ABSTRACT
Bacteria capable of reduction of nitrous oxide (N2O) to N2 separate into clade I and clade II organisms on the basis of nos operon structures and nosZ sequence features. To explore the possible ecological consequences of distinct nos clusters, the growth of bacterial isolates with either clade I (Pseudomonas stutzeri strain DCP-Ps1, Shewanella loihica strain PV-4) or clade II (Dechloromonas aromatica strain RCB, Anaeromyxobacter dehalogenans strain 2CP-C) nosZ with N2O was examined. Growth curves did not reveal trends distinguishing the clade I and clade II organisms tested; however, the growth yields of clade II organisms exceeded those of clade I organisms by 1.5- to 1.8-fold. Further, whole-cell half-saturation constants (Kss) for N2O distinguished clade I from clade II organisms. The apparent Ks values of 0.324 ± 0.078 μM for D. aromatica and 1.34 ± 0.35 μM for A. dehalogenans were significantly lower than the values measured for P. stutzeri (35.5 ± 9.3 μM) and S. loihica (7.07 ± 1.13 μM). Genome sequencing demonstrated that Dechloromonas denitrificans possessed a clade II nosZ gene, and a measured Ks of 1.01 ± 0.18 μM for N2O was consistent with the values determined for the other clade II organisms tested. These observations provide a plausible mechanistic basis for why the relative activity of bacteria with clade I nos operons compared to that of bacteria with clade II nos operons may control N2O emissions and determine a soil's N2O sink capacity.
IMPORTANCE Anthropogenic activities, in particular fertilizer application for agricultural production, increase N2O emissions to the atmosphere. N2O is a strong greenhouse gas with ozone destruction potential, and there is concern that nitrogen may become the major driver of climate change. Microbial N2O reductase (NosZ) catalyzes N2O reduction to environmentally benign dinitrogen gas and represents the major N2O sink process. The observation that bacterial groups with clade I nosZ versus those with clade II nosZ exhibit distinct affinities to N2O has implications for N2O flux models, and these distinct characteristics may provide opportunities to curb N2O emissions from relevant soil ecosystems.
INTRODUCTION
Nitrous oxide (N2O) is a potent greenhouse gas with a strong potential to drive climate change (1, 2) and will continue to be the largest contributor to ozone depletion in the stratosphere (2, 3). Anthropogenic activities, predominantly, fertilizer application in agricultural production, have contributed to a steady increase in atmospheric N2O concentrations, and a continued upward trend is expected (4–6). Particularly troublesome are the findings of a recent study that concluded that without solutions for the N2O problem, carbon dioxide (CO2) emission reductions even greater than those already proposed will be required to avoid climate change (7). Due to its environmental impact, the pathways leading to the generation and consumption of N2O have received heightened interest.
In the environment, N2O is predominantly formed as an intermediate of denitrification and a nitrification by-product (8). Denitrification is the stepwise reduction of NO3−/NO2− to gaseous products (i.e., N2O, N2), with each step being mediated by distinct enzyme systems (9). A kinetic imbalance in the rates of reactions producing and consuming N2O during denitrification leads to the release of N2O to the atmosphere (8, 10). In nitrification, N2O is generated by nitrifier denitrification and as a by-product of ammonia oxidation (8, 11, 12). A recent report indicated that nitrifiers, rather than denitrifiers, may be the primary source of N2O in agricultural soils (12). Other processes contributing to N2O formation include respiratory ammonification (also known as dissimilatory nitrate/nitrite reduction to ammonium [DNRA]) and chemodenitrification (i.e., the abiotic reaction of NO2− with ferrous iron) (13, 14). In contrast to the diverse pathways of N2O generation, the only known major biological pathway for the removal of N2O is by reduction to N2, catalyzed by the enzyme nitrous oxide reductase (NosZ) (8, 15–17). Recent efforts have identified a new cluster of atypical nosZ genes, now designated clade II nosZ genes (15, 16). Interestingly, many clade II nosZ genes were found on genomes lacking the nirS and/or nirK gene, suggesting that nondenitrifiers contribute to N2O reduction (18). Growth experiments with Anaeromyxobacter dehalogenans, an organism carrying the clade II nosZ, demonstrated that clade II NosZ functions as a respiratory oxidoreductase (15). PCR and metagenome analyses demonstrated the clade II nosZ gene abundance in different soils and suggested a correlation between the abundance and diversity of microorganisms carrying the clade II nosZ and a soil's N2O sink capacity (16, 18, 19).
The clade II NosZ enzymes have <50% amino acid sequence similarity with the clade I NosZ enzymes, but both NosZ enzymes catalyze the same reaction. While clade I NosZ enzymes have been studied in detail (17, 20, 21), only the clade II NosZ enzyme of Wolinella succinogenes has been characterized to some extent biochemically (22, 23). The detailed kinetic parameters of the clade I and II NosZ enzymes have not been determined in vivo. Thus, the kinetic properties of clade I NosZ and those of clade II NosZ may differ, and these different kinetic properties may reflect distinguishing phenotypic characteristics of the N2O-consuming hosts. To explore whether organisms carrying clade I nosZ and organisms carrying clade II nosZ exhibit distinct responses to N2O, the growth characteristics of N2O-respiring pure cultures representing both clades were examined. The measurement of growth yields and whole-cell N2O consumption kinetics demonstrated that bacteria with clade I nosZ and bacteria with clade II nosZ exhibit distinct properties, suggesting niche differentiation and a relevant role for clade II organisms for controlling N2O emissions to the atmosphere.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Five different bacterial isolates harboring nosZ were selected for a comparison of growth characteristics with N2O as the electron acceptor. Included in the analysis were Anaeromyxobacter dehalogenans, a nondenitrifying deltaproteobacterium with clade II nosZ; the denitrifying betaproteobacterium Dechloromonas aromatica with clade II nosZ; and the denitrifying gammaproteobacteria Pseudomonas stutzeri and Shewanella loihica, both of which carry a clade I nosZ. Also included in the initial analysis was Dechloromonas denitrificans, an organism that shares physiological features with Dechloromonas aromatica but that apparently possesses a clade I nosZ (Table 1). D. aromatica strain RCB was kindly provided by John D. Coates, and D. denitrificans strain ED-1 was acquired from the American Type Culture Collection (ATCC BAA-841T). S. loihica strain PV-4 was grown in saline (2.0% [wt/vol] NaCl) phosphate-buffered basal salts medium as described previously (24). The same medium with a lower NaCl concentration of 0.05% was used to grow D. aromatica strain RCB, D. denitrificans strain ED-1, and P. stutzeri strain DCP-Ps1. A. dehalogenans strain 2CP-C was grown in 30 mM bicarbonate-buffered basal salts medium with an N2-CO2 headspace (80:20, vol/vol) (25). The final pH was 7.0 in all media used. Culture bottles were prepared using the Hungate technique and sealed with black butyl rubber stoppers (Geo-Microbial Technologies, Inc., Ochelata, OK, USA) to maintain anoxic conditions. Cultures were initiated from freezer stocks and maintained in 100 ml of the respective medium in 160-ml glass serum bottles amended with 5 mM acetate and 1 mM NO3−. NH4Cl was used as a nitrogen source and was added to a final concentration of 0.5 mM. All culture vessels were incubated horizontally at 30°C with shaking at 110 rpm.
TABLE 1.
Bacteria harboring nosZ used to obtain kinetic data during growth with N2O provided as the electron acceptor and acetate as the electron donor
| Organism | Type culture collection accession no. | nosZ clade (GenBank accession no.) | GenBank accession no. of genome sequence |
|---|---|---|---|
| Dechloromonas aromatica strain RCB | ATCC BAA-1848 | II (YP_284794.1) | CP000089.1 |
| Dechloromonas denitrificans strain ED-1 | ATCC BAA-841, DSM 15892 | II (KT592356.1)a | GCA_001551835.1b |
| Anaeromyxobacter dehalogenans strain 2CP-C | ATCC BAA-259 | II (YP_002491966.1) | CP000251.1 |
| Pseudomonas stutzeri strain DCP-Ps1 | Not deposited | I (JQ513867) | Not sequenced |
| Shewanella loihica strain PV-4 | ATCC BAA-1088, DSM 17748 | I (YP_001095525.1) | CP000606.1 |
Dechloromonas denitrificans strain ED-1 was initially characterized as a clade I bacterium harboring the typical nosZ (GenBank accession number AJ704212.1). Genome sequencing performed in this study revealed that strain ED-1 harbors a clade II nosZ.
GenBank assembly accession number (draft genome).
Bacterial growth and N2O consumption were monitored following inoculation of 0.5 ml (0.5%, vol/vol) of a cell suspension from cultures that had consumed all NO3− and NO2− to fresh medium amended with 2 ml (∼83 μmol) of N2O gas (>99%; Sigma-Aldrich, St. Louis, MO). Acetate (500 μmol) was added in 24-fold excess in terms of electron equivalents (i.e., acetate− + 2H2O → 2CO2 + 8e− +7H+; N2O + 2H+ + 2e− → N2 + H2O) to ensure that N2O reduction activity was not electron donor limited. The optical density at 600 nm (OD600) values of the cultures and N2O concentrations were monitored until all N2O was consumed and growth ceased. N2O-negative controls exhibited no measurable growth.
Analytical procedures.
N2O consumption in growing cultures was monitored by measuring culture headspace samples with an Agilent 3000A MicroGC gas chromatograph (Palo Alto, CA) equipped with a Plot Q column and a thermal conductivity (TCD) detector (26). This instrument has a detection limit of ∼50 ppmv N2O. For measurement of the kinetic parameters half-saturation constant (Ks) and Vmax, N2O was quantified using an Agilent (HP) 7890A gas chromatograph equipped with an HP-Plot Q column (30 m by 0.320 mm in diameter; film thickness, 20 μm) and a micro-electron capture detector (μECD). This analytical system, which had a detection limit of ∼0.3 ppmv N2O, offered more sensitive quantification than the 3000A MicroGC instrument. The injector, oven, and detector temperatures were set to 200°C, 100°C, and 250°C, respectively. Helium (>99.999% purity) and N2 (>99.999% purity) (Airgas, Knoxville, TN) served as the carrier and makeup gases, respectively. The split ratio of the injector was set to 20 when the initial N2O concentration was below 200 ppmv and was increased to 100 when the initial N2O concentrations exceeded 200 ppmv. For each measurement, 0.1 ml headspace gas was withdrawn from the culture vessel and manually injected into the gas chromatograph. A calibration curve with the appropriate split ratio for the experimental samples was generated immediately before each experiment. The aqueous concentration (in micromolar) of N2O was calculated from the headspace concentrations using equation 1 as follows and a dimensionless Henry's constant of 1.94 for N2O at 30°C (27):
| (1) |
where Caq, Cg, and H are the aqueous concentration (in moles liter−1), the headspace concentration (in moles liter−1), and Henry's constant (dimensionless), respectively. A Henry’s constant of 2.21 was used to correct for the ionic strength effects in the saline medium used for culturing S. loihica (28–30). OD600 measurements were performed with a Spectronic 20D+ spectrophotometer (Thermo Scientific, Waltham, MA) using disposable plastic cuvettes with a 1-cm path length.
Growth yield determination.
The growth yields were measured for the bacteria listed in Table 1. Triplicate 160-ml serum bottles with 100 ml fresh medium received 1.0 ml of inoculum from the respective precultures grown with 5 mM acetate and 2 ml N2O (>99%). After inoculation, each vessel was amended with 4 ml N2O (165.8 μmol), and the initial amount of N2O was quantified following equilibration at 30°C. Immediately after N2O depletion, 40-ml cell suspensions were withdrawn from each bottle and the biomass was collected on 0.22-μm-pore-size membrane filters (Millipore, Billerica, MA) by vacuum filtration. The filters were dried at 100°C for 24 h and placed in a desiccator to cool to room temperature, and the weight was recorded after weight consistency was achieved. The biomass dry weight was calculated by subtraction of the premeasured weight of the dry membrane filter from the weight of the dried filter holding the biomass. The measurements were adjusted for the weight of the salts in the culture medium that stayed on the membrane filter after filtration. This adjustment was necessary because media with different ionic strengths were used for the cultivation of the microorganisms (30).
Determination of Michaelis-Menten parameters.
The rates of N2O consumption by D. aromatica strain RCB, D. denitrificans strain ED-1, A. dehalogenans strain 2CP-C, P. stutzeri strain DCP-Ps1, and S. loihica strain PV-4 cultures were measured. The bacterial cultures tested exhibited different growth characteristics and N2O reduction rates. In order to determine Michaelis-Menten parameters, extensive preliminary experiments were performed to elucidate conditions that maintained pseudo-steady-state substrate and biomass concentrations. Initially, cells were grown with 4 ml of N2O and 5 mM acetate in 160-ml serum bottles containing 100 ml of medium. Immediately following the consumption of 4 ml of N2O, 0.4 ml of 5% (vol/vol) N2O in N2 was added to the vessels and its consumption was monitored. All culture bottles were incubated at 30°C on a shaker platform set to 110 rpm. The concentrations of dissolved N2O were plotted against the rate of N2O consumption to determine whether saturation of N2O consumption activity (i.e., Vmax) was observed within the examined concentration range. When saturation in the Michaelis-Menten curve was not observed, the experiments were repeated with increased amounts of N2O so that consumption rates could be determined at higher N2O concentrations. Since the added amount of N2O was small (<1%) relative to the amount already consumed by the cultures, additional growth was considered negligible and confirmed by constant OD600 values. Thus, the amount of catalyst (i.e., the amount of NosZ per cell) was assumed to be constant. This approach was applied to all cultures to determine the optimal N2O concentration ranges for the kinetic measurements. In addition, the preliminary experiments determined the appropriate cell densities to avoid N2O concentration decreases exceeding 5% between consecutive measurements.
For each experiment, 0.5 ml of culture grown with 1 mM NO3− was inoculated into vessels containing fresh medium. Subsequently, N2O (2 ml for D. aromatica strain RCB and D. denitrificans strain ED-1, 4 ml for A. dehalogenans strain 2CP-C and S. loihica strain PV-4) was added to the headspace and the culture bottles were incubated at 30°C on a shaker platform set to 110 rpm until the N2O concentration reached the detection limit of the Agilent 7890 gas chromatograph. The D. aromatica strain RCB and D. denitrificans strain ED-1 cultures used for rate determination were prepared by transferring 5-ml aliquots from N2O-depleted culture bottles with these organisms to autoclaved 160-ml serum bottles filled with N2 gas at atmospheric pressure. Each vessel received 0.4 ml of 5% (vol/vol) N2O (N2O gas diluted with N2). The remaining cell suspensions were used for the dry weight measurements described below. After A. dehalogenans strain 2CP-C and S. loihica strain PV-4 cultures had consumed the initial amount of N2O, additional N2O (0.4 ml of 5% N2O for A. dehalogenans strain 2CP-C cultures, 0.3 ml of 99% N2O for S. loihica strain PV-4 cultures) was provided for construction of progression curves for N2O consumption. Following a 10-min equilibration period, gas chromatography measurements were taken at 2- to 5-min intervals. The total amounts of N2O in the bottles at these time points were calculated from the headspace N2O concentrations. Progression curves were prepared from three independent experiments, each of which was performed with a single culture bottle for each measurement. From each progression curve, 5 to 6 linear sections, each with at least three datum points, were selected. The slope of each linear section (i.e., a rate value) generated a datum point in the Michaelis-Menten plot. For each linear region, the averages of the aqueous-phase N2O concentrations served as the representative N2O concentrations, and the N2O consumption rates were determined from the slope of the linear regression lines. Immediately following the completion of the kinetic measurements, the culture suspensions were used for cell dry weight determinations.
Due to the fast growth and rapid consumption of N2O in P. stutzeri strain DCP-Ps1 cultures, the determination of the Michaelis-Menten parameters required a different approach. Following growth in 160-ml serum bottles containing 100 ml of medium with 5 mM acetate and 4 ml of N2O (i.e., the N2O was completely consumed), aliquots of 5-ml cell suspensions were transferred to serum bottles filled with N2 gas and sealed with black butyl rubber stoppers. After adjustment to atmospheric pressure, 0.5 ml of 5% N2O was injected, and after 10 min of equilibration at 30°C and agitation at 110 rpm, up to six gas chromatography measurements were taken at 4-min intervals. The gas chromatography measurements were repeated with various initial quantities of N2O (1.0 ml of 5% N2O; 0.1, 0.2, 0.5, and 1.0 ml of >99% N2O) and various intervals between each measurement. The Michaelis-Menten plot was generated as described above. This entire procedure was independently repeated three times. The data from each individual experiment were combined after normalization by the total biomass (dry weight) of the culture and used in the nonlinear regression analysis.
The volumes of cell suspensions remaining after the rate measurements (i.e., 30 ml for the D. aromatica strain RCB, D. denitrificans strain ED-1, and P. stutzeri strain DCP-Ps1 cultures and 15 ml for the A. dehalogenans strain 2CP-C and S. loihica strain PV-4 cultures) were used to determine the dry weight of the biomass.
The Michaelis-Menten parameters Ks and Vmax were determined from the nonlinear regression analysis of the plotted data using SPSS 22 for Mac (IBM Corp., Armonk, NY, USA).
Identification of nosZ in D. denitrificans strain ED-1.
D. denitrificans strain ED-1 was grown in a 160-ml serum bottle amended with 5 mM acetate and 2 mM NO3− and incubated until the NO3− was consumed. From a 50-ml cell suspension, genomic DNA was isolated using the cetyltrimethylammonium bromide (CTAB) method (http://jgi.doe.gov/collaborate-with-jgi/pmo-overview/protocols-sample-preparation-information/). RNA was removed with RNase I (Thermo Scientific, Waltham, MA), and the quantity and purity of the extracted genomic DNA were confirmed with a Qubit fluorometer (Life Technologies, Carlsbad, CA) and a NanoDrop 1100 spectrophotometer (NanoDrop Technologies, Wilmington, DE), respectively. The genomic DNA was sequenced using the Illumina MiSeq platform, and 2,193,064 paired-end sequences (total length, 293 Mbp) were obtained. To confirm that the sequenced organism was D. denitrificans strain ED-1 and to identify the nosZ gene(s) in the genomic fragments, CLC Genomic Workbench software (CLC bio, Aarhus, Denmark) was used to map the sequence reads onto the 16S rRNA gene sequence of D. denitrificans strain ED-1 (GenBank accession number AJ318917.1) and nosZ sequences of D. aromatica strain RCB (Daro_1571 and Daro_1575 in CP000089.1) and D. denitrificans strain ED-1 (AJ704212.1) obtained from the NCBI database.
Nucleotide sequence accession numbers.
The nosZ consensus sequence of D. denitrificans and the D. denitrificans draft genome sequence have been submitted to the NCBI GenBank database (accession numbers KT592356.1 and GCA_001551835.1, respectively).
RESULTS
Growth with N2O as the electron acceptor.
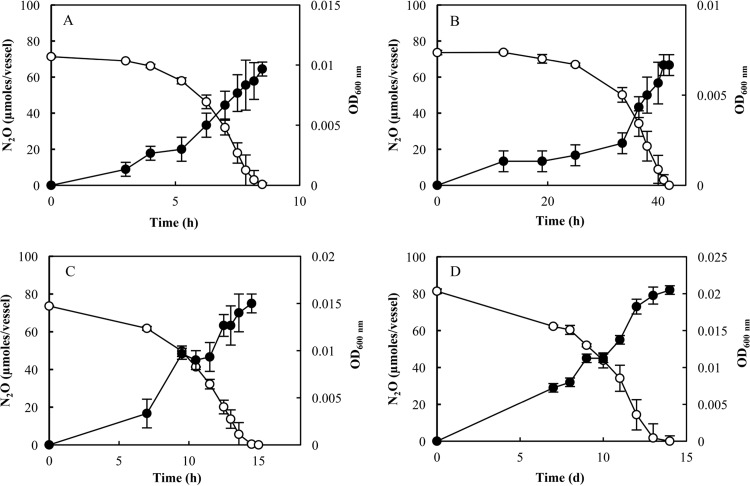
All tested cultures grew with N2O provided as the electron acceptor, whereas no growth was observed in the same medium without N2O (data not shown). The bacterial cultures exhibited distinct growth characteristics with N2O (Fig. 1; Table 2). P. stutzeri strain DCP-Ps1 cultures consumed 71.4 ± 0.6 μmol N2O and grew to a final OD600 of 0.009 ± 0.001. S. loihica strain PV-4 grew to a final OD600 value of 0.007 ± 0.001 after 42 h of incubation with the concomitant consumption of 73.6 ± 1.2 μmol of N2O. D. aromatica strain RCB and A. dehalogenans strain 2CP-C grew to significantly higher OD600 values but consumed similar amounts of N2O. D. aromatica strain RCB consumed 73.6 ± 0.1 μmol N2O in <15 h, growing to a final cell density of OD600 of 0.015 ± 0.001. A. dehalogenans strain 2CP-C required 14 days to consume 81.3 ± 2.4 μmol N2O and reach OD600 readings of 0.021 ± 0.001. The highest rate of N2O consumption was observed for P. stutzeri strain DCP-Ps1, with the maximum consumption rate being 282 nmol h−1 per ml of cell culture. The minimum rate of 8.3 nmol h−1 per ml of cell culture was observed for A. dehalogenans strain 2CP-C. No consistency in terms of the growth rates between bacteria with the same type of NosZ was observed; however, the final OD600 values suggested that the clade II N2O reducers consistently yielded more biomass than clade I N2O reducers (Table 2).
FIG 1.
Growth (●) and N2O consumption (○) in cultures of P. stutzeri strain DCP-Ps1 (clade I) (A), S. loihica strain PV-4 (clade I) (B), D. aromatica strain RCB (clade II) (C), and A. dehalogenans strain 2CP-C (clade II) (D). The error bars represent the standard deviations of measurements made with samples extracted from three separate culture vessels.
TABLE 2.
Growth characteristics and Ks values of bacterial species with clade I or clade II nosZ in batch culture experimentsa
| Organism | nosZ clade | Exponential growth rate (h−1) | Final OD600 | Biomass yield (mg [dry wt]/mmol N2O consumed) | Ks (μM) for N2O | Maximum N2O consumption rate (μmol/min/mg biomass) |
|---|---|---|---|---|---|---|
| Pseudomonas stutzeri strain DCP-1 | I | 0.326 (0.033) | 0.009 (0.001) | 7.24 (0.45) | 35.5 (9.3) | 4.16 (0.44) |
| Shewanella loihica strain PV-4 | I | 0.0892 (0.0131) | 0.007 (0.001) | 6.31 (1.31) | 7.07 (1.13) | 0.446 (0.024) |
| Dechloromonas aromatica strain RCB | II | 0.182 (0.030) | 0.015 (0.001) | 10.2 (0.7) | 0.324 (0.078) | 0.461 (0.034) |
| Anaeromyxobacter dehalogenans strain 2CP-C | II | 0.00762 (0.00072) | 0.021 (0.001) | 11.2 (0.5) | 1.34 (0.35) | 0.0171 (0.0024) |
For yield measurements, cell biomass was determined following the complete consumption of 0.16 mmol N2O. The electron donor acetate was provided in excess (0.5 mmol). The numbers in parentheses represent the standard errors. The standard errors of the growth rates measured during exponential growth, half-saturation constants, and maximum rates of N2O consumption were determined from the linear or nonlinear regression analysis using SPSS 22 for Mac with data obtained from triplicate experiments. The standard errors of the final OD600 and biomass yields were determined from the standard deviations of data for triplicate samples.
The biomass yield measurements were within the range of values previously reported for cultures grown with N2O as the sole electron acceptor (15, 31); however, higher yields of clade II organisms suggest differences in energy conservation efficiencies during growth with N2O (Table 2). The biomass yields of A. dehalogenans strain 2CP-C (11.2 ± 0.5 mg cell dry weight mmol N2O−1) and D. aromatica strain RCB (10.2 ± 0.7 mg cell dry weight mmol N2O−1) were significantly greater (P < 0.05) than the biomass yields of P. stutzeri strain DCP-Ps1 (7.24 ± 0.45 mg cell dry weight mmol N2O−1) and S. loihica strain PV-4 (6.31 ± 1.31 mg cell dry weight mmol N2O−1) (Table 2). The clade II N2O reducers with the clade II nosZ had significantly higher biomass yields than the clade I N2O reducers with the clade I nosZ, suggesting that the clade II NosZ allows more efficient energy conservation.
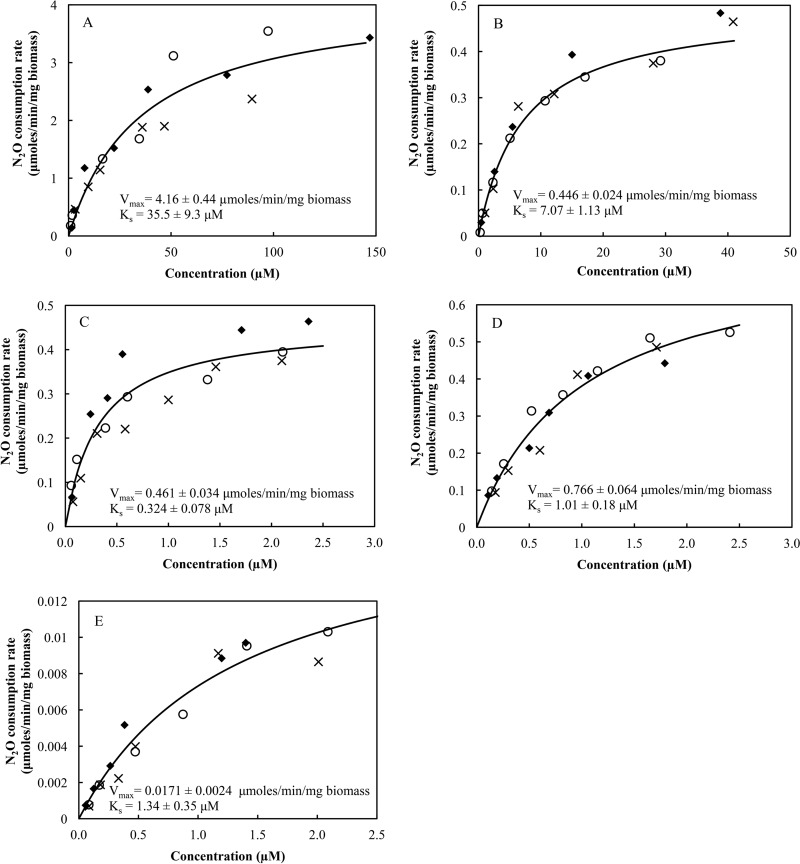
Michaelis-Menten parameters of N2O consumption.
Whole-cell Michaelis-Menten parameters Ks and Vmax were determined for the bacterial strains listed in Table 1 (Fig. 2). P. stutzeri strain DCP-Ps1 exhibited the highest Vmax of 4.16 ± 0.44 μmol min−1 mg biomass−1, a value at least five times greater than the Vmax values of any other organism tested. As implicated from its low growth rate on N2O, the lowest Vmax value (0.0171 ± 0.0024 μmol min−1 mg biomass−1) was observed for A. dehalogenans strain 2CP-C. The lowest Ks value of 0.324 ± 0.078 μM was observed for D. aromatica strain RCB; that value was ∼100 times lower than the Ks value observed for P. stutzeri strain DCP-Ps1 (35.5 ± 9.3 μM). D. denitrificans strain ED-1 and A. dehalogenans strain 2CP-C demonstrated statistically significantly similar Ks values of 1.01 ± 0.18 μM and 1.34 ± 0.35 μM, respectively (P > 0.05). The Ks value of S. loihica strain PV-4 was 7.07 ± 1.13 μM, which fell between the Ks values of N2O reducers harboring a clade II nosZ and the Ks value of P. stutzeri strain DCP-Ps1 harboring a clade I nosZ (P < 0.05).
FIG 2.
Whole-cell Michaelis-Menten kinetics curves of P. stutzeri strain DCP-Ps1 (clade I) (A), S. loihica strain PV-4 (clade I) (B), D. aromatica strain RCB (clade II) (C), D. denitrificans strain ED-1 (clade II) (D), and A. dehalogenans strain 2CP-C (clade II) (E). The different symbols (○, ×, and ◆) represent the results from independent experiments.
Identification of nosZ in D. denitrificans.
On the basis of nosZ amplicon sequence analysis, the genome of D. denitrificans strain ED-1 was originally reported to encode a clade I NosZ with 92% amino acid sequence identity to the NosZ of Bradyrhizobium japonicum strain USDA110 (32). Unexpectedly, we determined similar Ks and Vmax values for D. denitrificans strain ED-1 (reported to possess a clade I nosZ) and D. aromatica strain RCB with a clade II nosZ. To verify the type of NosZ produced by strain ED-1, the genome of this organism was sequenced. Assuming that D. denitrificans strain ED-1 and D. aromatica strain RCB share similarly sized genomes of 4.5 Mbp, coverage of the D. denitrificans strain ED-1 genome of ∼50 times was obtained. No reads mapped to the D. denitrificans nosZ nucleotide sequence in the NCBI database (GenBank accession number AJ704212.1), suggesting that the nosZ sequence previously reported does not belong to strain ED-1. Instead, 1,044 sequence reads mapped onto the D. aromatica strain RCB nosZ gene, and the 2,292-bp consensus sequence (GenBank accession number KT592356.1) shared 89% nucleotide sequence identity with the nosZ sequence of strain RCB. Mapping of the reads onto the 16S rRNA gene sequence of D. denitrificans strain ED-1 (GenBank accession number AJ318917.1) resulted in a consensus sequence with only one mismatch over the 1,487-bp gene length, which confirmed that strain ED-1 was sequenced. The genome sequencing efforts indicated that D. denitrificans strain ED-1 does not harbor a clade I nosZ gene but instead possesses a clade II nosZ gene.
DISCUSSION
The whole-cell Michaelis-Menten N2O reduction kinetics measurements performed with five proteobacterial isolates revealed differences between organisms harboring a clade I nosZ and organisms harboring a clade II nosZ. The organisms with a clade II nosZ, D. aromatica strain RCB and A. dehalogenans strain 2CP-C, had Ks values up to 2 orders of magnitude lower than those of the organisms harboring a clade I nosZ, P. stutzeri strain DCP-Ps1 and S. loihica strain PV-4. D. denitrificans strain ED-1, chosen as a Dechloromonas strain with a clade I nosZ (32), exhibited an unexpectedly low Ks that was similar to the values measured for the organisms with a clade II nosZ. Sequencing of strain ED-1 genomic DNA revealed that this organism actually possesses a clade II nosZ with 89% nucleotide sequence identity (94% amino acid sequence identity) to the nosZ of D. aromatica strain RCB. Taken together, these findings suggest that clade II organisms exhibit lower Ks values for N2O than clade I organisms. The notion that clade II organisms have a higher affinity to N2O than clade I organisms is also supported by previous observations. Betlach and Tiedje (33) observed a Ks value for N2O reduction by a Flavobacterium sp. of ∼0.4 μM. Although the nosZ gene of the Flavobacterium sp. used in that experiment has not been sequenced, all Flavobacterium nosZ sequences deposited in the NCBI database belong to clade II and share >89% translated amino acid sequence similarity with each other. On the other hand, the N2O consumption curves of soybean nodules inoculated with Bradyrhizobium japonicum strain USDA110, possessing a clade I nosZ, indicated a Ks value for N2O of greater than 15 μM (34). Thus, the substrate affinity constants reported in prior work support the hypothesis that clade II organisms exhibit a higher affinity to N2O and play a crucial role in attenuating N2O emissions. Of course, a broader diversity of organisms harboring nosZ should be investigated to conclusively demonstrate that differences in NosZ kinetic properties distinguish the ecophysiology of clade I and clade II organisms.
The distinguishing Ks values, growth yields with N2O, and N2O consumption rates may have implications for the competition between the clade I and clade II organismal groups when N2O is used as the electron acceptor. N2O concentrations rarely exceed 5 ppm (∼0.10 μM in pore water at 30°C and atmospheric pressure) in soils, except for agricultural soils following intensive fertilizer or manure applications (35, 36). Thus, under most environmental conditions, clade II organisms should outcompete clade I N2O reducers with higher Ks values. The analyses of soil metagenomes support this conclusion, and clade II nosZ dominated the nosZ gene pool in temperate grassland and tropical forest soils where no fertilizer was applied and elevated N2O concentrations were not likely (19, 37). In a fertilized agricultural soil ecosystem, N2O concentrations reaching 1,200 ppmv were observed following the application of cattle urine, and elevated N2O concentrations exceeding 100 ppmv were sustained for at least 10 days (38). Nitrogen fertilizer application can also increase N2O flux for prolonged time periods (39). Under such conditions, organisms with high Vmax values, such as P. stutzeri, would have a competitive advantage, regardless of the NosZ substrate affinity, and are predicted to be the dominant N2O consumers.
Along with the higher affinity to N2O, the tested clade II organisms produced at least 1.5 times more biomass per mole of N2O consumed than the tested clade I organisms. Growth yield is a measure of metabolic efficiency (40), and higher growth yields indicate tight coupling between catabolism and energy conservation. A mechanistic understanding about the differences in electron transfer and energy conservation between clade I NosZ and clade II NosZ is lacking, and detailed structural studies are warranted to explain the observed growth yield differences (15, 23, 41, 42). Organisms with more efficient energy conservation mechanisms can direct a greater fraction of electrons toward biomass synthesis (i.e., increased fs), which provides a competitive advantage, particularly in oligotrophic environments.
Quantitative PCR data and metagenomic analyses revealed that nosZ genes affiliated with the Anaeromyxobacter genus are abundant in agricultural soils of the U.S. Midwest corn belt (15, 19). The results of Anaeromyxobacter genome analyses suggest that members of this genus do not possess NO-forming nitrite reductase genes (nirK or nirS) but share a clade II nosZ gene (15). Members of the Anaeromyxobacter genus are versaphiles but have apparently adapted to utilize the N2O released in N2O-producing biogeochemical reactions occurring in the soil environment, as suggested by the high growth yield of strain 2CP-C (∼1.8-fold higher than that of P. stutzeri) and the low Ks of strain 2CP-C (∼22-fold lower than that of P. stutzeri). Despite the lower Ks, the N2O consumption rate calculated with experimentally acquired Vmax and Ks values was orders of magnitudes lower for strain 2CP-C than for P. stutzeri strain DCP-Ps1 at any N2O concentration. Vmax is less relevant than Ks for survival and growth under famine (i.e., low N2O concentration) conditions in the environment, as would be expected in natural, nonfertilized soils. Observations made with high-affinity methanotrophs support this conclusion, and the half-saturation constants and threshold concentrations for methane were more relevant for sustenance of methane oxidation than Vmax or catalytic efficiency (i.e., Vmax/Ks) (43). Fast-growing r-strategists, such as Pseudomonas spp., generally have high Vmax values for substrate consumption when the substrate is plentiful and, thus, an advantage in the competition for shared resources; however, a trade-off for fast growth is the loss of efficiency and a high maintenance energy (44, 45). Indeed, P. stutzeri had a growth yield 1.8-fold lower than the value measured for Anaeromyxobacter dehalogenans strain 2CP-C. Fast-growing r-strategists are rarely abundant in nutrient-limited soil environments, and the slow-growing K strategists (e.g., Anaeromyxobacter, Opitutus, and Gemmatimonas populations) are generally more robust under such conditions (19, 46, 47). Competition experiments performed under controlled N2O flux conditions will provide further insights and directly reveal the relevance of clade I versus clade II organisms for modulating soil N2O emissions.
Several studies have reported on the N2O sink capacities of natural soils as well as agricultural soils managed under various conditions (18, 48–50). The reason why soils act as net N2O sources or sinks is a controversial topic (10, 18, 49, 51–55). To provide an explanation in an ecological context, the abundance of denitrifiers lacking nosZ genes was used to explain N2O emission from soil (10, 54, 56). A recent study correlated the soil N2O sink capacity with the abundance and phylogenetic diversity of clade II organisms (18); however, causality was not demonstrated. The findings reported here provide a plausible mechanistic explanation for the observed correlation between clade II nosZ abundance and a soil's N2O sink capacity. The observation that NosZ kinetic properties distinguish groups of N2O-consuming bacteria is an example of how a detailed understanding of the microbiology can provide relevant information to advance models of N2O emission from diverse soil biomes. The specific stimulation of bacteria with clade II nosZ may provide opportunities to curb N2O emissions from relevant soil ecosystems.
Funding Statement
This work was supported by the U.S. Department of Energy, Office of Biological and Environmental Research, Genomic Science Program, award DE-SC0006662, and in part by the National Research Foundation of Korea, award 2014R1A1A2058543.
REFERENCES
- 1.Lashof DA, Ahuja DR. 1990. Relative contributions of greenhouse gas emissions to global warming. Nature 344:529–531. doi: 10.1038/344529a0. [DOI] [Google Scholar]
- 2.Ravishankara AR, Daniel JS, Portmann RW. 2009. Nitrous oxide (N2O): the dominant ozone-depleting substance emitted in the 21st century. Science 326:123–125. doi: 10.1126/science.1176985. [DOI] [PubMed] [Google Scholar]
- 3.Portmann RW, Daniel JS, Ravishankara AR. 2012. Stratospheric ozone depletion due to nitrous oxide: influences of other gases. Philos Trans R Soc Lond B Biol Sci 367:1256–1264. doi: 10.1098/rstb.2011.0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aneja VP, Schlesinger WH, Erisman JW. 2009. Effects of agriculture upon the air quality and climate: research, policy, and regulations. Environ Sci Technol 43:4234–4240. doi: 10.1021/es8024403. [DOI] [PubMed] [Google Scholar]
- 5.Foley JA, DeFries R, Asner GP, Barford C, Bonan G, Carpenter SR, Chapin FS, Coe MT, Daily GC, Gibbs HK, Helkowski JH, Holloway T, Howard EA, Kucharik CJ, Monfreda C, Patz JA, Prentice IC, Ramankutty N, Snyder PK. 2005. Global consequences of land use. Science 309:570–574. doi: 10.1126/science.1111772. [DOI] [PubMed] [Google Scholar]
- 6.Kroeze C. 1994. Nitrous oxide and global warming. Sci Total Environ 143:193–209. doi: 10.1016/0048-9697(94)90457-X. [DOI] [PubMed] [Google Scholar]
- 7.Pinder RW, Davidson EA, Goodale CL, Greaver TL, Herrick JD, Liu L. 2012. Climate change impacts of US reactive nitrogen. Proc Natl Acad Sci U S A 109:7671–7675. doi: 10.1073/pnas.1114243109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomson AJ, Giannopoulos G, Pretty J, Baggs EM, Richardson DJ. 2012. Biological sources and sinks of nitrous oxide and strategies to mitigate emissions. Philos Trans R Soc Lond B Biol Sci 367:1157–1168. doi: 10.1098/rstb.2011.0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zumft WG. 1997. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61:533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Philippot L, Andert J, Jones CM, Bru D, Hallin S. 2011. Importance of denitrifiers lacking the genes encoding the nitrous oxide reductase for N2O emissions from soil. Glob Change Biol 17:1497–1504. doi: 10.1111/j.1365-2486.2010.02334.x. [DOI] [Google Scholar]
- 11.Law Y, Ni B-J, Lant P, Yuan Z. 2012. N2O production rate of an enriched ammonia-oxidising bacteria culture exponentially correlates to its ammonia oxidation rate. Water Res 46:3409–3419. doi: 10.1016/j.watres.2012.03.043. [DOI] [PubMed] [Google Scholar]
- 12.Zhu X, Burger M, Doane TA, Horwath WR. 2013. Ammonia oxidation pathways and nitrifier denitrification are significant sources of N2O and NO under low oxygen availability. Proc Natl Acad Sci U S A 110:6328–6333. doi: 10.1073/pnas.1219993110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stremińska MA, Felgate H, Rowley G, Richardson DJ, Baggs EM. 2012. Nitrous oxide production in soil isolates of nitrate-ammonifying bacteria. Environ Microbiol Rep 4:66–71. doi: 10.1111/j.1758-2229.2011.00302.x. [DOI] [PubMed] [Google Scholar]
- 14.Bremner JM. 1997. Sources of nitrous oxide in soils. Nutr Cycl Agroecosyst 49:7–16. doi: 10.1023/A:1009798022569. [DOI] [Google Scholar]
- 15.Sanford RA, Wagner DD, Wu Q, Chee-Sanford JC, Thomas SH, Cruz-Garcia C, Rodríguez G, Massol-Deyá A, Krishnani KK, Ritalahti KM, Nissen S, Konstantinidis KT, Löffler FE. 2012. Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. Proc Natl Acad Sci U S A 109:19709–19714. doi: 10.1073/pnas.1211238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones CM, Graf DRH, Bru D, Philippot L, Hallin S. 2013. The unaccounted yet abundant nitrous oxide-reducing microbial community: a potential nitrous oxide sink. ISME J 7:417–426. doi: 10.1038/ismej.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coyle CL, Zumft WG, Kroneck PMH, Körner H, Jakob W. 1985. Nitrous oxide reductase from denitrifying Pseudomonas perfectomarina. Purification and properties of a novel multicopper enzyme. Eur J Biochem 153:459–467. doi: 10.1111/j.1432-1033.1985.tb09324.x. [DOI] [PubMed] [Google Scholar]
- 18.Jones CM, Spor A, Brennan FP, Breuil M-C, Bru D, Lemanceau P, Griffiths B, Philippot L. 2014. Recently identified microbial guild mediates soil N2O sink capacity. Nat Clim Change 4:801–805. doi: 10.1038/nclimate2301. [DOI] [Google Scholar]
- 19.Orellana LH, Rodriguez-R LM, Higgins S, Chee-Sanford JC, Sanford RA, Ritalahti KM, Löffler FE, Konstantinidis KT. 2014. Detecting nitrous oxide reductase (nosZ) genes in soil metagenomes: method development and implications for the nitrogen cycle. mBio 5:e01193-14. doi: 10.1128/mBio.01193-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kristjansson JK, Hollocher TC. 1980. First practical assay for soluble nitrous oxide reductase of denitrifying bacteria and a partial kinetic characterization. J Biol Chem 255:704–707. [PubMed] [Google Scholar]
- 21.Brown K, Tegoni M, Prudencio M, Pereira AS, Besson S, Moura JJ, Moura I, Cambillau C. 2000. A novel type of catalytic copper cluster in nitrous oxide reductase. Nat Struct Mol Biol 7:191–195. doi: 10.1038/73288. [DOI] [PubMed] [Google Scholar]
- 22.Zhang C, Jones AM, Hollocher TC. 1992. An apparently allosteric effect involving N2O with the nitrous oxide reductase from Wolinella succinogenes. Biochem Biophys Res Commun 187:135–139. doi: 10.1016/S0006-291X(05)81469-X. [DOI] [PubMed] [Google Scholar]
- 23.Simon J, Einsle O, Kroneck PMH, Zumft WG. 2004. The unprecedented nos gene cluster of Wolinella succinogenes encodes a novel respiratory electron transfer pathway to cytochrome c nitrous oxide reductase. FEBS Lett 569:7–12. doi: 10.1016/j.febslet.2004.05.060. [DOI] [PubMed] [Google Scholar]
- 24.Yoon S, Sanford RA, Löffler FE. 2013. Shewanella spp. use acetate as an electron donor for denitrification but not ferric iron or fumarate reduction. Appl Environ Microbiol 79:2818–2822. doi: 10.1128/AEM.03872-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas SH, Sanford RA, Amos BK, Leigh MB, Cardenas E, Löffler FE. 2010. Unique ecophysiology among U(VI)-reducing bacteria as revealed by evaluation of oxygen metabolism in Anaeromyxobacter dehalogenans strain 2CP-C. Appl Environ Microbiol 76:176–183. doi: 10.1128/AEM.01854-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoon S, Cruz-Garcia C, Sanford R, Ritalahti KM, Löffler FE. 2015. Denitrification versus respiratory ammonification: environmental controls of two competing dissimilatory NO3−/NO2− reduction pathways in Shewanella loihica strain PV-4. ISME J 9:1093–1104. doi: 10.1038/ismej.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sander R. 1999. Compilation of Henry's law constants for inorganic and organic species of potential importance in environmental chemistry. Max Plank Institute of Chemistry, Mainz, Germany: http://www.henrys-law.org/henry-3.0.pdf. [Google Scholar]
- 28.Schumpe A. 1993. The estimation of gas solubilities in salt solutions. Chem Eng Sci 48:153–158. doi: 10.1016/0009-2509(93)80291-W. [DOI] [Google Scholar]
- 29.Schumpe A, Quicker G, Deckwer W-D. 1982. Gas solubilities in microbial culture media. Adv Biochem Eng Biotechnol 24:1–38. [Google Scholar]
- 30.Yoon S, Sanford R, Löffler FE. 2015. Nitrite control over dissimilatory nitrate/nitrite reduction pathways in Shewanella loihica strain PV-4. Appl Environ Microbiol 81:3510–3517. doi: 10.1128/AEM.00688-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bryan BA, Jeter RM, Carlson CA. 1985. Inability of Pseudomonas stutzeri denitrification mutants with the phenotype of Pseudomonas aeruginosa to grow in nitrous oxide. Appl Environ Microbiol 50:1301–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horn MA, Drake HL, Schramm A. 2006. Nitrous oxide reductase genes (nosZ) of denitrifying microbial populations in soil and the earthworm gut are phylogenetically similar. Appl Environ Microbiol 72:1019–1026. doi: 10.1128/AEM.72.2.1019-1026.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Betlach MR, Tiedje JM. 1981. Kinetic explanation for accumulation of nitrite, nitric oxide, and nitrous oxide during bacterial denitrification. Appl Environ Microbiol 42:1074–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sameshima-Saito R, Chiba K, Hirayama J, Itakura M, Mitsui H, Eda S, Minamisawa K. 2006. Symbiotic Bradyrhizobium japonicum reduces N2O surrounding the soybean root system via nitrous oxide reductase. Appl Environ Microbiol 72:2526–2532. doi: 10.1128/AEM.72.4.2526-2532.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanai Y, Hirota T, Iwata Y, Nemoto M, Nagata O, Koga N. 2011. Accumulation of nitrous oxide and depletion of oxygen in seasonally frozen soils in northern Japan—snow cover manipulation experiments. Soil Biol Biochem 43:1779–1786. doi: 10.1016/j.soilbio.2010.06.009. [DOI] [Google Scholar]
- 36.Metay A, Oliver R, Scopel E, Douzet J-M, Aloisio Alves Moreira J, Maraux F, Feigl BJ, Feller C. 2007. N2O and CH4 emissions from soils under conventional and no-till management practices in Goiânia (Cerrados, Brazil). Geoderma 141:78–88. doi: 10.1016/j.geoderma.2007.05.010. [DOI] [Google Scholar]
- 37.Vitousek P, Howarth R. 1991. Nitrogen limitation on land and in the sea: how can it occur? Biogeochemistry 13:87–115. [Google Scholar]
- 38.Boon A, Robinson JS, Chadwick DR, Cardenas LM. 2014. Effect of cattle urine addition on the surface emissions and subsurface concentrations of greenhouse gases in a UK peat grassland. Agric Ecosyst Environ 186:23–32. doi: 10.1016/j.agee.2014.01.008. [DOI] [Google Scholar]
- 39.Mosier A, Schimel D, Valentine D, Bronson K, Parton W. 1991. Methane and nitrous oxide fluxes in native, fertilized and cultivated grasslands. Nature 350:330–332. doi: 10.1038/350330a0. [DOI] [Google Scholar]
- 40.Russell JB, Cook GM. 1995. Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol Rev 59:48–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kern M, Simon J. 23 September 2015. Three transcription regulators of the Nss family mediate the adaptive response induced by nitrate, nitric oxide or nitrous oxide in Wolinella succinogenes. Environ Microbiol doi: 10.1111/1462-2920.13060. [DOI] [PubMed] [Google Scholar]
- 42.Pauleta SR, Dell'Acqua S, Moura I. 2013. Nitrous oxide reductase. Coord Chem Rev 257:332–349. doi: 10.1016/j.ccr.2012.05.026. [DOI] [Google Scholar]
- 43.Knief C, Dunfield PF. 2005. Response and adaptation of different methanotrophic bacteria to low methane mixing ratios. Environ Microbiol 7:1307–1317. doi: 10.1111/j.1462-2920.2005.00814.x. [DOI] [PubMed] [Google Scholar]
- 44.Novak M, Pfeiffer T, Lenski RE, Sauer U, Bonhoeffer S. 2006. Experimental tests for an evolutionary trade-off between growth rate and yield in E. coli. Am Nat 168:242–251. doi: 10.1086/506527. [DOI] [PubMed] [Google Scholar]
- 45.Klappenbach JA, Dunbar JM, Schmidt TM. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol 66:1328–1333. doi: 10.1128/AEM.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janssen P. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol 72:1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohini-Kumar M, Osborne JW, Saravanan VS. 2013. Comparison of soil bacterial communities of Pinus patula of Nilgiris, Western Ghats with other biogeographically distant pine forest clone libraries. Microb Ecol 66:132–144. doi: 10.1007/s00248-012-0167-y. [DOI] [PubMed] [Google Scholar]
- 48.Goldberg SD, Gebauer G. 2009. Drought turns a Central European Norway spruce forest soil from an N2O source to a transient N2O sink. Glob Change Biol 15:850–860. doi: 10.1111/j.1365-2486.2008.01752.x. [DOI] [Google Scholar]
- 49.Berger S, Jang I, Seo J, Kang H, Gebauer G. 2013. A record of N2O and CH4 emissions and underlying soil processes of Korean rice paddies as affected by different water management practices. Biogeochemistry 115:317–332. doi: 10.1007/s10533-013-9837-1. [DOI] [Google Scholar]
- 50.Chapuis-Lardy L, Wrage N, Metay A, Chotte J-L, Bernoux M. 2007. Soils, a sink for N2O? A review. Glob Change Biol 13:1–17. doi: 10.1111/j.1365-2486.2006.01280.x. [DOI] [Google Scholar]
- 51.Henault C, Devis X, Page S, Justes E, Reau R, Germon JC. 1998. Nitrous oxide emissions under different soil and land management conditions. Biol Fertil Soils 26:199–207. doi: 10.1007/s003740050368. [DOI] [Google Scholar]
- 52.Weslien P, Kasimir Klemedtsson Å, Börjesson G, Klemedtsson L. 2009. Strong pH influence on N2O and CH4 fluxes from forested organic soils. Eur J Soil Sci 60:311–320. doi: 10.1111/j.1365-2389.2009.01123.x. [DOI] [Google Scholar]
- 53.Čuhel J, Šimek M, Laughlin RJ, Bru D, Chèneby D, Watson CJ, Philippot L. 2010. Insights into the effect of soil pH on N2O and N2 emissions and denitrifier community size and activity. Appl Environ Microbiol 76:1870–1878. doi: 10.1128/AEM.02484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu B, Frostegård Å, Bakken LR. 2010. Denitrification gene pools, transcription and kinetics of NO, N2O and N2 production as affected by soil pH. FEMS Microbiol Ecol 72:407–417. doi: 10.1111/j.1574-6941.2010.00856.x. [DOI] [PubMed] [Google Scholar]
- 55.Morales SE, Cosart T, Holben WE. 2010. Bacterial gene abundances as indicators of greenhouse gas emission in soils. ISME J 4:799–808. doi: 10.1038/ismej.2010.8. [DOI] [PubMed] [Google Scholar]
- 56.Henderson SL, Dandie CE, Patten CL, Zebarth BJ, Burton DL, Trevors JT, Goyer C. 2010. Changes in denitrifier abundance, denitrification gene mRNA levels, nitrous oxide emissions, and denitrification in anoxic soil microcosms amended with glucose and plant residues. Appl Environ Microbiol 76:2155–2164. doi: 10.1128/AEM.02993-09. [DOI] [PMC free article] [PubMed] [Google Scholar]