ABSTRACT
The E7 oncoprotein of the high-risk human papillomavirus (HPV) plays a major role in HPV-induced carcinogenesis. E7 abrogates the G1 cell cycle checkpoint and induces genomic instability, but the mechanism is not fully understood. In this study, we performed RNA sequencing (RNA-seq) to characterize the transcriptional profile of keratinocytes expressing HPV 16 (HPV-16) E7. At the transcriptome level, 236 genes were differentially expressed between E7 and vector control cells. A subset of the differentially expressed genes, most of them novel to E7-expressing cells, was further confirmed by real-time PCR. Of interest, the activities of multiple transcription factors were altered in E7-expressing cells. Through bioinformatics analysis, pathways altered in E7-expressing cells were investigated. The upregulated genes were enriched in cell cycle and DNA replication, as well as in the DNA metabolic process, transcription, DNA damage, DNA repair, and nucleotide metabolism. Specifically, we focused our studies on the gene encoding WDHD1 (WD repeat and high mobility group [HMG]-box DNA-binding protein), one of the genes that was upregulated in E7-expressing cells. WDHD1 is a component of the replisome that regulates DNA replication. Recent studies suggest that WDHD1 may also function as a DNA replication initiation factor as well as a G1 checkpoint regulator. We found that in E7-expressing cells, the steady-state level of WDHD1 protein was increased along with the half-life. Moreover, downregulation of WDHD1 reduced E7-induced G1 checkpoint abrogation and rereplication, demonstrating a novel function for WDHD1. These studies shed light on mechanisms by which HPV induces genomic instability and have therapeutic implications.
IMPORTANCE The high-risk HPV types induce cervical cancer and encode an E7 oncoprotein that plays a major role in HPV-induced carcinogenesis. However, the mechanism by which E7 induces carcinogenesis is not fully understood; specific anti-HPV agents are not available. In this study, we performed RNA-seq to characterize transcriptional profiling of keratinocytes expressing HPV-16 E7 and identified more than 200 genes that were differentially expressed between E7 and vector control cells. Through bioinformatics analysis, pathways altered in E7-expressing cells were identified. Significantly, the WDHD1 gene, one of the genes that is upregulated in E7-expressing cells, was found to play an important role in E7-induced G1 checkpoint abrogation and rereplication. These studies shed light on mechanisms by which HPV induces genomic instability and have therapeutic implications.
INTRODUCTION
Human papillomaviruses (HPVs) are small DNA viruses that replicate in squamous epithelia. Specific types of HPV (high-risk HPVs) are the causative agents for cervical and several other cancers (1). The transforming properties of high-risk HPVs such as HPV 16 (HPV-16) primarily depend on E7 as well as E6 oncogenes (1, 2). HPV E6 and E7 proteins promote the degradation of p53 and pRb, respectively (3, 4). E7 from the high-risk HPV types can abrogate cell cycle checkpoints and induces genomic instability. Although several transcription profiling studies for E7 have been conducted using DNA microarray analysis (3, 5–7), the HPV E7 activities downstream from, or independent of, pRb responsible for deregulation of cell cycle and induction of genomic instability are not fully understood.
Cell cycle progression is regulated by cyclins and by cyclin-dependent kinases (Cdks) and their regulatory proteins at several checkpoints (8). Once the checkpoint becomes abnormal, genomic instability may occur (8). Genomic instability is a hallmark of cancer progression (9). Polyploidy is a type of genomic instability where cells have more than two sets of chromosomes and has been recognized as a causal factor for tumorigenesis (10). Significantly, polyploidy can be detected in the early stage of cervical carcinogenesis (11). Polyploidy can be formed via rereplication, a process of successive rounds of host DNA replication without entering mitosis (12). Rereplication may lead to not only polyploidy but also gene amplification, DNA fragmentation, DNA breaks, and cellular DNA damage response (13–15). We recently demonstrated that HPV-16 E7 induces rereplication and that the cellular DNA replication initiation factor Cdt1 plays a role in this process (16).
DNA replication is regulated by sequential and interactive mechanisms to ensure that the genome is accurately replicated only once per cell cycle. The process of replication initiation is divided into two steps, pre-replicative complex (pre-RC) assembly and activation; the latter leads to generation of replication forks. Pre-RC starts with the association of the origin recognition complex (ORC), which then promotes the recruitment of two proteins, Cdc6 and Cdt1, onto origins. This is followed by recruitment of minichromosome maintenance 2-7 (MCM2-7) onto chromatin as a result of concerted actions of Cdc6 and Cdt1 (9). Prior to the S phase, origins are licensed by the binding of components of the replicative DNA helicase MCMs in eukaryotes (17). Afterward, licensing proteins are downregulated or inhibited such that no more origins can be licensed and rereplication of DNA is prevented. Cells employ a licensing checkpoint to monitor that sufficient origins are licensed, inhibiting S-phase entry until that state is established (18). The G1 arrest observed in cells that have engaged in the licensing checkpoint is associated with low levels of G1 Cdk-cyclin activity and pRb hypophosphorylation.
WDHD1 (WD repeat and high mobility group [HMG]-box DNA-binding protein 1) contains multiple N-terminal WD40 domains and a C-terminal HMG box. WD40 domains are found in a variety of eukaryotic proteins and may function as adaptor/regulatory modules in signal transduction, pre-mRNA processing, and cytoskeleton assembly. HMG boxes are found in many eukaryotic proteins involved in chromatin assembly, transcription, and replication. In addition to its established role in pre-RC activation (19–21), WDHD1 is also involved in pre-RC assembly (19). The WDHD1 gene is localized adjacent to replication foci, interacts with human primase-DNA polymerase/MCM10, and is required for DNA synthesis (20, 22, 23). A role for WDHD1 in G1 checkpoint control has recently been suggested (23). In addition, depletion of WDHD-1 increases DNA damage, leading to the accumulation of late S- and/or G2-phase cells (24).
In this study, we performed high-throughput RNA sequencing (RNA-seq) to characterize the transcriptional profile of keratinocytes expressing HPV-16 E7. In E7-expressing cells, the expression levels of hundreds of genes were found to be differentially regulated, the activity of multiple transcription factors was altered, and multiple molecular pathways were changed. The WDHD1 gene is among the genes upregulated in E7-expressing cells. Significantly, downregulation of the WDHD1 gene reduced E7-induced G1 checkpoint abrogation and rereplication. These results should help provide insights into the cellular pathways targeted during tumor development caused by HPV.
MATERIALS AND METHODS
Cell culture.
Primary human keratinocytes (PHKs) and spontaneously immortalized human foreskin keratinocytes (NIKS cells) were cultured on mitomycin C-treated J2-3T3 feeder cells with E medium composed of 3 parts Dulbecco's modified Eagle medium (DMEM) and 1 part Ham's F12 medium plus 5% fetal bovine serum (FBS). Cells of the human telomerase reverse transcriptase-expressing human retinal pigment epithelium cell line RPE1 were maintained in a 1:1 dilution of DMEM-Ham's F-12 medium plus 10% FBS. PHK and NIKS cells expressing HPV-16 E7 and RPE1 cells expressing HPV-16 E7 or HPV-6 E7 were established using a pBabe retroviral system as described previously (16). Populations of infected cells were pooled and expanded. PHKs, NIKS cells, and RPE1-derived cell lines were maintained in puromycin and used within 15 passages.
RNA-seq.
Total RNA from NIKS cells was used to construct cDNA libraries for RNA-seq. First, the rRNA was removed by the use of a RiboMinus kit (Life Technologies) combined with custom-designed DNA probes for rRNAs. The processed total RNA was then used to construct RNA-seq libraries with a NEBNext mRNA library preparation kit (New England BioLabs). In this way, double-stranded cDNA was synthesized from rRNA-depleted total RNA. RNA was then end repaired, dA tailed, and ligated to standard Illumina adaptor oligonucleotides. Adaptor-ligated cDNA libraries were amplified with Phusion PCR master mix. The cDNA libraries were then loaded into a HiSeq 2000 system (Illumina) for sequencing at the Washington University Genome Technology Access Center. The resulting raw sequence reads were first analyzed using a custom bioinformatics pipeline to remove low-quality reads and then clustered before sequentially mapping to the human transcriptome and genome with Bowtie was performed (25). In this way, about 90% of all sequence reads were mapped to known human sequences. Sequence reads mapping to the same transcript were combined and normalized on the basis of the length of the transcript and the number of total reads from each sample (reads per kilobase per million [RPKM]). Normalized read counts were compared across samples to identify changes in transcript abundance. Transcripts from the same gene locus in the human genome were combined for evaluation of expression changes at the gene level.
Bioinformatics analysis.
The official gene designations for the genes that showed significant expression differences between E7 and vector cells were submitted to the Database for Annotation, Visualization and Integrated Discovery (DAVID, v6.7) Bioinformatics Resources. Homo sapiens were selected as the annotation species. Functional classification of differentially expressed genes was analyzed by gene ontology (GO) via the use of the Web-based Gene Set Analysis Toolkit. Pathway analysis was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) PATHWAY database via Web Gestalt. To identify the statistical significance of the cluster results, we set a P value of ≤0.05 as the cluster filter. We then used the gene counts and terms to make the column chart separately for each category.
RT-PCR.
Total RNA from RPE1, NIKS, and PHK control cells and the corresponding E7-expressing cells was isolated using an RNeasy kit (Qiagen) according to the manufacturer's instruction. cDNA was synthesized with a Superscript VILO cDNA synthesis kit (Invitrogen). iTaq Universal SYBR green Supermix (Bio-Rad) was used in a Bio-Rad CFX96 Touch Real-Time PCR detection system for quantitative real-time PCR (qRT-PCR). Data were analyzed using the threshold cycle (2−ΔΔCT) method. The primer sequences are listed in Table 1.
TABLE 1.
The primer sequences
| Gene | Forward primer | Reverse primer |
|---|---|---|
| WDHD1 | GCTTCAGGTCGTCCTAGACAG | CCTTTGGGATGTTACAAGTGGT |
| UHRF1 | AGGTGGTCATGCTCAACTCAC | ACAGTTGGCGTAGAGTTCCC |
| CHAF1B | CGGAGCAGATCGCTTTTCAG | CCCAGGTTACTCCTTGGACATAA |
| ASF1B | TCCGGTTCGAGATCAGCTTC | GTCGGCCTGAAAGACAAACA |
| MRE11A | GGGGCAGATGCACTTTGTG | GAAGCAAAACCGGACTAATGTCT |
| MELK | TCTCCCAGTAGCATTCTGCTT | TGATCCAGGGATGGTTCAATAGA |
| HMMR | ATGATGGCTAAGCAAGAAGGC | TTTCCCTTGAGACTCTTCGAGA |
| DEK | AACTGCTTTACAACAGGCCAG | ATGGTTTGCCAGAAGGCTTTG |
| FN1 | CGGTGGCTGTCAGTCAAAG | AAACCTCGGCTTCCTCCATAA |
| UBE2L6 | TGGACGAGAACGGACAGATTT | GGCTCCCTGATATTCGGTCTATT |
| TIMP1 | AGAGTGTCTGCGGATACTTCC | CCAACAGTGTAGGTCTTGGTG |
| PTHLH | ATTTACGGCGACGATTCTTCC | GCTTGGAGTTAGGGGACACC |
| c-MYC | AAGCTGAATTGTGCAGGCATC | CTCACCCAAAGGCATTTTAAG |
| HPV16E7 | AGTGTGACTCTACGCTTCGGTTG | CTGAGAACAGATGGGGCACAC |
| HPV6E7 | GACGAAGTGGACGGACAAGA | TCCGCCATCGTTGTTAGGTC |
| MCM3 | GAAGACCAGGGAATTT | AGGCAACCAGCTCCTACCAG |
ChIP assay.
The chromatin immunoprecipitation (ChIP) assay was performed using a ChIP assay kit from Millipore, following the supplied protocol. Immunoprecipitations were performed using anti-MCM3 or control IgG antibodies. PCR was performed with the primers designed from the sequences of the human c-MYC gene as follows: sense, AAGCTGAATTGTGCAGTGCATC; antisense, CTCACCCAAAGGCATTTTAAG. It was shown that the MCM protein complex binds to the DNA replication initiation zone upstream of the c-MYC gene (26).
Flow cytometry.
For cell cycle and polyploidy analysis, asynchronous cultured cells expressing HPV E7 or vector alone were treated with phosphate-buffered saline (PBS) or bleomycin (Alexis Biochemicals) (4 μg/ml in PBS). At 24 h later, cells were fixed in 70% ethanol, treated with 50 μg/ml RNase A plus 50 μg/ml propidium iodide, and analyzed by fluorescence-activated cell sorting (FACS). For the bromodeoxyuridine (BrdU) labeling experiment, BrdU (final concentration, 20 μM) was added to the medium 2 h before collection of cells. After fixation, cells were permeabilized with 2 N HCl–0.5% Triton X-100, neutralized with 0.1 M sodium tetraborate, stained with monoclonal anti-BrdU (BD Biosciences) followed by treatment with anti-mouse IgG F(ab)2-fluorescein isothiocyanate (FITC) (Sigma), and counterstained with PBS–7-aminoactinomycin D (7-AAD)–RNase A. Flow cytometric analysis was performed on a BD FACSAria III sorter instrument equipped with BD FACSDiva 7.0 software (BD Biosciences, NJ, USA). FITC 490-nm fluorescence was acquired in logarithmic amplification in FL1, and 7-AAD 650-nm fluorescence was acquired in linear amplification in FL3. Cell cycle analysis was done using a Cytomics FC500 Flow Cytometry CXP 2.0 system.
siRNAs and transfection.
Cells were transfected with a final concentration of 20 nM small interfering RNA (siRNA) per target gene using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's instructions. For gene knockdown analysis, cells were harvested 48 h posttransfection and specific protein levels were analyzed by immunoblotting. For cell cycle analysis, 24 h after transfection, cells were treated with bleomycin (4 μg/ml) for an additional 24 h. For polyploidy analysis, cells were blocked in G1 phase with thymidine for 16 h after transfection with 20 nM siRNA for 24 h, and then cells were treated with bleomycin (4 μg/ml) for an additional 48 h. The siRNA duplexes were as follows: for the small interfering WDHD1-1 (si-WDHD1-1) sense strand, 5′-GCAUGUACCCUAAGAAUAA-3′; for the si-WDHD1-2 sense strand, 5′-GCAAAGUUAUGGAAAGUAU-3′; for the si-MCM3 sense strand, 5′-GCATTGTCACTAAATGTTCTCTAGT-3′ (27); and for the negative-control (NC) siRNA sense strand, 5′-UUCUCCGAACGUGUCACGU-3′.
Immunoblotting.
Total cellular protein was prepared in lysis buffer (10 mM Tris [pH 7.4], 1% SDS, 1.0 mM sodium orthovanadate). To obtain cytoplasmic and nuclear proteins, cells were extracted with a nuclear extract kit (Active motif; 100946). The protein concentration was measured by the use of bicinchoninic acid (BCA) protein assay reagent (Pierce) and confirmed by Coomassie blue staining of membranes after blotting. Equal amounts of protein from each cell lysate were separated in an SDS polyacrylamide gel (PAGE) and transferred onto a nitrocellulose filter membrane (NC) membrane. Membranes were blotted with antibodies against WDHD1 (abcam; ab72436), MCM3 (abcam; ab4460), SP1 (Cell Signaling; catalog no. 9389), and tubulin (Sigma; T-4026). Protein bands were detected using an Odyssey infrared imaging system (Li-COR, Lincoln, NE) and quantified using ImageJ (NIH). WDHD1 half-life was measured following a cycloheximide (25 μg/ml) chase procedure and calculated using Half Life Calculator (www.calculator.net).
Statistical analysis.
Data are presented as means and standard deviations (SDs). The Student t test was used to evaluate the differences between means. P values of <0.05 were considered significant.
RESULTS
Gene expression profiling of HPV-16 E7-expressing cells.
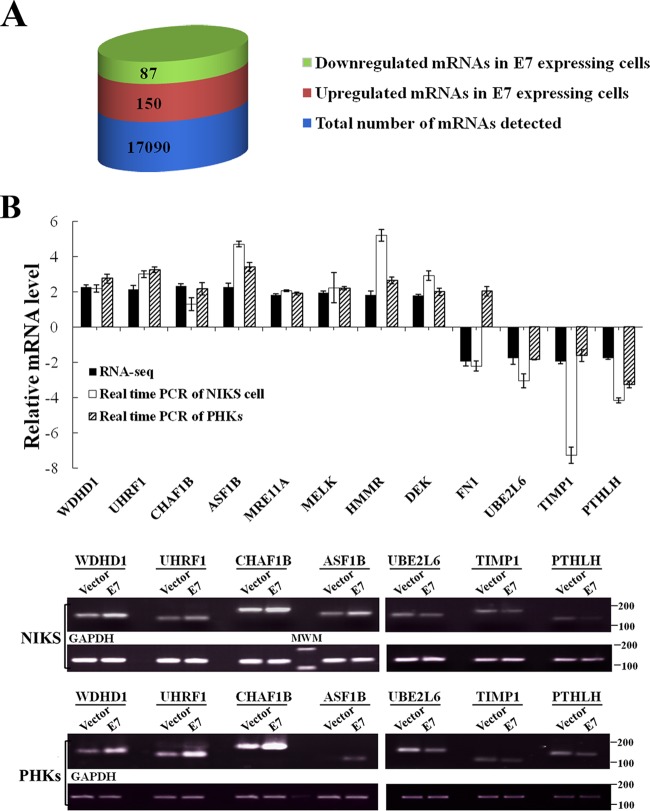
To identify genes differentially expressed between cells with and without E7, we used NIKS cells (28). NIKS cells exhibit many characteristics of early-passage human keratinocytes, the natural host cells for HPV, including stratification, differentiation, and the ability to sustain the HPV life cycle (29–31). The use of NIKS cells for HPV oncogene studies can avoid comparisons being made between senescing (vector) and proliferating (E7) cells, when the experimental goal is to explore the biological activities of E7. In addition, while PHKs do not proliferate efficiently, NIKS cells grow relatively well in culture. For gene expression profiling analysis, the RNA-seq approach was used. From RNA-seq data, we detected a total of 20,537 transcripts that included 17,090 mRNAs. After normalization, 237 genes were identified that were differentially expressed between E7-expressing and vector control NIKS cells (Fig. 1A). Among these, 150 genes were upregulated (between 1.7-fold and 4.3-fold) and 87 genes were downregulated (between 1.7-fold and 4.3-fold) in NIKS cells expressing E7 (Fig. 1A; see also Table S1 in the supplemental material).
FIG 1.
Genes differentially expressed in HPV-16 E7-expressing cells. (A) Numbers of genes differentially expressed between NIKS cells expressing E7 and a vector control, as determined by RNA-seq, are presented as columns. Blue, total number of protein-coding mRNAs detected; red, upregulated genes in E7-expressing cells; green, downregulated genes in E7-expressing cells. (B) Real-time PCR assay for selected genes in keratinocytes. Upper panel, relative expression levels of mRNAs in E7-expressing NIKS cells or PHKs compared with vector, measured by real-time PCR and RNA-seq. Lower panels, images of DNA gel electrophoresis. MWM, molecular weight marker.
To verify the RNA-seq results, we performed real-time PCR assay for nearly a dozen genes selected on the basis of their potential E7-related biological functions. Among these genes, consistent with RNA-seq analysis results, the WDHD1, UHRF1, CHAF1B, ASF1B, MRE11, MELK, HMMR, and DEK genes were shown to be upregulated whereas the FN1 (fibronectin), UBE2L6, and TIMP1 genes were shown to be downregulated in E7-expressing NIKS cells (Fig. 1B). For example, the WDHD1 (WD repeat and HMG-box DNA-binding protein 1) gene was shown to be upregulated by more than 2-fold in E7-expressing NIKS cells by RNA-seq and it was verified to be similarly upregulated by real-time PCR; the UBE2L6 (ubiquitin/ISG15-conjugating enzyme E2 L6) gene was shown to be downregulated by 1.77-fold in E7-expressing NIKS cells by RNA-seq, and it was verified to be similarly downregulated by real-time PCR. Although the fold changes as demonstrated by real-time PCR for some genes such as those encoding CHAF1B and FN1 were not dramatic, the differences are statistically significant. These are newly described differentially expressed genes in E7-expressing cells, with the exception of the DEK and FN1 genes. Thus, the RNA-seq results provided genome-wide gene expression profiling in E7-expressing cells that can be verified by real-time PCR.
Using real-time PCR, we also examined the expression of the selected genes described above in primary human keratinocytes (PHKs) expressing HPV-16 E7 or containing a vector. The expression profiles of the selected genes were similar to what was observed in NIKS cells (Fig. 1B). However, the opposite results were obtained for FN1 expression. While FN1 was downregulated in E7-expressing NIKS cells, it was upregulated in E7-expressing PHKs. It was previously reported that FN1 was downregulated in three immortal nontumorigenic cell lines that expressed the high-risk HPV E7 protein (32). These results show that the expression profiles for the selected genes are similar, with the exception of the expression profiles of NIKS cells and PHKs expressing HPV E7.
Functional implications of differentially expressed genes in HPV E7-expressing cells.
Some of the differentially expressed genes in E7-expressing cells identified in this study have previously been shown to be regulated by HPV E7 at the transcription level. These genes are summarized in Table 2. E7 is known to abrogate cell cycle checkpoints. Consistent with this notion, a group of cell cycle-related genes are differentially expressed in E7-expressing cells. These include the CCNA2, Cdc25A, and MYBL2 genes. Expression of the CCNA2 gene, which encodes cyclin A2, is upregulated in E7-expressing cells (51). The tyrosine phosphatase Cdc25A gene is an E2F1 target gene (52) and is involved in the regulation of the G1/S-phase transition (53). The Cdc25A gene is upregulated in E7-expressing cells (54, 55, 57). The MYBL2 (B-Myb) gene is an E2F-responsive gene and a component of the DREAM complex that promotes expression of genes during the G2/M phase of the cell cycle (48). B-Myb has been implicated in a positive regulatory role for Cdk1 expression (49, 56). We and others have shown that E7 upregulates B-Myb expression (7, 46). Moreover, downregulation of B-Myb-induced G1 arrest in E7-expressing cells upon DNA damage has been described previously (59).
TABLE 2.
Differentially expressed genes in E7-expressing cells
| Gene designation | Ratio of E7/vector by RNA-seq | Regulation categorya (reference[s]) | Product description |
|---|---|---|---|
| S100P | 4.27 | ↑ (50, 52) | S100 calcium binding protein P |
| DHFR | 3.54 | ↑ (53) | Dihydrofolate reductase |
| CDC6 | 3.00 | ↑ (54) | Cell division cycle 6 |
| RAD51AP1 | 1.98 | ↑ (55) | RAD51-associated protein 1 |
| APOBEC3B | 1.97 | ↑ (47) | Apolipoprotein B mRNA editing enzyme catalytic polypeptide-like 3B |
| DEK | 1.80 | ↑ (48, 49) | DEK oncogene |
| CDC25A | 1.82 | ↑ (56) | Cell division cycle 25A |
| MSH6 | 1.78 | ↑ (57) | MutS homolog 6 |
| CCNA2 | 1.78 | ↑ (57) | Cyclin A2 |
| BARD1 | 1.77 | ↑ (59) | BRCA1-associated RING domain 1 |
| MYBL2 | 1.79 | ↑ (7, 46) | V-myb myeloblastosis viral oncogene homolog (avian)-like 2 |
| S100A8 | 1.70 | ↑ (58) | S100 calcium binding protein A8 |
| FN1 | 0.50 | ↓ (59) | Fibronectin 1 |
Upward-pointing arrows represent upregulation; downward-pointing arrows represent downregulation.
Some other genes such as the APOBEC-3B, BARD1, DEK, DHFR, MSH6, RAD51AP1, S100P, and S100A8 genes are upregulated in E7-expressing cells and may play critical roles in cervical carcinogenesis. Among these genes, the APOBEC-3B gene, a member of the cytidine deaminase gene family, is activated by HPV infection and increases genome instability (46). The nuclear protein DEK has shown to be upregulated in E7-expressing cells as well as cervical intraepithelial neoplasia (CIN) and cervical cancers. DEK was reported to be required for cellular immortalization by HPV E7 (47, 48). DEK upregulation may be a common event in human carcinogenesis and may reflect its senescence inhibitory function (47). The RAD51AP1 gene (a DNA repair gene) is an S-phase cell cycle checkpoint gene that is directly induced by E2F1 and becomes overexpressed when pRB is inactivated by E7 (49).
Bioinformatic analysis of genes differentially expressed in E7-expressing cells.
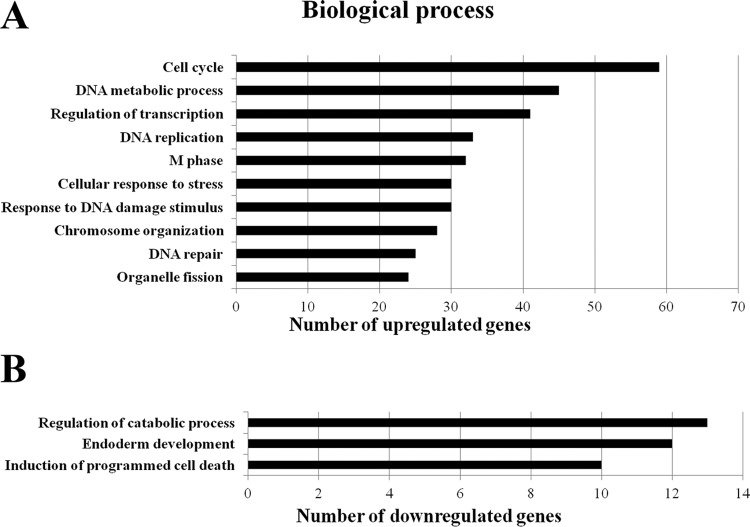
We next performed bioinformatics analysis on genes differentially expressed in E7-expressing cells. First we used the Gene Ontology (GO) system, a gene function classification system that provides a comprehensive description of gene properties (60). Based on the GO annotation, the 150 upregulated genes could be classified into multiple functional groups (Fig. 2A). In the “biological processes” category, more than 50 genes belonged to the “cell cycle” functional group. Some additional cell cycle-related genes were in the “M phase,” “cell division,” and “cell proliferation” functional groups. Taking the data together, after eliminating overlapping genes, a total of 59 genes were shown to be cell cycle related. These results are consistent with the known cell cycle regulatory functions of HPV E7 (61). Interestingly, more than 40 upregulated genes in E7-expressing cells were in the “DNA metabolic process” (such as DNA repair and replication) group. In addition, 33 upregulated genes belonged to the “DNA replication” functional group, which is consistent with the known function of E7 to establish an S-phase-like environment in otherwise differentiating epithelial cells to support HPV replication (62). There were also 41 genes and 28 genes, respectively, in the “regulation of transcription” and “transcription” functional groups. It is known that E7 is involved in transcriptional regulation, partly through degradation of pRb and release of E2F.
FIG 2.
GO categories of differentially expressed genes in NIKS cells expressing HPV-16 E7. (A) Biological process category. Ten groups (with more than 22 genes each) of upregulated genes (n = 157) are listed. (B) Biological process category. Three groups (with more than 10 genes each) of downregulated genes (n = 67) are listed.
E7 has been known to regulate the DNA damage response (16). In fact, E7 also induces DNA damage (63, 64). Consistently, a total of 30 genes that were upregulated in E7-expressing cells belonged to the “cellular response to stress,” “response to DNA damage stimulus,” and “DNA repair” groups. In addition, a total of 30 genes belonged to the “chromosome organization” group and 24 genes belonged to the “organelle fission” group (the members of which are employed to obtain a variety of organelles in cell division). The significance of the latter two groups to E7 function remains to be established.
Interestingly, genes in a group related to “regulation of catabolic process” were downregulated in E7-expressing cells (Fig. 2B). Genes for “endoderm development” were also downregulated in E7-expressing cells. However, the significance of these findings is not known. Notably, antiapoptotic genes in the “induction of programmed cell death” functional group are also downregulated in E7-expressing cells. Cells expressing HPV-16 E7 are predisposed to undergo apoptosis (65, 66), and the observation is therefore consistent with this finding.
Next, we searched the differentially expressed genes using the KEGG PATHWAY database, a collection of pathway maps representing knowledge on the molecular interaction and reaction networks. Nine biological pathways that were significantly upregulated (6 or more genes with altered expression) in E7-expressing NIKS cells are listed in Table 3. Consistent with results of GO analysis and the known E7 functions, the “cell cycle” and “DNA replication” pathways were significantly regulated in E7-expressing cells. As expected, the members of the “pathways in cancer” group were changed. Up to 24% of upregulated genes were associated with DNA repair (“mismatch repair,” “nucleotide excision repair,” and “homologous recombination” groups), and 12% of upregulated genes were associated with nucleotide metabolism (“purine metabolism” and “pyrimidine metabolism” groups). In addition, 6% of upregulated genes are in the “p53 signaling pathway” group (data not shown).
TABLE 3.
KEGG pathway analysis of differentially expressed genes in E7-expressing cells
| Pathway IDa | KEGG pathway(s) | Total no. of genes | Highly expressed genes |
|---|---|---|---|
| 04110 | Cell cycle | 20 | E2F1, CDC6/7/25A/45, CDK1/2, RBL1, TTK, MCM27, CCNE2, CDKN2A, PCNA, MDM2, CCNA2 |
| 03030 | DNA replication | 15 | POLE, POLA1, MCM2–7, RPA1, RFC5, POLD3, PRIM1, RPA2, RFC3, PCNA |
| 05200 | Cancer (several pathways) | 9 | E2F1, CCNE2, MSH6, CDKN2A, MSH2, MDM2, BRCA2, BIRC3, CDK2 |
| 03430 | Mismatch repair | 8 | POLD3, RFC5, RPA1, MSH6, RPA2, RFC3, MSH2, PCNA |
| 03420 | Nucleotide excision repair | 7 | POLD3, RFC5, RPA1, RPA2, RFC3, POLE, PCNA |
| 04115 | p53 signaling | 6 | CCNE2, CDK1, CDKN2A, RRM2, MDM2, CDK2 |
| 03440 | Homologous recombination | 6 | POLD3, RPA1, RPA2, MRE11A, BRCA2, RAD54B |
| 00230 | Purine metabolism | 6 | POLD3, PRIM1, RRM2, RRM1, POLE, POLA1 |
| 00240 | Pyrimidine metabolism | 6 | POLD3, PRIM1, RRM2, RRM1, POLE, POLA1 |
ID, identifier.
Targets of specific transcription factors are differentially expressed in HPV E7-expressing cells.
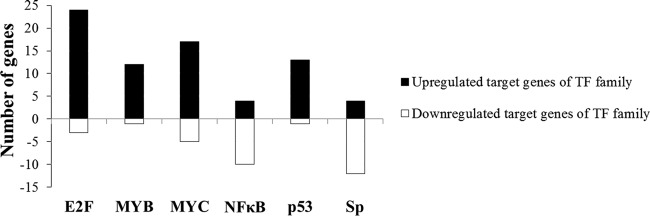
We reasoned that, on the basis of the results seen with differentially expressed genes, we might be able to identify transcription factors whose activities are differentially regulated in E7-expressing cells. For this, we analyzed 41 of the cancer-related transcription factor families described in TRED (https://cb.utdallas.edu/cgi-bin/TRED/tred.cgi?process=searchTFGeneForm) for their predicted target gene expression characteristics. Significantly, targets of the transcription family that includes E2F, MYB, MYC, NF-κB, p53, and SP are significantly differentially expressed (with at least 10 target genes that were up- or downregulated) in E7-expressing cells (Fig. 3).
FIG 3.
Differentially expressed transcription factor target gene families in HPV E7-expressing cells. Genes differentially expressed between NIKS-E7 and NIKS-vector cells are categorized as targets of transcription factors. y axis, numbers of differentially expressed genes. x axis, names of transcription factor gene families.
E7 targets the degradation of pRb and release of E2F1. E7 may also activate E2F2 transcription through interaction with HDACs (67). In contrast, E7 can associate with and inactivate the transcriptional repression activity of E2F6 (68). Consistent with these observations, as many as 24 target genes of the E2F family are upregulated in E7-expressing cells.
The MYB gene family consists of three members, namely, A-, B-, and C-Myb. Multiple lines of evidence suggest that B-Myb plays an important role in HPV-associated carcinogenesis (7, 42, 69–71). We recently showed that HPV E7 is able to activate the B-Myb gene (42, 44, 45).
Up to 17 MYC targets were upregulated in E7-expressing cells. This is consistent with the observation that HPV-18 E7 binds and augments c-Myc transcriptional activity (72). We have also demonstrated that activation of c-Myc contributes to bovine papillomavirus type 1 E7-induced cell proliferation (73). Nonetheless, the results cannot rule out the possibility that upregulation of MYC targets is an indirect effect of E2F, as most (9 of 17) of the upregulated MYC targets are also E2F targets.
Interestingly, most (10 of 14) of the targets of NF-κB family were downregulated in E7-expressing cells. NF-κB plays a key role in regulating the immune response to infection. Abnormal regulation of NF-κB has been linked to cancer, viral infection, and improper immune development. Inhibition of NF-κB may reflect the function of HPV to downregulate the immune system. Consistently, HPV-16 E7 inhibited NF-κB DNA binding activity (74) and attenuated NF-κB activation (75, 76). However, and in contrast, it was observed that E7, together with E6, upregulated NF-κB-responsive genes (77). Regulation of NF-κB activity by E7 can be complicated, depending on the cellular context.
Results from several studies indicate that in HPV E7-expressing cells, the activity of p53 is inhibited, although its steady-state level is high (78–81). However, results of one study suggest that p53 remains active in E7-expressing cells (82). The p53 family targets include p53, p63, and p73. Some were not confirmed (data not shown). Among the experimentally confirmed p53 targets, all three genes that are positively regulated by p53 (the MDM2, MSH2, and S100A9 genes) were upregulated in E7-expressing cells whereas the one gene negatively regulated by p53, the PTHLH gene (encoding parathyroid hormone-related protein), was downregulated. These results favor the notion that p53 functions in E7-expressing cells.
The Sp family transcription factors are widely expressed in human tissues and involved in the regulation of multiple cellular processes and responses to cellular microenvironment. We know little about the functional interaction of HPV with Sp family members other than SP1 binding to the HPV early promoter (83). Interestingly, we found that most of the transcriptional targets of Sp family members were downregulated in E7-expressing cells (Fig. 3).
Expression and localization of WDHD1 in E7-expressing cells.
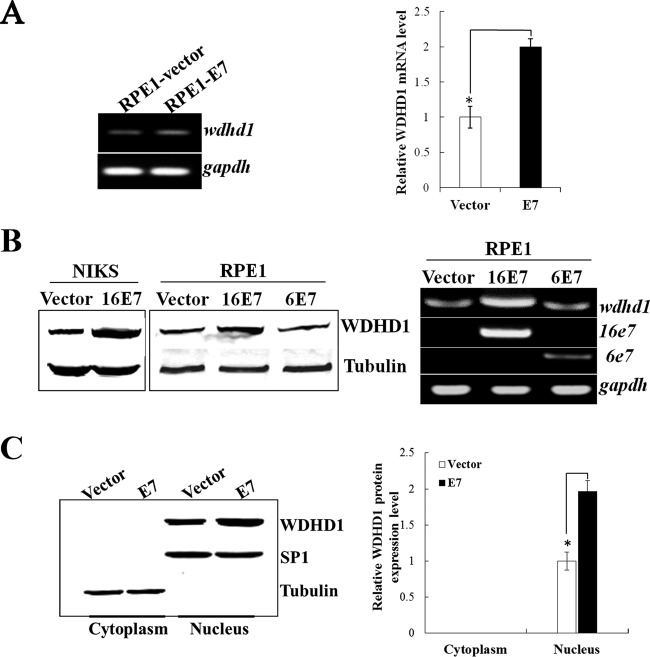
Given the fact that cell cycle and DNA replication pathways and their related genes have shown significant disregulation in E7-expressing cells, we selected WDHD1, which is known to be involved in both DNA replication and potentially G1 checkpoint regulation, for further analysis. As transfection efficiencies in keratinocytes are not very high, we included RPE1 cells. RPE1 cells expressing the wild-type E7 (RPE1-E7) and vector control (RPE1-vector) have been used in our recent HPV-related functional studies (16, 84, 85). Similarly to what was observed in keratinocytes, WDHD1 mRNA levels were increased (∼2-fold) in E7-expressing RPE1 cells (Fig. 4A). We then further examined the steady-state level of WDHD1 protein. As shown in Fig. 4B, the levels of WDHD1 protein were upregulated in both RPE1-E7 cells (1.9-fold) and NIKS-E7 cells (2.2-fold). As an initial step toward understanding the mechanism by which E7 regulates WDHD1, we examined the steady-state level of WDHD1 protein in RPE1 cells expressing HPV-6 E7, which does not degrade pRb (86). As shown in Fig. 4B, the level of WDHD1 protein was comparable to that seen with the vector control cells, suggesting that pRb degradation is important for WDHD1 upregulation by HPV E7.
FIG 4.
Upregulation and nuclear localization of WDHD1 in E7-expressing cells. (A) WDHD1 mRNA levels in RPE1 cells determined by real-time PCR analysis. (B) WDHD1 protein levels in RPE1 and NIKS cells expressing E7 or vector examined by Western blotting (left panel). WDHD1 mRNA levels in RPE1 cells expressing HPV-6 E7 or −16 E7 (right panel) are indicated. gapdh, GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene. (C) Cellular localization of WDHD1 in E7-expressing cells. Right panel, quantification of WDHD1 protein expression in different cellular compartment. Error bars reflect the standard deviations of the means. Data from an experiment representative of 3 are shown. *, P < 0.05.
WDHD1 is usually localized in the nucleus adjacent to replication foci and is required for efficient DNA replication (24, 87). We examined cellular localization of WDHD1 in E7-expressing RPE1 cells. Our study demonstrated that WDHD1 was mainly located in the nucleus, where it was present in levels that were about 2-fold higher in E7-expressing cells than in control cells (Fig. 4C).
WDHD1 depletion induces G1 arrest.
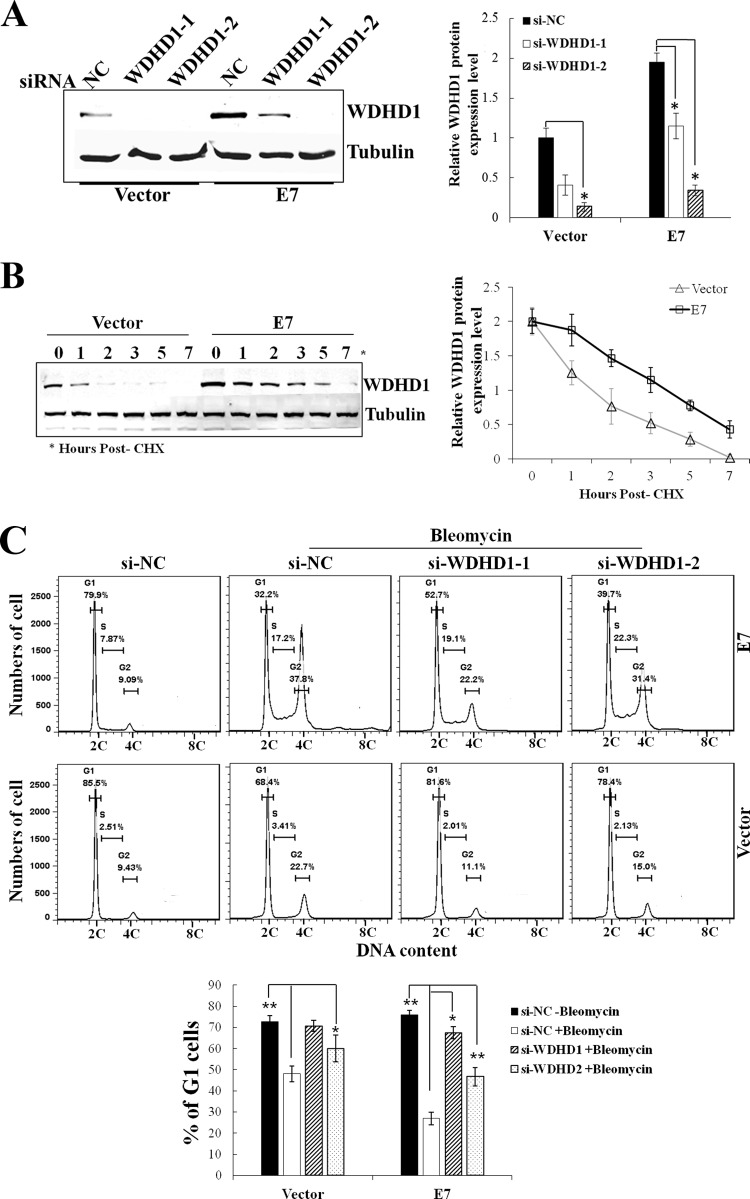
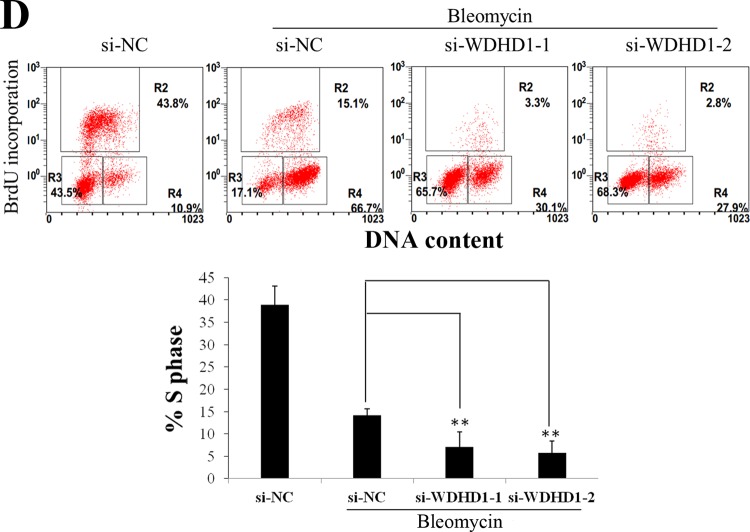
Many HMG proteins facilitate the assembly of nucleoprotein complexes to influence cell cycle (23–25). DNA replication initiation factors may also be involved in the licensing checkpoint to modulate S-phase entry (18). As an HMG box-containing protein and a DNA replication initiation factor, WDHD1 may play a role in cell cycle control in E7-expressing cells. To test this possibility, we employed the RNA interference (RNAi) approach by using two independent siRNAs. The steady-state level of WDHD1 protein was downregulated (to 0.2-fold) after transfection with si-WDHD1-2 and (to 0.5-fold) after transfection with si-WDHD1-1 in RPE1-E7 cells (Fig. 5A). We also determined the protein stability of WDHD1 in E7-expressing cells. Two hours after cycloheximide treatment, the steady-state level of WDHD1 in the vector-containing RPE1 cells had dropped by more than 50%. In contrast, ∼75% of WDHD1 was maintained in E7-expressing cells (Fig. 5B). The protein half-life of WDHD1 in E7-expressing cells was greatly increased compared with that in the vector control cells (3.4 h versus 1.6 h). Next, we examined changes of cell cycle profiles of E7-expressing and vector-containing RPE1 cells after siRNA knockdown of WDHD1. Consistent with what we have recently observed, upon treatment using the DNA-damaging agent bleomycin, fewer cells were arrested at the G1 phase in the nonsilencing siRNA control-transfected E7-expressing cells than in the vector control cells (Fig. 5C), indicating abrogation of the G1 checkpoint in E7-expressing cells. Notably, knockdown of WDHD1 with siRNAs led to an increase in the G1 peak (from 26.9% to 67.5% for si-WDHD1-1 and 46.7% for si-WDHD1-2) in E7-expressing cells. To demonstrate the role of WDHD1 in promoting S-phase entry of cells more directly, we transfected siRNAs targeting WDHD1 into E7-expressing cells and measured BrdU incorporation upon bleomycin treatment. Significantly, knockdown of WDHD1 by siRNAs led to a significant reduction of BrdU incorporation (from 13.7% to 5.3% for si-WDHD1-1 and 5.7% for si-WDHD1-2) in RPE1-E7 cells (Fig. 5D). These results demonstrate an important role of WDHD1 in the G1 cell cycle control and S-phase entry of E7-expressing cells.
FIG 5.
WDHD1 plays a role at the G1/S transition in E7-expressing cells. (A) RPE1 cells expressing E7 or containing a vector were transfected with siRNAs targeting WDHD1. The steady-state levels of WDHD1 were measured by Western blotting. Tubulin was used as a loading control. Right panel, quantification of relative WDHD1 levels from 3 independent experiments. (B) RPE1 cells were incubated with cycloheximide (CHX) and harvested at the indicated times. The stability of WDHD1 was monitored by immunoblotting analyses (left panel). Data are summarized in the right panel. (C) After siRNA transfection, cells were treated with bleomycin, stained with propidium iodide (PI), and analyzed by flow cytometry. Results of 1 experiment representative of 3 are shown (upper panel). The percentages of cells with 2C and 4C DNA content are indicated. Data are summarized in the lower panel. (D) After siRNA transfection, cells were treated with bleomycin, stained with BrdU, and analyzed by flow cytometry. Data of 3 independent experiments are summarized in the lower panel. *, P < 0.05; **, P < 0.01. NC, negative control.
WDHD1 depletion reduces rereplication in E7-expressing cells.
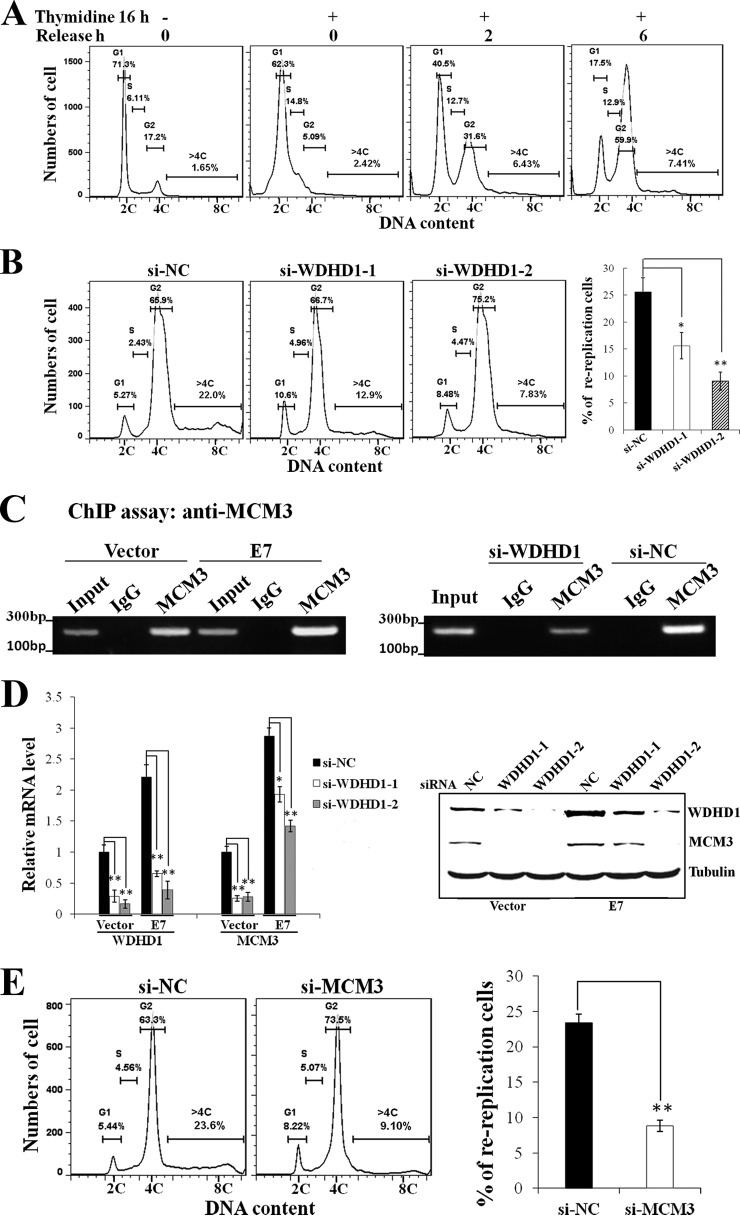
We have recently demonstrated that cells expressing HPV-16 E7 undergo rereplication upon DNA damage and that the DNA replication-initiating factor Cdt1 plays an important role in this process (16). Since WDHD1 has been implicated as a DNA replication-initiating factor (22), we examined its potential role in E7-induced rereplication. As WDHD1 depletion may block cells at the G1/S phase, we synchronized cells with thymidine and then released them into S phase and G2 phase (Fig. 6A). As shown in Fig. 6A, after thymidine release for 2 h, 30% of the cells synchronized in G1 bypassed S phase and entered G2 phase and 6% became polyploid. As time passed, 60% of cells entered G2 phase and 8% became polyploid by 6 h, suggesting that rereplication had occurred. We then used siRNAs to assess the role of WDHD1 in E7-induced rereplication upon DNA damage. Notably, the percentage of polyploidy was significantly reduced in E7-expressing cells after WDHD1 knockdown (Fig. 6B). Our recent study demonstrated that, under those conditions, polyploidy in E7-expressing cells was due to rereplication, where DNA replication occurs once again in the G2 phase of the cell cycle (88). These results indicate that WDHD1 plays an important role in E7-induced rereplication.
FIG 6.
WDHD1 depletion reduces rereplication and decreases MCM3 chromatin loading and expression. (A) RPE1-E7 cells were treated with thymidine for 16 h, harvested at different times after release from thymidine, and analyzed with flow cytometry. (B) RPE1-E7 cells were transfected with siRNAs targeting WDHD1 and treated with thymidine for 16 h followed by bleomycin treatment for 48 h after release for 2 h from thymidine. Cells were analyzed with flow cytometry. Cells with >4C DNA content were quantified (right panel). (C) RPE1 cells were subjected to ChIP assay using antibody against MCM3 (left panel). RPE1-E7 cells were transfected with siRNA targeting WDHD1 and cells were subjected to ChIP assay (right panel). (D) RPE1-E7 cells were transfected with siRNAs targeting WDHD1, and real-time PCR analysis was performed. Another set of cells (right panel) was analyzed for steady-state levels of MCM3 by Western blotting. (E) RPE1-E7 cells were transfected with siRNAs targeting MCM3. Cells with >4C DNA content were quantified (right panel). Data from an experiment representative of 3 are shown. *, P < 0.05; **, P < 0.01.
As an initial step toward understanding the mechanism by which WDHD1 knockdown causes a reduction in rereplication, we examined the loading of MCM3 onto chromatin by ChIP assay. It is known that DNA replication-initiating factors recruit MCMs to the origin of replication (89). Consistent with this notion, more MCM3 (∼2.5-fold) bound to chromatin in E7-expressing cells than in vector control cells (Fig. 6C, left panel). WDHD1 knockdown by siRNA significantly reduced MCM3 loading to the chromatin (Fig. 6C, right panel). Next, we examined the effect of knocking down WDHD1 on MCM3 expression. As shown in Fig. 6D, both MCM3 mRNA levels (left panel) and the steady-state levels of MCM3 went down after WDHD1 knockdown. In addition, WDHD1 knockdown significantly reduced polyploidy (Fig. 6E). These results indicate that WDHD1 facilitates rereplication in E7-expressing cells by modulating MCM3, via either expression or loading. The extent to which the reduced MCM3 chromatin levels seen after WDHD1 knockdown are due to loading per se or to its expression remains to be determined.
DISCUSSION
Previous transcriptional profiles of E7 were mostly determined in the presence of E6. Nonetheless, several gene expression profiling studies have been conducted on HPV E7-expressing cells using microarray analysis. The cells employed included mouse cells, cervical cancer cell lines (C33A and CaSki), and PHKs (3, 5–7, 90). However, there are major limitations with the DNA microarray approach, such as the relatively high background noise resulting from solution-based DNA probe hybridization as well as the limited scope of profiling analysis, which is confined by the DNA probe set. RNA-seq is a more sensitive method for reliably identifying differentially expressed genes, especially low-abundance genes, than traditional methods for transcriptome profiling such as microarray analyses (91). Additionally, RNA-seq profiling is more comprehensive than microarray analyses, as all expressed transcripts in the cells are included in the analysis. Our study was the first to profile the gene expression of HPV E7-expressing cells using RNA-seq. With this new method, we have identified many E7-induced gene expression changes that were not reported previously from microarray studies.
It was observed that although the steady-state level of p53 was high in HPV E7-expressing cells, its activity was low (78, 80, 92). It was therefore proposed that p53 activity is inhibited by HPV E7. However, one study found the opposite phenomenon (82). Is p53 functioning in E7-expressing cells? This is an open question and remains to be resolved. Using an unbiased approach, we showed that among the experimentally confirmed p53 targets, all genes positively regulated by p53 were upregulated in E7-expressing cells while the one negatively regulated by p53 (the PTHLH gene) (93) was downregulated. These results favor the notion that p53 is functional in E7-expressing cells. Consistently, up to 6% of the upregulated genes identified in this study have been shown to participate in the p53 signaling pathway (data not shown).
While the established cellular function is pre-RC activation for DNA synthesis, WDHD1 has also been implicated in a role in pre-RC assembly, functioning as a DNA replication initiation factor (19). The known DNA replication initiation factors, Cdt1 and Cdc6, have been shown to be required for rereplication (9, 10, 14, 34, 44, 63). However, no such function has been reported for WDHD1. The current study was the first to demonstrate that WDHD1 plays a role in rereplication. The mechanism by which WDHD1 contributes to rereplication is likely that of loading MCMs. Consistent with this notion and with a previous observation, we have shown that WDHD1 knockdown reduced chromatin loading of MCM3. Interestingly, WDHD1 knockdown reduced both MCM3 expression and its chromatin loading. It is therefore not clear whether the reduced chromatin loading of MCM3 after WDHD1 knockdown is a result of reduced MCM3 expression or of loading to chromatin or of a combination of the two. The detailed mechanism by which WDHD1 regulates MCM3 expression is also not known. Future studies will address this question.
DNA replication initiation factors have been shown to play a role in G1 checkpoint regulation. Consistently, we have shown that knockdown of WDHD1 led to G1 arrest in E7-expressing cells. It is believed that G1 arrest induced by knockdown of DNA replication-initiating factors is due to their requirement for DNA replication initiation. This is normally associated with reduced cyclin D and cyclin E activities (94). Interestingly, our study showed that cells still arrested in the G1 phase even with partial knockdown of WDHD1 (Fig. 5). In this case, the steady-state level of WDHD1 after knockdown in E7-expressing cells was similar to that seen with the NC-transfected control cells, where normal DNA replication is expected, suggesting that WDHD1 regulates the G1 checkpoint in E7-expressing cells by a mechanism independent of DNA replication initiation. Consistently, mimosine can prevent WDHD1 binding to chromosomes and arrest the cell cycle in the G1 phase by activating HIF1- (23). How WDHD1 precisely regulates the G1 checkpoint requires further studies.
(23). How WDHD1 precisely regulates the G1 checkpoint requires further studies.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of our laboratories for helpful discussions and Wesley Haynes for English editing.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00513-16.
REFERENCES
- 1.zur Hausen H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer 2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 2.Doorbar J. 2006. Molecular biology of human papillomavirus infection and cervical cancer. Clin Sci (Lond) 110:525–541. doi: 10.1042/CS20050369. [DOI] [PubMed] [Google Scholar]
- 3.Lee KA, Shim JH, Kho CW, Park SG, Park BC, Kim JW, Lim JS, Choe YK, Paik SG, Yoon DY. 2004. Protein profiling and identification of modulators regulated by the E7 oncogene in the C33A cell line by proteomics and genomics. Proteomics 4:839–848. doi: 10.1002/pmic.200300626. [DOI] [PubMed] [Google Scholar]
- 4.Kuner R, Vogt M, Sultmann H, Buness A, Dymalla S, Bulkescher J, Fellmann M, Butz K, Poustka A, Hoppe-Seyler F. 2007. Identification of cellular targets for the human papillomavirus E6 and E7 oncogenes by RNA interference and transcriptome analyses. J Mol Med (Berl) 85:1253–1262. doi: 10.1007/s00109-007-0230-1. [DOI] [PubMed] [Google Scholar]
- 5.Cortés-Malagón EM, Bonilla-Delgado J, Díaz-Chávez J, Hidalgo-Miranda A, Romero-Cordoba S, Uren A, Celik H, McCormick M, Munguía-Moreno JA, Ibarra-Sierra E, Escobar-Herrera J, Lambert PF, Mendoza-Villanueva D, Bermudez-Cruz RM, Gariglio P. 2013. Gene expression profile regulated by the HPV16 E7 oncoprotein and estradiol in cervical tissue. Virology 447:155–165. doi: 10.1016/j.virol.2013.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim E, Kang J, Cho M, Lee S, Seo E, Choi H, Kim Y, Kim J, Kang KY, Kim KP, Han J, Sheen Y, Yum YN, Park SN, Yoon DY. 2009. Profiling of transcripts and proteins modulated by the E7 oncogene in the lung tissue of E7-Tg mice by the omics approach. Mol Med Rep 2:129–137. [DOI] [PubMed] [Google Scholar]
- 7.Pang CL, Toh SY, He P, Teissier S, Ben Khalifa Y, Xue Y, Thierry F. 2014. A functional interaction of E7 with B-Myb-MuvB complex promotes acute cooperative transcriptional activation of both S- and M-phase genes. (129 c). Oncogene 33:4039–4049. doi: 10.1038/onc.2013.426. [DOI] [PubMed] [Google Scholar]
- 8.Murray AW. 2004. Recycling the cell cycle: cyclins revisited. Cell 116:221–234. doi: 10.1016/S0092-8674(03)01080-8. [DOI] [PubMed] [Google Scholar]
- 9.Bochman ML, Schwacha A. 2009. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol Mol Biol Rev 73:652–683. doi: 10.1128/MMBR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. 2005. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 11.Olaharski AJ, Sotelo R, Solorza-Luna G, Gonsebatt ME, Guzman P, Mohar A, Eastmond DA. 2006. Tetraploidy and chromosomal instability are early events during cervical carcinogenesis. Carcinogenesis 27:337–343. doi: 10.1093/carcin/bgi218. [DOI] [PubMed] [Google Scholar]
- 12.Porter AC. 2008. Preventing DNA over-replication: a Cdk perspective. Cell Div 3:3. doi: 10.1186/1747-1028-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davidson IF, Li A, Blow JJ. 2006. Deregulated replication licensing causes DNA fragmentation consistent with head-to-tail fork collision. Mol Cell 24:433–443. doi: 10.1016/j.molcel.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaziri C, Saxena S, Jeon Y, Lee C, Murata K, Machida Y, Wagle N, Hwang DS, Dutta A. 2003. A p53-dependent checkpoint pathway prevents rereplication. Mol Cell 11:997–1008. doi: 10.1016/S1097-2765(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 15.Mariani BD, Schimke RT. 1984. Gene amplification in a single cell cycle in Chinese hamster ovary cells. J Biol Chem 259:1901–1910. [PubMed] [Google Scholar]
- 16.Fan X, Liu Y, Heilman SA, Chen JJ. 2013. Human papillomavirus E7 induces rereplication in response to DNA damage. J Virol 87:1200–1210. doi: 10.1128/JVI.02038-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parker MJ, Gillespie LD, Gillespie WJ. 2001. Hip protectors for preventing hip fractures in the elderly. Cochrane Database Syst Rev 2001:CD001255. doi: 10.1002/14651858. [DOI] [PubMed] [Google Scholar]
- 18.Shreeram S, Sparks A, Lane DP, Blow JJ. 2002. Cell type-specific responses of human cells to inhibition of replication licensing. Oncogene 21:6624–6632. doi: 10.1038/sj.onc.1205910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Xiao H, de Renty C, Jaramillo-Lambert A, Han Z, DePamphilis ML, Brown KJ, Zhu W. 2012. The involvement of acidic nucleoplasmic DNA-binding protein (And-1) in the regulation of prereplicative complex (pre-RC) assembly in human cells. J Biol Chem 287:42469–42479. doi: 10.1074/jbc.M112.404277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu W, Ukomadu C, Jha S, Senga T, Dhar SK, Wohlschlegel JA, Nutt LK, Kornbluth S, Dutta A. 2007. Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Dev 21:2288–2299. doi: 10.1101/gad.1585607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato N, Koinuma J, Fujita M, Hosokawa M, Ito T, Tsuchiya E, Kondo S, Nakamura Y, Daigo Y. 2010. Activation of WD repeat and high-mobility group box DNA binding protein 1 in pulmonary and esophageal carcinogenesis. Clin Cancer Res 16:226–239. doi: 10.1158/1078-0432.CCR-09-1405. [DOI] [PubMed] [Google Scholar]
- 22.Esposito F, Tornincasa M, Federico A, Chiappetta G, Pierantoni GM, Fusco A. 2014. Retraction. High-mobility group A1 protein inhibits p53-mediated intrinsic apoptosis by interacting with Bcl-2 at mitochondria. Cell Death Dis 5:e1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park SY, Im JS, Park SR, Kim SE, Wang HJ, Lee JK. 2012. Mimosine arrests the cell cycle prior to the onset of DNA replication by preventing the binding of human Ctf4/And-1 to chromatin via Hif-1alpha activation in HeLa cells. Cell Cycle 11:761–766. doi: 10.4161/cc.11.4.19209. [DOI] [PubMed] [Google Scholar]
- 24.Yoshizawa-Sugata N, Masai H. 2009. Roles of human AND-1 in chromosome transactions in S phase. J Biol Chem 284:20718–20728. doi: 10.1074/jbc.M806711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinoshita Y, Johnson EM. 2004. Site-specific loading of an MCM protein complex in a DNA replication initiation zone upstream of the c-MYC gene in the HeLa cell cycle. J Biol Chem 279:35879–35889. doi: 10.1074/jbc.M401640200. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Deng M, Wei Q, Liu T, Tong X, Ye X. 2011. Phosphorylation of MCM3 protein by cyclin E/cyclin-dependent kinase 2 (Cdk2) regulates its function in cell cycle. J Biol Chem 286:39776–39785. doi: 10.1074/jbc.M111.226464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambert PF, Ozbun MA, Collins A, Holmgren S, Lee D, Nakahara T. 2005. Using an immortalized cell line to study the HPV life cycle in organotypic “raft” cultures. Methods Mol Med 119:141–155. [DOI] [PubMed] [Google Scholar]
- 29.Zehbe I, Lichtig H, Westerback A, Lambert PF, Tommasino M, Sherman L. 2011. Rare human papillomavirus 16 E6 variants reveal significant oncogenic potential. Mol Cancer 10:77. doi: 10.1186/1476-4598-10-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Genther SM, Sterling S, Duensing S, Munger K, Sattler C, Lambert PF. 2003. Quantitative role of the human papillomavirus type 16 E5 gene during the productive stage of the viral life cycle. J Virol 77:2832–2842. doi: 10.1128/JVI.77.5.2832-2842.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zehbe I, Richard C, DeCarlo CA, Shai A, Lambert PF, Lichtig H, Tommasino M, Sherman L. 2009. Human papillomavirus 16 E6 variants differ in their dysregulation of human keratinocyte differentiation and apoptosis. Virology 383:69–77. doi: 10.1016/j.virol.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mannhardt B, Weinzimer SA, Wagner M, Fiedler M, Cohen P, Jansen-Durr P, Zwerschke W. 2000. Human papillomavirus type 16 E7 oncoprotein binds and inactivates growth-inhibitory insulin-like growth factor binding protein 3. Mol Cell Biol 20:6483–6495. doi: 10.1128/MCB.20.17.6483-6495.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mansour M, Touka M, Hasan U, Bellopede A, Smet A, Accardi R, Gabet AS, Sylla BS, Tommasino M. 2007. E7 properties of mucosal human papillomavirus types 26, 53 and 66 correlate with their intermediate risk for cervical cancer development. Virology 367:1–9. doi: 10.1016/j.virol.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Zhang W, Chen H, Chen Y, Liu J, Wang X, Yu X, Chen JJ, Zhao W. 2015. Cancerous inhibitor of protein phosphatase 2A contributes to human papillomavirus oncoprotein E7-induced cell proliferation via E2F1. Oncotarget 6:5253–5262. doi: 10.18632/oncotarget.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blomberg I, Hoffmann I. 1999. Ectopic expression of Cdc25A accelerates the G(1)/S transition and leads to premature activation of cyclin E- and cyclin A-dependent kinases. Mol Cell Biol 19:6183–6194. doi: 10.1128/MCB.19.9.6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu L, Goodwin EC, Naeger LK, Vigo E, Galaktionov K, Helin K, DiMaio D. 2000. E2F-Rb complexes assemble and inhibit cdc25A transcription in cervical carcinoma cells following repression of human papillomavirus oncogene expression. Mol Cell Biol 20:7059–7067. doi: 10.1128/MCB.20.19.7059-7067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen T, Huang S. 2012. The role of Cdc25A in the regulation of cell proliferation and apoptosis. Anticancer Agents Med Chem 12:631–639. doi: 10.2174/187152012800617678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moon MS, Lee CJ, Um SJ, Park JS, Yang JM, Hwang ES. 2001. Effect of BPV1 E2-mediated inhibition of E6/E7 expression in HPV16-positive cervical carcinoma cells. Gynecol Oncol 80:168–175. doi: 10.1006/gyno.2000.6053. [DOI] [PubMed] [Google Scholar]
- 39.Sadasivam S, DeCaprio JA. 2013. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer 13:585–595. doi: 10.1038/nrc3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sala A, Calabretta B. 1992. Regulation of BALB/c 3T3 fibroblast proliferation by B-myb is accompanied by selective activation of cdc2 and cyclin D1 expression. Proc Natl Acad Sci U S A 89:10415–10419. doi: 10.1073/pnas.89.21.10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu W, Giangrande PH, Nevins JR. 2004. E2Fs link the control of G1/S and G2/M transcription. EMBO J 23:4615–4626. doi: 10.1038/sj.emboj.7600459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fan X, Chen JJ. 2014. Role of Cdk1 in DNA damage-induced G1 checkpoint abrogation by the human papillomavirus E7 oncogene. Cell Cycle 13:3249–3259. doi: 10.4161/15384101.2014.953879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johung K, Goodwin EC, DiMaio D. 2007. Human papillomavirus E7 repression in cervical carcinoma cells initiates a transcriptional cascade driven by the retinoblastoma family, resulting in senescence. J Virol 81:2102–2116. doi: 10.1128/JVI.02348-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Armstrong DJ, Roman A. 1997. The relative ability of human papillomavirus type 6 and human papillomavirus type 16 E7 proteins to transactivate E2F-responsive elements is promoter- and cell-dependent. Virology 239:238–246. doi: 10.1006/viro.1997.8885. [DOI] [PubMed] [Google Scholar]
- 45.Lam EW, Morris JD, Davies R, Crook T, Watson RJ, Vousden KH. 1994. HPV16 E7 oncoprotein deregulates B-myb expression: correlation with targeting of p107/E2F complexes. EMBO J 13:871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohba K, Ichiyama K, Yajima M, Gemma N, Nikaido M, Wu Q, Chong P, Mori S, Yamamoto R, Wong JE, Yamamoto N. 2014. In vivo and in vitro studies suggest a possible involvement of HPV infection in the early stage of breast carcinogenesis via APOBEC3B induction. PLoS One 9:e97787. doi: 10.1371/journal.pone.0097787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wise-Draper TM, Allen HV, Thobe MN, Jones EE, Habash KB, Munger K, Wells SI. 2005. The human DEK proto-oncogene is a senescence inhibitor and an upregulated target of high-risk human papillomavirus E7. J Virol 79:14309–14317. doi: 10.1128/JVI.79.22.14309-14317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams AK, Hallenbeck GE, Casper KA, Patil YJ, Wilson KM, Kimple RJ, Lambert PF, Witte DP, Xiao W, Gillison ML, Wikenheiser-Brokamp KA, Wise-Draper TM, Wells SI. 2015. DEK promotes HPV-positive and -negative head and neck cancer cell proliferation. Oncogene 34:868–877. doi: 10.1038/onc.2014.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou X, Han S, Wang S, Chen X, Dong J, Shi X, Xia Y, Wang X, Hu Z, Shen H. 2009. Polymorphisms in HPV E6/E7 protein interacted genes and risk of cervical cancer in Chinese women: a case-control analysis. Gynecol Oncol 114:327–331. doi: 10.1016/j.ygyno.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 50.Jakubickova L, Barathova M, Pastorekova S, Pastorek J, Gibadulinova A. 2005. Expression of S100P gene in cervical carcinoma cells is independent of E7 human papillomavirus oncogene. Acta Virol 49:133–137. http://www.elis.sk/index.php?page=shop.product_details&flypage=flypage.tpl&product_id=99&category_id=4&option=com_virtuemart&vmcchk=1&Itemid=1. [PubMed] [Google Scholar]
- 51.Hellung Schonning B, Bevort M, Mikkelsen S, Andresen M, Thomsen P, Leffers H, Norrild B. 2000. Human papillomavirus type 16 E7-regulated genes: regulation of S100P and ADP/ATP carrier protein genes identified by differential-display technology. J Gen Virol 81:1009–1015. doi: 10.1099/0022-1317-81-4-1009. [DOI] [PubMed] [Google Scholar]
- 52.Aisner DL, Nguyen TT, Paskulin DD, Le AT, Haney J, Schulte N, Chionh F, Hardingham J, Mariadason J, Tebbutt N, Doebele RC, Weickhardt AJ, Varella-Garcia M. 2014. ROS1 and ALK fusions in colorectal cancer, with evidence of intratumoral heterogeneity for molecular drivers. Mol Cancer Res 12:111–118. doi: 10.1158/1541-7786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yeager TR, Reznikoff CA. 1998. Methotrexate resistance in human uroepithelial cells with p53 alterations. J Urol 159:581–585. doi: 10.1016/S0022-5347(01)63988-0. [DOI] [PubMed] [Google Scholar]
- 54.Martin CM, Astbury K, McEvoy L, O'Toole S, Sheils O, O'Leary JJ. 2009. Gene expression profiling in cervical cancer: identification of novel markers for disease diagnosis and therapy. Methods Mol Biol 511:333–359. doi: 10.1007/978-1-59745-447-6_15. [DOI] [PubMed] [Google Scholar]
- 55.Martinez I, Wang J, Hobson KF, Ferris RL, Khan SA. 2007. Identification of differentially expressed genes in HPV-positive and HPV-negative oropharyngeal squamous cell carcinomas. Eur J Cancer 43:415–432. doi: 10.1016/j.ejca.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nguyen DX, Westbrook TF, McCance DJ. 2002. Human papillomavirus type 16 E7 maintains elevated levels of the cdc25A tyrosine phosphatase during deregulation of cell cycle arrest. J Virol 76:619–632. doi: 10.1128/JVI.76.2.619-632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Myklebust MP, Bruland O, Fluge O, Skarstein A, Balteskard L, Dahl O. 2011. MicroRNA-15b is induced with E2F-controlled genes in HPV-related cancer. Br J Cancer 105:1719–1725. doi: 10.1038/bjc.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li JZ, Pan HY, Zheng JW, Zhou XJ, Zhang P, Chen WT, Zhang ZY. 2008. Benzo (a) pyrene induced tumorigenesity of human immortalized oral epithelial cells: transcription profiling. Chin Med J (Engl) 121:1882–1890. [PubMed] [Google Scholar]
- 59.Rey O, Lee S, Park NH. 2000. Human papillomavirus type 16 E7 oncoprotein represses transcription of human fibronectin. J Virol 74:4912–4918. doi: 10.1128/JVI.74.10.4912-4918.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang X, Chen H, Wang X, Zhao W, Chen JJ. 2014. Expression and transcriptional profiling of the LKB1 tumor suppressor in cervical cancer cells. Gynecol Oncol 134:372–378. doi: 10.1016/j.ygyno.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moody CA, Laimins LA. 2010. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer 10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 62.Banerjee NS, Genovese NJ, Noya F, Chien WM, Broker TR, Chow LT. 2006. Conditionally activated E7 proteins of high-risk and low-risk human papillomaviruses induce S phase in postmitotic, differentiated human keratinocytes. J Virol 80:6517–6524. doi: 10.1128/JVI.02499-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park JW, Shin MK, Lambert PF. 2014. High incidence of female reproductive tract cancers in FA-deficient HPV16-transgenic mice correlates with E7's induction of DNA damage response, an activity mediated by E7's inactivation of pocket proteins. Oncogene 33:3383–3391. doi: 10.1038/onc.2013.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park JW, Shin MK, Pitot HC, Lambert PF. 2013. High incidence of HPV-associated head and neck cancers in FA deficient mice is associated with E7's induction of DNA damage through its inactivation of pocket proteins. PLoS One 8:e75056. doi: 10.1371/journal.pone.0075056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Howes KA, Ransom N, Papermaster DS, Lasudry JG, Albert DM, Windle JJ. 1994. Apoptosis or retinoblastoma: alternative fates of photoreceptors expressing the HPV-16 E7 gene in the presence or absence of p53. Genes Dev 8:1300–1310. doi: 10.1101/gad.8.11.1300. [DOI] [PubMed] [Google Scholar]
- 66.Pan H, Griep AE. 1995. Temporally distinct patterns of p53-dependent and p53-independent apoptosis during mouse lens development. Genes Dev 9:2157–2169. doi: 10.1101/gad.9.17.2157. [DOI] [PubMed] [Google Scholar]
- 67.Longworth MS, Wilson R, Laimins LA. 2005. HPV31 E7 facilitates replication by activating E2F2 transcription through its interaction with HDACs. EMBO J 24:1821–1830. doi: 10.1038/sj.emboj.7600651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McLaughlin-Drubin ME, Huh KW, Munger K. 2008. Human papillomavirus type 16 E7 oncoprotein associates with E2F6. J Virol 82:8695–8705. doi: 10.1128/JVI.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen Y, Miller C, Mosher R, Zhao X, Deeds J, Morrissey M, Bryant B, Yang D, Meyer R, Cronin F, Gostout BS, Smith-McCune K, Schlegel R. 2003. Identification of cervical cancer markers by cDNA and tissue microarrays. Cancer Res 63:1927–1935. [PubMed] [Google Scholar]
- 70.Santin AD, Zhan F, Bignotti E, Siegel ER, Cane S, Bellone S, Palmieri M, Anfossi S, Thomas M, Burnett A, Kay HH, Roman JJ, O'Brien TJ, Tian E, Cannon MJ, Shaughnessy J Jr, Pecorelli S. 2005. Gene expression profiles of primary HPV16- and HPV18-infected early stage cervical cancers and normal cervical epithelium: identification of novel candidate molecular markers for cervical cancer diagnosis and therapy. Virology 331:269–291. doi: 10.1016/j.virol.2004.09.045. [DOI] [PubMed] [Google Scholar]
- 71.Rampias T, Sasaki C, Weinberger P, Psyrri A. 2009. E6 and e7 gene silencing and transformed phenotype of human papillomavirus 16-positive oropharyngeal cancer cells. J Natl Cancer Inst 101:412–423. doi: 10.1093/jnci/djp017. [DOI] [PubMed] [Google Scholar]
- 72.Wang YW, Chang HS, Lin CH, Yu WC. 2007. HPV-18 E7 conjugates to c-Myc and mediates its transcriptional activity. Int J Biochem Cell Biol 39:402–412. doi: 10.1016/j.biocel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 73.Fan X, Liu Y, Chen JJ. 2003. Activation of c-Myc contributes to bovine papillomavirus type 1 E7-induced cell proliferation. J Biol Chem 278:43163–43168. doi: 10.1074/jbc.M306008200. [DOI] [PubMed] [Google Scholar]
- 74.Perea SE, Massimi P, Banks L. 2000. Human papillomavirus type 16 E7 impairs the activation of the interferon regulatory factor-1. Int J Mol Med 5:661–666. [DOI] [PubMed] [Google Scholar]
- 75.Spitkovsky D, Hehner SP, Hofmann TG, Moller A, Schmitz ML. 2002. The human papillomavirus oncoprotein E7 attenuates NF-kappa B activation by targeting the Ikappa B kinase complex. J Biol Chem 277:25576–25582. doi: 10.1074/jbc.M201884200. [DOI] [PubMed] [Google Scholar]
- 76.Vandermark ER, Deluca KA, Gardner CR, Marker DF, Schreiner CN, Strickland DA, Wilton KM, Mondal S, Woodworth CD. 2012. Human papillomavirus type 16 E6 and E 7 proteins alter NF-kB in cultured cervical epithelial cells and inhibition of NF-kB promotes cell growth and immortalization. Virology 425:53–60. doi: 10.1016/j.virol.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nees M, Geoghegan JM, Hyman T, Frank S, Miller L, Woodworth CD. 2001. Papillomavirus type 16 oncogenes downregulate expression of interferon-responsive genes and upregulate proliferation-associated and NF-kappaB-responsive genes in cervical keratinocytes. J Virol 75:4283–4296. doi: 10.1128/JVI.75.9.4283-4296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shaikh F, Sanehi P, Rawal R. 2012. Molecular screening of compounds to the predicted protein-protein interaction site of Rb1-E7 with p53-E6 in HPV. Bioinformation 8:607–612. doi: 10.6026/97320630008607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nair P, Somasundaram K, Krishna S. 2003. Activated Notch1 inhibits p53-induced apoptosis and sustains transformation by human papillomavirus type 16 E6 and E7 oncogenes through a PI3K-PKB/Akt-dependent pathway. J Virol 77:7106–7112. doi: 10.1128/JVI.77.12.7106-7112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Massimi P, Banks L. 1997. Repression of p53 transcriptional activity by the HPV E7 proteins. Virology 227:255–259. doi: 10.1006/viro.1996.8315. [DOI] [PubMed] [Google Scholar]
- 81.Wang Y, Okan I, Pokrovskaja K, Wiman KG. 1996. Abrogation of p53-induced G1 arrest by the HPV 16 E7 protein does not inhibit p53-induced apoptosis. Oncogene 12:2731–2735. [PubMed] [Google Scholar]
- 82.Eichten A, Westfall M, Pietenpol JA, Munger K. 2002. Stabilization and functional impairment of the tumor suppressor p53 by the human papillomavirus type 16 E7 oncoprotein. Virology 295:74–85. doi: 10.1006/viro.2002.1375. [DOI] [PubMed] [Google Scholar]
- 83.Dong XP, Pfister H. 1999. Overlapping YY1- and aberrant SP1-binding sites proximal to the early promoter of human papillomavirus type 16. J Gen Virol 80(Pt 8):2097–2101. doi: 10.1099/0022-1317-80-8-2097. [DOI] [PubMed] [Google Scholar]
- 84.Liu Y, Heilman SA, Illanes D, Sluder G, Chen JJ. 2007. p53-independent abrogation of a postmitotic checkpoint contributes to HPV E6-induced polyploidy. Cancer Res 67:2603–2610. doi: 10.1158/0008-5472.CAN-06-3436. [DOI] [PubMed] [Google Scholar]
- 85.Heilman SA, Nordberg JJ, Liu Y, Sluder G, Chen JJ. 2009. Abrogation of the postmitotic checkpoint contributes to polyploidization in human papillomavirus E7-expressing cells. J Virol 83:2756–2764. doi: 10.1128/JVI.02149-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Demers GW, Foster SA, Halbert CL, Galloway DA. 1994. Growth arrest by induction of p53 in DNA damaged keratinocytes is bypassed by human papillomavirus 16 E7. Proc Natl Acad Sci U S A 91:4382–4386. doi: 10.1073/pnas.91.10.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hsieh CL, Lin CL, Liu H, Chang YJ, Shih CJ, Zhong CZ, Lee SC, Tan BC. 2011. WDHD1 modulates the post-transcriptional step of the centromeric silencing pathway. Nucleic Acids Res 39:4048–4062. doi: 10.1093/nar/gkq1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Banerjee NS, Wang HK, Broker TR, Chow LT. 2011. Human papillomavirus (HPV) E7 induces prolonged G2 following S phase reentry in differentiated human keratinocytes. J Biol Chem 286:15473–15482. doi: 10.1074/jbc.M110.197574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Braun TE, Poole E, Sinclair J. 2012. Depletion of cellular pre-replication complex factors results in increased human cytomegalovirus DNA replication. PLoS One 7:e36057. doi: 10.1371/journal.pone.0036057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Uetake Y, Sluder G. 2004. Cell cycle progression after cleavage failure: mammalian somatic cells do not possess a “tetraploidy checkpoint”. J Cell Biol 165:609–615. doi: 10.1083/jcb.200403014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang C, Gong B, Bushel PR, Thierry-Mieg J, Thierry-Mieg D, Xu J, Fang H, Hong H, Shen J, Su Z, Meehan J, Li X, Yang L, Li H, Labaj PP, Kreil DP, Megherbi D, Gaj S, Caiment F, van Delft J, Kleinjans J, Scherer A, Devanarayan V, Wang J, Yang Y, Qian HR, Lancashire LJ, Bessarabova M, Nikolsky Y, Furlanello C, Chierici M, Albanese D, Jurman G, Riccadonna S, Filosi M, Visintainer R, Zhang KK, Li J, Hsieh JH, Svoboda DL, Fuscoe JC, Deng Y, Shi L, Paules RS, Auerbach SS, Tong W. 2014. The concordance between RNA-seq and microarray data depends on chemical treatment and transcript abundance. Nat Biotechnol 32:926–932. doi: 10.1038/nbt.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mathevet P, Frappart L, Hittelman W. 2000. Cervix dysplasias: study of Rb and p53 gene expression and correlation with mitotic activity. Gynecol Obstet Fertil 28:44–50. (In French.) [PubMed] [Google Scholar]
- 93.Foley J, Wysolmerski JJ, Broadus AE, Philbrick WM. 1996. Parathyroid hormone-related protein gene expression in human squamous carcinoma cells is repressed by mutant isoforms of p53. Cancer Res 56:4056–4062. [PubMed] [Google Scholar]
- 94.Ma Q, Fonseca A, Liu W, Fields AT, Pimsler ML, Spindola AF, Tarone AM, Crippen TL, Tomberlin JK, Wood TK. 2012. Proteus mirabilis interkingdom swarming signals attract blow flies. ISME J 6:1356–1366. doi: 10.1038/ismej.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.