ABSTRACT
Lactobacillus rhamnosus GG is a lactic acid bacterium widely marketed by the food industry. Its genomic analysis led to the identification of a gene cluster encoding mucus-binding SpaCBA pili, which is located in a genomic island enriched in insertion sequence (IS) elements. In the present study, we analyzed by genome-wide resequencing the genomic integrity of L. rhamnosus GG in four distinct evolutionary experiments conducted for approximately 1,000 generations under conditions of no stress or salt, bile, and repetitive-shearing stress. Under both stress-free and salt-induced stress conditions, the GG population (excluding the mutator lineage in the stress-free series [see below]) accumulated only a few single nucleotide polymorphisms (SNPs) and no frequent chromosomal rearrangements. In contrast, in the presence of bile salts or repetitive shearing stress, some IS elements were found to be activated, resulting in the deletion of large chromosomal segments that include the spaCBA-srtC1 pilus gene cluster. Remarkably, a high number of SNPs were found in three strains obtained after 900 generations of stress-free growth. Detailed analysis showed that these three strains derived from a founder mutant with an altered DNA polymerase subunit that resulted in a mutator phenotype. The present work confirms the stability of the pilus production phenotype in L. rhamnosus GG under stress-free conditions, highlights the possible evolutionary scenarios that may occur when this probiotic strain is extensively cultured, and identifies external factors that affect the chromosomal integrity of GG. The results provide mechanistic insights into the stability of GG in regard to its extensive use in probiotic and other functional food products.
IMPORTANCE Lactobacillus rhamnosus GG is a widely marketed probiotic strain that has been used in numerous clinical studies to assess its health-promoting properties. Hence, the stability of the probiotic functions of L. rhamnosus GG is of importance, and here we studied the impact of external stresses on the genomic integrity of L. rhamnosus GG. We studied three different stresses that are relevant for understanding its robustness and integrity under both ex vivo conditions, i.e., industrial manufacturing conditions, and in vivo conditions, i.e., intestinal tract-associated stress. Overall, our findings contribute to predicting the genomic stability of L. rhamnosus GG and its ecological performance.
INTRODUCTION
Lactobacillus rhamnosus GG is a human intestinal isolate (1) that has been extensively studied over the last 2 decades for its probiotic properties and impact on human health (2, 3). In numerous clinical trials, L. rhamnosus was shown to display beneficial properties in humans, such as the reduction of diarrhea (4–7), atopic eczema (2), and respiratory infections (8, 9). Its implementation in a large variety of applications, food products, and clinical trials was possible, thanks to the remarkably high adaptability and resilience of L. rhamnosus GG to adverse culturing, handling, or storage conditions that actually differ significantly from those in the human intestinal tract. Coping with these environmental changes over time may impact the performance, probiotic properties, metabolic capabilities, and, therefore, the coding capacities of L. rhamnosus GG. This may be important to consider specifically in the interpretation of trials with L. rhamnosus GG that showed limited reproducibility (10).
Bacterial genomes have the propensity to change over time, and a number of experimental evolution studies have been conducted on different bacterial species to assess the plasticity of bacterial genomes over a long time period under various culturing conditions (11–14). Excluding the acquisition of foreign mobile DNA, such as plasmids, conjugative transposons, or phages, bacterial genomes would mostly evolve through the loss, decay, or duplication of genes that result from either small mutational events, i.e., single nucleotide polymorphisms (SNPs) and insertions/deletions (indels), or larger genomic rearrangement events that are typically mediated by insertion sequence (IS) elements. In addition to causing large chromosomal deletions or inversions (15, 16), IS elements can also have some more localized impacts on coding capacities and gene expression by inactivating or activating gene(s) (17). Thus, we identified that the activation of the pilus gene cluster spaCBA-srtC1 was caused by the insertion of an iso-IS30 element that created an active and constitutive promoter upstream of the spaC gene, from which we determined the transcriptional start site experimentally (18). The occurrence of spontaneous mutagenesis mediated by IS elements can be in some cases relatively high (19) and is generally considered harmful for the integrity of the bacteria due to the deleterious effects they can have. Nonetheless, IS elements can also lead to positive and beneficial effects for the bacterial cells with respect to the organism's niche/host, as illustrated by the promoter reconstitution upstream of the spaCBA-srtC1 gene cluster in L. rhamnosus GG (18). A total of 69 IS-like elements were annotated throughout the L. rhamnosus GG chromosome (20). It is noteworthy that the genomic island GGISL2 containing the pilus gene cluster spaCBA-srtC1 and numerous phosphotransferase systems (PTSs) is highly enriched in IS elements (17 IS elements within an ∼125-kb region), explaining why this region has high variability in L. rhamnosus isolates as well as in derivatives of L. rhamnosus GG (16, 21, 22).
Studies have addressed the genomic stability of lactic acid bacteria under various experimental conditions (23). In a recent study, the propagation of a Lactococcus lactis plant strain in a milk environment for 1,000 generations demonstrated the adaptation processes occurring in ecological niche transition (24). Of interest, the same study described the emergence of a mutation-prone lineage (defective mutL gene) (24). All of these adaptations result in non-genetically modified organism (GMO) derivatives (based on European Union regulations) that could be instrumental for cause-effect studies as these strains can be consumed without specific regulations. Such a non-GMO strategy has also been carried out for L. rhamnosus GG, where a set of mutations were identified that affected the production of the mucus-binding pili (16). In the present study, we focus on L. rhamnosus GG that was grown for 1,000 generations under four conditions, including the presence of various stressors, to provide insights into the genomic dynamics and plasticity and the impact of IS elements. Specifically, we examined the integrity of the genomic island encoding the SpaCBA mucus-binding pili since they are associated with host interaction and immuno-signaling (25, 26). In combination with genomic resequencing and functional analysis, we identified stresses that are deleterious for the genomic integrity of L. rhamnosus GG and its associated probiotic functions. Moreover, we provide an understanding of the molecular mechanisms underlying the stability of L. rhamnosus GG and report a mutator strain that could be useful for future non-GMO improvements of this globally used probiotic strain.
MATERIALS AND METHODS
Bacterial strain, growth conditions, and experimental evolution setup.
The reference L. rhamnosus strain GG was cultured in Man-Rogosa-Sharpe (MRS) (Difco BD, NJ, USA) broth and agar plates under anaerobic and static conditions at 37°C. It is noteworthy that the L. rhamnosus strain GG used in the present study was obtained from a glycerol stock derived from ATCC 53103 and was found to contain three SNPs compared to the sequence of the reference genome (20), as further detailed below. Four distinct experimental evolution setups were used: a control (growth in MRS), one with salt-induced osmotic stress (MRS broth with 3% [wt/vol] NaCl), one with bile salt-induced stress (0.25% [wt/vol] ox gall), and one with mechanical shear force-induced stress (30s/day vortex treatment). The generation time of L. rhamnosus GG under all four conditions was determined (see Table S1 in the supplemental material). Next, L. rhamnosus GG was serially transferred for approximately 1,000 generations using consecutive 1,000-fold dilutions of the cultures and the estimated 10 generations that would be needed to obtain full growth under these conditions after 24 h of incubation. As a slightly higher growth rate was obtained in the control incubation (15 generations in 24 h) than in the other incubations (the expected 10 generations in 24 h), in some cases a 100-fold dilution (approximately 5 generations) was applied during the day. Cell aliquots were collected at different time points (designated by the approximate number of generations) throughout the experiment and stored in 20% (vol/vol) glycerol at −80°C for further analyses.
Colony PCR screening.
For a given time point, aliquots of the L. rhamnosus GG cultures from the different incubations were diluted and plated on MRS agar plates and further incubated overnight at 37°C anaerobically. A selected number of isolates from single-colony plates were examined by colony PCR, as previously described (27). To obtain a snapshot of the piliation percentage within a given population of L. rhamnosus GG, 20 single colonies from a given time point were picked, transferred into strip PCR tubes, and heated in a microwave oven at full power for 3 min. DreamTaq master mix (Thermo Scientific) was added according to the manufacturer's instructions, and the PCR mixtures were run in a PCR thermal cycler. Subsequently, the PCR mixture was analyzed by DNA gel electrophoresis. Subsequently, 10 of the colonies screened by PCR for each condition were further analyzed by MiSeq genome resequencing, as described below.
Immunoelectron microscopy.
L. rhamnosus GG samples were grown overnight and then analyzed by transmission electron microscopy. Polyclonal anti-SpaA rabbit antibodies were used for detection of pilus structures. Samples were prepared as previously described (28). Grids were visualized using a JEOL 1400 transmission electron microscope (JEOL, Ltd., Japan).
Mucus-binding assay.
Mucus-binding assays of the three L. rhamnosus isolates (N1, N3, and N8), the pilus-negative strain PB12 (16), and the reference strain GG were carried out, as described previously (16). The relative binding capacities of N1, N3, N8, and PB12 were calculated compared to the capacity of the reference strain GG. The results were obtained from 12 technical replicates of three biological replicates.
qPCR.
Genomic DNA from L. rhamnosus GG overnight cultures corresponding to different time points was extracted as per the manufacturer's instructions (Wizard Genomic DNA purification kit; Promega, WI, USA). Quantitative PCRs (qPCRs) were performed as recommended by the manufacturer in a total volume of 25 μl (5 μl of ∼50 ng/μl gDNA, 1 μl 50 pmol/μl of each oligonucleotide primer, 5 μl of 5× Hot FirePol EvaGreen qPCR Mix Plus [Solis Biodyne, Estonia]). qPCRs were carried out and subsequently analyzed using a Stratagene Mxp3005p thermocycler and Stratagene Mxp3005p software (Agilent Technologies, CA, USA). Three technical replicates from three biological samples were performed for each time point. Two sets of primers were employed, the pair srtC1-for (5′-AGTGCGACTATTAGCTTTA-3′) and srtC1-rev (5′-GGATCTTGTGACCTTAATG-3′) and a GG-specific primer pair (29).
Genome sequencing.
Forty colonies (10 for each evolutionary experiment as described above) were randomly picked after 1,000 generations and resequenced. Briefly, genomic DNA of all 40 colonies/isolates was extracted using a Wizard Genomic DNA purification kit (Promega), as per the manufacturer's instructions. Genomes of the 40 isolates were sequenced at the DNA Sequencing and Genomics Laboratory, Institute of Biotechnology, University of Helsinki, and further compared to the sequence of the L. rhamnosus GG genome using the MiSeq platform (Illumina) as previously described (16). Briefly, paired fastq reads were mapped against the L. rhamnosus GG genome sequence (GenBank accession number FM179322.1, GI:257146922) with SHRiMP, version 2.2.2 software (30), using the command “gmapper-ls –qv-offset 33.” Samtools, version 0.1.18 (31), commands “view -bS,” “sort,” and “index” were used to generate sorted and indexed BAM files. The Samtools command “faidx” was applied to the L. rhamnosus GG reference genome fasta file, and consensus sequences of the mappings were generated by “mpileup -C30 -uf,” “bcftools view -cg-,” and “vcfutils.pl vcf2fq” commands in that order. SNP calling on the consensus sequences were performed with MUMmer software, version 3.23 (32), using commands “nucmer –maxmatch -c 100” and “show-snps -C.” From the output results obtained from the MUMmer, version 3.0, software package, we analyzed only nonambiguous and nonrepetitive regions of the GG genome using an approach similar to that defined in a previous study in Salmonella enterica serovar Typhi by Holt et al. (33) since these regions may be subject to read mismappings. Specifically, and based on the chosen experimental design, we therefore excluded ambiguous single base insertions, deletions, and SNPs (single nucleotide polymorphisms) located in variable-number tandem repeats (VNTRs; >90% identical repeats of >40 bp, determined using http://minisatellites.u-psud.fr) (34–36), CRISPR array, phage regions, and IS elements from the chromosome of L. rhamnosus GG. The excluded region represented a total of 4.7% of the L. rhamnosus GG genome (see Table S2 in the supplemental material). Tree reconstructions of the 40 resequenced L. rhamnosus colonies (see Fig. 5) were done manually and iteratively by comparison of shared SNPs among isolates. The relationship between isolates relies on the assumption that two or more isolates sharing the same SNP derive from a common ancestor. All sequences of IS elements were recovered manually from the L. rhamnosus GG annotated genome sequence and were analyzed using T-Coffee (37) and Genesis (38) to generate the heat map shown in Fig. 7. The mutations of interest were further verified by DNA sequencing, as specified in the main text.
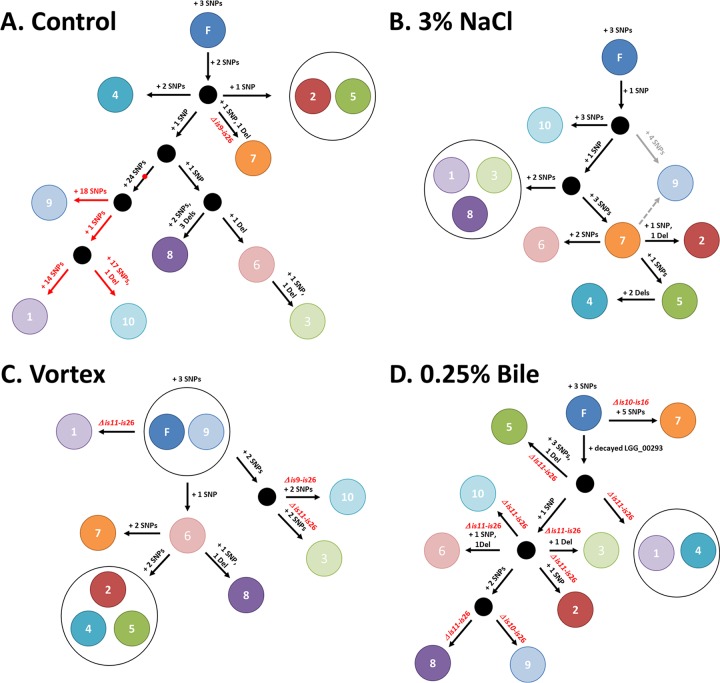
FIG 5.
Tree reconstructions of the 40 resequenced L. rhamnosus colonies recovered after culturing for 1,000 generations. The proposed trees were constructed using the output analysis of the resequenced isolates and highlight possible scenarios that may have occurred toward the diversification of the L. rhamnosus population under each condition tested. For each experiment, isolates are shown as a circle with its corresponding number. (A) The red dot highlights a SNP within the gene encoding the DNA polymerase III, δ unit, of isolates C1, C9, and C10. The subsequent mutator lineage is shown with red arrows. (B) The phylogeny of isolate N9 is unclear, and two possible scenarios (gray arrows) are plausible based on the genomic data. IS-mediated chromosomal deletions are indicated in red on the schematics. (C) Three of 10 isolates underwent IS-mediated chromosomal deletions. (D) All 10 isolates lost a large chromosomal region containing the spaCBA-srtC1 pilus gene cluster and also had a decayed gene (LGG_00293). F, founder.
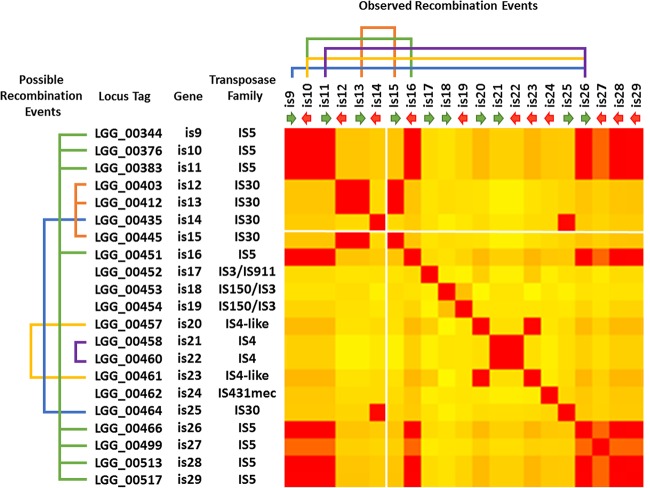
FIG 7.
BLAST analysis of the different IS mobile elements located in the vicinity of the spaCBA-srtC1 pilus gene cluster. On the left-hand side are indicated all possible recombination events that may be potentially occurring based on IS element sequence homologies. On the top of the heat map are shown the recombination events (chromosomal deletions) observed in the present study and also in previous work (16, 21). The position of the spaCBA-srtC1 cluster is located between IS14 and IS15 and marked with two white lines across the heat map. The orientation of the IS elements within the chromosome is also indicated by green and red arrows. The figure is based on data generated with T-Coffee (37) and Genesis (38).
Nucleotide sequence accession number.
The genome sequences of all 40 GG derivatives were deposited in NCBI's Sequence Read Archive (SRA). All accession numbers for sample, experiment, and run are grouped under the project number SRP067389 and are further detailed in Table S3 in the supplemental material.
RESULTS AND DISCUSSION
IS elements remained dormant in L. rhamnosus GG when it was propagated in a stress-free environment.
Based on both colony PCR and qPCR analysis, we observed that the L. rhamnosus GG population remained piliated after culturing for approximately 1,000 generations in a stress-free environment (Table 1 and Fig. 1 and 2). No significant changes in piliation of the GG population were observed over time, suggesting that the genomic island harboring the spaCBA-srtC1 pilus gene cluster remains intact. L. rhamnosus GG retained its piliation phenotype under in vitro conditions although it was originally isolated from the human intestinal tract, where such a phenotypic trait is more relevant for the ecological fitness of GG. When colony PCR screening was performed, 1 colony out of 20 was found to be negative for the presence of the spaCBA-srtC1 pilus genes. Using dot blot analysis, 1 out of 96 single colonies lacked pili (data not shown). Data from qPCR, PCR, and immunoblotting analysis suggested that only a minor fraction of the GG population underwent chromosomal deletion events after 1,000 generations. In an effort to further identify what genomic alterations or changes occurred during the control (stress-free) experimental evolution setup, we selected 10 colonies (or isolates), including the one pilus-less colony identified by colony PCR. All 10 single-colony isolates were resequenced and compared to the L. rhamnosus GG reference genome sequence (20). Genomic data were in agreement with the PCR screening initially performed on the srtC1 gene (Table 1). Only isolate C7 was shown to be devoid of the spaCBA-srtC1 pilus gene cluster (Fig. 3). In isolate C7, the deletion of an ∼125-kb-long chromosomal segment was mediated by IS elements IS9 and IS26. Such an event appeared to be marginal in the control sample, and we hypothesized that collateral mutations might have triggered such a genomic rearrangement. In isolate C7, we identified one SNP (coordinate 1424629) that is located within the open reading frame of LGG_01420, encoding a DNA topoisomerase IV, A subunit. Recent work in Saccharomyces cerevisiae by Yadav et al. showed that DNA topoisomerase I reduces genome instability in hot spots for chromosomal recombination (39). Another study has indicated a role for type 1A topoisomerases in genome stability in Escherichia coli (40). Although we have only in silico data, a similar mechanism in strain C7 may be a plausible explanation for the loss of the genomic island containing the pilus gene cluster. However, we also found three other piliated isolates to carry a similar SNP. Therefore, we still cannot exclude the possibility that the activation of the IS elements may be due to other external factors or collateral mutations. In earlier generations, no measurable loss of the genomic island containing the pilus gene cluster could be observed, indicating that such probiotic properties remained intact over an extended amount of time in the absence of stresses.
TABLE 1.
Analysis of L. rhamnosus colonies for the presence of the spa CBA-srtC1 pilus gene cluster
| Estimated no. of generations and growth condition | srtC1 PCR screening (no. of positive isolates/total no. of isolates)a |
|---|---|
| 100 generations | |
| Control (MRS only) | 10/10 |
| 3% (wt/vol) NaCl | 10/10 |
| 30-s vortex treatment | 10/10 |
| 0.25% (wt/vol) bile salts | 10/10 |
| 1,000 generations | |
| Control (MRS only) | 19/20 |
| 3% (wt/vol) NaCl | 20/20 |
| 30-s vortex treatment | 13/20 |
| 0.25% (wt/vol) bile salts | 0/20 |
Isolates were screened after approximately 100 and 1,000 generations by conventional endpoint PCR using srtC1-specific primers (see Materials and Methods).
FIG 1.
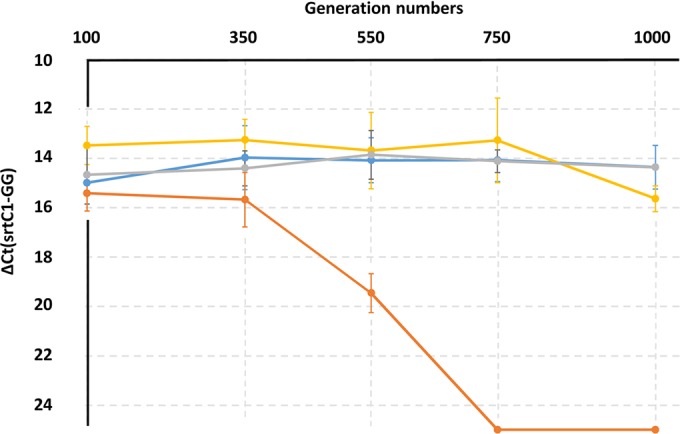
Piliation of L. rhamnosus GG populations in all four evolutionary experiments using quantitative real-time PCR. For each experiment, the change in threshold cycle (ΔCT) was calculated as follows: CT (srtC1-specific primers) − CT(GG-specific primers). The experiment was performed in triplicates. For two bile salt-induced stress samples (750 and 1,000 generations), no amplification of the srtC1 gene was detected, indicating a complete loss of the pilus gene clusters in these two samples. The value for the change in threshold cycle was therefore arbitrarily set at 25.00 as a baseline for the pilus-less bacterial samples. Colony PCRs using the srtC1 primers were also performed for the first (100 generations) and last (1,000 generations) time points as detailed in Table 1. When results for the time points (100 generations 1,000 generations) were compared, differences were significant for the bile-induced stress samples (P < 0.0001) and shearing force-induced stress samples (P = 0.016) only. Blue, control; gray, salt-induced stress; yellow, shearing force-induced stress; orange, bile-induced stress.
FIG 2.
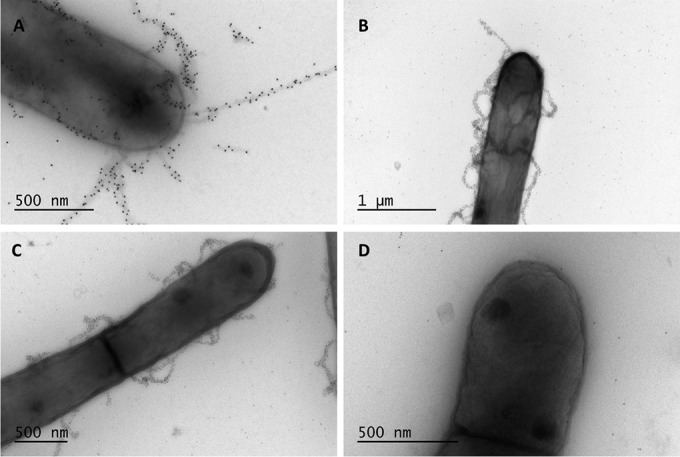
Transmission electron microscopy observations of samples after 1,000 generations. For each condition, one representative bacterial cell of the dominant pilus phenotype is shown. Pili were labeled using anti-SpaA polyclonal antibodies and gold particle-labeled protein A (10 nm). Images represent the following conditions: control (stress-free) (A), high salinity (B), shearing stress (C), and bile salt-induced stress (D).
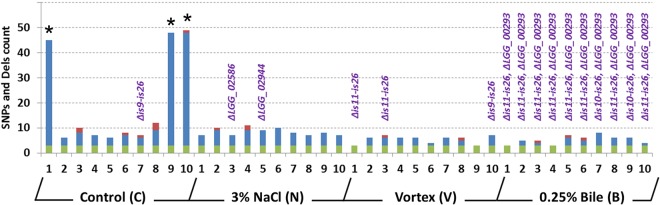
FIG 3.
SNP and deletion counts based on MiSeq genome resequencing of 10 colonies from each evolutionary experiment after 1,000 generations. Founder SNPs, SNPs, and deletions are shown in green, blue, and red, respectively. For each isolate, genomic deletions and decayed/missed genes are indicated (purple labels). The mutator strains are indicated with an asterisk (*). Dels, deletions.
Low mutation rate in stress-free and also stress-induced environments.
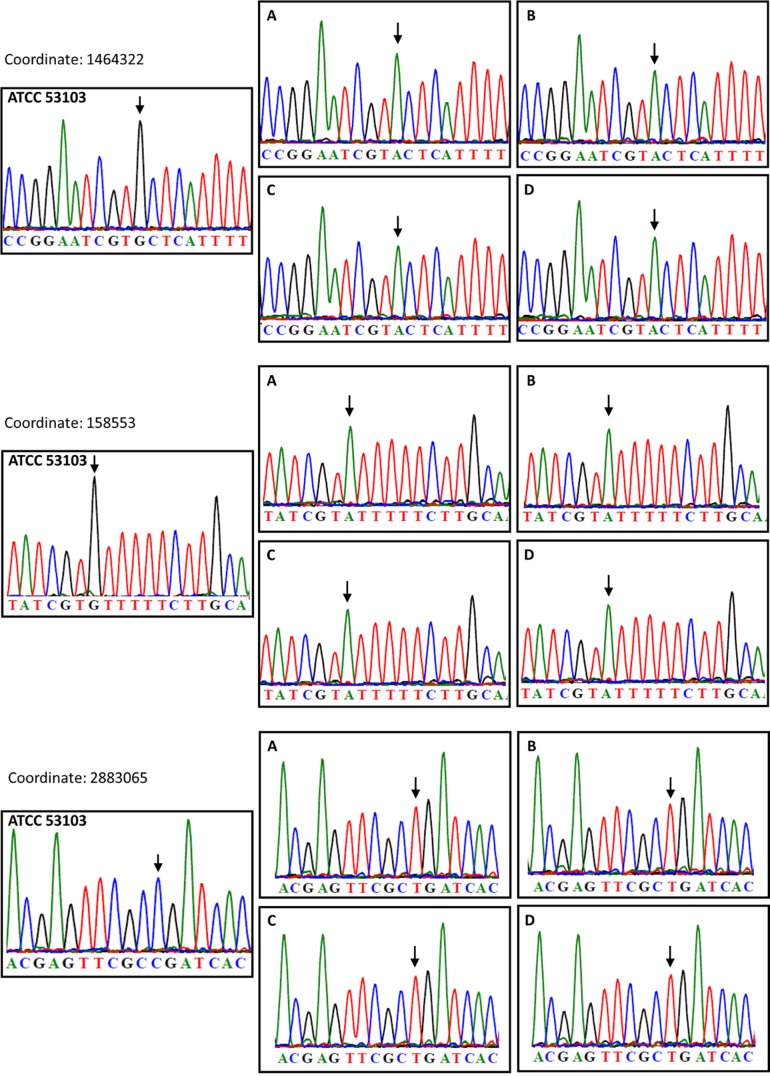
In 37 out of the 40 resequenced L. rhamnosus isolates, low numbers of SNPs or single-base deletions were found, and the mutation (or evolution) rate was at similar levels under all four conditions, i.e., control (estimate of 2.65 × 10−9 SNPs or single base deletions per nucleotide per generation), bile salt-induced stress (estimate of 3.98 × 10−9 SNPs or single base deletions per nucleotide per generation), mechanical shear force-induced stress (estimate of 1.79 × 10−9 SNPs or single base deletions per nucleotide per generation), and salt-induced stress (estimate of 2.79 × 10−9 SNPS and single base deletions per nucleotide per generation) as opposed to levels in three other isolates from the control sample (estimate of 1.37 × 10−7 SNPs or single base deletions per nucleotide per generation, based on the hypothesis that the mutator lineage emerged, at the earliest, after 900 generations, as further discussed in the next section and shown on Fig. 3). Overall, the observed mutation rates are slightly higher than the rate of spontaneous mutations reported in E. coli (41). The reported single base mutations were scattered throughout the chromosome and did not show clear regions more prone for single-base mutations (see Fig. S1 in the supplemental material). The accumulation rate of SNPs and deletions was therefore not dramatically exacerbated by external stresses over time. Interestingly, we identified three founder SNPs that were present in all 40 isolates (Fig. 3; see also Table S4). These were initially found in 38 isolates, and we further confirmed these SNPs in the two remaining isolates using Sanger sequencing (see Fig. S2 in the supplemental material). Additional Sanger sequencing confirmed that these three founder SNPs were not present in the original L. rhamnosus GG strain (ATCC 53103) but only in the one particular aliquot used for the present experimental evolution (glycerol stock derived from ATCC 53103) (Fig. 4). Here, the power of resequencing typically illustrates the need for good practices in strain management and raises questions about the dissemination/distribution of strains (strain integrity).
FIG 4.
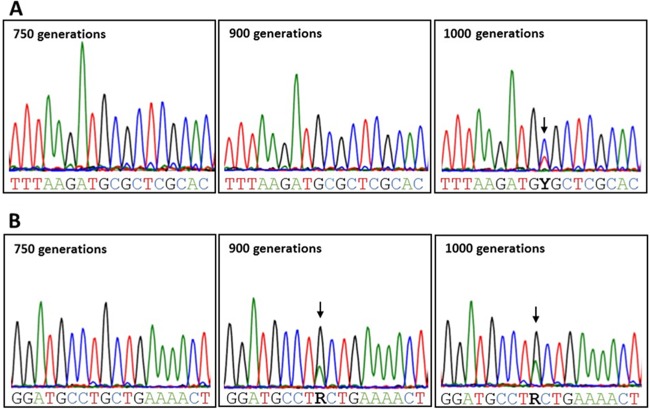
Sanger sequence analysis of founder SNPs. For all three founder SNPS, sequencing on gDNA from the original ATCC 53103 (GG) strain and also from the cultures at 100 generations was performed. SNPs are indicated with black arrows. Graphs represent the following conditions: control (stress-free) (A), salt-induced stress (B), shearing stress (C), and bile salt-induced stress (D).
Identification of a mutator lineage.
Remarkably, 3 out of 10 isolates in the control sample actually had a much higher number of SNPs (>40) (Fig. 3), and further analysis revealed that these three isolates (called C1, C9, and C10) shared a striking number of 30 SNPs, including one point mutation at coordinate 2327174. This mutation is located within the open reading frame of gene LGG_02260, coding for a DNA polymerase III, δ unit, and introduces an amino acid substitution (CGC → CAC, R70H). Alteration of LGG_02260 is likely to impact its intrinsic function (assembly of factor β) within the DNA polymerase III holoenzyme and may explain the higher mutation rates in all three isolates sharing this SNP. Reconstruction of the phylogeny of the 10 control isolates clearly indicated the emergence of a mutator lineage (Fig. 5A). To determine when the mutation of the DNA polymerase III, δ unit, occurred, we extracted the total genomic DNA (gDNA) at different time points (generations) of the overall L. rhamnosus GG population (control sample) and then amplified by PCR a 400-bp region which comprises the SNP (coordinate 2327174). Sanger sequencing allowed us to estimate that the mutation of the DNA polymerase III holoenzyme occurred rather late, i.e., not earlier than 900 generations (Fig. 6A). This subsequently triggered a much higher mutation rate in the related L. rhamnosus GG lineage (Fig. 5 and 6). The mutator lineage constitutes a significant proportion of the L. rhamnosus bacterial population after 1,000 generations, based on genome resequencing (3 out of 10) and Sanger sequencing (Fig. 6A, peak area). We further tested the ability of isolates C1, C9, and C10 to acquire resistance to rifampin (see Fig. S3 in the supplemental material) and demonstrated that all three isolates were more prone to acquire a mutant phenotype (rifampin-resistant phenotype; ∼8-fold higher) than GG. This confirms the mutation-prone properties of this lineage. It is noteworthy that we did not observe a similar mutation (coordinate 2327174) in any of the other 37 resequenced isolates.
FIG 6.
Sequence analysis of the emergence of two SNPs of interest over time. (A) Sanger sequencing of the DNA polymerase III, δ unit, mutation (LGG_02260; coordinate 2327174, C → T) from genomic DNA of the total population (control sample) at different time points. The mutation was initially identified in colonies C1, C9, and C10 (Fig. 3 and 5). (B) Sanger sequencing of the spaC mutation (LGG_00444; coordinate 446541, G → A) from genomic DNA of the total population (salt-induced stress) at different time points. The mutation was previously identified in colonies N1, N3, and N8 (Fig. 3 and 5).
Salt-induced stress does not trigger an IS-mediated recombination event in L. rhamnosus GG.
In lactobacilli, osmotic stresses are usually caused by large amounts of salts (NaCl or KCl) in the environment. Since salts are typically used in cured or fermented food products, it is of industrial relevance to assess the long-term impact of osmotic stress in L. rhamnosus GG. A number of studies in lactobacilli and lactococci have been conducted to look at the transcriptome, proteome, and other “omes” under such environmental conditions (42–45). Here, we aimed at determining possible genomic events triggered by long-term osmotic stress. Similar to the results observed in the control sample, there was no loss of piliation in the L. rhamnosus GG population after propagation for 1,000 generations (Fig. 1 and 2), and this was confirmed when 10 L. rhamnosus GG isolates were resequenced (Fig. 3). Both qPCR and resequencing results indicated that IS elements were not activated and did not result in the loss of any chromosomal segments. The mutation rate in the salt-induced stress sample also remained low and at a level similar to that of the control sample. In N3 and N5, instances of gene decay were observed but could not be related to any adaptation to high-saline conditions. Among SNPs and deletions examined in all 10 isolates, one mutation was common to all 10 resequenced isolates (coordinate 1276820, LGG_01280; FtsI cell division protein). In L. acidophilus, it was demonstrated that the inactivation of the cell division protein CdpA lowered its resistance to osmotic stresses by possibly having an impact on the cell wall architecture (46). Although no sequence similarity was found between CdpA and FtsI, a similar mechanism involving FtsI (LGG_01280) and associated with the cell division process may possibly enhance the tolerance of L. rhamnosus GG to NaCl. Among other reported mutated genes, there were the ABC transporter, PTS, Mg transporter, lysine-specific permease, and putative proteins. It remained difficult, however, to determine whether these mutations related to an adaptation to a high-NaCl condition. Remarkably, three isolates (N1, N3, and N8) from the same lineage (Fig. 5B) also had a substitution mutation within the spaC gene that introduced a stop codon (coordinate 446541; CAG → TAG; Q → STOP). SpaC is the tip pilin and has been shown to be associated with mucus binding (20, 47, 48). Such a mutation is predicted to truncate the SpaC protein and therefore to abolish the mucus-binding properties of L. rhamnosus N1, N3, and N8. This was confirmed by a mucus adhesion assay (see Fig. S4 in the supplemental material). The spaC mutation was not reported under other conditions, indicating that it is not a hot spot for mutations. Sanger sequencing showed that the spaC substitution mutation arose after 750 generations (Fig. 6B).
Activation of IS elements in L. rhamnosus GG upon long-term treatment to mechanical shear forces and bile salts.
Since L. rhamnosus GG is marketed as a probiotic strain in the food industry, we investigated the effects of two additional stresses on the GG genome. In one case, bacterial cells were vortexed for 30 s daily throughout the evolutionary experiment to exaggeratedly mimic possible shearing forces applied to the cells during manufacturing or handling processes. In another case, bacterial cells were propagated in MRS broth in the presence of bile salts (stress encountered in the intestinal tract). In both cases, we found a notable impact on the piliation of the L. rhamnosus GG cells (Fig. 1 and Table 1). For the shearing force-induced stress, a decrease in piliation was observed after 1,000 generations. When 10 isolates were resequenced, 3 out of 10 had lost a chromosomal segment containing the pilus gene cluster (samples V1, V3, and V10). In all three, the deleted segment was flanked by an IS element (Fig. 3). The numbers of SNPs and deletions, however, remained at rates similar to those in the control samples, suggesting that shearing forces activated IS elements, resulting in sudden chromosomal deletion, but did not impact the mutation rate. More strikingly, when L. rhamnosus GG was propagated in MRS broth supplemented with bile salts, the whole population was pilus-less after 750 generations. The loss of piliation was already quantifiable after 350 generations (Fig. 1). At 100 generations, we did not see significant changes (Fig. 1 and Table 1). Such dramatic deletion was mediated by IS elements, as seen in the genomic data (Fig. 3). The deleted region was also in all cases flanked by IS elements. On one end of the chromosomal deletion, the IS26 element was constantly involved, as seen also under other conditions. Excluding IS-mediated deletions, the mutation rate remained at a rate comparable to rates under other tested conditions. SNPs and deletions were found in genes encoding a cell division protein (LGG_00901), the intergenic region of the membrane protein (LGG_01708), ABC transporter (LGG_01853), PTS protein (LGG_00603), and other putative/conserved proteins. The reconstruction of the isolate tree (Fig. 5D) also illustrates that the IS-mediated deletion was not just one event occurring in the culture but that it was recurrent and involved different IS elements, indicating that the IS elements were highly active under that particular condition.
The IS elements identified in the L. rhamnosus GG genome mostly belong to distinct transposase families, i.e., IS5 and IS30 (Fig. 7), and are abundant in the vicinity of the region harboring the spaCBA-srtC1 pilus gene cluster (20, 21). Although in a stress-free environment IS elements remain dormant for a relatively long period of time, a number of stresses clearly induce them, resulting in large chromosomal deletions. Based on the present study and also on previous genomic work, it can be noted that, with the exception of one reported case (21), IS26 plays a dominant role in the deletion event. A BLAST analysis of IS elements in the vicinity of the spaCBA-srtC1 pilus gene cluster showed that the recombination event occurred between highly similar IS elements and that it is not affected by their orientation within the chromosome (Fig. 7). As far as is known, five distinct IS-mediated deletion events have been reported in L. rhamnosus GG even though bioinformatics sequence analysis would suggest additional possible recombination events (Fig. 7).
Concluding remarks.
The evolutionary experiment using L. rhamnosus GG in a nutrient-rich medium for 1,000 generations did not cause any dramatic metabolic simplifications on a large scale within the L. rhamnosus population. The resequencing of L. rhamnosus GG isolates did, however, reveal some phenotypes of interest, i.e., mutator lineage and piliated mutants. The number of SNPs remained low and did not show any clear adaptation pattern to in vitro conditions under all four tested conditions, i.e., stress-free, high salinity, shearing force-induced stress, or bile salt stress. The heterogeneity of the GG population after 1,000 generations also indicated that no beneficial mutations resulted in a single clonal population. Although SNP and deletion accumulation rates were comparable in all four experiments, larger chromosomal deletions could be clearly associated with specific stresses. Shearing forces did trigger to some degree the loss of a large chromosomal segment by IS elements, resulting in pilus-less isolates. The IS-mediated events did, however, emerge relatively late in our experimental setup but still highlight the potential risks associated with such stress when L. rhamnosus GG is being used. We hypothesize that stronger shearing forces may further increase IS element activity. More surprisingly, bile salts strongly induced the IS-mediated loss of the islands harboring the pilus gene cluster. After 100 generations in bile salts, all screened L. rhamnosus cells were still shown to be piliated; but, as observed by qPCRs, the loss of piliation appeared at around 350 generations, and after 1,000 generations, no piliated cells could be detected by qPCRs. Bile salts are known to trigger a strong response in bacterial cells, and we showed that they can also induce IS elements and lead to large recombination events. In the present case, it is relevant since these rearrangements were associated with the loss of some colonization-associated genes, e.g., a mucus-binding pilus gene cluster.
To conclude, the IS elements play an important role in the coding capacity and ecological performance of L. rhamnosus GG with regard to its probiotic applications and should therefore be carefully monitored when experiments are conducted in which IS element-inducing stresses are involved. This study also illustrates the pivotal role of IS elements in shaping the genomes of L. rhamnosus along with contributing to the diversification of the species.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Daragh Hill for technical assistance during the present study. We thank Kirsi Lipponen, Eeva-Marja Turkki, and Pia Laine for assistance in DNA sequencing. We are, finally, grateful to the personnel of the Electron Microscopy Unit (Viikki Campus, University of Helsinki).
Funding Statement
This study was supported by the ERC grant Microbes Inside (grant ERC 250172 to WMdV), the Academy of Finland (grants 141140 to AP, 252123 to FPD and 141130 to WMdV) and the University of Helsinki.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00255-16.
REFERENCES
- 1.Silva M, Jacobus NV, Deneke C, Gorbach SL. 1987. Antimicrobial substance from a human Lactobacillus strain. Antimicrob Agents Chemother 31:1231–1233. doi: 10.1128/AAC.31.8.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalliomäki M, Salminen S, Poussa T, Arvilommi H, Isolauri E. 2003. Probiotics and prevention of atopic disease: 4-year follow-up of a randomised placebo-controlled trial. Lancet 361:1869–1871. doi: 10.1016/S0140-6736(03)13490-3. [DOI] [PubMed] [Google Scholar]
- 3.Szajewska H, Mrukowicz JZ. 2001. Probiotics in the treatment and prevention of acute infectious diarrhea in infants and children: a systematic review of published randomized, double-blind, placebo-controlled trials. J Pediatr Gastroenterol Nutr 33(Suppl 2):S17–S25. doi: 10.1097/00005176-200110002-00004. [DOI] [PubMed] [Google Scholar]
- 4.Canani RB, Cirillo P, Terrin G, Cesarano L, Spagnuolo MI, De Vincenzo A, Albano F, Passariello A, De Marco G, Manguso F, Guarino A. 2007. Probiotics for treatment of acute diarrhoea in children: randomised clinical trial of five different preparations. BMJ 335:340. doi: 10.1136/bmj.39272.581736.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Österlund P, Ruotsalainen T, Korpela R, Saxelin M, Ollus A, Valta P, Kouri M, Elomaa I, Joensuu H. 2007. Lactobacillus supplementation for diarrhoea related to chemotherapy of colorectal cancer: a randomised study. Br J Cancer 97:1028–1034. doi: 10.1038/sj.bjc.6603990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Majamaa H, Isolauri E, Saxelin M, Vesikari T. 1995. Lactic acid bacteria in the treatment of acute rotavirus gastroenteritis. J Pediatr Gastroenterol Nutr 20:333–338. doi: 10.1097/00005176-199504000-00012. [DOI] [PubMed] [Google Scholar]
- 7.Arvola T, Laiho K, Torkkeli S, Mykkänen H, Salminen S, Maunula L, Isolauri E. 1999. Prophylactic Lactobacillus GG reduces antibiotic-associated diarrhea in children with respiratory infections: a randomized study. Pediatrics 104:e64. doi: 10.1542/peds.104.5.e64. [DOI] [PubMed] [Google Scholar]
- 8.Hojsak I, Snovak N, Abdović S, Szajewska H, Mišak Z, Kolaček S. 2010. Lactobacillus GG in the prevention of gastrointestinal and respiratory tract infections in children who attend day care centers: a randomized, double-blind, placebo-controlled trial. Clin Nutr 29:312–316. doi: 10.1016/j.clnu.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Hatakka K, Savilahti E, Pönkä A, Meurman JH, Poussa T, Näse L, Saxelin M, Korpela R. 2001. Effect of long term consumption of probiotic milk on infections in children attending day care centres: double blind, randomised trial. BMJ 322:1327. doi: 10.1136/bmj.322.7298.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nylund L, Satokari R, Nikkilä J, Rajilić-Stojanović M, Kalliomäki M, Isolauri E, Salminen S, de Vos WM. 2013. Microarray analysis reveals marked intestinal microbiota aberrancy in infants having eczema compared to healthy children in at-risk for atopic disease. BMC Microbiol 13:12. doi: 10.1186/1471-2180-13-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elena SF, Lenski RE. 2003. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet 4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 12.Finkel SE, Kolter R. 1999. Evolution of microbial diversity during prolonged starvation. Proc Natl Acad Sci U S A 96:4023–4027. doi: 10.1073/pnas.96.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jerison ER, Desai MM. 2015. Genomic investigations of evolutionary dynamics and epistasis in microbial evolution experiments. Curr Opin Genet Dev 35:33–39. doi: 10.1016/j.gde.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF. 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 15.Schneider D, Duperchy E, Coursange E, Lenski RE, Blot M. 2000. Long-term experimental evolution in Escherichia coli. IX. Characterization of insertion sequence-mediated mutations and rearrangements. Genetics 156:477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasinkangas P, Reunanen J, Douillard FP, Ritari J, Uotinen V, Palva A, de Vos WM. 2014. Genomic characterization of non-mucus-adherent derivatives of Lactobacillus rhamnosus GG reveals genes affecting pilus biogenesis. Appl Environ Microbiol 80:7001–7009. doi: 10.1128/AEM.02006-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saedler H, Reif HJ, Hu S, Davidson N. 1974. IS2, a genetic element for turn-off and turn-on of gene activity in E. coli. Mol Gen Genet 132:265–289. doi: 10.1007/BF00268569. [DOI] [PubMed] [Google Scholar]
- 18.Douillard FP, Ribbera A, Järvinen HM, Kant R, Pietilä TE, Randazzo C, Paulin L, Laine PK, Caggia C, von Ossowski I, Reunanen J, Satokari R, Salminen S, Palva A, de Vos WM. 2013. Comparative genomic and functional analysis of Lactobacillus casei and Lactobacillus rhamnosus strains marketed as probiotics. Appl Environ Microbiol 79:1923–1933. doi: 10.1128/AEM.03467-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider D, Lenski RE. 2004. Dynamics of insertion sequence elements during experimental evolution of bacteria. Res Microbiol 155:319–327. doi: 10.1016/j.resmic.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 20.Kankainen M, Paulin L, Tynkkynen S, von Ossowski I, Reunanen J, Partanen P, Satokari R, Vesterlund S, Hendrickx APA, Lebeer S, De Keersmaecker SCJ, Vanderleyden J, Hämäläinen T, Laukkanen S, Salovuori N, Ritari J, Alatalo E, Korpela R, Mattila-Sandholm T, Lassig A, Hatakka K, Kinnunen KT, Karjalainen H, Saxelin M, Laakso K, Surakka A, Palva A, Salusjärvi T, Auvinen P, de Vos WM. 2009. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc Natl Acad Sci U S A 106:17193–17198. doi: 10.1073/pnas.0908876106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sybesma W, Molenaar D, van IW, Venema K, Kort R. 2013. Genome instability in Lactobacillus rhamnosus GG. Appl Environ Microbiol 79:2233–2239. doi: 10.1128/AEM.03566-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Douillard FP, Ribbera A, Kant R, Pietilä TE, Järvinen HM, Messing M, Randazzo CL, Paulin L, Laine P, Ritari J, Caggia C, Lähteinen T, Brouns SJ, Satokari R, von Ossowski I, Reunanen J, Palva A, de Vos WM. 2013. Comparative genomic and functional analysis of 100 Lactobacillus rhamnosus strains and their comparison with strain GG. PLoS Genet 9:e1003683. doi: 10.1371/journal.pgen.1003683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Douillard FP, de Vos WM. 2014. Functional genomics of lactic acid bacteria: from food to health. Microb Cell Fact 13(Suppl 1):S8. doi: 10.1186/1475-2859-13-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bachmann H, Starrenburg MJC, Molenaar D, Kleerebezem M, van Hylckama Vlieg JET. 2012. Microbial domestication signatures of Lactococcus lactis can be reproduced by experimental evolution. Genome Res 22:115–124. doi: 10.1101/gr.121285.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lebeer S, Verhoeven TLA, Perea Velez M, Vanderleyden J, De Keersmaecker SCJ. 2007. Impact of environmental and genetic factors on biofilm formation by the probiotic strain Lactobacillus rhamnosus GG. Appl Environ Microbiol 73:6768–6775. doi: 10.1128/AEM.01393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lebeer S, Claes I, Tytgat HLP, Verhoeven TLA, Marien E, von Ossowski I, Reunanen J, Palva A, Vos WM, De Keersmaecker SCJ, Vanderleyden J. 2012. Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl Environ Microbiol 78:185–193. doi: 10.1128/AEM.06192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Keersmaecker SCJ, Braeken K, Verhoeven TLA, Perea Velez M, Lebeer S, Vanderleyden J, Hols P. 2006. Flow cytometric testing of green fluorescent protein-tagged Lactobacillus rhamnosus GG for response to defensins. Appl Environ Microbiol 72:4923–4930. doi: 10.1128/AEM.02605-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Douillard FP, Rasinkangas P, von Ossowski I, Reunanen J, Palva A, de Vos WM. 2014. Functional identification of conserved residues involved in Lactobacillus rhamnosus strain GG sortase specificity and pilus biogenesis. J Biol Chem 289:15764–15775. doi: 10.1074/jbc.M113.542332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahlroos T, Tynkkynen S. 2009. Quantitative strain-specific detection of Lactobacillus rhamnosus GG in human faecal samples by real-time PCR. J Appl Microbiol 106:506–514. doi: 10.1111/j.1365-2672.2008.04018.x. [DOI] [PubMed] [Google Scholar]
- 30.David M, Dzamba M, Lister D, Ilie L, Brudno M. 2011. SHRiMP2: sensitive yet practical SHort Read Mapping. Bioinformatics 27:1011–1012. doi: 10.1093/bioinformatics/btr046. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delcher AL, Phillippy A, Carlton J, Salzberg SL. 2002. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res 30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holt KE, Parkhill J, Mazzoni CJ, Roumagnac P, Weill FX, Goodhead I, Rance R, Baker S, Maskell DJ, Wain J, Dolecek C, Achtman M, Dougan G. 2008. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat Genet 40:987–993. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denoeud F, Vergnaud G. 2004. Identification of polymorphic tandem repeats by direct comparison of genome sequence from different bacterial strains: a web-based resource. BMC Bioinformatics 5:4. doi: 10.1186/1471-2105-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Fleche P, Hauck Y, Onteniente L, Prieur A, Denoeud F, Ramisse V, Sylvestre P, Benson G, Ramisse F, Vergnaud G. 2001. A tandem repeats database for bacterial genomes: application to the genotyping of Yersinia pestis and Bacillus anthracis. BMC Microbiol 1:2. doi: 10.1186/1471-2180-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grissa I, Bouchon P, Pourcel C, Vergnaud G. 2008. On-line resources for bacterial micro-evolution studies using MLVA or CRISPR typing. Biochimie 90:660–668. doi: 10.1016/j.biochi.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 37.Notredame C, Higgins DG, Heringa J. 2000. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol 302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 38.Sturn A, Quackenbush J, Trajanoski Z. 2002. Genesis: cluster analysis of microarray data. Bioinformatics 18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 39.Yadav P, Harcy V, Argueso JL, Dominska M, Jinks-Robertson S, Kim N. 2014. Topoisomerase I plays a critical role in suppressing genome instability at a highly transcribed G-quadruplex-forming sequence. PLoS Genet 10:e1004839. doi: 10.1371/journal.pgen.1004839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Usongo V, Drolet M. 2014. Roles of type 1A topoisomerases in genome maintenance in Escherichia coli. PLoS Genet 10:e1004543. doi: 10.1371/journal.pgen.1004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drake JW, Charlesworth B, Charlesworth D, Crow JF. 1998. Rates of spontaneous mutation. Genetics 148:1667–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prasad J, McJarrow P, Gopal P. 2003. Heat and osmotic stress responses of probiotic Lactobacillus rhamnosus HN001 (DR20) in relation to viability after drying. Appl Environ Microbiol 69:917–925. doi: 10.1128/AEM.69.2.917-925.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Louesdon S, Charlot-Rougé S, Juillard V, Tourdot-Maréchal R, Béal C. 2014. Osmotic stress affects the stability of freeze-dried Lactobacillus buchneri R1102 as a result of intracellular betaine accumulation and membrane characteristics. J Appl Microbiol 117:196–207. doi: 10.1111/jam.12501. [DOI] [PubMed] [Google Scholar]
- 44.Taïbi A, Dabour N, Lamoureux M, Roy D, LaPointe G. 2011. Comparative transcriptome analysis of Lactococcus lactis subsp. cremoris strains under conditions simulating cheddar cheese manufacture. Int J Food Microbiol 146:263–275. doi: 10.1016/j.ijfoodmicro.2011.02.034. [DOI] [PubMed] [Google Scholar]
- 45.van de Guchte M, Serror P, Chervaux C, Smokvina T, Ehrlich SD, Maguin E. 2002. Stress responses in lactic acid bacteria. Antonie Van Leeuwenhoek 82:187–216. doi: 10.1023/A:1020631532202. [DOI] [PubMed] [Google Scholar]
- 46.Altermann E, Buck LB, Cano R, Klaenhammer TR. 2004. Identification and phenotypic characterization of the cell-division protein CdpA. Gene 342:189–197. doi: 10.1016/j.gene.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 47.Ardita CS, Mercante JW, Kwon YM, Luo L, Crawford ME, Powell DN, Jones RM, Neish AS. 2014. Epithelial adhesion mediated by pilin SpaC is required for Lactobacillus rhamnosus GG-induced cellular responses. Appl Environ Microbiol 80:5068–5077. doi: 10.1128/AEM.01039-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reunanen J, von Ossowski I, Hendrickx APA, Palva A, de Vos WM. 2012. Characterization of the SpaCBA pilus fibers in the probiotic Lactobacillus rhamnosus GG. Appl Environ Microbiol 78:2337–2344. doi: 10.1128/AEM.07047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.