shared among the three batches, Lb. acidophilus characterized the ripening time; S. thermophilus was found during the manufacturing and the early-stage ripening in production E and randomly in production F, while it became persistent during ripening in production D. Lactococcus lactis was detected only in few samples during the ripening time, and only in batches D and E. Regarding the contaminant species, Bacillus subtilis was detected mainly in the F samples, while Acinetobacter baumannii, present randomly in the milk in productions D and E, was not found in F samples. Propionibacterium sp. was randomly detected in the three productions (Table 2).
FIG 1.
(a) Partial least-squares discriminant analysis model built on the similarity distance matrix based on RT-PCR-DGGE fingerprint profiles. (b) Principal coordinates analysis of weighted UniFrac distances for 16S rRNA (cDNA) gene sequencing data. Samples are color coded as a function of the batch: D, blue; F, red; and E, yellow.
TABLE 2.
Results of the identification of selected RT-PCR-DGGE band sequencing
Species identified from sequencing of bands in the profiles of each sample. The presence of the black box indicates the presence of the bands in the DGGE profiles. The sequences obtained were aligned with those in GenBank with BLAST program.
Sample descriptions are reported in Table 1.
16S rRNA gene pyrosequencing.
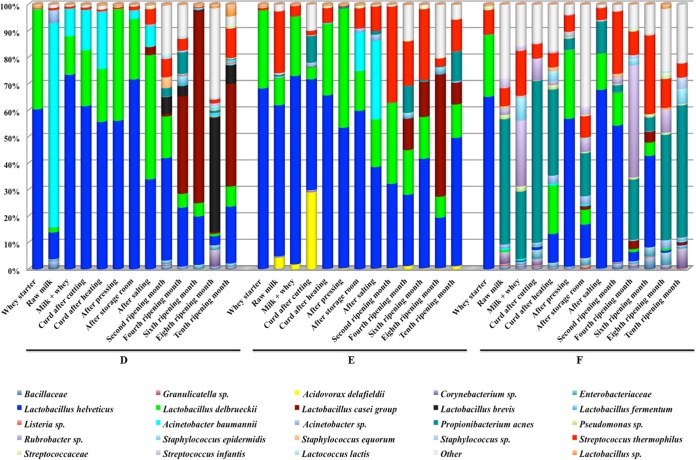
A total of 198,276 raw reads were obtained after the 454 processing; 148,944 reads passed the filters applied through QIIME, with an average value of 3,819 reads/sample and an average length of 469 bp. The rarefaction analysis and the estimated sample coverage (Table 3) indicated that there was satisfactory coverage for all the samples (estimated sample coverage, >99%). Whey starters used in the three manufactures showed similar qualitative and quantitative microbiota compositions, characterized by almost the same relative abundance of Lb. helveticus and Lb. delbrueckii (Fig. 2).
FIG 2.
Abundance of the major taxonomic groups detected by pyrosequencing. Only OTUs with an incidence above 0.5% in at least 2 samples are shown. Samples are grouped according to batch.
Alpha-diversity indices (Table 3) showed that raw milk samples from batches F and E had generally higher levels of complexity than samples from batch D. On the contrary, cheese from batch D showed a higher level of complexity during the ripening.
In Fig. 2, only OTUs with a relative abundance of 0.5% in at least two samples are shown. The milk sample from production D was characterized by Lb. helveticus (10%) and by the predominance of Acinetobacter baumannii (76%), which survived throughout the ripening and appeared in most of the samples of the same batch. Lb. helveticus (55%) was the dominant species in milk E, followed by S. thermophilus (22%) and Lb. delbrueckii (10%). Propionibacterium acnes (38%) characterized milk from batch F, while S. thermophilus and genera belonging to Acinetobacter and Streptococcaceae were also present as minor OTUs. The main differences between the three cheese batches were the presence of several contaminant OTUs, such as P. acnes, Acidovorax, Acinetobacter, and Pseudomonas. Cheese in production D was characterized by the presence of Lb. casei group (37%), Lb. brevis (6.8%), Lb. fermentum (0.6%), and S. thermophilus (10.8%), while cheese from batch F showed low levels of NSLAB (<1%). Moreover, P. acnes and Staphylococcus were present at 42% and 5% of abundance, respectively. Interestingly, starter species were absent in cheeses from production F during ripening. S. thermophilus and P. acnes were found to be significantly more abundant (G test, P < 0.001) in samples from batch F. Acinetobacter baumannii, Lb. delbrueckii, and Lb. casei group discriminated samples D, while Lb. helveticus characterized batch E (P < 0.001).
Nevertheless, OTUs characteristic of each production drove the sample clustering according to the batch (Fig. 1B), and sample differentiation according to the batch was supported by Adonis and Anosim statistical tests (P < 0.001).
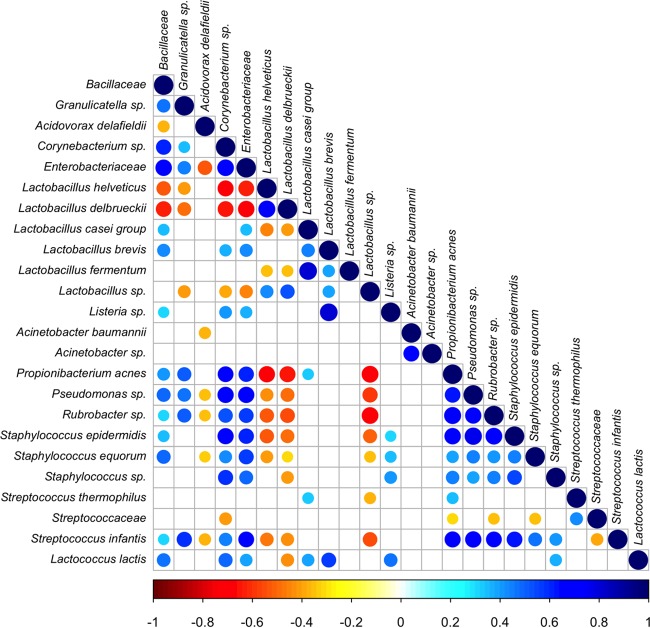
The OTU cooccurrence/exclusion pattern is shown in Fig. 3, where only significant correlations are reported (at a false discovery rate [FDR] of <0.05). Lb. delbrueckii, Lb. helveticus, and Lb. casei group showed the highest number of negative correlations, including a strong exclusion with Pseudomonas and other contaminants.
FIG 3.
Significant cooccurrence and coexclusion relationships between bacterial OTUs. The figure presents a Spearman's rank correlation matrix of OTUs with >0.5% abundance in at least 2 samples. Strong correlations are indicated by large circles, whereas weak correlations are indicated by small circles. The color of the scale bar denotes the nature of the correlation, with 1 indicating a perfect positive correlation (dark blue) and −1 indicating a perfect negative correlation (dark red). Only significant correlations (FDR < 0.05) are shown.
DISCUSSION
Insights into the microbial ecology and active bacterial communities during the manufacturing and ripening of a Piedmont hard cheese were provided in this study. The experimental approach used exploited different culture-independent methods based on RNA. Culture-independent methods have rapidly been recognized as a valuable tool for the study of biodiversity and the identification of microbial species in food samples (31). The results of qPCR were able to give more precise information about the total active bacteria counts and showed higher values than those observed by culture-dependent methods on selective media for enumeration of LAB (22), emphasizing the presence of other, non-LAB populations.
By using RT-PCR-DGGE and pyrosequencing, Lb. rhamnosus (belonging to Lb. casei group) and Lb. helveticus were the dominant taxa during the whole manufacturing and ripening process. On the contrary, Bautista-Gallego et al. (22) showed the predominance of Lb. helveticus only in the early stages of cheese production, while Lb. rhamnosus dominated in the following months. As reported from other studies (32), Lb. helveticus can contribute to the ripening process due to its autolytic properties, which could increase the proteolysis in aged cheese and, consequently, the flavor formation. Moreover, members of the Lb. casei group are able to utilize products of Lb. helveticus lysis as a unique energy source (33). Lc. lactis was also detected. This evidence is in agreement with recent data concerning the viability of Lc. lactis throughout the manufacturing and ripening of cheese (34, 35).
The microbial cooccurrence/exclusion patterns suggested that the presence of Lb. helveticus and Lb. delbrueckii coexcluded the presence of other populations. Those results confirmed that the core microbial genera of the starter tend to dominate the cheese microbiota and to limit the development of spoilage bacteria and contaminants (36). On the other hand, the 16S data revealed that P. acnes, present as main contaminant in milk from production F, showed a cooccurrence pattern with other contaminant taxa. Moreover, it remained metabolically active until the end of the ripening, confirming the impact of raw milk quality in the development of the microbiota during ripening. As previously demonstrated, Propionibacterium is normally isolated from milk and cheese and may contribute to the formation of distinctive flavors arising from its metabolism (37, 38).
PCoA and PLS-DA analyses clearly showed a separation of the samples from the three production batches, probably driven by the different milk samples used. The 16S data highlighted a higher level of complexity in the raw milk used in production F, with the presence of minor OTUs, such as Corynebacterium sp., A. baumannii, P. acnes, Pseudomonas sp., and Streptococcaceae, besides Lb. helveticus and S. thermophilus. The contaminants observed in the raw milk samples were previously detected in milk and cheese and were suggested to cause spoilage (39–41). The differences in microbiota composition of the raw milk used for the three productions can be attributed to the hygiene practices in the farm, including the teat surface hygiene, air, dust, stable conditions, and milking parlor environment, which are responsible for milk contamination (7). It is well demonstrated that the microbiota involved in cheese production are commonly found on the processing surfaces (36), highlighting the importance of appropriate hygienic measures to avoid contamination from the production environment. They are also critical for minimizing contamination and preventing the growth of spoilage microorganism. In addition, milk supplied to the farm is not always immediately processed. In particular, the storage conditions and storage time before the processing can favor the growth and the development of minor microbiota which can become dominant, leading to unacceptable quality of the final dairy product (42, 43).
This study highlights and confirms that the microbial quality of raw milk has an important effect on the development of the contaminant microbiota during ripening of a Grana-like cheese. The results showed that the core genera of the starter tended to limit the development of the spoilage bacteria. On the other hand, in the case of batch F, the whey starter was not competitive enough against the contaminant species, which impacted the microbial composition of the final products. For this production, qPCR also revealed a small amount of active bacterial cells in the milk and in the manufacturing and ripening processes.
This study underlines the influence of different factors that can affect the final microbiota composition. Further studies based on metatranscriptomics and metabolomics are needed for verifying the effect of the subdominant microbiota of the raw milk on the organoleptic and sensorial properties of the cheeses.
Funding Statement
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
REFERENCES
- 1.Giraffa G, Mucchetti G, Addeo F, Neviani E. 1997. Evolution of lactic acid microflora during Grana cheesemaking and ripening. Microbiol Aliment Nutr 15:115–122. [Google Scholar]
- 2.Neviani E, Bottari B, Lazzi C, Gatti M. 2013. New developments in the study of the microbiota of raw-milk, long-ripened cheeses by molecular methods: the case of Grana Padano and Parmigiano Reggiano. Front Microbiol 4:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lazzi C, Rossetti L, Zago M, Neviani E, Giraffa G. 2004. Evaluation of bacterial communities belonging to natural whey starters for Grana Padano cheese by length heterogeneity PCR. J Appl Microbiol 96:481–490. doi: 10.1111/j.1365-2672.2004.02180.x. [DOI] [PubMed] [Google Scholar]
- 4.Rossi F, Gatto V, Sabattini G, Torriani S. 2012. An assessment of factors characterizing the microbiology of Grana Trentino cheese, a Grana-type cheese. Int J Dairy Technol 65:401–409. doi: 10.1111/j.1471-0307.2012.00844.x. [DOI] [Google Scholar]
- 5.Montel M-C, Buchin S, Mallet A, Delbes-Paus C, Vuitton DA, Desmasures N, Berthier F. 2014. Traditional cheeses: rich and diverse microbiota with associated benefits. Int J Food Microbiol 177:136–154. doi: 10.1016/j.ijfoodmicro.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 6.Holzapfel WH. 2002. Appropriate starter culture technologies for small-scale fermentation in developing countries. Int J Food Microbiol 75:197–212. doi: 10.1016/S0168-1605(01)00707-3. [DOI] [PubMed] [Google Scholar]
- 7.Gatti M, Bottari B, Lazzi C, Neviani E, Mucchetti G. 2014. Invited review: microbial evolution in raw-milk, long-ripened cheeses produced using undefined natural whey starters. J Dairy Sci 97:573–591. doi: 10.3168/jds.2013-7187. [DOI] [PubMed] [Google Scholar]
- 8.Quigley L, O'Sullivan O, Stanton C, Beresford TP, Ross RP, Fitzgerald GF, Cotter PD. 2013. The complex microbiota of raw milk. FEMS Microbiol Rev 37:664–698. doi: 10.1111/1574-6976.12030. [DOI] [PubMed] [Google Scholar]
- 9.Settanni L, Moschetti G. 2010. Nonstarter lactic acid bacteria used to improve cheese quality and provide health benefits. Food Microbiol 27:691–697. doi: 10.1016/j.fm.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 10.Coppola S, Blaiotta G, Ercolini D. 2007. Dairy products, p 31–90. In Cocolin L, Ercolini D (ed), Molecular techniques in the microbial ecology of fermented foods. Springer, New York, NY. [Google Scholar]
- 11.Cocolin L, Ercolini D. 2015. Zooming into food-associated microbial consortia: a cultural evolution. Curr Opin Food Sci 2:43–50. doi: 10.1016/j.cofs.2015.01.003. [DOI] [Google Scholar]
- 12.Ercolini D. 2013. High-throughput sequencing and metagenomics: moving forward in the culture-independent analysis of food microbial ecology. Appl Environ Microbiol 79:3148–3155. doi: 10.1128/AEM.00256-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke SF, Murphy EF, O'Sullivan O, Ross RP, O'Toole PW, Shanahan F, Cotter PD. 2013. Targeting the microbiota to address diet-induced obesity: a time dependent challenge. PLoS One 8:e65790. doi: 10.1371/journal.pone.0065790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lessard M-H, Viel C, Boyle B, St-Gelais D, Labrie S. 2014. Metatranscriptome analysis of fungal strains Penicillium camemberti and Geotrichum candidum reveal cheese matrix breakdown and potential development of sensory properties of ripened Camembert-type cheese. BMC Genomics 15:235. doi: 10.1186/1471-2164-15-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dugat-Bony E, Straub C, Teissandier A, Onésime D, Loux V, Monnet C, Irlinger F, Landaud S, Leclercq-Perlat M-N, Bento P, Fraud S, Gibrat J-F, Aubert J, Fer F, Guédon E, Pons N, Kennedy S, Beckerich J-M, Swennen D, Bonnarme P. 2015. Overview of a surface-ripened cheese community functioning by meta-omics analyses. PLoS One 10:e0124360. doi: 10.1371/journal.pone.0124360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Filippis F, Genovese A, Ferranti P, Gilbert JA, Ercolini D. 2016. Metatranscriptomics reveals temperature-driven functional changes in microbiome impacting cheese maturation rate. Sci Rep 6:21871. doi: 10.1038/srep21871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dolci P, Zenato S, Pramotton R, Barmaz A, Alessandria V, Rantsiou K, Cocolin L. 2013. Cheese surface microbiota complexity: RT-PCR-DGGE, a tool for a detailed picture? Int J Food Microbiol 162:8–12. doi: 10.1016/j.ijfoodmicro.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 18.De Angelis M, Montemurno E, Vannini L, Cosola C, Cavallo N, Gozzi G, Maranzano V, Di Cagno R, Gobbetti M, Gesualdo L. 2015. Effect of whole-grain barley on the human fecal microbiota and metabolome. Appl Environ Microbiol 81:7945–7956. doi: 10.1128/AEM.02507-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rantsiou K, Alessandria V, Urso R, Dolci P, Cocolin L. 2008. Detection, quantification and vitality of Listeria monocytogenes in food as determined by quantitative PCR. Int J Food Microbiol 121:99–105. doi: 10.1016/j.ijfoodmicro.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Vauterin L, Vauterin P. 1992. Computer-aided objective comparison of electrophoresis patterns for grouping and identification of microorganisms. Eur Microbiol 1:37–42. [Google Scholar]
- 21.Alessandria V, Dolci P, Rantsiou K, Pattono D, Dalmasso A, Civera T, Cocolin L. 2010. Microbiota of the Planalto de Bolona: an artisanal cheese produced in uncommon environmental conditions in the Cape Verde Islands. World J Microbiol Biotechnol 26:2211–2221. doi: 10.1007/s11274-010-0406-7. [DOI] [Google Scholar]
- 22.Bautista-Gallego J, Alessandria V, Fontana M, Bisotti S, Taricco S, Dolci P, Cocolin L, Rantsiou K. 2014. Diversity and functional characterization of Lactobacillus spp. isolated throughout the ripening of a hard cheese. Int J Food Microbiol 181:60–66. doi: 10.1016/j.ijfoodmicro.2014.04.020. [DOI] [PubMed] [Google Scholar]
- 23.Higuchi R, Fockler C, Dollinger G, Watson R. 1993. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Nat Biotechnol 11:1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- 24.Ercolini D, De Filippis F, La Storia A, Iacono M. 2012. Remake by high-throughput sequencing of the microbiota involved in the production of water buffalo mozzarella cheese. Appl Environ Microbiol 78:8142–8145. doi: 10.1128/AEM.02218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greppi A, Ferrocino I, La Storia A, Rantsiou K, Ercolini D, Cocolin L. 2015. Monitoring of the microbiota of fermented sausages by culture independent rRNA-based approaches. Int J Food Microbiol 212:67–75. doi: 10.1016/j.ijfoodmicro.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 27.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 28.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dixon P. 2003. Vegan, a package of R functions for community ecology. J Veg Sci 14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 30.Friendly M. 2002. Corrgrams: exploratory displays for correlation matrices. Am Stat 56:316–324. doi: 10.1198/000313002533. [DOI] [Google Scholar]
- 31.Ercolini D, Moschetti G, Blaiotta G, Coppola S. 2001. The potential of a polyphasic PCR-DGGE approach in evaluating microbial diversity of natural whey cultures for water-buffalo mozzarella cheese production: bias of culture-dependent and culture-independent analyses. Syst Appl Microbiol 24:610–617. doi: 10.1078/0723-2020-00076. [DOI] [PubMed] [Google Scholar]
- 32.Zambonelli C, Chiavari C, Benevelli M, Coloretti F. 2002. Effects of lactic acid bacteria autolysis on sensorial characteristics of fermented foods. Food Technol Biotechnol 40:347–351. [Google Scholar]
- 33.Sgarbi E, Bottari B, Gatti M, Neviani E. 2014. Investigation of the ability of dairy nonstarter lactic acid bacteria to grow using cell lysates of other lactic acid bacteria as the exclusive source of nutrients. Int J Dairy Technol 67:342–347. doi: 10.1111/1471-0307.12132. [DOI] [Google Scholar]
- 34.Dolci P, De Filippis F, La Storia A, Ercolini D, Cocolin L. 2014. rRNA-based monitoring of the microbiota involved in fontina PDO cheese production in relation to different stages of cow lactation. Int J Food Microbiol 185:127–135. doi: 10.1016/j.ijfoodmicro.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 35.Ruggirello M, Dolci P, Cocolin L. 2014. Detection and viability of Lactococcus lactis throughout cheese ripening. PLoS One 9:e114280. doi: 10.1371/journal.pone.0114280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stellato G, De Filippis F, La Storia A, Ercolini D. 2015. Coexistence of lactic acid bacteria and potential spoilage microbiota in a dairy-processing environment. Appl Environ Microbiol 81:7893–7904. doi: 10.1128/AEM.02294-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meile L, Le Blay G, Thierry A. 2008. Safety assessment of dairy microorganisms: Propionibacterium and Bifidobacterium. Int J Food Microbiol 126:316–320. doi: 10.1016/j.ijfoodmicro.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 38.Thierry A, Maillard M-B, Bonnarme P, Roussel E. 2005. The addition of Propionibacterium freudenreichii to raclette cheese induces biochemical changes and enhances flavor development. J Agric Food Chem 53:4157–4165. doi: 10.1021/jf0481195. [DOI] [PubMed] [Google Scholar]
- 39.Ercolini D, Russo F, Ferrocino I, Villani F. 2009. Molecular identification of mesophilic and psychrotrophic bacteria from raw cow's milk. Food Microbiol 26:228–231. doi: 10.1016/j.fm.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 40.Giannino ML, Aliprandi M, Feligini M, Vanoni L, Brasca M, Fracchetti F. 2009. A DNA array based assay for the characterization of microbial community in raw milk. J Microbiol Methods 78:181–188. doi: 10.1016/j.mimet.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 41.Masoud W, Takamiya M, Vogensen FK, Lillevang S, Al-Soud WA, Sørensen SJ, Jakobsen M. 2011. Characterization of bacterial populations in Danish raw milk cheeses made with different starter cultures by denaturating gradient gel electrophoresis and pyrosequencing. Int Dairy J 21:142–148. doi: 10.1016/j.idairyj.2010.10.007. [DOI] [Google Scholar]
- 42.O'Sullivan DJ, Cotter PD, O'Sullivan O, Giblin L, McSweeney PLH, Sheehan JJ. 2015. Temporal and spatial differences in microbial composition during the manufacture of a continental-type cheese. Appl Environ Microbiol 81:2525–2533. doi: 10.1128/AEM.04054-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gargouri A, Hamed H, Elfeki A. 2013. Analysis of raw milk quality at reception and during cold storage: combined effects of somatic cell counts and psychrotrophic bacteria on lipolysis. J Food Sci 78:M1405–M1411. doi: 10.1111/1750-3841.12188. [DOI] [PubMed] [Google Scholar]