ABSTRACT
Several essential viral proteins are proposed to participate in genome encapsidation of human cytomegalovirus (HCMV), among them pUL77 and pUL93, which remain largely uncharacterized. To gain insight into their properties, we generated an HCMV mutant expressing a pUL77-monomeric enhanced green fluorescent protein (mGFP) fusion protein and a pUL93-specific antibody. Immunoblotting demonstrated that both proteins are incorporated into capsids and virions. Conversely to data suggesting internal translation initiation sites within the UL93 open reading frame (ORF), we provide evidence that pUL93 synthesis commences at the first start codon. In infected cells, pUL77-mGFP was found in nuclear replication compartments and dot-like structures, colocalizing with capsid proteins. Immunogold labeling of nuclear capsids revealed that pUL77 is present on A, B, and C capsids. Pulldown of pUL77-mGFP revealed copurification of pUL93, indicating interaction between these proteins, which still occurred when capsid formation was prevented. Correct subnuclear distribution of pUL77-mGFP required pUL93 as well as the major capsid protein (and thus probably the presence of capsids), but not the tegument protein pp150 or the encapsidation protein pUL52, demonstrating that pUL77 nuclear targeting occurs independently of the formation of DNA-filled capsids. When pUL77 or pUL93 was missing, generation of unit-length genomes was not observed, and only empty B capsids were produced. Taken together, these results show that pUL77 and pUL93 are capsid constituents needed for HCMV genome encapsidation. Therefore, the task of pUL77 seems to differ from that of its alphaherpesvirus orthologue pUL25, which exerts its function subsequent to genome cleavage-packaging.
IMPORTANCE The essential HCMV proteins pUL77 and pUL93 were suggested to be involved in viral genome cleavage-packaging but are poorly characterized both biochemically and functionally. By producing a monoclonal antibody against pUL93 and generating an HCMV mutant in which pUL77 is fused to a fluorescent protein, we show that pUL77 and pUL93 are capsid constituents, with pUL77 being similarly abundant on all capsid types. Each protein is required for genome encapsidation, as the absence of either pUL77 or pUL93 results in a genome packaging defect with the formation of empty capsids only. This distinguishes pUL77 from its alphaherpesvirus orthologue pUL25, which is enriched on DNA-filled capsids and exerts its function after the viral DNA is packaged. Our data for the first time describe an HCMV mutant with a fluorescent capsid and provide insight into the roles of pUL77 and pUL93, thus contributing to a better understanding of the HCMV encapsidation network.
INTRODUCTION
The life cycle of human cytomegalovirus (HCMV), the prototype member of the betaherpesviruses, comprises a nuclear phase that includes transcription of viral genes, replication of the double-stranded DNA genome, assembly of procapsids, packaging of the viral DNA into the preformed capsids, and maturation of the DNA-filled capsids, promoting their egress into the cytoplasm, where they undergo secondary envelopment (1). Capsid formation commences by assembly of the major capsid protein (MCP; encoded by the UL86 open reading frame [ORF]) around a protein scaffold formed by the assembly protein precursor (UL80.5) and the protease precursor (UL80a), followed by stabilization of the MCP capsomers through the triplex proteins, which consist of two copies of the minor capsid protein (mCP; UL85) and one copy of the mCP-binding protein (mCP-BP; UL46) (2, 3), resulting in spherical procapsids. The MCP assembles into hexons and pentons, the latter being restricted to the vertices of the eventually icosahedral capsid. The small capsid protein (SCP; UL48.5) may be present already in procapsids (4), and in mature capsids, it decorates the tips of the hexons but not of the pentons. Procapsids are believed to constitute the substrate for viral genome packaging, during which the scaffold is cleaved and expelled from the capsids. Successful genome packaging generates DNA-filled C capsids. The two other nuclear capsid forms are empty shells, with B capsids probably arising from spontaneous angularization of procapsids and A capsids originating from abortive packaging events without retention of the genomes within the capsids (1, 3, 5, 6). B capsids were recently discussed to be intermediate capsid forms during the genome packaging process rather than dead end products (7). While both A and B capsids lack DNA, they are distinguished by the presence (B capsids) or absence (A capsids) of the scaffold protein.
The capsids of alpha-, beta- and gammaherpesviruses share many characteristics, but differences also exist. Although HCMV has the largest genome of all mammalian DNA viruses (∼240 kbp); the diameter of its capsid is similar to that of the herpes simplex virus 1 capsid (HSV-1; genome size, 150 kbp). As a consequence, encapsidated HCMV DNA is more densely packed (8), resulting in DNA-filled capsids that are under higher pressure and may thus need additional stabilization (9). Furthermore, the structure of the HCMV inner tegument is distinct from that of HSV-1, insofar as the HCMV tegument contacts both hexons and pentons, whereas in HSV-1, the inner tegument is attached to pentons only (10, 11). Moreover, HCMV encodes betaherpesvirus-specific tegument proteins: e.g., pp150 (UL32) and pUL96. Recently it was shown that pp150 is the most inner tegument protein forming a netlike layer around the capsids (12). A follow-up study disclosed the SCP as being necessary for recruitment of pp150 to capsids (13), thus providing an explanation of why in betaherpesviruses SCP is essential (14), while in HSV-1 it is dispensable for viral growth. Taken together, these features of betaherpesviruses may point to divergent mechanisms in capsid maturation and stabilization following genome packaging.
HCMV genome encapsidation requires at least five gene products, in addition to the proteins needed for procapsid assembly. These are as follows: (i) the terminase subunits pUL56 and pUL89, which bind to the concatemeric viral DNA and cleave it into unit-length genomes (15–17); (ii) pUL51, which interacts with pUL56 and pUL89 and may represent a third subunit of the terminase complex (18); (iii) the portal protein pUL104, present at one capsid vertex (19, 20); and (iv) pUL52, whose function is elusive, yet essential for genome cleavage-packaging (21). For only two of those proteins, pUL51 and pUL52, are HCMV mutants available that allow analysis of the phenotypic consequences caused by the lack of either of these proteins (18, 21). This is in part due to the difficulties in generating complementing systems for essential HCMV proteins (22). Studies with alphaherpesvirus mutants, for which complementing cells can be more easily established, revealed that two additional viral proteins (pUL17 and pUL25) are involved in encapsidation of genomes and their stable retention within capsids. pUL17 and pUL25 form a heterodimer that is located exclusively around pentons (23–26) and was therefore named the “capsid vertex-specific component” (CVSC) (27). Loading of the CVSC on DNA-filled capsids is assumed to label mature capsids as ready for nuclear egress, which may be initiated by interaction of the CVSC with the nuclear egress complex (NEC).
Orthologous proteins to HSV-1 pUL25 exist throughout the herpesviruses, and in HCMV, the orthologue is the UL77 protein. However, the similarity of the amino acid sequences of pUL77 and HSV-1 pUL25 is only moderate (37%). Based on the findings obtained with alphaherpesviruses, pUL77 was proposed to represent an additional HCMV gene product involved in DNA encapsidation. So far, only one publication on pUL77 is available, reporting that it interacts with DNA and components of the HCMV genome packaging machinery (28), but its role during HCMV infection is unknown. HCMV does not encode a protein sharing substantial sequence similarity with HSV pUL17; however, due to a similar position within the viral genome, the HCMV UL93 ORF was defined as a “positional homologue” of HSV-1 UL17. Until now, the protein encoded by UL93 is completely uncharacterized.
The aim of this study was to investigate the HCMV proteins pUL77 and pUL93 and their putative role during the infection cycle. To this end, we generated an HCMV mutant expressing a UL77-monomeric enhanced green fluorescent protein (EGFP) fusion protein (UL77-mGFP) and produced a pUL93-specific monoclonal antibody. The usage of an efficient transfection protocol for HCMV bacterial artificial chromosomes (BACs) (29) made it possible to examine mutant HCMV genomes in which either UL77 or UL93 is disrupted directly in BAC-transfected cells. pUL77 and pUL93 were expressed as late proteins and were found to be associated with nuclear capsids and virus particles. In cells transfected with HCMV BACs lacking the UL77 or UL93 ORF, we did not detect unit-length genomes, and solely B capsids were observed. Our data provide a characterization of pUL77 and pUL93 in infected cells and assign a role to either protein in genome cleavage and packaging.
MATERIALS AND METHODS
Viruses and cells.
The recombinant viruses and genomes generated in this study are based on HCMV strain AD169 cloned as a bacterial artificial chromosome (BAC) in Escherichia coli (30). pHB5 denotes the original AD169 BAC, and pHG represents an enhanced green fluorescent protein (EGFP)-expressing pHB5 derivative (31). hTERT-RPE-1 cells (Clontech) were propagated as described previously (29). Cultivation of human foreskin fibroblasts (HFFs), preparation of HCMV stocks, and analysis of viral growth by standard plaque assay were done as reported before (30).
BAC mutagenesis, transfection by adenofection, and analysis of viral DNA.
Insertion of the monomeric EGFP (mGFP) sequence into ORF UL77 of pHB5 was performed by en passant mutagenesis (32). The mGFP ORF together with a kanamycin resistance cassette (Knr) and an I-SceI site was amplified with primer pairs UL77-GFP2.for and UL77-GFP2.rev or UL77-GFP3.for and UL77-GFP3.rev (Table 1) using plasmid pEP-mGFP-in as the template (kindly provided by Beate Sodeik). The resulting PCR products were recombined with pHB5 by red-α, -β, -γ-mediated homologous recombination in E. coli, resulting in pHB5-UL77-mGFP-2 and pHB5-UL77-mGFP-3. HCMV BACs carrying deletions in ORF UL32 (pp150), UL93, or UL86 (MCP) were constructed in an analogous manner with pHB5-UL77-mGFP-3 as the backbone, pEP-Kan-S2 as the template, and the primer pairs given in Table 1. Deletion of the UL52 ORF from pHB5-UL77-mGFP-3 was done as described previously (21), and construction of pHG-ΔUL77 lacking the UL77 ORF was reported elsewhere (33). pHG-ΔUL93 was generated by disrupting the UL93 ORF utilizing the respective primers shown in Table 1. Successful mutagenesis was verified for each BAC through restriction analysis and sequencing. Isolation of BACs from bacterial cultures was done by applying an alkaline lysis procedure as described before (34). To reconstitute virus progeny from HCMV BAC genomes, HFFs were transfected with defective adenovirus particles as carriers (“adenofection”) as published recently (29). For investigation of recombinant HCMV genomes in which essential genes were disrupted, hTERT-RPE-1 cells (3 × 106) were adenofected accordingly with 3 μg of BAC DNA prepared by a protocol yielding endotoxin-free DNA (Nucleobond PC 500 EF kit; Macherey & Nagel, Düren, Germany). To analyze viral DNA by pulsed-field gel electrophoresis (PFGE), 3 × 106 adenofected hTERT-RPE-1 cells were harvested on day 5 posttransfection and cast into agarose blocks. The conditions of PFGE, Southern blotting, and hybridization to the probe specific for the HCMV b-repeat region were as delineated earlier (21).
TABLE 1.
Oligonucleotides used in this study to generate mutant HCMV BAC genomes
| Primer name | Sequence (5′→3′) |
|---|---|
| UL77-GFP2.for | CCCGCGGACGCTGCTACAACCACCGTGTCGTCCGCGTTCTGTGAGCAAGGGCGAGGAGCTGTT |
| UL77-GFP2.rev | GGACGGCAGAAGCTCCTGCCACGCCGGTCCCTGGGGACGACTTGTACAGCTCGTCCATGCCGA |
| UL77-GFP3.for | CGTGGTCGCCCCGTCTGACGCGGTCGCCGCGTCAGCGGCCGTGAGCAAGGGCGAGGAGCTGTT |
| UL77-GFP3.rev | GCTCGGCGCACTGCGCCAGCCAGGTAGAAGAAGCACCGGCCTTGTACAGCTCGTCCATGCCGA |
| UL93-KO.for | AAGTCGAACTATACCGGGCGTTGGACGCTTATCGGGCGTAGGACTGATTCGCGCCGTGCGCAAGGATGACGACGATAAG |
| UL93-KO.rev | AGTACAGCGTAGATCTCGTCGCGCACGGCGCGAATCAGTCACTACGCCCGATAAGCGTCCAGCCAGTGTTACAACCAATT |
| UL32-ko.for | CCGCGCTTACGAGCGCGCGTTCCACGATCTCGGCATCGTGCTAGTTACCTATTAACGGTCTAGCACCAAGGATGACGACGATAAG |
| UL32-ko.rev | CGTGGTAAGGCGCGTGACCACCTGGCTGTGCTAGACCGTTAATAGGTAACTAGCACGATGCCGAGAGCCAGTGTTACAACCAATT |
| MCP-KO.for | GCAGTGTCTGTAAAAAGCTGTGAATCAAGCCGCGCTCCATCTAGTTACCTATTACGCGCGCAGCACCCAAGGATGACGACGATAAG |
| MCP-KO.rev | GATCCTCAACGTGGAAGCCATGCACACGGTGCTGCGCGCGTAATAGGTAACTAGATGGAGCGCGGCTGCCAGTGTTACAACCAATT |
Protein biochemistry techniques and generation of a UL93-specific MAb.
To produce mouse monoclonal antibodies (MAb) directed against pUL93, two different parts of UL93 were expressed as recombinant proteins in E. coli. To this end, PCR products were generated with the primers UL93-N.for (5′-CGCGGATCCTTCTATGCCGTCTTCACTACG-3′) and UL93-N.rev (5′-CGCAAGCTTGCGACTGCGCCAAAAGGAATT-3′) or UL93-C.for (5′-CGCGGATCCGAGCTGAGCTACGATGACCAC-3′) and UL93-C.rev (5′-CGCAAGCTTAAGATCGTCGAACGGCAAGCG-3′) and pHB5 as the template and were cloned into plasmid pQE-30 (Qiagen, Hilden, Germany), giving rise to pQE-UL93-N and pQE-UL93-C. Expression of the recombinant proteins pUL93-N and pUL93-C and purification, immunization of mice, and generation of hybridoma cultures was done as reported previously (18). Antibodies reactive with the recombinant proteins were finally evaluated with HCMV-infected cells by immunoblotting (IB) and immunofluorescence (IF) microscopy done as described previously (18, 21, 29). Hybridoma cultures secreting pUL93-specific antibodies were only obtained after immunization with pUL93-N, but not with pUL93-C. Other antibodies used were GFP mouse MAb (Santa Cruz; catalog no. sc-9996), UL44 and pp150 mouse MAb (kindly provided by Bodo Plachter, University of Mainz, Germany), IE1 mouse MAb (NEA-9221; PerkinElmer), and anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) (rabbit MAb 14C10; Cell Signaling; catalog no. 2118). The MCP-specific mouse MAb was obtained from Klaus Radsak (University of Marburg, Germany), the pUL50 and pUL53 Abs were received from Manfred Marschall (University of Erlangen, Germany), and the MAb directed against the HCMV SCP was a kind gift from William Britt (University of Alabama, Birmingham). Polyclonal rabbit sera raised against the HCMV mCP or the mCP-BP were generously provided by Wade Gibson (Johns Hopkins University, Baltimore, MD). Generation of MAbs specific for UL52, UL56, and UL89 was described previously (18). The kinetic class of HCMV protein expression was determined by applying inhibitors of early or late gene expression (cycloheximide/actinomycin D or phosphonoacetic acid) as given elsewhere (21).
To pull down the UL77-mGFP fusion from infected HFFs, 4 × 106 cells were infected with either HB5 or HB5-UL77-mGFP-3 at a multiplicity of infection (MOI) of 3, and on day 5 postinfection (p.i.) lysed in 1 ml of immunoprecipitation (IP) buffer (50 mM Tris-HCl [pH 7.4], 300 mM KCl, 5 mM EDTA, 0.5% NP-40, containing protease inhibitors [protease inhibitor cocktail set II; Calbiochem]). After freezing at −80°C, lysates were thawed on ice, and insoluble material was removed by centrifugation for 15 min at 4°C and 16,000 × g. Each sample was precleared with protein G Sepharose before being incubated for 2 h at 4°C on a tumbling wheel with 50 μl of GFP-Trap_A beads (ChromoTek, Martinsried, Germany). Beads were washed five times with IP buffer, and bound proteins were eluted with 150 μl of reducing buffer RotiLoad-1 (Carl Roth, Karlsruhe, Germany). One percent of the cell lysates before IP and 8% of the eluted material were separated by SDS-PAGE, transferred to nitrocellulose membranes, and examined by IB with the Ab indicated. Pulldown of pUL77-mGFP from adenofected hTERT-RPE-1 cells was performed in the same way using 3 × 106 cells per sample.
Preparation of nuclear capsids and extracellular virions.
For capsid preparation, HFFs (1.6 × 107 cells) were infected at an MOI of 1. On day 5 p.i., cells were trypsinized, collected by centrifugation at 230 × g for 7 min at 4°C, and washed once with MNT buffer (30 mM 2-[N-morpholino]-ethanesulfonic acid, 100 mM NaCl, 20 mM Tris-HCl [pH 7.4]). Cells were resuspended in 4 volumes of sterile water containing 10 mM dithiothreitol (DTT) and protease inhibitors, incubated on ice for 1 min, and snap-frozen in liquid nitrogen. After thawing at 37°C, cells were homogenized by passing 10 times through a hollow needle (0.6 × 30 mm), and nuclei were pelleted (230 × g, 7 min, 4°C) and resuspended in 4 pellet volumes of TNE–1% Triton X-100–10 mM DTT. (TNE is 20 mM Tris [pH 7.5], 0.5 M NaCl, and 1 mM EDTA.) Following snap-freezing in liquid nitrogen and thawing at 37°C, nuclei were sonicated at 4°C (amplitude of 60% with 10-s pulses until the sum of the applied energy was 4 kJ). MgCl2 (final concentration, 1 mM) and 100 U of Benzonase were added, and samples were incubated overnight on a tumbling wheel at 4°C. Benzonase was inactivated by adding EDTA (pH 8.0) to a final concentration of 20 mM, and debris was removed by centrifugation for 50 min at 8,000 × g at 4°C. The supernatant was layered on 2 ml of 35% sucrose in TNE–10 mM DTT, and capsids were pelleted at 70,000 × g for 90 min at 4°C. Capsids were resuspended by adding 100 μl of 20 mM Tris (pH 7.5)–150 mM NaCl–1 mM EDTA–10 mM DTT to the pellet, followed by incubation on ice for 10 min and careful pipetting using a cut pipette tip. Finally, capsids were snap-frozen in liquid nitrogen and stored at −80°C. All steps were done at 4°C using ice-cold buffer solutions containing protease inhibitors. Gradient purification of capsids was done by following previously described protocols with slight modifications (35, 36). HFFs (5 × 107 cells) were infected as described above and harvested on day 4 p.i. by trypsinization. After being washed in phosphate-buffered saline (PBS), cells were resuspended in 20 mM Tris-HCl (pH 7.5) and incubated on ice for 20 min, followed by addition of Triton X-100 to a final concentration of 1.5% and further incubation on ice for another 30 min. Nuclei were spun down at 950 × g for 10 min and resuspended in TNE, and subsequent sonication, Benzonase treatment, and removal of debris were performed as delineated above. The resulting supernatant was layered on a 20 to 50% sucrose gradient (prepared in TNE) followed by centrifugation at 100,000 × g for 60 min at 4°C and collection of the light-scattering bands.
To purify virions from the supernatant of infected HFFs, medium was harvested from cell cultures exhibiting 100% cytopathic effect (CPE), and cell debris was removed by centrifugation at 3,500 × g for 10 min at 4°C. The sample was layered on a 15% sucrose solution in virus standard buffer (50 mM Tris-HCl [pH 7.8], 12 mM KCl, 5 mM EDTA) and centrifuged for 90 min at 4°C at 70,000 × g. The virus particles in the pellet were resuspended in 150 μl of Roti-Load 1.
EM.
For analysis of intracellular capsids by transmission electron microscopy (EM), glass coverslips were seeded with adenofected hTERT-RPE-1 cells and processed 5 days later essentially as described previously (18). Cells were fixed for 1 h at room temperature with 2% (wt/vol) glutaraldehyde in 130 mM cacodylate buffer at pH 7.4 containing 2 mM CaCl2 and 10 mM MgCl2. Then the cells were washed and postfixed with 1% (wt/vol) OsO4 in 165 mM cacodylate buffer at pH 7.4 containing 1.5% (wt/vol) K3[FeIII(CN)6] for 1 h, followed by incubation in 0.5% (wt/vol) uranyl acetate in 50% (vol/vol) ethanol overnight. The cells were flat embedded in Epon, and 50-nm sections were cut parallel to the substrate. Images were taken with an FEI Morgagni electron microscope at a magnification of 14,000×, which allowed the identification of nuclear capsid types. These images were stitched to larger images covering the entire nuclear area using the “photomerge” function of the Adobe Photoshop software. Nuclear A, B, and C capsids were counted, and the sectioned nuclear areas were measured. For negative staining and immunogold EM, preparations of nuclear capsids were adsorbed onto carbon and Formvar film-coated 400 mesh copper grids (Stork Veco). As a negative control, capsids of HB5-UL77-mGFP-3 were additionally treated with 2 M guanidinium hydrochloride (GuHCl) for 1 h at room temperature (RT). The samples were washed with PBS, followed either directly by negative staining or immunogold staining. For immunogold staining, grids were blocked with 10 mg/ml bovine serum albumin (BSA) in PBS, incubated for 30 min with rabbit GFP antiserum (Invitrogen) at a dilution of 1:100, and washed with PBS, followed by a 15-min incubation with protein A-labeled gold particles (10-nm diameter; Cell Microscopy Center, Utrecht School of Medicine, The Netherlands). After being washed with PBS and distilled water, preparations were negative stained using 2% (wt/vol) uranyl acetate (Merck) and analyzed with a Tecnai G20 (FEI) at 200 kV. Images of capsids were randomly taken at a magnification of 55,000×. Gold particles were counted.
RESULTS
Generation of HCMV mutants expressing a UL77-mGFP fusion protein.
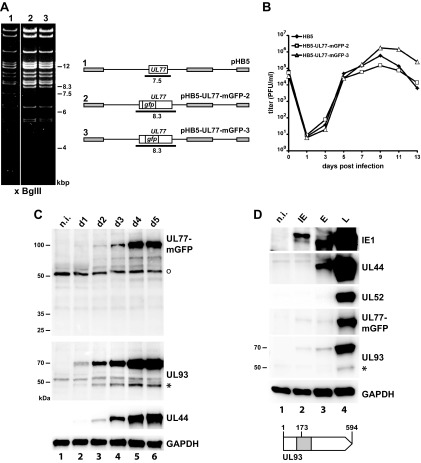
To investigate the UL77 protein in infected cells, we aimed at generating an HCMV mutant that encodes pUL77 fused to a fluorescent protein. Adding tags to essential viral proteins can potentially impair their function. To minimize such effects, we chose a monomeric EGFP (mGFP) that does not induce dimer formation (37). From previous experiments, we knew that tagging of pUL77 at the N terminus is not compatible with virus growth, but small epitope tags were successfully added to the C terminus (33). Unexpectedly, insertion of the mGFP sequence after the last codon of the UL77 open reading frame (ORF) was not compatible with the production of viral progeny (data not shown). Instead, we inserted mGFP after amino acid (aa) 114 or 154 of pUL77 because the respective domains are nonconserved compared to the orthologous proteins of other herpesviruses (38) and presumably form loops on the protein surface as suggested by structure prediction of pUL77 (www.robetta.org [39]; data not shown). Restriction analysis of the resulting HCMV bacterial artificial chromosomes (BACs) HB5-UL77-mGFP-2 and HB5-UL77-mGFP-3 displayed the expected shift of a 7.5-kbp BglII fragment (containing the UL77 ORF) to 8.3 kbp (Fig. 1A) due to insertion of the mGFP ORF. Following transfection of fibroblasts, both BACs gave rise to infectious virus, and the virus mutants exhibited growth kinetics similar to that of the parental HB5 virus (Fig. 1B). The integration of the mGFP ORF between codons 114 and 115 of the UL77 ORF in the HB5-UL77-mGFP-2 BAC disrupts the partially overlapping UL76 ORF. Although nonessential for replication of HCMV in cell culture (as shown in reference 40 and demonstrated here) or perhaps even inhibitory (41), several functions have been assigned to UL76 (42, 43), including regulation of UL77 expression (44). Thus, we decided to perform the further experiments with the mutant HB5-UL77-mGFP-3, in which UL76 remains intact.
FIG 1.
Characteristics of HCMV mutants expressing a UL77-mGFP fusion protein. (A) Construction of the recombinant HCMV BACs pHB5-UL77-mGFP-2 and pHB5-UL77-mGFP-3, which differ in the position of the mGFP sequence inserted into the UL77 open reading frame (ORF). The schematic drawing of the respective BACs is given to the right (pHB5, parental genome), with the expected BglII restriction fragments indicated as black lines. Gray boxes represent repeat regions of the HCMV genome. The left part shows the BglII restriction pattern of the parental BAC (lane 1) compared to that of the mutant genomes (lanes 2 and 3). (Please note the shift of a 7.5-kbp fragment in pHB5 to 8.3 kbp in the UL77-mGFP mutants.) (B) Growth kinetics of the parental virus HB5 (black diamonds) and the HCMV mutants expressing the UL77-mGFP fusion proteins (open triangles and squares, respectively). Fibroblasts were infected at a multiplicity of infection (MOI) of 0.1, supernatants were taken at the time points indicated postinfection, and titers were determined by plaque assay. The values for time point 0 indicate the inoculation doses. (C) Expression of the UL77-mGFP fusion protein and the UL93 protein after infection of fibroblasts with HB5-UL77-mGFP-3 (MOI of 1). Cell lysates were prepared on days 1 to 5 p.i. and analyzed for the indicated proteins by immunoblotting. pUL77-mGFP was detected with a GFP-specific antibody. The nonspecific reactivity with a cellular protein is marked with an open circle. The HCMV early protein pUL44 served as control for viral gene expression and GAPDH as a loading control. Molecular mass markers (in kildodaltons) are given to the left. n.i., noninfected fibroblasts. The asterisk denotes the putative 48-kDa version of pUL93. (D) Expression kinetics of the UL77-mGFP and UL93 proteins. IE, immediate early (i.e., treatment with cycloheximide/actinomycin D); E, early (i.e., treatment with phosphonoacetic acid); L, late (i.e., protein expression in the absence of inhibitors). Fibroblasts were infected with HB5-UL77-mGFP-3 at an MOI of 1, kept with or without the respective inhibitors, followed by preparation of cell lysates on day 3 p.i. and analysis by immunoblotting. IE1, UL44, and UL52 are representative IE, E, and L viral proteins. The open arrow depicts the full-length UL93 ORFs (codons 1 to 594). A putative internal initiation codon at position 173 would give rise to the synthesis of a 48-kDa protein. The part of the UL93 ORF marked in gray was used to express a recombinant protein employed for generation of the specific antibody.
Characterization of the UL77-mGFP and UL93 proteins.
We first analyzed the expression of the UL77-mGFP fusion protein in fibroblasts from day 1 to day 5 following infection with HB5-UL77-mGFP-3 by immunoblotting with a GFP antibody (Fig. 1C). A protein of ∼100 kDa was detected that was expressed mainly at late times of infection (day 3 to day 5), which is consistent with the calculated molecular mass of the UL77-mGFP fusion protein. To characterize the protein(s) encoded by ORF UL93, we generated specific mouse monoclonal antibodies (MAbs) by immunizing mice with a bacterially produced recombinant protein, comprising aa 125 to 339 of the predicted UL93 protein (gray box of the UL93 scheme in Fig. 1D). When tested by immunoblotting, two of the obtained MAbs reacted well with lysates of HCMV-infected cells but performed poorly in immunofluorescence (IF) experiments (not shown). Both MAbs detected a viral protein of ∼70 kDa and also reacted with a weaker band migrating at ∼48 kDa (e.g., see Fig. 1C, lane 6). When testing the lysates of HB5-UL77-mGFP-3-infected cells harvested on day 1 to day 5 by immunoblotting with one of the pUL93-specific MAbs, the 70-kDa protein was the prominent form, which considerably increased in amount during the viral infection cycle, whereas the 48-kDa protein (Fig. 1C, asterisk) was present in small amounts only and did not change much over time. The ∼70-kDa band probably represents the expected 68-kDa protein encoded by the full-length UL93 ORF as first annotated by Chee et al. (45). This is an interesting finding because a recent study did not find evidence for usage of the most 5′-located start codon of the UL93 ORF when applying ribosome profiling analysis of mRNAs extracted from HCMV-infected cells (46). Their data rather suggested that translation of the UL93 transcript starts at downstream initiation codons, giving rise to a protein of 422 aa (ORFL221W, 48 kDa) and two smaller ones. The observed 48-kDa band would be in line with alternative initiation of protein synthesis (possibly at codon 173 of the UL93 ORF, as proposed [c.f., UL93 scheme in Fig. 1D]); however, the main initiation site appears to be the first ATG.
To determine whether pUL77 and pUL93 are immediate early (IE), early (E), or late (L) proteins, infected cells were either left untreated or were incubated with inhibitors typically used to arrest HCMV gene expression in the respective phase of the infection cycle. IE1, UL44, and UL52 served as controls for immediate early, early, and late viral proteins, respectively. As can be seen in Fig. 1D, both pUL77-mGFP and pUL93 were expressed efficiently only in the absence of inhibitors (lane 4, L), classifying them as late proteins.
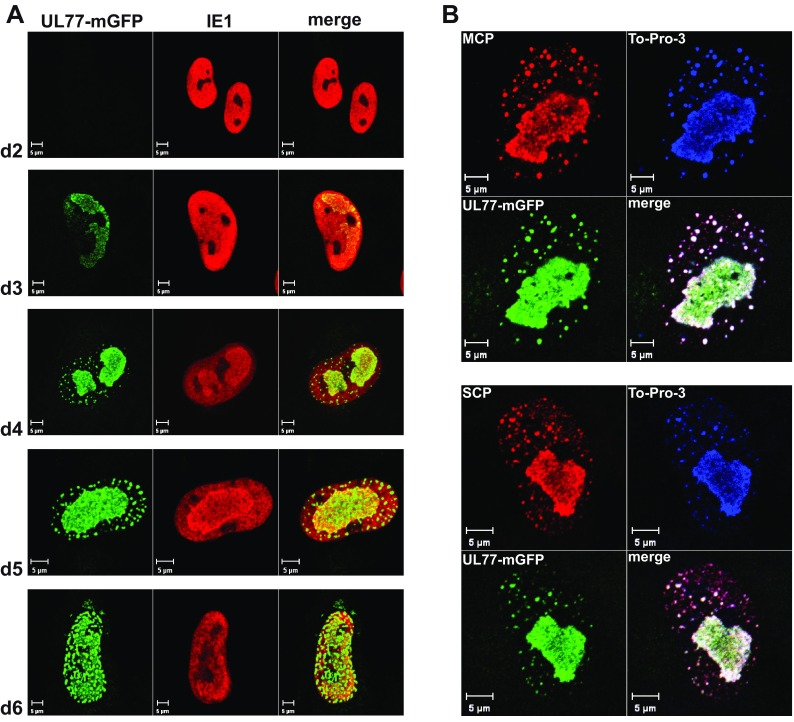
Next we analyzed the subcellular localization of the UL77-mGFP fusion protein during the course of HCMV infection by confocal laser scanning microscopy. In nuclei of infected cells (labeled by IE1), pUL77-mGFP became detectable on day 3 p.i. in patches reminiscent of replication compartments (Fig. 2A). Additionally, green fluorescent dots were visible outside the replication compartments later (Fig. 2A, d4 and d5) and finally throughout the whole nucleus (Fig. 2A, d6). To check whether these dots also contain capsid proteins, cells infected with HB5-UL77-mGFP-3 were labeled on day 5 p.i. with antibodies specific for the major or small capsid protein (Fig. 2B). The green fluorescent dots overlapped almost completely with the signals for the capsid proteins, especially with the ones for the major capsid protein (MCP) (Fig. 2B, upper panel). The larger dots with sizes of 800 to 1,000 nm may result from accumulation of several capsids, whereas the smaller dots (∼400 nm) may constitute individual fluorescent capsids. Taken together, these results suggest that pUL77-mGFP associates with capsid proteins, possibly with complete capsid shells.
FIG 2.
Subcellular localization of pUL77-mGFP in infected cells. (A) Fibroblasts were infected with HB5-UL77-mGFP-3 (MOI, 0.1) and fixed on days 2 to 6 p.i. The UL77-mGFP fusion protein was detected by fluorescence microscopy, and labeling of the viral IE1 protein with a specific Ab was used to delineate the nuclei of infected cells. (B) Fibroblasts infected with HB5-UL77-mGFP-3 at an MOI of 0.1 were fixed on day 5 p.i. The UL77-mGFP fusion protein was detected via the GFP fluorescence, To-Pro-3 was used to stain nucleic acid, and capsid proteins were visualized with Abs directed against the major capsid protein (MCP; upper panel) or the small capsid protein (SCP; lower panel). Scale bars, 5 μm.
pUL77-mGFP and pUL93 are present on nuclear capsids and in extracellular virions.
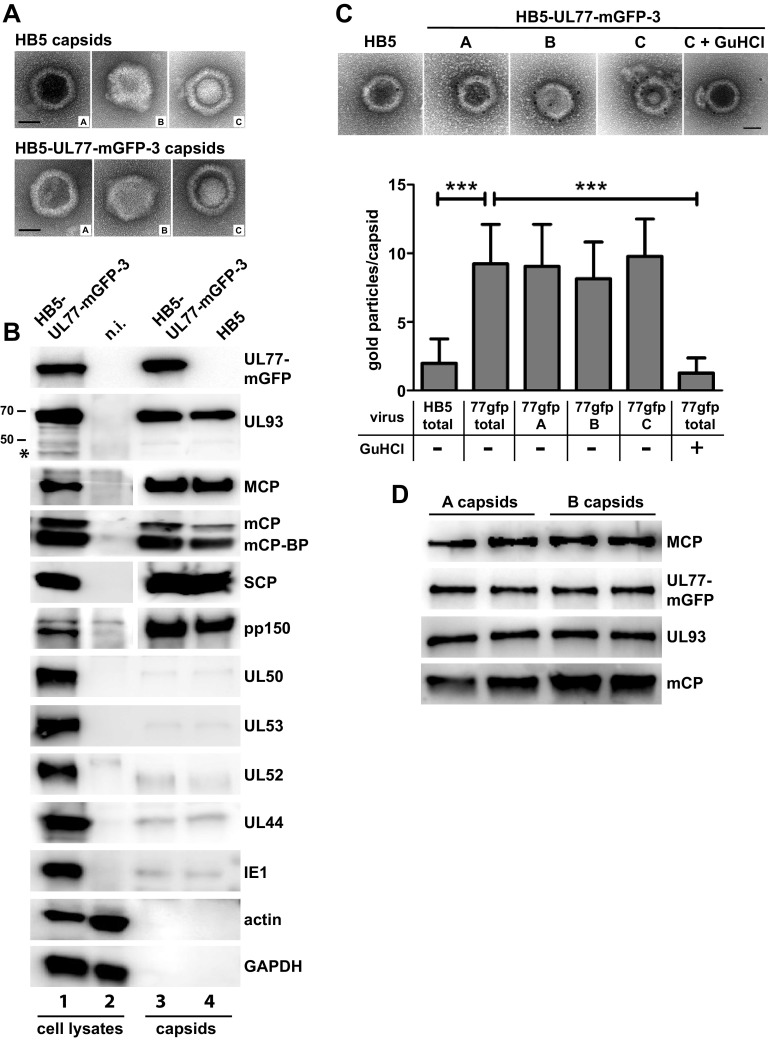
To gain further insight into the capsid association of pUL77-mGFP and pUL93, we isolated capsids from the nuclei of infected cells. At first, capsids were pelleted by ultracentrifugation, which allowed us to isolate intact capsids of all three types (A, B, and C capsids) from nuclei of fibroblasts infected with either the parental HB5 virus or HB5-UL77-mGFP-3 (Fig. 3A). Analysis of these capsid preparations by immunoblotting showed the presence of pUL77-mGFP in capsids derived from cells infected with the mutant (Fig. 3B, top panel, lane 3). pUL93 was present as well in nuclear capsids, with only the 68-kDa full-length form being detected (Fig. 3B, second panel, lanes 3 and 4). Other known capsid proteins (MCP, mCP, mCP-BP, and SCP) were readily identified as expected, and also the betaherpesvirus-specific tegument protein pp150 was found to be associated with capsids. This is in line with data reporting that attachment of pp150 to capsids starts already in the cell nucleus, presumably stabilizing C capsids during nuclear egress (12, 13, 47, 48). However, we cannot exclude that a portion of pp150 detected here originates from some cytoplasmic capsids copurifying with the nuclear capsids and not solely from C capsids. To evaluate the quality of the capsid preparations, several viral nuclear as well as cellular proteins served as controls. pUL52, an HCMV late protein required for genome cleavage and packaging, the viral polymerase processivity factor pUL44, and the viral regulatory protein IE1 were detected in minimal amounts only, and actin and GAPDH were absent (Fig. 3B, bottom panels). This confirmed the purity of the capsids. Moreover, the viral nuclear egress proteins pUL50 and pUL53 were present in marginal amounts only, similar to the background signals obtained for nonstructural viral control proteins (Fig. 3B, panels 7 and 8).
FIG 3.
Detection of pUL77-mGFP and pUL93 in nuclear capsids. Capsids isolated from the nuclei of fibroblasts infected with HB5-UL77-mGFP-3 or the parental virus HB5 (MOI of 1) were visualized by negative staining and electron microscopy (scale bars, 50 nm) (A) and by immunoblotting (B) for the presence of the indicated proteins. Lysates of HB5-UL77-mGFP-3-infected fibroblasts and of noninfected cells served as controls. The 48-kDa UL93 product is marked with an asterisk. Two percent of the cell lysates and 8% of the capsid preparations were loaded. (C) Immunogold labeling of isolated nuclear capsids using a GFP antibody followed by EM (top). The gold particles attached to the different capsid types were quantified as shown in the lower part (“total,” label on all capsid types). GuHCl, treatment with guanidinium hydrochloride. The numbers of capsids analyzed are as follows: HB5 total, 41; 77GFP total, 56; 77GFP A, 26; 77GFP B, 7; 77GFP C, 23; and 77GFP total plus GuHCl (right), 15. For statistical analysis, the unpaired t test was applied. ***, P < 0.0001. Scale bars, 50 nm. (D) Comparison of pUL77-mGFP and pUL93 amounts of gradient-purified A and B capsids by immunoblotting. Following adjustment to equal MCP levels, the samples were loaded in duplicates onto the gel.
Next we asked to which types of capsids pUL77-mGFP is attached by performing immunogold labeling with a GFP antibody followed by electron microscopy (EM). As can be seen in Fig. 3C, there was specific labeling of HB5-UL77-mGFP A, B, and C capsids compared to the background signal obtained for HB5 capsids, yet we did not observe preferential labeling of A or C capsids as described for alphaherpesviruses. Upon treatment of HB5-UL77-mGFP capsids with guanidinium hydrochloride (GuHCl), the occupancy of capsids with gold particles was reduced to background level, thus corroborating the localization of pUL77-mGFP on the capsid surface.
Since the comparable amounts of pUL77-mGFP on all three capsid types contrast with the findings obtained for alphaherpesviruses, we further investigated this aspect by using gradient-purified capsids. Albeit well established with alphaherpesviruses, the procedure is challenging for HCMV because of the much lower number of nuclear capsids produced and their reduced stability (35, 36). Upon rate velocity sedimentation of nuclear capsids in a sucrose gradient, the light-scattering bands were collected and analyzed by negative staining and EM. The upper two bands exhibited efficient enrichment of either A or B capsids—namely, 85% A capsids and 80% B capsids, respectively. The band below was very faint—an observation also made by others (36)—and contained only 5% C capsids besides numerous A and B capsids, and also many damaged capsids (data not shown). This is probably due to the larger and more densely packed HCMV genome compared to those of other herpesviruses, resulting in a high internal capsid pressure more easily leading to capsid rupture. The purified A and B capsids were examined by immunoblotting for the presence of pUL77-mGFP after normalization of the samples to equal amounts of MCP (Fig. 3D). Comparable levels of pUL77-mGFP were detected on both capsid types, and the same was found for pUL93. We could not check the latter finding by immunogold labeling and EM until now because the UL93-specific antibody does not work when the native protein has to be recognized. These results provide further evidence that pUL77-mGFP is not preferentially associated with one type of capsids, as is the case with the alphaherpesvirus orthologue pUL25, and moreover indicate that also pUL93 is present in A and B capsids in similar amounts.
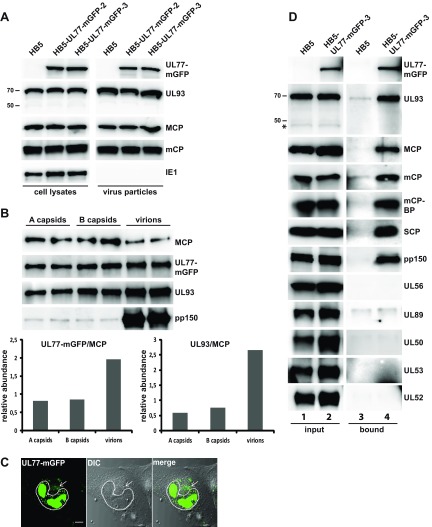
Some viral proteins interact only transiently with capsids and dissociate after accomplishing their task (e.g., the terminase subunits or the proteins of the nuclear egress complex), whereas others are retained as structural components. Therefore, we tested whether pUL77-mGFP and pUL93 are present in extracellular virions. Virus particles were purified from the supernatant of fibroblasts infected with either the parental virus HB5 or the HB5-UL77-mGFP mutants and analyzed by immunoblotting (Fig. 4A). As is obvious from the figure, pUL77-mGFP was detected in the virions of the mutants, and pUL93 was found in all virion preparations. Major and minor capsid proteins (MCP and mCP, respectively) were used as positive controls for viral proteins associated with virions, and the nuclear, nonstructural IE1 protein served as a negative control, which was present in cell lysates, but not in virus particles, underlining the purity of the virion preparations. Next we assessed the amounts of pUL77-mGFP and pUL93 in virus particles in comparison to A and B capsids using immunoblotting and determined the ratios of the pUL77-mGFP and pUL93 signals to the MCP signals (Fig. 4B). Again, this experiment also did not point to a preference of either pUL77-mGFP or pUL93 for A capsids, yet virions contained larger amounts of both pUL77-mGFP and pUL93 than A or B capsids (Fig. 4B; factor 2 for pUL77-mGFP and factor 2.7 for pUL93). As expected, large amounts of pp150 were associated with virus particles only (Fig. 4B, lowest panel).
FIG 4.
Detection of pUL77-mGFP and pUL93 in virus particles and identification of viral proteins associated with pUL77-mGFP. (A) Virus particles purified from the supernatant of fibroblasts infected with the parental virus HB5 or the HB5-UL77-mGFP mutants (MOI of 1) were analyzed by immunoblotting (right). The lysates of cells the virions were derived from were used for comparison (left). Two percent of the cell lysates and 10% of the virus preparations were analyzed. (B) Investigation into pUL77-mGFP and pUL93 levels in gradient-purified A and B capsids and virions by immunoblotting. Each sample was loaded twice onto the gel, and signals were quantified with the ImageJ 1.46r software, using membranes exposed for a few seconds only. The lower part depicts the relative abundance of pUL77-mGFP and pUL93 as obtained after dividing the respective signals by the MCP signal. (C) Live-cell image of HFFs infected with HB5-UL77-mGFP-3 on day 5 p.i. The dotted line marks the cell nucleus, and the arrow points to the viral assembly compartment. (D) Viral proteins associated with the UL77-mGFP fusion protein in infected cells. Fibroblasts were infected with HB5 or HB5-UL77-mGFP-3 at an MOI of 1 and lysed on day 5 p.i. pUL77-mGFP was pulled down using GFP-trap beads and copurifying proteins were analyzed by immunoblotting. One percent of the cell lysates before pulldown (input) and 8% of the material eluted from the beads (bound) were loaded onto the gels. An asterisk indicates the 48-kDa version of pUL93.
Having shown that pUL77-mGFP is present in virions, we expected also a fluorescence signal of particles in the cytoplasm, where they undergo further tegumentation and secondary envelopment. In fixed and permeabilized cells, we did not detect a distinct cytoplasmic localization of pUL77-mGFP (c.f. Fig. 2) and therefore investigated additionally living cells. As is shown in Fig. 4C, pUL77-mGFP was found in the cytoplasm of infected fibroblasts late in infection, mostly in a juxtanuclear region reminiscent of the HCMV assembly compartment.
In summary, these results show that pUL77 and pUL93 are structural proteins that remain associated with capsids and are ultimately incorporated into mature extracellular virus particles.
Viral proteins interacting with pUL77-mGFP.
We then examined which of the HCMV capsid proteins are interacting with pUL77-mGFP and may thus mediate its binding to capsids. For this, fibroblasts were infected with either HB5 or HB5-UL77-mGFP-3, cell lysates were prepared, and pUL77-mGFP was pulled down using GFP-Trap beads. This approach utilizes fragments of Camelidae GFP Abs (nanobodies), which combine the characteristics of high-affinity binding and small size (∼10 kDa compared to 150 kDa for conventional Abs). This enables the nanobodies to gain access to GFP even when hidden in small cavities (as might be the case for pUL77-mGFP when incorporated into viral capsid structures). Viral protein levels in whole-cell lysates (Fig. 4D, left panels, input) of parental and mutant virus-infected cells demonstrated comparable infection efficiency. As expected, pUL77-mGFP could be precipitated from lysates of cells infected with HB5-UL77-mGFP-3 (Fig. 4D, right panels, bound). Several viral proteins copurified with pUL77-mGFP, namely, pUL93 and the capsid proteins MCP, mCP, mCP-BP, and SCP, as well as the tegument protein pp150. No evidence of association of the putative 48-kDa pUL93 version was obtained as exclusively the full-length 68-kDa protein was visible. The terminase subunits pUL56 and pUL89 were not detected in the fraction of proteins pulled down with pUL77-mGFP, and also the pUL50 and pUL53 components of the nuclear egress machinery (49, 50), as well as the encapsidation protein pUL52 (21), were not observed. Few proteins exhibited some unspecific attachment to the beads, as can be seen by the weak signals obtained for the HB5 sample (i.e., pUL93, mCP, and pUL89 [Fig. 4D, lane 3]). Here, the differences in signal strength between the HB5 and HB5-UL77-mGFP-3 samples (Fig. 4D, right panels, bound) indicated that pUL93 and mCP do associate with the pUL77-mGFP fusion protein, whereas the signals for pUL89 were similar, reflecting background binding.
These results suggested a number of potential viral interaction partners for pUL77. An alternative explanation would be that rather whole capsids than individual proteins were coprecipitated. When examining the lysates of infected cells used for the GFP-trap pulldown assays by EM, we could indeed find capsids as well as structures resembling capsid fragments (data not shown).
Requirements for subnuclear localization and for interactions of pUL77.
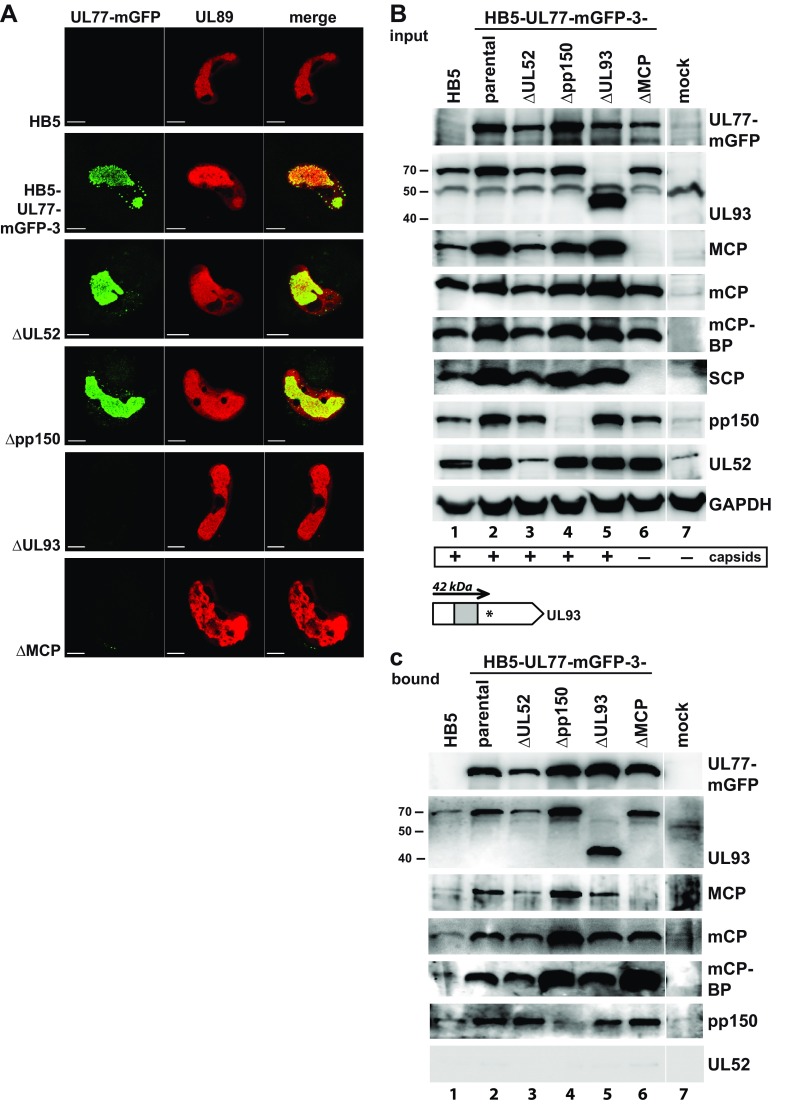
To further dissect the observed pUL77 protein interactions, we asked which viral proteins are necessary for these interactions to occur and disrupted the ORFs encoding the major capsid protein (MCP), pUL52, pUL93, or pp150 in the BAC HB5-UL77-mGFP-3. HCMV BACs with mutations in essential genes are not infectious, and no complementing cells were available for propagation of most of these mutants. To overcome these limitations, we applied a transfection protocol that allows the study of essential viral proteins directly in cells transfected with the respective HCMV deletion genomes (29). Using this method, which is based on defective adenovirus particles as carriers and was therefore termed “adenofection,” we transfected HCMV-permissive RPE-1 cells with the parental BAC HB5 or HB5-UL77-mGFP-3, as well as with the deletion genomes derived from HB5-UL77-mGFP-3. There was no difference in the localization of pUL77-mGFP between cells transfected with HB5-UL77-mGFP-3 or with genomes lacking intact ORFs for pUL52 or pp150 (Fig. 5A). Overlap of the pUL77-mGFP and pUL89 signals—the latter being a marker for the replication centers—confirmed that pUL77-mGFP is mainly found in this compartment. When pUL93 or the MCP was missing, pUL77-mGFP was no longer detectable, possibly because it was dispersed throughout the cell and hence was below the detection limit of the microscope settings applied. These results indicate that pUL93 is crucial for the correct localization of pUL77-mGFP within nuclei. The fact that MCP was necessary as well indicates that it is either directly involved in pUL77-mGFP nuclear targeting or that correct localization of the fusion protein is dependent on the presence of assembled capsids. Furthermore, viral genome encapsidation is not a prerequisite for the proper subcellular distribution of pUL77-mGFP, because the cleavage-packaging protein pUL52 proved to be dispensable for its nuclear localization.
FIG 5.
Localization and interaction of pUL77-mGFP in the absence of individual proteins of the encapsidation network. (A) Subnuclear distribution of the UL77-mGFP fusion protein in RPE-1 cells adenofected with the parental HB5 BAC, the HB5-UL77-mGFP-3 BAC, or mutant BAC genomes derived thereof. pUL77-mGFP was detected by GFP fluorescence, and viral replication compartments were visualized with an antibody specific for the terminase subunit pUL89. For all images, the same microscope settings were used. Scale bars, 10 μm. (B) Immunoblot analysis of total lysates of RPE-1 cells 5 days after adenofection with the indicated BACs. Please note that the pUL52 and pp150 MAbs exhibit some cross-reactivity with cellular proteins of RPE-1 cells migrating with similar mobility to the respective viral protein. The presence of capsids in the respective cell lysates as assessed by negative staining and EM is depicted at the bottom. The scheme at the bottom shows the UL93 ORF of the ΔUL93 mutant carrying an inserted stop codon (asterisk), which results in the synthesis of a truncated 42-kDa protein. (The gray box depicts the part used to generate the pUL93-specific MAb.) (C) Pulldown of pUL77-mGFP from the given cell lysates by GFP-trap followed by investigation of viral proteins associated with pUL77-mGFP by immunoblotting.
Another reason why pUL77-mGFP was not detectable by fluorescence microscopy anymore could be that it was degraded when pUL93 or MCP were missing. Immunoblot analysis demonstrated, however, that the fusion protein was present in all RPE-1 cells adenofected with HB5-UL77-mGFP-3 or the derivatives of it (Fig. 5B, top panel). For the genomes that do not express UL52, pp150, or MCP, the absence of the respective proteins was confirmed (Fig. 5B). Interestingly, upon knockout of the ORF for MCP, the SCP became undetectable in the cell lysate as well (Fig. 5B, SCP, lane 6). In cells transfected with the UL93-null genome, the UL93-specific MAb detected an ∼42-kDa protein (Fig. 5B, second panel, lane 5). The size of this protein is compatible with translation commencing at the first start codon of the UL93 ORF and terminating at the introduced stop codon (c.f. scheme at the bottom of Fig. 5B), supporting our other data that point to the use of the first ATG for pUL93 synthesis.
To investigate which interactions of pUL77-mGFP can still occur when one of the mentioned proteins is missing, the fusion protein was precipitated from the respective cell lysates (Fig. 5C, first panel). Examination of the cell lysates by EM confirmed that in the absence of MCP, capsid formation did not occur, whereas it was not affected when one of the other proteins was missing (indicated at the bottom of Fig. 5B) (data not shown). For detection of interaction partners coprecipitating with pUL77-mGFP from lysates of transfected RPE-1 cells, long exposure times were needed. This led to high background signals, resulting from nonspecific attachment of proteins to the beads, as can be seen in the lane for the HB5 control lysate (Fig. 5C, lane 1). Despite this technical limitation, we concluded the following: (i) in the absence of pUL52—a protein that is not a capsid component and does not interact with pUL77-mGFP (c.f. Fig. 3B and 4D)—association of pUL77-mGFP with the viral proteins tested was not affected (Fig. 5C, lane 3), (ii) the same was observed when the tegument protein pp150 was missing (Fig. 5C, lane 4), and (iii) in lysates of cells transfected with the ΔUL93 BAC, the truncated 42-kDa UL93 protein was coprecipitated with pUL77-mGFP, and also capsid proteins and pp150 did copurify. Apparently, the truncated UL93 protein can still mediate interaction to some extent, although this is not sufficient for correct localization of pUL77-mGFP and the generation of infectious progeny. Since capsids are made when either the UL52, UL32 (encoding pp150), or UL93 ORF is disrupted, we cannot make a conclusion about individual protein-protein interactions as coprecipitation with pUL77-mGFP could also reflect pulldown of whole capsids. However, when MCP was missing and capsid assembly was prevented, we still observed interaction with pUL93, mCP, mCP-BP, and pp150 (Fig. 5C, lane 6). This hints at the formation of higher-order protein complexes of the structural viral proteins even in the absence of capsid formation.
Functional consequences of UL77 or UL93 deletion.
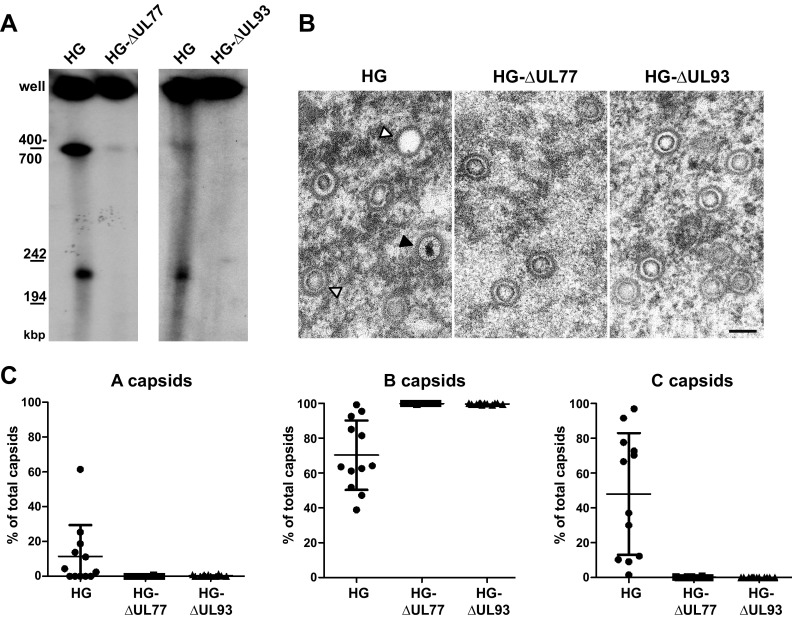
Having shown that pUL77-mGFP and pUL93 are capsid constituents, we next asked whether these proteins may play a role in HCMV genome encapsidation. To this end, either UL77 or UL93 was deleted from the HG genome, an AD169 strain-based EGFP-expressing HCMV BAC (31). We first tested if genome cleavage is still carried out after disruption of UL77 or UL93. The mutated genomes were used for adenofection of RPE-1 cells, and total DNA was analyzed 5 days posttransfection by pulsed-field gel electrophoresis and Southern blotting with a probe specific for the viral genome (Fig. 6A). For both HG-ΔUL77 and HG-ΔUL93, similar amounts of newly replicated concatemeric viral DNA were obtained compared to the parental HG genome (Fig. 6A, signal labeled “well”). Concatemeric DNA is of high molecular weight and is believed to adopt branched and complex structures, and thus is retained within the wells of the gel. Conversely, when genome cleavage takes place, unit-length genomes can be resolved in pulsed-field gels and are detected at approximately 240 kbp (Fig. 6A). (The 400- to 700-kbp DNA species also seen in Fig. 6A presumably represents intermediate products of the cleavage process.) As shown in Fig. 6A, after deletion of UL77 or UL93, unit-length genomes were not detectable any longer, implying that genome cleavage does not (or does not efficiently) occur in the absence of these viral proteins.
FIG 6.
Analysis of genome cleavage and capsid formation in the absence of pUL77 or pUL93. (A) Total DNA of RPE-1 cells adenofected with the parental HG BAC or the ORF UL77- or UL93-deleted genomes was analyzed on day 5 posttransfection by pulsed-field gel electrophoresis followed by Southern blotting using an HCMV-specific probe. (B) The RPE-1 cells were adenofected as in panel A and examined by EM for the presence of different capsid species in the nucleus. Open arrowhead, A capsid; gray arrowhead, B capsid; black arrowhead, DNA-filled C capsid. Scale bar, 100 nm. (C) Quantification of capsid types shown in panel B. The number of capsids per cell nucleus (n = 12 cells, with each dot representing a section through one nucleus) was determined, and the percentage of the capsid types (A, B, and C) is depicted (percentage of total capsids).
Since viral genome packaging and cleavage occur concomitantly, we also investigated which capsid types were present in cells adenofected with the UL77 or UL93 mutant genomes (Fig. 6B and C). In cells transfected with the parental HG genome and analyzed by transmission EM, all three capsid types were identified (Fig. 6B, first panel), whereas upon transfection of the mutated genomes, only B capsids were found (Fig. 6B, second and third panels). The percentages of the different capsid types were quantified (Fig. 6C), corroborating the absence of A and C capsids in cells transfected with HG-ΔUL77 or HG-ΔUL93. This is in line with our observation that viral genomes remain uncleaved when pUL77 or pUL93 is missing. The exclusive occurrence of B capsids, which are thought to be dead end products arising from spontaneous angularization of procapsids, together with the lack of A capsids that originate from abortive DNA packaging attempts, suggested that genome packaging is not even initiated in the absence of pUL77 or pUL93. In summary, these data assign an essential role to both pUL77 and pUL93 for HCMV genome encapsidation.
DISCUSSION
In this work, we analyzed the properties and functions of the essential HCMV UL77 and UL93 proteins. Our results show that both pUL77 and pUL93 are expressed with late kinetics and are structural components incorporated into capsids as well as virions. The pUL77-mGFP fusion protein was detected on all three capsid types (A, B, and C) in similar amounts. In the absence of pUL77 or pUL93, viral genome encapsidation did not ensue, and only empty B capsids were produced. pUL77-mGFP was found to interact with pUL93, as well as with capsid proteins and also with the betaherpesvirus-specific tegument protein pp150, and these interactions even occurred when capsid assembly was prevented. However, the localization of pUL77-mGFP to nuclear replication compartments required the presence of pUL93 as well as of the major capsid protein (i.e., intact capsids), but was independent of the tegument protein pp150 or the cleavage-packaging protein pUL52, demonstrating that C capsid formation is not a prerequisite for correct subnuclear distribution of pUL77. Taken together, our results indicate that pUL77 and pUL93 are important components of the HCMV encapsidation network.
While in alphaherpesviruses, several mutants encoding fluorescent capsid proteins have been generated successfully (23, 51–54), similar HCMV mutants were not available. Our previous attempts to label the CMV small capsid protein (SCP) with GFP did not result in viable progeny, because it turned out that—in contrast to alphaherpesviruses—the SCP is essential in both mouse CMV (MCMV) and HCMV (14). A major step forward was the recent generation of an MCMV recombinant in which EGFP is fused to SCP at a different position than before, although this MCMV mutant has to incorporate the native SCP in addition to the EGFP fusion into its capsids to be viable (55). For HCMV, addition of the EGFP to pp150 (as well as tagging of the envelope protein gM with mCherry) provided a valuable tool to study virus entry and intracellular particle movement (47, 56). pp150 was recently defined as the innermost tegument protein of HCMV (12), and although its addition to capsids commences in the nucleus, the majority of pp150 is acquired in the cytoplasm. Thus, the UL77-mGFP virus described here represents the first HCMV mutant with a fluorescently labeled capsid. In this study, we utilized the GFP tag mainly as a means to pull down interacting proteins of pUL77; however, in future studies, this mutant may be a versatile tool to investigate capsid assembly, nuclear capsid trafficking, or nuclear egress, for instance, by live cell imaging.
By examining the expression of pUL77-mGFP and pUL93 during HCMV infection, we found that both proteins are of late kinetics. This is in accordance with a recent proteomics study of HCMV-infected cells, indicating that pUL77 and pUL93 are late proteins expressed simultaneously (57). In our immunoblotting experiments, pUL93 was seen as a major band migrating with a molecular mass of 68 kDa and as a minor form of 48 kDa. This was unexpected, since a recent study reporting on ribosome profiling with HCMV-infected cells did not find evidence for usage of the first ATG of the UL93 ORF, which would yield a 68-kDa protein (46). Instead, it was proposed that translation starts at internal initiation codons, giving rise to smaller UL93 proteins or peptides. For the following reasons, we believe that the full-length pUL93 is the biologically relevant protein species: (i) the 68-kDa UL93 protein was expressed at high levels, whereas only small amounts of the 48-kDa form were found, (ii) predominantly the 68-kDa UL93 protein was detected in capsids and virions and was also the one interacting with pUL77-mGFP, and (iii) the interruption of the UL93 ORF yielded a nonfunctional, C-terminally truncated UL93 protein with a size that exactly matched a product whose synthesis starts at the first ATG.
Inspection of capsids isolated from the nuclei of infected cells and of extracellular virions demonstrated that pUL77 and pUL93 are capsid constituents that stay associated with capsids and are incorporated into virus particles. The latter finding is in line with proteomic analyses of HCMV particles, in which peptides originating from pUL77 and pUL93 were detected (58, 59). Also, capsid-association of pUL77 was previously seen after fractionation of extracellular HCMV particles into envelope, tegument, and capsid (28). The association of pUL77 and pUL93 with nuclear capsids is described here for the first time. Further EM experiments revealed that pUL77-mGFP is present on A, B, and DNA-filled C capsids in comparable amounts, and quantitative immunoblotting showed equal amounts of pUL77 as well as of UL93 on A and B capsids. This is different from the UL25 protein of alphaherpesviruses, whose abundance increases from B to A to C capsids (24, 25, 60–64). In fact, the HSV-1 UL25/UL17 heterodimer was initially considered to be attached exclusively to C capsids and was thus termed the “C capsid-specific component” (CCSC) (25), yet its subsequent detection in A and B capsids led to its renaming as the “capsid vertex-specific component” (CVSC) (27). The enrichment of CVSC on C capsids described by several groups gave rise to the hypothesis that addition of CVSC concomitant with viral DNA packaging creates a signature indicating that mature capsids are ready for nuclear egress. At first glance, the finding that pUL77-mGFP is apparently present in similar amounts on all three capsid types—which needs to be corroborated by means of other techniques such as cryo-electron microscopy (cryo-EM)—argues against pUL77 providing the signal that triggers nuclear egress. In this respect, one has to point out that the organizations of the tegument of alpha- and betaherpesviruses differ (10, 11), and moreover, HCMV encodes additional tegument proteins which are specific for betaherpesviruses and may take over this task. On the other hand, it might be rather the joint binding of pUL77 and pUL93 to C capsids concomitant with conformational changes that could lead to the exposure of domains providing the signal for nuclear egress. Assessment of pUL93 association with different capsid types by immunogold labeling is therefore one of the immediate next tasks. For this, we are currently generating additional antibodies that will recognize as well the native protein. Furthermore, we consider constructing an HCMV mutant that expresses a UL93 protein tagged in a similar way as done for pUL77. Future cryo-EM experiments will disclose whether pUL77 is indeed located at the capsid vertices and will elucidate the precise localization of pUL77 and pUL93 on HCMV capsids and their positions relative to other capsid and tegument proteins.
We could copurify pUL77-mGFP and pUL93, which is reminiscent of the interaction of the alphaherpesvirus CVSC proteins pUL25 and pUL17. In addition, capsid proteins and pp150 were identified in the immunoprecipitated material, which was not unexpected, because a recent high-resolution cryo-EM study of HCMV capsids showed spatial proximity of MCP, SCP, triplex proteins, and pp150 (13). However, based on our data, one cannot conclude that pUL77-mGFP and pUL93 interact directly with these proteins. Since the cell lysates also contained capsid fragments and even whole capsids, pulldown of one of the proteins is sufficient to coprecipitate the other components. We did not observe an interaction of pUL77 with the terminase proteins pUL89 or pUL56, although the latter was reported before (28). This discrepancy may be due to different experimental conditions. Even so, the terminase is supposed to detach from capsids after cleavage-packaging is completed, and thus its capsid association may be hard to discover. Likewise, the nuclear egress complex (NEC) components pUL50 and pUL53 neither coprecipitated with pUL77-mGFP nor were detected in nuclear capsids in substantial amounts. In contrast, for alphaherpesviruses, an association of the HCMV pUL53 homologue pUL31 with capsids was reported (65–67). Interaction of herpesvirus capsids with the NEC is transient and probably of low affinity, because the capsids have to dissociate from the NEC to get released into the cytosol. This may hamper the detection of NEC components on HCMV capsids. Upon overexpression of isolated proteins, interactions among the CVSCs of different herpesviruses and terminase or NEC components were seen (68–71). To assess whether during infection pUL77 or pUL93 is required for NEC binding, a similar approach to that described for HSV-1 by Leelawong et al. (66) could be applied, using mutants lacking either UL50 or UL53. This would lead to inhibition of nuclear egress, stabilizing the capsid association with the remaining component of the NEC and thus facilitating the detection of the interaction.
We also investigated whether the subcellular localization of pUL77 or its interaction with pUL93 depends on the presence of other proteins of the HCMV encapsidation network. Correct localization of pUL77-mGFP was not impaired in the absence of pp150 (UL32) or pUL52. The latter finding is contrary to that of the HSV-1 pUL32 orthologue, which influences HSV-1 pUL25 nuclear localization (72). This may be due to the recently described ability of pUL32 to affect the disulfide bond profile of pUL25 (73, 74); however, it is not known whether HCMV pUL52 exerts a similar function. In any case, the results presented here indicate that pUL52 is neither a capsid constituent nor an interaction partner of pUL77. The dependence of pUL77-mGFP localization on pUL93 or MCP is similar to data obtained with HSV-1, for which pUL17 and also capsid proteins are necessary for nuclear targeting of pUL25 (72). Although pUL77-mGFP and pUL93 could still interact when capsid formation was abrogated—for instance, upon disruption of the MCP ORF—proper nuclear localization of pUL77 seems to depend on additional factors. Whether pUL77 and pUL93 interact directly, as reported for the corresponding proteins of alpha- and gammaherpesviruses (23, 24, 75), remains to be tested by expressing the proteins independent of viral infection. Interestingly, in the accompanying article by P. Köppen-Rung and colleagues (84), direct interaction of pUL77 and pUL93 was confirmed by using such an approach, and the domain mediating the binding to UL93 was mapped to the N terminus of pUL77.
Following disruption of either the UL77 or UL93 ORF, viral concatemeric genomes remained uncleaved, and exclusively B capsids were detected, demonstrating that both proteins are essential for HCMV genome encapsidation. The phenotype of the UL93 mutant is comparable to that of alphaherpesvirus UL17 deletion mutants (76, 77). Moreover, a recent study using a UL93 mutant in which the initiation codon was mutated to a stop codon also established a pivotal role for pUL93 in genome cleavage and packaging (36). Thus, our data provide evidence that UL93 indeed represents a functional orthologue of HSV-1 UL17, albeit the two proteins do not share sequence similarity.
In contrast, the phenotypic consequences resulting from deletion of UL77 are different from those of UL25-null mutants of alphaherpesviruses. pUL25 exerts its function after the viral DNA has been cut (78, 79) and rather is involved in retention of genomes within capsids. The conformational changes in capsid structure that accompany genome packaging are believed to expose pUL25 binding sites, resulting in preferential attachment of pUL25 to C capsids and their stabilization (25, 64, 80). In cells infected with UL25 deletion mutants, DNA-filled C capsids were detected (27, 63), whereas upon capsid purification, a larger amount of A capsids, but no C capsids, was seen (24, 52). These findings are not necessarily contradictory, since the isolation of nuclear capsids by gradient purification can lead to the loss of capsid-associated proteins and of the packaged DNA (12, 13, 24). The increased number of alphaherpesvirus A capsids suggested that DNA packaging was initiated, yet the genomes were not stably retained within capsids in the absence of pUL25. In support of this scenario, small amounts of terminal genomic fragments indicative of low-level genome cleavage in the absence of pUL25 were observed (27, 78). We searched for genomic termini in RPE-1 cells transfected with the UL77-deleted genome. However, the portion of free genomic ends is already low in HCMV-infected fibroblasts (18, 36) and was near the detection limit of the Southern blot analysis of viral DNA isolated from RPE-1 cells transfected with the parental HCMV BAC (data not shown). Therefore, a fractional amount of terminal fragments possibly produced in the absence of pUL77 cannot be evaluated in this setting. Still, the lack of C (and A) capsids in ΔUL77-transfected cells distinguishes pUL77 from its alphaherpesvirus orthologues, overall pointing to a somehow different function in betaherpesviruses. Notably, even the roles of the CVSC proteins of alphaherpesviruses seem to be slightly divergent, as functional complementation of pUL17 or pUL25 between PRV and HSV-1 (and vice versa) was not or was only partially possible (81, 82).
Another unsettled issue—also in alpha- and gammaherpesviruses—is whether the CVSC is also present at the portal vertex. If this was the case in HCMV, our finding that DNA packaging is not even attempted in the absence of pUL77 or pUL93 hints at an effect of the CVSC on the portal protein or the interaction of the portal with the terminase complex. Such virus-specific protein-protein interactions may constitute an Achilles heel of the virus and could serve as targets for the development of novel antiviral compounds (83). Further knowledge about the role and structure of pUL77 and pUL93 may thus provide the basis for the design of custom-tailored inhibitors.
ACKNOWLEDGMENTS
We are grateful to all colleagues generously providing antibodies, namely, William Britt, Wade Gibson, Manfred Marschall, Bodo Plachter, and Klaus Radsak. We also thankfully acknowledge the support of the Research Core Units for Laser Microscopy and for Electron Microscopy at Hannover Medical School.
E.M.B., R.B., A.B., T.M.S., S.N., K.W., B.S. and L.S. performed experiments and analyzed data. T.L.R. and S.J. conducted monoclonal antibody generation and production. E.M.B. and M.M. conceived the study and wrote the manuscript.
Funding Statement
This work was funded by Deutsche Forschungsgemeinschaft (DFG; individual grant BO4196/1-2 to Eva Maria Borst) and in part by Deutsches Zentrum für Infektionsforschung (DZIF; grant IICH07.0802 to Martin Messerle). The work of Beate Sodeik was funded by the Excellence Cluster REBIRTH EXC62/2, and the work of Stipan Jonjić was funded by EC | European Research Council (ERC) (322693) and by Ministry of Science, Education and Sports Croatia (CERVir-Vac).
Footnotes
For a companion article on this topic, see doi:10.1128/JVI.00351-16.
REFERENCES
- 1.Mocarski ES, Shenk T, Griffiths PD, Pass RF. 2013. Cytomegaloviruses, p 1960–2014. In Knipe DM, Howley PM (ed), Fields virology 6th ed Lippincott, Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Britt WJ, Boppana S. 2004. Human cytomegalovirus virion proteins. Hum Immunol 65:395–402. doi: 10.1016/j.humimm.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Gibson W, Bogner E. 2013. Morphogenesis of the cytomegalovirus virion and subviral particles, p 230–246. In Reddehase MJ. (ed), Cytomegaloviruses: from molecular pathogenesis to intervention, vol 1 Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 4.Lai L, Britt WJ. 2003. The interaction between the major capsid protein and the smallest capsid protein of human cytomegalovirus is dependent on two linear sequences in the smallest capsid protein. J Virol 77:2730–2735. doi: 10.1128/JVI.77.4.2730-2735.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardone G, Heymann JB, Cheng N, Trus BL, Steven AC. 2012. Procapsid assembly, maturation, nuclear exit: dynamic steps in the production of infectious herpesvirions. Adv Exp Med Biol 726:423–439. doi: 10.1007/978-1-4614-0980-9_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tandon R, Mocarski ES. 2012. Viral and host control of cytomegalovirus maturation. Trends Microbiol 20:392–401. doi: 10.1016/j.tim.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tandon R, Mocarski ES, Conway JF. 2015. The A, B, Cs of herpesvirus capsids. Viruses 7:899–914. doi: 10.3390/v7030899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhella D, Rixon FJ, Dargan DJ. 2000. Cryomicroscopy of human cytomegalovirus virions reveals more densely packed genomic DNA than in herpes simplex virus type 1. J Mol Biol 295:155–161. doi: 10.1006/jmbi.1999.3344. [DOI] [PubMed] [Google Scholar]
- 9.Bauer DW, Li D, Huffman J, Homa FL, Wilson K, Leavitt JC, Casjens SR, Baines J, Evilevitch A. 2015. Exploring the balance between DNA pressure and capsid stability in herpes and phage. J Virol 89:9288–9298. doi: 10.1128/JVI.01172-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen DH, Jiang H, Lee M, Liu F, Zhou ZH. 1999. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology 260:10–16. doi: 10.1006/viro.1999.9791. [DOI] [PubMed] [Google Scholar]
- 11.Zhou ZH, Chen DH, Jakana J, Rixon FJ, Chiu W. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J Virol 73:3210–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu X, Shah S, Lee M, Dai W, Lo P, Britt W, Zhu H, Liu F, Zhou ZH. 2011. Biochemical and structural characterization of the capsid-bound tegument proteins of human cytomegalovirus. J Struct Biol 174:451–460. doi: 10.1016/j.jsb.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai X, Yu X, Gong H, Jiang X, Abenes G, Liu H, Shivakoti S, Britt WJ, Zhu H, Liu F, Zhou ZH. 2013. The smallest capsid protein mediates binding of the essential tegument protein pp150 to stabilize DNA-containing capsids in human cytomegalovirus. PLoS Pathog 9:e1003525. doi: 10.1371/journal.ppat.1003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borst EM, Mathys S, Wagner M, Muranyi W, Messerle M. 2001. Genetic evidence of an essential role for cytomegalovirus small capsid protein in viral growth. J Virol 75:1450–1458. doi: 10.1128/JVI.75.3.1450-1458.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bogner E, Radsak K, Stinski MF. 1998. The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity. J Virol 72:2259–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheffczik H, Savva CG, Holzenburg A, Kolesnikova L, Bogner E. 2002. The terminase subunits pUL56 and pUL89 of human cytomegalovirus are DNA-metabolizing proteins with toroidal structure. Nucleic Acids Res 30:1695–1703. doi: 10.1093/nar/30.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bogner E. 2002. Human cytomegalovirus terminase as a target for antiviral chemotherapy. Rev Med Virol 12:115–127. doi: 10.1002/rmv.344. [DOI] [PubMed] [Google Scholar]
- 18.Borst EM, Kleine-Albers J, Gabaev I, Babic M, Wagner K, Binz A, Degenhardt I, Kalesse M, Jonjic S, Bauerfeind R, Messerle M. 2013. The human cytomegalovirus UL51 protein is essential for viral genome cleavage-packaging and interacts with the terminase subunits pUL56 and pUL89. J Virol 87:1720–1732. doi: 10.1128/JVI.01955-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dittmer A, Bogner E. 2005. Analysis of the quaternary structure of the putative HCMV portal protein pUL104. Biochemistry 44:759–765. doi: 10.1021/bi047911w. [DOI] [PubMed] [Google Scholar]
- 20.Dittmer A, Drach JC, Townsend LB, Fischer A, Bogner E. 2005. Interaction of the putative human cytomegalovirus portal protein pUL104 with the large terminase subunit pUL56 and its inhibition by benzimidazole-d-ribonucleosides. J Virol 79:14660–14667. doi: 10.1128/JVI.79.23.14660-14667.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borst EM, Wagner K, Binz A, Sodeik B, Messerle M. 2008. The essential human cytomegalovirus gene UL52 is required for cleavage-packaging of the viral genome. J Virol 82:2065–2078. doi: 10.1128/JVI.01967-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruzsics Z, Borst EM, Bosse JB, Brune W, Messerle M. 2013. Manipulating cytomegalovirus genomes by BAC mutagenesis: strategies and applications, p 37–57. In Reddehase MJ. (ed), Cytomegaloviruses: from molecular pathogenesis to intervention, vol I Caister Academic Press, Hethersett, Norwich, United Kingdom. [Google Scholar]
- 23.Toropova K, Huffman JB, Homa FL, Conway JF. 2011. The herpes simplex virus 1 UL17 protein is the second constituent of the capsid vertex-specific component required for DNA packaging and retention. J Virol 85:7513–7522. doi: 10.1128/JVI.00837-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Homa FL, Huffman JB, Toropova K, Lopez HR, Makhov AM, Conway JF. 2013. Structure of the pseudorabies virus capsid: comparison with herpes simplex virus type 1 and differential binding of essential minor proteins. J Mol Biol 425:3415–3428. doi: 10.1016/j.jmb.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trus BL, Newcomb WW, Cheng N, Cardone G, Marekov L, Homa FL, Brown JC, Steven AC. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol Cell 26:479–489. doi: 10.1016/j.molcel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conway JF, Cockrell SK, Copeland AM, Newcomb WW, Brown JC, Homa FL. 2010. Labeling and localization of the herpes simplex virus capsid protein UL25 and its interaction with the two triplexes closest to the penton. J Mol Biol 397:575–586. doi: 10.1016/j.jmb.2010.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cockrell SK, Huffman JB, Toropova K, Conway JF, Homa FL. 2011. Residues of the UL25 protein of herpes simplex virus that are required for its stable interaction with capsids. J Virol 85:4875–4887. doi: 10.1128/JVI.00242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meissner CS, Köppen-Rung P, Dittmer A, Lapp S, Bogner E. 2011. A “coiled-coil” motif is important for oligomerization and DNA binding properties of human cytomegalovirus protein UL77. PLoS One 6:e25115. doi: 10.1371/journal.pone.0025115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elbasani E, Gabaev I, Steinbrück L, Messerle M, Borst EM. 2014. Analysis of essential viral gene functions after highly efficient adenofection of cells with cloned human cytomegalovirus genomes. Viruses 6:354–370. doi: 10.3390/v6010354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borst EM, Hahn G, Koszinowski UH, Messerle M. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J Virol 73:8320–8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borst EM, Messerle M. 2005. Analysis of human cytomegalovirus oriLyt sequence requirements in the context of the viral genome. J Virol 79:3615–3626. doi: 10.1128/JVI.79.6.3615-3626.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless Red recombination system. Methods Mol Biol 634:421–430. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 33.Glass M, Busche A, Wagner K, Messerle M, Borst EM. 2009. Conditional and reversible disruption of essential herpesvirus proteins. Nat Methods 6:577–579. doi: 10.1038/nmeth.1346. [DOI] [PubMed] [Google Scholar]
- 34.Borst EM, Benkartek C, Messerle M. 2007. Use of bacterial artificial chromosomes in generating targeted mutations in human and mouse cytomegaloviruses, p 10.32.1-10.32.30. In Coligan JE, Bierer B, Margulies DH, Shevach EM, Strober W, Coico R (ed), Current protocols in immunology. John Wiley & Sons, New York, NY. [DOI] [PubMed] [Google Scholar]
- 35.Irmiere A, Gibson W. 1985. Isolation of human cytomegalovirus intranuclear capsids, characterization of their protein constituents, and demonstration that the B-capsid assembly protein is also abundant in noninfectious enveloped particles. J Virol 56:277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeRussy BM, Tandon R. 2015. Human cytomegalovirus pUL93 is required for viral genome cleavage and packaging. J Virol 89:12221–12225. doi: 10.1128/JVI.02382-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Müller-Taubenberger A, Anderson KI. 2007. Recent advances using green and red fluorescent protein variants. Appl Microbiol Biotechnol 77:1–12. doi: 10.1007/s00253-007-1131-5. [DOI] [PubMed] [Google Scholar]
- 38.Bowman BR, Welschhans RL, Jayaram H, Stow ND, Preston VG, Quiocho FA. 2006. Structural characterization of the UL25 DNA-packaging protein from herpes simplex virus type 1. J Virol 80:2309–2317. doi: 10.1128/JVI.80.5.2309-2317.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim DE, Chivian D, Baker D. 2004. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res 32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu D, Silva MC, Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc Natl Acad Sci U S A 100:12396–12401. doi: 10.1073/pnas.1635160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang SK, Duh CY, Wu CW. 2004. Human cytomegalovirus UL76 encodes a novel virion-associated protein that is able to inhibit viral replication. J Virol 78:9750–9762. doi: 10.1128/JVI.78.18.9750-9762.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin SR, Jiang MJ, Wang HH, Hu CH, Hsu MS, Hsi E, Duh CY, Wang SK. 2013. Human cytomegalovirus UL76 elicits novel aggresome formation via interaction with S5a of the ubiquitin proteasome system. J Virol 87:11562–11578. doi: 10.1128/JVI.01568-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costa H, Nascimento R, Sinclair J, Parkhouse RM. 2013. Human cytomegalovirus gene UL76 induces IL-8 expression through activation of the DNA damage response. PLoS Pathog 9:e1003609. doi: 10.1371/journal.ppat.1003609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Isomura H, Stinski MF, Murata T, Nakayama S, Chiba S, Akatsuka Y, Kanda T, Tsurumi T. 2010. The human cytomegalovirus UL76 gene regulates the level of expression of the UL77 gene. PLoS One 5:e11901. doi: 10.1371/journal.pone.0011901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chee MS, Bankier AT, Beck S, Bohni R, Brown CM, Cerny R, Horsnell T, Hutchison CA, Kouzarides T, Martignetti JA, Preddie E, Satchwell SC, Tomlinson P, Weston KM, Barrell BG. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol 154:125–169. [DOI] [PubMed] [Google Scholar]
- 46.Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, Huang SX, Ma M, Shen B, Qian SB, Hengel H, Mann M, Ingolia NT, Weissman JS. 2012. Decoding human cytomegalovirus. Science 338:1088–1093. doi: 10.1126/science.1227919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sampaio KL, Cavignac Y, Stierhof YD, Sinzger C. 2005. Human cytomegalovirus labeled with green fluorescent protein for live analysis of intracellular particle movements. J Virol 79:2754–2767. doi: 10.1128/JVI.79.5.2754-2767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tandon R, Mocarski ES. 2011. Cytomegalovirus pUL96 is critical for the stability of pp150-associated nucleocapsids. J Virol 85:7129–7141. doi: 10.1128/JVI.02549-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Milbradt J, Auerochs S, Sticht H, Marschall M. 2009. Cytomegaloviral proteins that associate with the nuclear lamina: components of a postulated nuclear egress complex. J Gen Virol 90:579–590. doi: 10.1099/vir.0.005231-0. [DOI] [PubMed] [Google Scholar]
- 50.Milbradt J, Kraut A, Hutterer C, Sonntag E, Schmeiser C, Ferro M, Wagner S, Lenac T, Claus C, Pinkert S, Hamilton ST, Rawlinson WD, Sticht H, Coute Y, Marschall M. 2014. Proteomic analysis of the multimeric nuclear egress complex of human cytomegalovirus. Mol Cell Proteomics 13:2132–2146. doi: 10.1074/mcp.M113.035782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Desai P, Person S. 1998. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J Virol 72:7563–7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cockrell SK, Sanchez ME, Erazo A, Homa FL. 2009. Role of the UL25 protein in herpes simplex virus DNA encapsidation. J Virol 83:47–57. doi: 10.1128/JVI.01889-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bohannon KP, Sollars PJ, Pickard GE, Smith GA. 2012. Fusion of a fluorescent protein to the pUL25 minor capsid protein of pseudorabies virus allows live-cell capsid imaging with negligible impact on infection. J Gen Virol 93:124–129. doi: 10.1099/vir.0.036145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lebrun M, Thelen N, Thiry M, Riva L, Ote I, Conde C, Vandevenne P, Di Valentin E, Bontems S, Sadzot-Delvaux C. 2014. Varicella-zoster virus induces the formation of dynamic nuclear capsid aggregates. Virology 454-455:311–327. doi: 10.1016/j.virol.2014.02.023. [DOI] [PubMed] [Google Scholar]
- 55.Bosse JB, Bauerfeind R, Popilka L, Marcinowski L, Taeglich M, Jung C, Striebinger H, von Einem J, Gaul U, Walther P, Koszinowski UH, Ruzsics Z. 2012. A beta-herpesvirus with fluorescent capsids to study transport in living cells. PLoS One 7:e40585. doi: 10.1371/journal.pone.0040585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sampaio KL, Jahn G, Sinzger C. 2013. Applications for a dual fluorescent human cytomegalovirus in the analysis of viral entry. Methods Mol Biol 1064:201–209. doi: 10.1007/978-1-62703-601-6_14. [DOI] [PubMed] [Google Scholar]
- 57.Weekes MP, Tomasec P, Huttlin EL, Fielding CA, Nusinow D, Stanton RJ, Wang EC, Aicheler R, Murrell I, Wilkinson GW, Lehner PJ, Gygi SP. 2014. Quantitative temporal viromics: an approach to investigate host-pathogen interaction. Cell 157:1460–1472. doi: 10.1016/j.cell.2014.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Varnum SM, Streblow DN, Monroe ME, Smith P, Auberry KJ, Pasa-Tolic L, Wang D, Camp DG, Rodland K, Wiley S, Britt W, Shenk T, Smith RD, Nelson JA. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J Virol 78:10960–10966. doi: 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reyda S, Buscher N, Tenzer S, Plachter B. 2014. Proteomic analyses of human cytomegalovirus strain AD169 derivatives reveal highly conserved patterns of viral and cellular proteins in infected fibroblasts. Viruses 6:172–188. doi: 10.3390/v6010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sheaffer AK, Newcomb WW, Gao M, Yu D, Weller SK, Brown JC, Tenney DJ. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J Virol 75:687–698. doi: 10.1128/JVI.75.2.687-698.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thurlow JK, Rixon FJ, Murphy M, Targett-Adams P, Hughes M, Preston VG. 2005. The herpes simplex virus type 1 DNA packaging protein UL17 is a virion protein that is present in both the capsid and the tegument compartments. J Virol 79:150–158. doi: 10.1128/JVI.79.1.150-158.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Newcomb WW, Homa FL, Brown JC. 2006. Herpes simplex virus capsid structure: DNA packaging protein UL25 is located on the external surface of the capsid near the vertices. J Virol 80:6286–6294. doi: 10.1128/JVI.02648-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klupp BG, Granzow H, Keil GM, Mettenleiter TC. 2006. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J Virol 80:6235–6246. doi: 10.1128/JVI.02662-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roos WH, Radtke K, Kniesmeijer E, Geertsema H, Sodeik B, Wuite GJ. 2009. Scaffold expulsion and genome packaging trigger stabilization of herpes simplex virus capsids. Proc Natl Acad Sci U S A 106:9673–9678. doi: 10.1073/pnas.0901514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang K, Baines JD. 2011. Selection of HSV capsids for envelopment involves interaction between capsid surface components pUL31, pUL17, and pUL25. Proc Natl Acad Sci U S A 108:14276–14281. doi: 10.1073/pnas.1108564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leelawong M, Guo D, Smith GA. 2011. A physical link between the pseudorabies virus capsid and the nuclear egress complex. J Virol 85:11675–11684. doi: 10.1128/JVI.05614-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang K, Wills E, Lim HY, Zhou ZH, Baines JD. 2014. Association of herpes simplex virus pUL31 with capsid vertices and components of the capsid vertex-specific complex. J Virol 88:3815–3825. doi: 10.1128/JVI.03175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fossum E, Friedel CC, Rajagopala SV, Titz B, Baiker A, Schmidt T, Kraus T, Stellberger T, Rutenberg C, Suthram S, Bandyopadhyay S, Rose D, von Brunn A, Uhlmann M, Zeretzke C, Dong YA, Boulet H, Koegl M, Bailer SM, Koszinowski U, Ideker T, Uetz P, Zimmer R, Haas J. 2009. Evolutionarily conserved herpesviral protein interaction networks. PLoS Pathog 5:e1000570. doi: 10.1371/journal.ppat.1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Uetz P, Dong YA, Zeretzke C, Atzler C, Baiker A, Berger B, Rajagopala SV, Roupelieva M, Rose D, Fossum E, Haas J. 2006. Herpesviral protein networks and their interaction with the human proteome. Science 311:239–242. doi: 10.1126/science.1116804. [DOI] [PubMed] [Google Scholar]
- 70.To A, Bai Y, Shen A, Gong H, Umamoto S, Lu S, Liu F. 2011. Yeast two hybrid analyses reveal novel binary interactions between human cytomegalovirus-encoded virion proteins. PLoS One 6:e17796. doi: 10.1371/journal.pone.0017796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vizoso Pinto MG, Pothineni VR, Haase R, Woidy M, Lotz-Havla AS, Gersting SW, Muntau AC, Haas J, Sommer M, Arvin AM, Baiker A. 2011. Varicella zoster virus ORF25 gene product: an essential hub protein linking encapsidation proteins and the nuclear egress complex. J Proteome Res 10:5374–5382. doi: 10.1021/pr200628s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Scholtes L, Baines JD. 2009. Effects of major capsid proteins, capsid assembly, and DNA cleavage/packaging on the pUL17/pUL25 complex of herpes simplex virus 1. J Virol 83:12725–12737. doi: 10.1128/JVI.01658-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Albright BS, Kosinski A, Szczepaniak R, Cook EA, Stow ND, Conway JF, Weller SK. 2015. The putative herpes simplex virus 1 chaperone protein UL32 modulates disulfide bond formation during infection. J Virol 89:443–453. doi: 10.1128/JVI.01913-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Szczepaniak R, Nellissery J, Jadwin JA, Makhov AM, Kosinski A, Conway JF, Weller SK. 2011. Disulfide bond formation contributes to herpes simplex virus capsid stability and retention of pentons. J Virol 85:8625–8634. doi: 10.1128/JVI.00214-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dai X, Gong D, Wu TT, Sun R, Zhou ZH. 2014. Organization of capsid-associated tegument components in Kaposi's sarcoma-associated herpesvirus. J Virol 88:12694–12702. doi: 10.1128/JVI.01509-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salmon B, Cunningham C, Davison AJ, Harris WJ, Baines JD. 1998. The herpes simplex virus type 1 U(L)17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J Virol 72:3779–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Klupp BG, Granzow H, Karger A, Mettenleiter TC. 2005. Identification, subviral localization, and functional characterization of the pseudorabies virus UL17 protein. J Virol 79:13442–13453. doi: 10.1128/JVI.79.21.13442-13453.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stow ND. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J Virol 75:10755–10765. doi: 10.1128/JVI.75.22.10755-10765.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McNab AR, Desai P, Person S, Roof LL, Thomsen DR, Newcomb WW, Brown JC, Homa FL. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J Virol 72:1060–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sae-Ueng U, Liu T, Catalano CE, Huffman JB, Homa FL, Evilevitch A. 2014. Major capsid reinforcement by a minor protein in herpesviruses and phage. Nucleic Acids Res 42:9096–9107. doi: 10.1093/nar/gku634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kuhn J, Leege T, Klupp BG, Granzow H, Fuchs W, Mettenleiter TC. 2008. Partial functional complementation of a pseudorabies virus UL25 deletion mutant by herpes simplex virus type 1 pUL25 indicates overlapping functions of alphaherpesvirus pUL25 proteins. J Virol 82:5725–5734. doi: 10.1128/JVI.02441-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kuhn J, Leege T, Granzow H, Fuchs W, Mettenleiter TC, Klupp BG. 2010. Analysis of pseudorabies and herpes simplex virus recombinants simultaneously lacking the pUL17 and pUL25 components of the C-capsid specific component. Virus Res 153:20–28. doi: 10.1016/j.virusres.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 83.Baines JD. 2011. Herpes simplex virus capsid assembly and DNA packaging: a present and future antiviral drug target. Trends Microbiol 19:606–613. doi: 10.1016/j.tim.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 84.Köppen-Rung P, Dittmer A, Bogner E. 2016. Intracellular distributions of capsid-associated pUL77 of human cytomegalovirus and interactions with packaging proteins and pUL93. J Virol 90:5876–5885. doi: 10.1128/JVI.00351-16. [DOI] [PMC free article] [PubMed] [Google Scholar]