ABSTRACT
HIV establishes reservoirs of infected cells that persist despite effective antiretroviral therapy (ART). In most patients, the virus begins to replicate soon after treatment interruption. However, a low frequency of infected cells at the time of treatment interruption has been associated with delayed viral rebound. Likewise, individuals who control the infection spontaneously, so-called HIV-1 controllers (HICs), carry particularly low levels of infected cells. It is unclear, however, whether and how this small number of infected cells contributes to durable viral control. Here we compared 38 HICs with 12 patients on effective combined antiretroviral therapy (cART) and found that the low frequency of infected cells in the former subjects was associated both with less efficient viral reactivation in resting CD4+ T cells and with less efficient virion production ex vivo. We also found that a potent HIV-specific CD8+ T cell response was present only in those HICs whose CD4+ T cells produced virus ex vivo. Long-term spontaneous control of HIV infection in HICs thus appears to be sustained on the basis of the inefficient reactivation of viruses from a limited number of infected cells and the capacity of HICs to activate a potent HIV-specific CD8+ T cell response to counteract efficient viral reactivation events.
IMPORTANCE There is a strong scientific interest in developing strategies to eradicate the HIV-1 reservoir. Very rare HIV-1-infected patients are able to spontaneously control viremia for long periods of time (HIV-1 controllers [HICs]) and are put forward as a model of HIV-1 remission. Here, we show that the low viral reservoirs found in HICs are a critical part of the mechanisms underlying viral control and result in a lower probability of HIV-1 reactivation events, resulting in limited HIV-1 release and spread. We found that those HICs in whom viral reactivation and spread from CD4+ T cells in vitro were the most difficult were those with diminished CD8+ T cell responses. These results suggest that, in some settings, low HIV-1 reservoirs decisively contribute to at least the temporary control of infection without antiretroviral therapy. We believe that this work provides information of relevance in the context of the search for HIV-1 remission.
INTRODUCTION
So-called human immunodeficiency virus type 1 (HIV-1) controllers (HICs) provide a valuable model of natural, durable control of HIV-1 infection (1). A better understanding of the mechanisms underlying this viral control could help with the development of therapeutic interventions capable of achieving HIV-1 remission in other patients. Numerous reports point to a prominent role of CD8+ T cells in the control of infection observed in HICs. Indeed, many HICs have high frequencies of CD8+ T cells that exert multiple effector functions in response to HIV-1 antigens (2–4). In particular, CD8+ T cells from many HICs efficiently eliminate infected CD4+ T cells ex vivo (4). Certain HLA class I alleles, such as B*57 and B*27, are overrepresented in HICs (4–7), but efficient anti-HIV CD8+ T cell responses are not restricted to individuals carrying these alleles (8). In addition, potent HIV-specific CD8+ T cell responses are not found in all HICs, at least during the chronic phase of infection (8, 9). We have found that HIV-specific CD8+ T cell responses in some HICs enrolled in the ANRS CO21 cohort wane over time, yet the plasma viral load remains undetectable (unpublished observations). Similar observations have been made in macaques spontaneously controlling simian immunodeficiency virus (SIV) SIVmac251 infection (10). In HICs, highly responsive CD8+ T cells tend to have an effector phenotype (4, 8, 11), whereas weakly responsive CD8+ T cells tend to have a resting memory phenotype (8, 9). Weakly responsive CD8+ T cells from HICs can regain their effector functions upon antigen stimulation (12), but their role in HIV-1 control in vivo is unclear. These results suggest that several factors probably contribute to long-term spontaneous HIV-1 control, acting in synergy or relieving each other during the period of control.
We and others have previously shown that despite the presence of replication-competent viruses (13–15), HICs are characterized by low levels of CD4+ T cell-associated HIV DNA (16, 17). Although this may be the consequence of viral control, different results indicate that the low frequencies of HIV-1-infected CD4+ T cells might also contribute to the maintenance of such control. The stochastic nature of HIV-1 reactivation from latency suggests that very low HIV-1 reservoirs might result in at least the temporary control of infection without therapy (18). Along this line, the control of HIV-1 viremia or a delayed viral rebound after the discontinuation of antiretroviral therapy (ART) has consistently been associated with low levels of cell-associated HIV DNA at the time of treatment interruption (19–22), even when a specific anti-HIV immune response was not present (23).
In the present study, we analyzed what the low frequency of HIV-1-infected CD4+ T cells found in HICs may represent in terms of virus reactivation and its contribution to the control of infection. We found that the low number of HIV-1-infected cells in HICs was associated with the less frequent and inefficient reactivation of HIV-1 infection in vitro and impaired viral spread. We also found that HICs whose CD4+ T cells did not produce HIV-1 proteins in vitro had a diminished HIV-specific CD8+ T cell response, suggesting that inefficient viral reactivation may suffice to maintain, at least temporarily, control of infection in the absence of antiretroviral treatment.
MATERIALS AND METHODS
Patients and samples.
We studied 38 HICs from the ANRS CO21 CODEX cohort and 12 patients receiving combined antiretroviral therapy (cART patients) from the Kremlin-Bicêtre University Hospital (France) and the Germans Trias i Pujol Hospital (Badalona, Spain). The HICs were patients who had been infected with HIV-1 for at least the previous 5 years and whose last five consecutive viral loads were below 400 HIV RNA copies/ml of plasma. Their median age at the time of the study was 49 years (interquartile range [IQR], 36 to 74 years), their median CD4+ T cell count was 786 cells/mm3 (IQR, 515 to 1,203 cells/mm3), and their median RNA load was <50 HIV RNA copies/ml (IQR, <50 to 213 HIV RNA copies/ml). The cART patients were individuals in whom ongoing antiretroviral treatment had suppressed their viral load (<50 copies HIV RNA/ml) for at least the past 2 years. Their median age at the time of the study was 56 years (IQR, 52 to 57 years), and their median CD4+ T cell count was 677 cells/mm3 (IQR, 433 to 1,165 cells/mm3). A detailed description of the patients is provided in Table 1.
TABLE 1.
Clinical and epidemiological characteristics of the study groupsa
| Subject identifier | Sex | Age (yr) | No. of CD4 cells/mm3 | VL (no. of RNA copies/ml) | No. of HIV DNA copies/106 PBMCs | Time with VL of <400 RNA copies/ml (yr) |
|---|---|---|---|---|---|---|
| 5002 | M | 54 | 1,314 | <50 | 11 | 13 |
| 18004 | M | 47 | 690 | <50 | 12 | 14 |
| 19001 | F | 52 | 702 | <50 | <10 | 13 |
| 23001 | F | 53 | 803 | <50 | 11 | 13 |
| 23002 | F | 52 | 649 | <50 | <10 | 14 |
| 30004 | F | 41 | 1,202 | <50 | 10 | 11 |
| 34003 | F | 45 | 857 | <50 | 16 | 12 |
| 34006 | M | 55 | 554 | <40 | NA | 19 |
| 34009 | M | 40 | 676 | <50 | <10 | 10 |
| 34014 | F | 44 | 931 | 79 | 22 | 10 |
| 34015 | M | 59 | NA | <50 | <10 | 2 |
| 34017 | F | 32 | 995 | <50 | <10 | 7.5 |
| 38002 | F | 42 | 1,060 | <50 | <10 | 11 |
| 41002 | M | 64 | 756 | <50 | <10 | 12 |
| 41003 | M | 55 | 776 | <50 | 45 | 11 |
| 47003 | F | 26 | 620 | <50 | 11 | 6 |
| 51001 | F | 44 | 520 | <50 | <10 | 12 |
| 54001 | F | 34 | 892 | 36 | 45 | 12.5 |
| 57002 | M | 48 | 1,150 | 42 | <10 | 13 |
| 58001 | F | 56 | 465 | <50 | <10 | 11 |
| 58003 | F | 36 | 649 | <50 | 17 | 11 |
| 60005 | M | 48 | 931 | <50 | <10 | 13.5 |
| 72002 | F | 45 | 1,154 | <50 | 34 | 15 |
| 72003 | M | 42 | 495 | <50 | <10 | 9 |
| 78001 | M | 55 | 934 | <50 | <10 | 12.5 |
| 81003 | F | 38 | 882 | <50 | <10 | 11 |
| 81004 | M | 50 | 942 | <50 | 25 | 11 |
| 85002 | M | 49 | 1,003 | 26 | 30 | 12 |
| 86001 | M | 72 | 575 | 72 | <10 | 11 |
| 86003 | F | 34 | 1,144 | <50 | NA | 8 |
| 86005 | F | 39 | 775 | <50 | 11 | 12 |
| 86006 | M | 52 | NA | <50 | 3 | 8 |
| 86007 | F | 36 | 667 | <50 | NA | 13 |
| 88001 | F | 64 | 752 | <50 | NA | 8 |
| 114002 | M | 65 | 979 | <50 | 11 | 10 |
| 134001 | M | 43 | 1,164 | <50 | 187 | 13 |
| 146001 | F | 39 | 464 | 81 | <10 | 13 |
| 171001 | M | 50 | 437 | <50 | <10 | 9 |
| ART1 | M | 65 | 281 | <50 | NA | 21 |
| ART2 | M | 58 | 774 | <50 | NA | 16 |
| ART3 | M | 33 | 580 | <50 | NA | 6 |
| ART4 | F | 53 | 1,151 | <50 | NA | 14 |
| ART5 | M | 60 | 437 | <50 | 317 | 6.5 |
| ART6 | M | 57 | 564 | <50 | 183 | 9.5 |
| ART7 | M | 56 | 1,058 | <50 | <13 | 6.5 |
| ART8 | M | 57 | 420 | <50 | 1,010 | 9.5 |
| ART9 | M | 56 | 385 | <50 | 120 | 12 |
| ART10 | M | 50 | 1,243 | <50 | 233 | 4.5 |
| ART11 | M | 55 | 1,419 | <50 | 103 | 7.5 |
| ART12 | M | 47 | 1,205 | <50 | 167 | 5.5 |
NA, data not available; M, male; F, female; VL, viral load.
Blood samples from HIV-seronegative donors used for viral amplifications experiments were obtained through the collaborative programs between the Institute Pasteur or the AIDS Research Institute-IrsiCaixa and the respective national blood banks.
Ethics statement.
All the subjects provided their written informed consent to participate in the study. The CO21 CODEX cohort and this substudy were funded and sponsored by ANRS and approved by the Ile de France VII Ethics Committee. The study was conducted according to the principles expressed in the Declaration of Helsinki.
HIV-1 reactivation from resting CD4+ T cells.
Peripheral blood mononuclear cells (PBMCs) were collected by Ficoll gradient centrifugation of 50 ml of fresh EDTA-treated blood. A first negative isolation step for total CD4+ T cells was performed with anti-CD8, -CD14, -CD16 (a and b), -CD19, -CD36, -CD56, -CD123, and -CD235a antibodies together with magnetic Dynabeads (Life Technologies, Foster City, CA, USA). A second negative isolation step was performed after CD25+ CD69+ HLADR+ cell depletion with unlabeled anti-CD25 (clone MA251), anti-CD69 (clone FN50), and anti-HLADR (clone L243) (all from BD Bioscience), followed by Dynabeads separation (Life Technologies, Foster City, CA, USA). The purity of the quiescent CD4+ CD25− CD69− HLADR− cells thus isolated was >97%.
The cells were resuspended in RPMI medium with 10% fetal calf serum (R10 medium) at 106 cells/ml, and 500,000 cells/well were cultured under the following conditions: prostratin (2.5 μM; Enzo Life Sciences), chaetocin (90 nM; Sigma-Aldrich), suberoylanilide hydroxamic acid (SAHA; 2.5 μM; Alexis Biochemicals), 5-aza-2´deoxicytidine (1 μM; Sigma-Aldrich), 10-[(3-hydroxy-4-methoxybenzylidene)]-9(10H)-anthracenone (HMBA; 5 mM; Sigma-Aldrich), and interleukin-7 (IL-7; 10 ng/ml; R&D Systems). As positive controls, we stimulated resting CD4+ T cells under two conditions: with anti-CD2/anti-CD28 antibodies (monoclonal antibody [MAb] 39C1.5 [rat IgG2a, anti-CD2] plus MAb 6F10.3 [mouse IgG1, anti-CD2] and MAb 28.2 [mouse IgG1, anti-CD28]) (24) and with phytohemagglutinin (PHA; 0.25 mg/ml) plus IL-2 (25 ng/ml). PHA-preactivated allogeneic CD8-depleted T cells from healthy volunteers were used for specific activation. On days 1, 3, and 6, 200 μl of supernatant was collected and replenished with medium alone on day 1 and with medium together with reactivating agents on days 3 and 6. At days 1, 3, 6, and 14, supernatants were collected and stored at −80°C. Viral RNA was extracted from the culture supernatant, and HIV RNA quantification was done in five PCR runs using the ANRS assay (HIV-1 Generic Charge Virale; Biocentric, Bandol, France). The PCR targets a conserved consensus region in the long terminal repeat (LTR) of the HIV-1 major group and has proved suitable for quantification of HIV-1 isolates of different clades (25). The sequences of the forward primer (primer NEC005) and the reverse primer (primer NEC131) were 5′-GCCTCAATAAAGCTTGCC-3′ and 5′-GGCGCCACTGCTAGAGATTTT-3′, respectively. The internal HIV-1 TaqMan probe (MLC1) MGB LTR (5′-AAGTRGTGTGTGCCC-3′) carried the 5′ reporter 6-carboxyfluorescein and the 3′ quencher 6-carboxytetramethylrhodamine (Applied Biosystems, Foster City, CA). Real-time PCR was performed on a CFX96 device (Bio-Rad), and experimental values were derived from the kit standards as described by the manufacturer.
HIV DNA quantification.
Total cell-associated HIV DNA in PBMCs from cART patients and in leukocytes or purified resting CD4+ T cells from HICs was quantified using an ultrasensitive ANRS real-time PCR assay (generic HIV DNA cell kit) as previously described (26). The primers and the probe were the same as those used in the real-time PCR for HIV-1 RNA quantitation. All runs were performed with a 50-ml volume containing DNA extract (1 mg of total DNA), primers and probe (a 200 nM concentration of each), 1× PCR buffer (Platinum 1 quantitative PCR SuperMix-UDG; Invitrogen, Cergy-Pontoise, France), and carboxy-X-rhodamine reference dye (500 nM; Invitrogen). Thermocycling conditions were 2 min at 50°C and 10 min at 95°C, followed by 50 cycles at 95°C for 15 s and 60°C for 1 min. Each entire DNA extract was tested in two to four PCR runs. The results are reported either as the actual HIV DNA copy number per million cells or as 50% of the detection limit for samples in which HIV DNA was undetectable. The detection limit varied according to cell availability (27).
UCA.
An ultrasensitive coculture assay (UCA) was applied to purified CD4+ T cells isolated by negative selection from total cryopreserved PBMCs from HICs and cART patients (Miltenyi, Spain). Purified CD4+ T cells were stimulated with a pool of allogeneic irradiated PBMCs at a ratio of 1:5 (20,000 CD4+ cells and 100,000 allogeneic PBMCs per well) in 96-well plates in the presence of PHA (1 μg/ml) and IL-2 (100 U/ml) (Novartis, Spain) for 72 h. Simultaneously, PBMCs from three HIV-seronegative donors were isolated and CD8+ T cells were depleted with RosetteSep human CD8+ depletion cocktail (Stemcell Technologies, France). Pooled CD8-depleted PBMCs were then stimulated under three different conditions (referred to here as 3×3 cells) as previously described (28, 29). After 72 h, the PHA was washed off and the 3×3 cells were cultured in R10 medium containing IL-2 (100 U/ml). On day 7, the culture was scaled up to 48-well plates with the addition of 200 μl of fresh medium. During the following 2 weeks, to maximize viral outgrowth, the cocultures were fed once a week with fresh medium and once a week with 3×3 cells. After 21 days in culture, the supernatants were tested for p24 by enzyme-linked immunosorbent assay (ELISA; PerkinElmer, Spain). p24-negative wells were given a value of zero. Supernatants and cells from positive wells were stored at −80°C.
HIV-1 spread, viral titration, and p24 input assay.
PBMCs from three HIV-1-seronegative donors were depleted of CD8+ T cells and stimulated as described above. After 72 h of activation, pellets of 1 × 106 cells were spinoculated for 2 h at 2,000 × g and 22°C with UCA supernatants containing ≥10 pg/ml p24 or with HIVNL43 as a positive control in the presence of dextran (25 μg/ml). After spinoculation, the cells were resuspended and cultured in R10 medium with IL-2 (100 U/ml; Novartis, Spain). Virus spread in the supernatants was monitored on days 0, 1, 7, and 10 postchallenge. Supernatants containing at least 1 × 106 pg/ml p24 were stored at −80°C for further analysis.
The numbers of 50% tissue culture infective doses (TCID50s) per milliliter of three primary viral isolates (two from cART patients and one from an HIC) and the HIVNL43 laboratory reference strain were measured in TZM-bl cells. In addition, 3×3 cells were spinoculated with 10-fold serial dilutions of virus (105 to 1 pg/ml p24) in the presence of dextran (25 μg/ml). After spinoculation, the cells were extensively washed and cultured in R10 medium with IL-2 (100 U/ml). Virus production in the supernatants was monitored by a p24 ELISA on days 3, 7, and 10 postchallenge.
Sequencing of HIV-1 nef.
Total DNA was extracted from UCA-positive wells with over 20 pg/ml of p24 for samples from HICs and patients receiving cART by using a QIAamp DNA minikit (Qiagen, Spain), and 721 bp of the nef gene was amplified with specific primers as previously described (56). Taking into account that we estimated that less than one cell per well carried proviral DNA in the UCA with CD4+ T cells from HICs and that we performed the PCRs in replicates (n = 8), we approximated that each viral sequence underwent a single genome amplification event. In addition, total viral RNA was extracted from virus expanded in the UCA by reverse transcription-PCR with the pairs of primers described above. The sequences were obtained by Sanger reactions using the Macrogen Service (Netherlands) and analyzed with the Sequencher program (version V; Gene Codes). Evolutionary analyses were conducted in MEGA (version 5) software. The number of APOBEC signatures for each patient was calculated with the Highlighter tool from the Los Alamos National Laboratory database, and the sequences were compared to HIV-1 consensus clade B sequences.
Analysis of the HIV-specific CD8+ T cell response.
The capacity of CD8+ T cells to suppress infection of autologous CD4+ T cells ex vivo was measured as previously described (30). The capacity of CD8+ T cells to suppress HIV-1 replication was calculated as the log of the decrease in the level of p24 production in the presence of CD8+ T cells relative to the level of p24 production in the presence of CD4+ T cells cultured alone.
The frequency of HIV-specific (gamma interferon [IFN-γ]-secreting) CD8+ T cells was estimated with an enzyme-linked immunosorbent spot assay after a brief stimulation with pools of optimal HIV peptides.
Statistical analysis.
P values were calculated by the Mann-Whitney test. Only significant values (P < 0.05) are shown in the figures. Spearman's correlation coefficient (r) and the associated two-tailed P value were also calculated.
RESULTS
A low frequency of viral reactivation correlates with a low number of latently infected resting CD4+ cells.
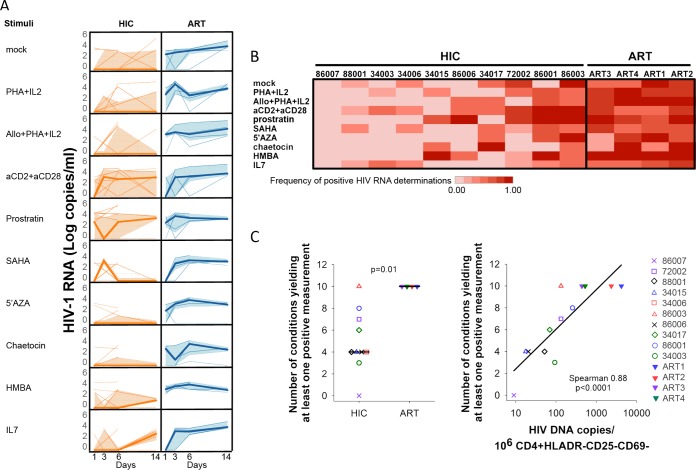
We first compared the capacity of resting CD4+ T cells from HICs and the capacity of resting CD4+ T cells from patients receiving antiretroviral treatment to reactivate HIV-1 replication. Because of the complex regulation of HIV-1 latency, the reactivation of all fully competent integrated provirus requires repeated stimulation of the cells (31) or the use of different stimuli (14, 32, 33). Therefore, we evaluated HIV-1 reactivation in isolated resting (HLADR− CD69− CD25−) CD4+ T cells from 10 HICs and 4 cART patients cultured under the following conditions: no stimulus (mock) or stimulus with PHA plus IL-2 with or without allogeneic CD8-depleted PBMCs, anti-CD2 plus anti-CD28, IL-7 (34), prostratin (24, 35), HMBA (36), 5-azacytidine (37), chaetocin (38), and SAHA (24, 39). All the stimuli were periodically replenished in the cultures. The efficiency of viral reactivation, measured in terms of positive HIV-1 RNA levels in the culture supernatants, varied among individuals and conditions. The total proportions of positive reactivations at days 1, 3, 6, and 14 were 0.48, 0.73, 0.9, and 0.93, respectively, for cART patients and 0.22, 0.23, 0.34, and 0.35, respectively, for HICs. Among the positive reactivations, higher median HIV-1 RNA levels per patient were measured at day 14 for cART patients (median number of HIV-1 RNA copies per milliliter at days 1, 3, 6, and 14, 1,126.5 [IQR, 412.5 to 1,530.8], 1,875 [IQR, 857 to 5,323.8], 2,120 [IQR, 618.8 to 13,285], and 5,023.5 [IQR, 1,601.7 to 5,0678.5], respectively; P = 0.012) and at day 6 for HICs (median number of HIV-1 RNA copies per milliliter at days 1, 3, 6, and 14, 1,044 [IQR, 311 to 2,536], 2,963[IQR, 2,002.8 to 17,151.8], 5,514.5 [IQR, 697.3 to 41,643.8], and 2,225 [IQR, 1,093.5 to 8,061.5], respectively; P = 0.3), although the differences over time were not statistically significantly different for HICs. These results are in agreement with reports showing that repeated stimulation is necessary to induce maximal reactivation (31). While HIV-1 RNA production in cultures of CD4+ T cells from cART patients was usually sustained and HIV-1 RNA accumulated over time, in the case of HICs, HIV-1 RNA was often detected at a single time point or the level of HIV-1 RNA production did not increase over time (Fig. 1A).
FIG 1.
Viral reactivation in resting CD4+ T cells from HICs and cART patients. (A) HIV RNA levels produced by CD4+ T cells from 10 HIV controllers and 4 cART-treated patients over time under the different culture conditions tested. Thin lines, data for individual subjects; thick lines and colored areas, the median and the IQR per group, respectively. (B) The heat map represents the frequency of positive HIV RNA determinations assessed in culture supernatants of CD4+ T cells from HICs and cART patients at different times of culture (days 1, 3, 6, and 14) in the presence of various stimuli. (C) Differences in the number of culture conditions resulting in at least one positive HIV RNA determination at any given time (days 1, 3, 6, and 14) with cells from HICs and cART patients (left) and correlation with the number of HIV DNA copies per million resting CD4+ T cells (right). Each symbol represents one patient (open symbols, HICs; filled symbols, cART patients). The results for HIC 34006 are not included in the correlation due to a technical problem during the quantification of the HIV DNA. Allo, allogeneic cells; 5′AZA, 5-azacytidine.
Viral reactivation was observed in cells from all the HICs (except HIC 86007) under at least one condition tested (Fig. 1B). The anti-CD2 plus anti-CD28 stimulus often resulted in viral reactivation events in HICs (median proportion of positive reactivations per HIC, 0.5 [IQR, 0.19 to 0.81; n = 10]; at least one reactivation event occurred in 8 of 10 HICs), but this difference was not statistically significantly higher than that under the other conditions tested. Viral reactivation occurred in resting CD4+ T cells from cART patients at least at one time point under all conditions tested (Fig. 1B). Overall, the number of conditions activating virus was much higher in cART patients than HICs (Fig. 1C, left) and was directly related to HIV DNA levels in resting CD4+ T cells for all subjects (Spearman r = 0.88, P < 0.0001) (Fig. 1C, right) and in the HIC subgroup (Spearman r = 0.783, P = 0.009).
HIV-1 outgrowth is reduced in HICs and limits viral spread.
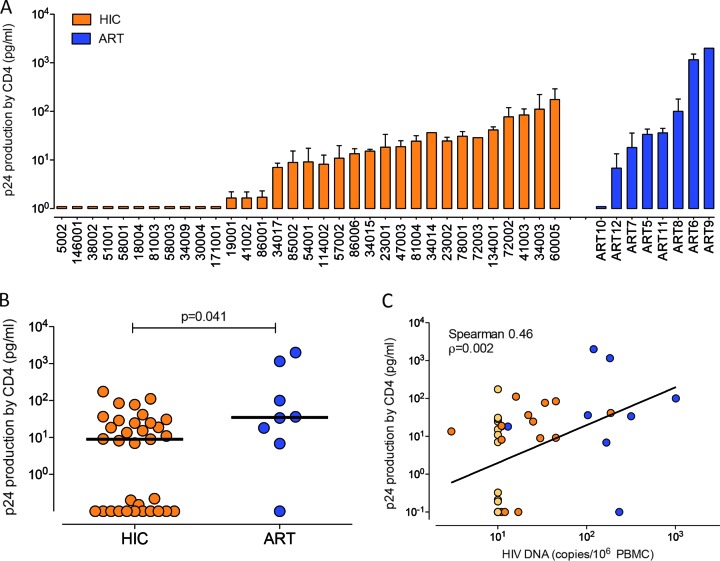
Besides the difference in the frequency of infected cells, additional factors might contribute to fewer reactivation events in HICs: (i) the differential capacity to produce viral particles, (ii) the resistance of HIC cells to HIV-1 infection (17, 40), or (iii) the lower replicative capacity of the newly produced virions. Although because of sample limitations we could not perform a standard limiting dilution assay, we used an ultrasensitive coculture assay (UCA) to monitor ex vivo HIV-1 production by CD4+ T cells from 33 HICs and 8 cART patients. We estimated that less than one cell per well in the UCA of CD4+ T cells from HICs carried HIV-1 DNA, based on the number of cells per well used (median, 0.8 CD4+ T cells carrying HIV-1 DNA per culture [IQR, 0.8 to 1.2 CD4+ T cells carrying HIV-1 DNA per culture], considering that all the HIV-1 DNA measured in the PBMCs was in CD4+ T cells, which made up ∼25% of the PBMCs, and that only one copy was present per infected cell). We detected the p24 antigen in the culture supernatants of CD4+ T cells from 22 HICs (66.6%) and 7 cART patients (87.6%) (Fig. 2A). Although fewer HIC CD4+ T cells than cART patient CD4+ T cells were analyzed in total (860,000 [IQR, 490,000 to 1,700,000) versus 1,880,000 [IQR, 1,700 000 to 2,240,000]), because of the limited availability of cells from HICs, no difference in the frequency of p24-positive cultures was found (medians, 4.8% [IQR, 0 to 9.1%] and 4.1% [IQR, 2.2 to 9.3%] for HIC and cART patients, respectively; P = 0.73). In contrast, cells from HICs produced significantly less p24 antigen than cells from cART patients (medians, 9.03 pg/ml p24 [IQR, 9.29 to 35.8 pg/ml p24] for HICs and 34.97 pg/ml p24 [IQR, 208.7 to 1,047 pg/ml p24] for cART patients; P = 0.041) (Fig. 2B). A trend toward a lower level of p24 production by cells from HICs was also observed when only p24-positive cultures were considered (medians, 18.6 pg/ml p24 [IQR, 8.7 to 37.7 pg/ml p24] and 36.2 pg/ml p24 [IQR, 18.1 to 1,158 pg/ml p24] for HICs [n = 22] and cART patients [n = 7], respectively; P = 0.1). A direct correlation between the PBMC HIV DNA load and the amount of p24 produced in culture was found (Spearman r = 0.46, P = 0.002) (Fig. 2C). These data show that HIV-1 can be reactivated from HIV-1 controllers' cells in the presence of heterologous cells but that virion production is often inefficient.
FIG 2.
UCA. The UCA was performed with CD4+ T cells isolated by negative selection from cryopreserved PBMCs from 33 HICs and 8 cART patients. (A) Each bar corresponds to one subject and represents the median level of HIV-1 p24 in positive wells. (B) Comparison of levels of p24 production in the UCA with cells from HICs (n = 33; orange) and cART patients (n = 8; blue). Each circle represents one subject, and the lines indicate mean values. The P value was calculated using the Mann-Whitney test. (C) Correlation between the PBMC HIV DNA load and the level of p24 production by CD4+ T cells. Light-colored symbols, cells with HIV DNA levels below the limit of detection, corresponding to HICs; line, data fit to a linear regression. The frequency of p24 positivity was calculated as follows: (number of p24-positive wells/total number of wells tested) × 100. Virus production in the UCA was calculated as the median level of p24 production in positive wells.
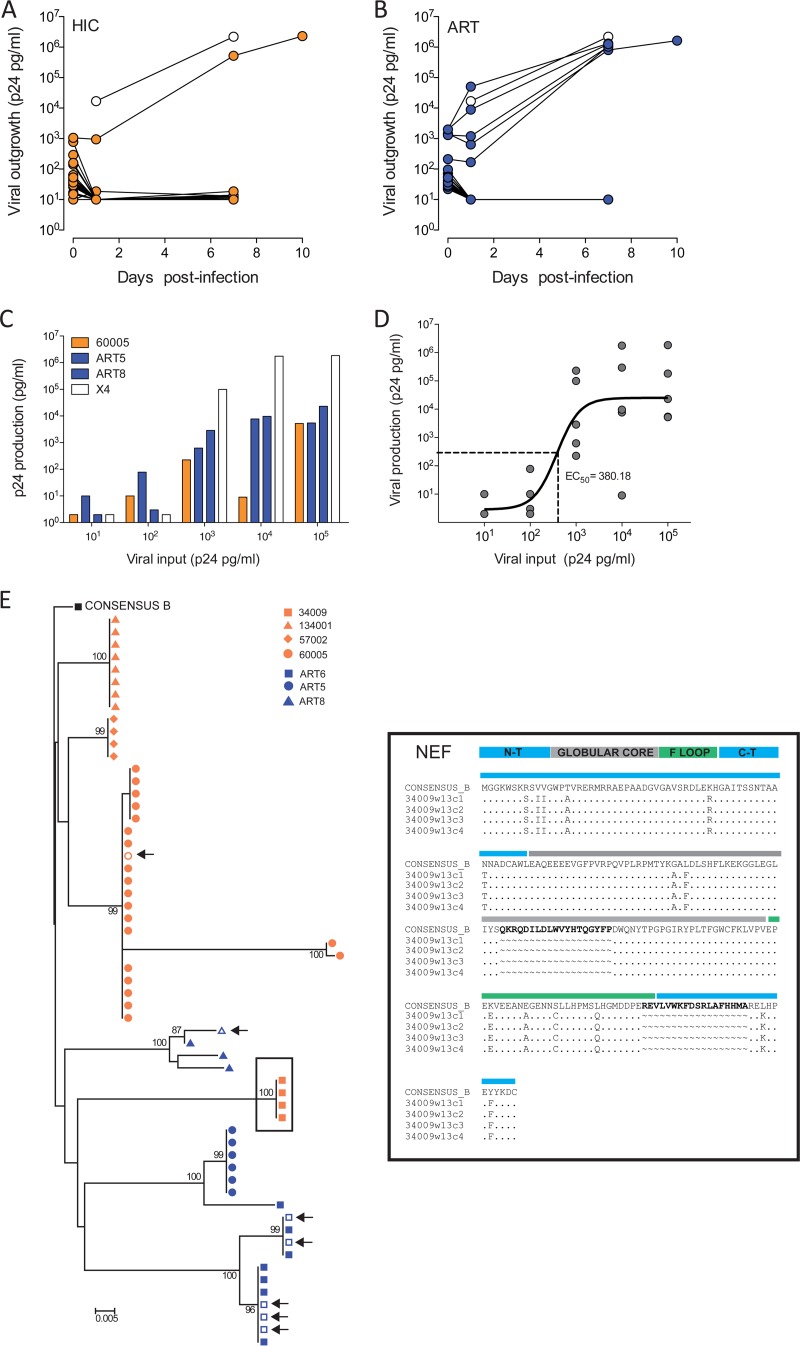
To further evaluate if differences in virus reactivation and production in HICs and cART patients affect viral spread, we infected preactivated primary allogeneic CD8 cell-depleted PBMCs with supernatants from the UCA containing ≥10 pg/ml of p24. Viral spread in cells that were in culture for a week was monitored by a p24 ELISA. Our data showed a lower frequency of spread of infections in samples from HICs than samples from cART patients (3% for HICs versus 21.4% for cART patients; P = 0.045) (Fig. 3A and B). To evaluate if viral spread was associated with a threshold of virus production from CD4+ T cells, we performed a p24 limiting dilution infectivity assay and tested the growth of virus from one HIC (HIC 60005) and two cART patients (patients ART5 and ART8) in cells in comparison with that of HIVNL43. We found a direct correlation between viral input and viral outgrowth (Spearman r = 0.77, P < 0.0001) (Fig. 3C), where sustained viral outgrowth was observed only at p24 values of >103 pg/ml, confirming the existence of a p24 threshold for successful viral spread. Additionally, by using a nonlinear fit of the data, we estimated that 380.18 pg/ml of p24 is the half-maximal effective concentration (EC50) required to achieve viral spread in 50% of cultures (Fig. 3D). This amount is well above the median level of production observed for CD4+ T cells in HICs (9.03 pg/ml) and further supports the suggestion that low viral spread from HIC CD4+ T cell cultures is limited by low levels of particle release after reactivation.
FIG 3.
HIV-1 spread from UCA wells and viral p24 limiting dilution input assay. (A and B) Viral spread in UCA wells containing >10 pg/ml of p24 from activated CD8-depleted PBMCs was monitored on days 0, 1, 7, and 10 postinfection by p24 ELISA of the supernatants of cultures of cells from HICs (A) and cART patients (B). (C) The outgrowth of HIV-1 from subjects 60005, ART5, and ART8 and the NL43 (X4) virus strain was measured in cells receiving 10-fold dilutions of input p24 ranging from 105 to 1 pg/ml. The viral outgrowth represented in the graphs was calculated as follows: (amount of p24 at day 7 postinfection) – (amount of p24 at day 3 postinfection), where the amount of p24 is given in picograms per milliliter. (D) Nonlinear regression of p24 input and production. Line, fit of the data; dashed line; half-maximal effective concentration (EC50). (E) (Left) Phylogenetic tree determined by the neighbor-joining method of nef sequences from HICs (HICs 34009, 134001, 57002, and 60005) and cART patients (cART patients ART5, ART6, and ART8). Sequences were obtained from proviral DNA (filled symbols) from UCA wells and viral RNA in successfully spreading infections, indicated by arrows (empty symbols). Sequences were aligned to the consensus clade B sequence using the BioEdit package, and the neighbor-joining tree was built with MEGA (version 5) software over 1,000 replicates. Mean evolutionary diversity analyses for the entire population were conducted using the maximum composite likelihood model. The analysis involved 33 nucleotide sequences for HICs and 22 nucleotide sequences for cART patients. Codon positions included were the 1st, 2nd, and 3rd positions and the noncoding sequence. All positions containing gaps and missing data were eliminated. There were a total of 610 positions in the final data set. The tree is rooted to the consensus clade B sequence. Only bootstrap values over 70 are represented. (Right) Nef amino acid sequence alignment of 34,009 clonal sequences shown at the top. Bold, deletions compared to the consensus sequence; colored regions, functional domains of the Nef protein (blue, N-terminal and C-terminal regions; gray, globular core; green, flexible [F] loop).
To evaluate whether differences in viral spread in HICs and cART patients could be due to the presence of defective virus, we extracted DNA and RNA from UCA wells containing cells from HIC 60005 and cART patients ART5 and ART8, whose viruses were able to infect allogeneic cells. We also analyzed cells from four additional subjects, three HICs (HICs 34009, 134001, and 57002) and one cART patient (patient ART8), whose virus did not spread. To increase the chances of amplification, we focused on the gene nef. As shown in Fig. 3E, we observed monophyletic groups of viruses with a lower mean population diversity in HICs than cART patients (0.022 for HICs versus 0.061 for cART patients). These results corroborate previous experimental data that identified in HICs monoclonal populations with low levels of diversity (41). Moreover, no defects in the nef reading frame were found in any of the study groups. An exception was HIC 34009, where two large deletions in nef (19 and 16 amino acids) were observed (Fig. 3E, right). No differences in APOBEC signatures were found between viruses from HICs and cART patients (not shown), dismissing the potential role of hypermutation in the differential spread potential of the viruses from both groups. Taken together, our data show that the production of fit viruses by CD4+ T cells is stochastic and highly dependent on the total frequency of infected cells. Viral reactivation in cells from HICs was difficult and resulted in only the transient release of viral particles that was not sufficient to ensure sustained viral spread.
The amplitude of CD8+ T cell responses is reduced in HIC virus nonproducers.
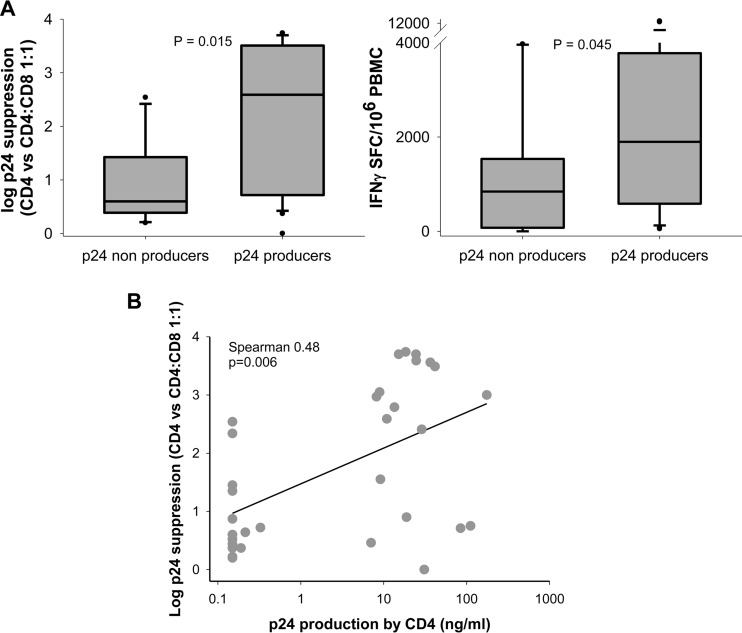
We then examined whether the different amplitudes of HIV-specific CD8+ T cell responses that we and others have observed in HICs (8, 9, 42) were related to the capacity of these patients' CD4+ T cells to produce viral particles ex vivo. Viral producers were defined as those HICs with p24 values in the supernatant over the limit of detection of the UCA. We found that the HIV-specific CD8+ T cell responses of HICs whose CD4+ T cells did not produce viral particles in the UCA did not differ from those of cART patients (9, 43). In particular, CD8+ T cells from the latter HICs did not efficiently suppress HIV-1 infection of autologous CD4+ T cells (Fig. 4A), a property considered characteristic of CD8+ T cells from HICs (4, 30, 44). Similarly, the frequency of circulating HIV-specific (IFN-γ-producing) CD8+ T cells in the same HICs was low (Fig. 4B). In contrast, HICs whose CD4+ T cells efficiently produced viral particles ex vivo had potent CD8+ T cell responses (Fig. 4A and B). Positive correlations were found between the capacity of CD8+ T cells from HICs to suppress HIV-1 infection and the level of p24 produced by their CD4+ T cells in vitro (Fig. 4B) and between the HIV-suppressive capacity or the frequency of HIV-specific CD8+ T cells and the frequency of p24-positive cultures (Spearman r = 0.48 [P = 0.006] and Spearman r = 0.46 [P = 0.007], respectively; not shown).
FIG 4.
HIC HIV-specific CD8+ T cell responses according to the amount of p24 produced by their CD4+ T cells in vitro. The responses of HIC CD8+ T cells tested in the UCA were analyzed ex vivo. HICs were classified as p24 producers or nonproducers depending on whether or not p24 was produced by their CD4+ T cells in the UCA. (A) (Left) CD8+ T cell capacity to suppress infection of autologous CD4+ T cells, as measured by the decrease in the level of p24 production in the presence of unstimulated autologous CD8+ T cells (n = 32; one value is missing, owing to unsuccessful CD4+ T cell infection). (Right) Frequency of IFN-γ-producing cells after PBMC stimulation with optimal HIV-1 peptides. The box plots represent the medians; 10%, 25%, 75%, and 90% ranges; and outliers. SFC, spot-forming cells. (B) Correlation between CD8+ T cell capacity to suppress HIV-1 infection and p24 production by CD4+ T cells. Each symbol represents one HIC. The line indicates data fit to a linear regression.
DISCUSSION
The results reported in this study provide functional evidence of the stochastic nature of HIV-1 reactivation and how it may contribute to the maintenance of virological control in the absence of antiretroviral treatment. In particular, we show that the lower that the frequency of infected resting CD4+ T cells in HICs is, the lower that the probability of viral reactivation, viral production, and viral spread in vitro compared to that in cART patients is.
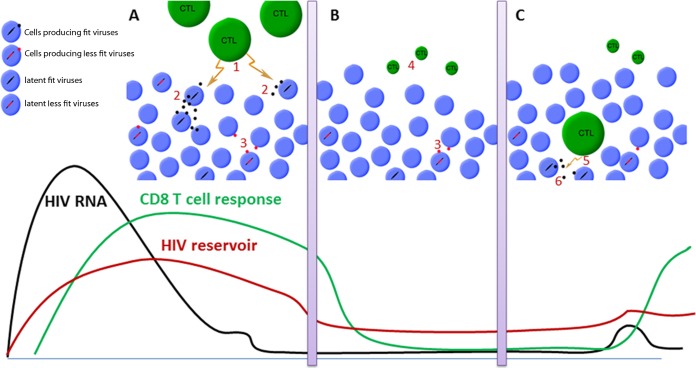
Our results suggest that natural control of HIV-1 infection can be maintained at least at two different levels. Strong but incomplete viral control would result in low-level virion release from infected cells, sustaining the strong CD8+ T cell responses observed in many HICs. This response, in turn, readily controls the infection, maintaining a steady state between infected cells and the immune system (Fig. 5A), which may result in residual systemic activation and mild progression in the absence of therapy (45–47). The virtuous circle between the virus and the host response would be interrupted when the frequency of latently infected CD4+ T cells falls to a very low level, hindering the stochastic reactivation of fit viruses (18) and leading to fewer viral amplification events. This would result in the contraction of CD8+ T cell responses, at least transiently, due to the absence of the antigenic boost (Fig. 5B), as has indeed been reported in some HICs after ART initiation (48). This implies that a low frequency of latently infected cells may be sufficient to transiently prevent viral outgrowth, even in the absence of a major contribution of the immune system, as recently reported in two patients in Boston, MA, and a baby in Mississippi (23, 49). This phenomenon may be particularly relevant and efficient in the long term in HICs, whose target cells are also strongly resistant to HIV-1 infection, particularly at low infectious doses (17, 40), providing an additional barrier to viral amplification. In addition, weakly responsive memory CD8+ T cells in HICs are ready to react and expand promptly in response to stochastic viral amplification events (12, 50) (Fig. 5C), thereby ensuring natural control of HIV-1 infection in the very long term.
FIG 5.
(A) Most HICs are able to generate potent HIV-specific effector CD8+ T cell responses (marker 1) that efficiently eliminate HIV-infected cells, allowing the control of viral replication and avoiding the replenishment of the viral reservoirs (marker 2). A virtuous circle between low-level viral replication and the CD8+ T cell response that ensures long-term control of infection is established. (B) We hypothesize that in some HICs most cells carrying fit viruses are intensely eliminated by a combination of the cytopathogenic effects of the viruses and the HICs' efficient CD8+ T cell responses. Considering the stochastic character of viral activation, the reactivation of fit viruses in these HICs would be rare, and more often, activation events would involve less fit viruses that result in a small viral burst insufficient to transmit infection (marker 3). In HICs, this phenomenon is reinforced by the intrinsic resistance of their cells to infection with low doses of viruses. In such a context, the control of infection would be related to rare and inefficient viral reactivation events and the CD8+ T cell response, which would no longer be required to maintain control, shrink, and deactivate (marker 4). (C) These cells, however, retain the capacity to expand and again exert effector functions (marker 5) if a viral amplification event occurs (marker 6). CTL, cytotoxic T lymphocyte.
The levels of cell-associated HIV-1 DNA have been shown to predict progression to disease and the capacity to control infection after treatment interruption (19, 22, 51–53). Although measurement of total HIV-1 DNA levels may take into account many copies of replication-deficient viruses, it is likely that under conditions of very low levels of viral replication, as in HICs, differences in the levels of HIV-1 DNA probably maintain proportionality between replication-competent and noncompetent proviruses, as recently observed in treated patients (54). This assumption is supported, despite limitations in the total numbers of CD4+ T cells tested, by the direct correlation between the levels of cell-associated HIV-1 DNA and HIV-1 reactivation, measured by determination of the levels of HIV-1 RNA or p24 production in culture supernatants, that was found when CD4+ T cells were split into parallel cultures and were subjected to different repeated stimuli (with polyclonal or allogeneic stimuli or the provision of latency-reversing agents).
We did not find significant differences in the hypermutation rates or defective sequences between the viruses from HICs and those from cART patients. However, the difference in the number of infected cells between HICs and cART patients resulted in different levels of virion production after activation, which ultimately would result in a lower probability of the spread of infections for HICs. The reduced cellular susceptibility to HIV-1 infection in HICs may contribute to the lower levels of infected cells found in these patients (17). Moreover, the block of HIV-1 replication in HIC CD4+ T cells has been shown to affect preintegration steps of the replication cycle (17, 40, 55) but also mRNA transcription from the integrated viruses (40), which could explain the reduced levels of viral particle production observed in HICs. Further mechanistic studies on HIV-1 transcriptional steps or particle production in HICs will contribute to a clarification of these findings.
Our study has some limitations. First, our analysis focused on CD4+ T cells not expressing activation markers ex vivo, which excluded other cell types that may contribute to the reservoir size. Second, our findings are limited to peripheral blood. The level/frequency of reactivatable competent viruses in the tissues of some HICs with low levels of viremia (i.e., HIC 146001) might be different from that in peripheral blood and might explain the absence of virus induction in vitro. Our analyses of viral sequences were limited to a small number of sequences and capture the diversity associated only with the nef region after ex vivo culture. Therefore, additional defects in other regions of the viral sequence were missed and potential artifacts due to in vitro viral expansion may have been introduced. However, other, deeper studies have not detected, in general, major defects in the viruses from HICs (15).
Overall, our results suggest that limiting the frequency of infected cells could suffice to achieve temporary control of infection in the absence of antiretroviral treatment. However, additional intrinsic and/or immune mechanisms are needed to reinforce this effect and ensure HIV-1 control in the long term.
ACKNOWLEDGMENTS
We thank Yves Colette for kindly contributing reagents (mouse IgG1, anti-CD28) and David Young for English editing of the manuscript.
We have no conflicts of interest to report.
Funding Statement
This work, including the efforts of Esther Jimenez, was funded by Redes Temáticas de Investigación en SIDA; Acción Estratégica en Salud, Plan Nacional de Investigación Científica, Desarrollo e Innovación Tecnológica 2008-2011; and Instituto de Salud Carlos III, Fondos FEDER (ISCIII RETIC RD12/0017/0002)
REFERENCES
- 1.Saez-Cirion A, Pancino G. 2013. HIV controllers: a genetically determined or inducible phenotype? Immunol Rev 254:281–294. doi: 10.1111/imr.12076. [DOI] [PubMed] [Google Scholar]
- 2.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, Roederer M, Koup RA. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Migueles SA, Laborico AC, Shupert WL, Sabbaghian MS, Rabin R, Hallahan CW, Van Baarle D, Kostense S, Miedema F, McLaughlin M, Ehler L, Metcalf J, Liu S, Connors M. 2002. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat Immunol 3:1061–1068. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- 4.Saez-Cirion A, Lacabaratz C, Lambotte O, Versmisse P, Urrutia A, Boufassa F, Barre-Sinoussi F, Delfraissy JF, Sinet M, Pancino G, Venet A. 2007. HIV controllers exhibit potent CD8 T cell capacity to suppress HIV infection ex vivo and peculiar cytotoxic T lymphocyte activation phenotype. Proc Natl Acad Sci U S A 104:6776–6781. doi: 10.1073/pnas.0611244104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flores-Villanueva PO, Yunis EJ, Delgado JC, Vittinghoff E, Buchbinder S, Leung JY, Uglialoro AM, Clavijo OP, Rosenberg ES, Kalams SA, Braun JD, Boswell SL, Walker BD, Goldfeld AE. 2001. Control of HIV-1 viremia and protection from AIDS are associated with HLA-Bw4 homozygosity. Proc Natl Acad Sci U S A 98:5140–5145. doi: 10.1073/pnas.071548198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Migueles SA, Sabbaghian MS, Shupert WL, Bettinotti MP, Marincola FM, Martino L, Hallahan CW, Selig SM, Schwartz D, Sullivan J, Connors M. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci U S A 97:2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M, Kadie CM, Carlson JM, Heckerman D, Graham RR, Plenge RM, Deeks SG, Gianniny L, Crawford G, Sullivan J, Gonzalez E, Davies L, Camargo A, Moore JM, Beattie N, Gupta S, Crenshaw A, Burtt NP, Guiducci C, Gupta N, Gao X, Qi Y, Yuki Y, Piechocka-Trocha A, Cutrell E, Rosenberg R, Moss KL, Lemay P, O'Leary J, Schaefer T, Verma P, Toth I, Block B, Baker B, Rothchild A, Lian J, Proudfoot J, Alvino DM, Vine S, Addo MM, Allen TM, et al. . 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lecuroux C, Saez-Cirion A, Girault I, Versmisse P, Boufassa F, Avettand-Fenoel V, Rouzioux C, Meyer L, Pancino G, Lambotte O, Sinet M, Venet A. 2014. Both HLA-B*57 and plasma HIV RNA levels contribute to the HIV-specific CD8+ T cell response in HIV controllers. J Virol 88:176–187. doi: 10.1128/JVI.02098-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saez-Cirion A, Sinet M, Shin SY, Urrutia A, Versmisse P, Lacabaratz C, Boufassa F, Avettand-Fenoel V, Rouzioux C, Delfraissy JF, Barre-Sinoussi F, Lambotte O, Venet A, Pancino G. 2009. Heterogeneity in HIV suppression by CD8 T cells from HIV controllers: association with Gag-specific CD8 T cell responses. J Immunol 182:7828–7837. doi: 10.4049/jimmunol.0803928. [DOI] [PubMed] [Google Scholar]
- 10.Bruel T, Hamimi C, Dereuddre-Bosquet N, Cosma A, Shin SY, Corneau A, Versmisse P, Karlsson I, Malleret B, Targat B, Barre-Sinoussi F, Le Grand R, Pancino G, Saez-Cirion A, Vaslin B. 2015. Long-term control of simian immunodeficiency virus (SIV) in cynomolgus macaques not associated with efficient SIV-specific CD8+ T-cell responses. J Virol 89:3542–3556. doi: 10.1128/JVI.03723-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Addo MM, Draenert R, Rathod A, Verrill CL, Davis BT, Gandhi RT, Robbins GK, Basgoz NO, Stone DR, Cohen DE, Johnston MN, Flynn T, Wurcel AG, Rosenberg ES, Altfeld M, Walker BD. 2007. Fully differentiated HIV-1 specific CD8+ T effector cells are more frequently detectable in controlled than in progressive HIV-1 infection. PLoS One 2:e321. doi: 10.1371/journal.pone.0000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ndhlovu ZM, Proudfoot J, Cesa K, Alvino DM, McMullen A, Vine S, Stampouloglou E, Piechocka-Trocha A, Walker BD, Pereyra F. 2012. Elite controllers with low to absent effector CD8+ T cell responses maintain highly functional, broadly directed central memory responses. J Virol 86:6959–6969. doi: 10.1128/JVI.00531-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blankson JN, Bailey JR, Thayil S, Yang HC, Lassen K, Lai J, Gandhi SK, Siliciano JD, Williams TM, Siliciano RF. 2007. Isolation and characterization of replication-competent human immunodeficiency virus type 1 from a subset of elite suppressors. J Virol 81:2508–2518. doi: 10.1128/JVI.02165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamine A, Caumont-Sarcos A, Chaix ML, Saez-Cirion A, Rouzioux C, Delfraissy JF, Pancino G, Lambotte O. 2007. Replication-competent HIV strains infect HIV controllers despite undetectable viremia (ANRS EP36 study). AIDS 21:1043–1045. doi: 10.1097/QAD.0b013e3280d5a7ac. [DOI] [PubMed] [Google Scholar]
- 15.Miura T, Brockman MA, Brumme CJ, Brumme ZL, Carlson JM, Pereyra F, Trocha A, Addo MM, Block BL, Rothchild AC, Baker BM, Flynn T, Schneidewind A, Li B, Wang YE, Heckerman D, Allen TM, Walker BD. 2008. Genetic characterization of human immunodeficiency virus type 1 in elite controllers: lack of gross genetic defects or common amino acid changes. J Virol 82:8422–8430. doi: 10.1128/JVI.00535-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewin SR, Rouzioux C. 2011. HIV cure and eradication: how will we get from the laboratory to effective clinical trials? AIDS 25:885–897. doi: 10.1097/QAD.0b013e3283467041. [DOI] [PubMed] [Google Scholar]
- 17.Saez-Cirion A, Hamimi C, Bergamaschi A, David A, Versmisse P, Melard A, Boufassa F, Barre-Sinoussi F, Lambotte O, Rouzioux C, Pancino G. 2011. Restriction of HIV-1 replication in macrophages and CD4+ T cells from HIV controllers. Blood 118:955–964. doi: 10.1182/blood-2010-12-327106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill AL, Rosenbloom DI, Fu F, Nowak MA, Siliciano RF. 2014. Predicting the outcomes of treatment to eradicate the latent reservoir for HIV-1. Proc Natl Acad Sci U S A 111:13475–13480. doi: 10.1073/pnas.1406663111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Assoumou L, Weiss L, Piketty C, Burgard M, Melard A, Girard P-M, Rouzioux C, Costagliola D, ANRS 116 SALTO Study Group . 2015. A low HIV-DNA level in PBMCs at antiretroviral treatment interruption predicts a higher probability of maintaining viral control. AIDS 29:2003–2007. doi: 10.1097/QAD.0000000000000734. [DOI] [PubMed] [Google Scholar]
- 20.Goujard C, Girault I, Rouzioux C, Lecuroux C, Deveau C, Chaix ML, Jacomet C, Talamali A, Delfraissy JF, Venet A, Meyer L, Sinet M. 2012. HIV-1 control after transient antiretroviral treatment initiated in primary infection: role of patient characteristics and effect of therapy. Antivir Ther 17:1001–1009. doi: 10.3851/IMP2273. [DOI] [PubMed] [Google Scholar]
- 21.Hocqueloux L, Prazuck T, Avettand-Fenoel V, Lafeuillade A, Cardon B, Viard JP, Rouzioux C. 2010. Long-term immunovirologic control following antiretroviral therapy interruption in patients treated at the time of primary HIV-1 infection. AIDS 24:1598–1601. doi: 10.1097/QAD.0b013e32833b61ba. [DOI] [PubMed] [Google Scholar]
- 22.Yerly S, Gunthard HF, Fagard C, Joos B, Perneger TV, Hirschel B, Perrin L. 2004. Proviral HIV-DNA predicts viral rebound and viral setpoint after structured treatment interruptions. AIDS 18:1951–1953. doi: 10.1097/00002030-200409240-00011. [DOI] [PubMed] [Google Scholar]
- 23.Henrich TJ, Hanhauser E, Marty FM, Sirignano MN, Keating S, Lee TH, Robles YP, Davis BT, Li JZ, Heisey A, Hill AL, Busch MP, Armand P, Soiffer RJ, Altfeld M, Kuritzkes DR. 2014. Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases. Ann Intern Med 161:319–327. doi: 10.7326/M14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reuse S, Calao M, Kabeya K, Guiguen A, Gatot JS, Quivy V, Vanhulle C, Lamine A, Vaira D, Demonte D, Martinelli V, Veithen E, Cherrier T, Avettand V, Poutrel S, Piette J, de Launoit Y, Moutschen M, Burny A, Rouzioux C, De Wit S, Herbein G, Rohr O, Collette Y, Lambotte O, Clumeck N, Van Lint C. 2009. Synergistic activation of HIV-1 expression by deacetylase inhibitors and prostratin: implications for treatment of latent infection. PLoS One 4:e6093. doi: 10.1371/journal.pone.0006093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rouet F, Chaix ML, Nerrienet E, Ngo-Giang-Huong N, Plantier JC, Burgard M, Peeters M, Damond F, Ekouevi DK, Msellati P, Ferradini L, Rukobo S, Marechal V, Schvachsa N, Wakrim L, Rafalimanana C, Rakotoambinina B, Viard JP, Seigneurin JM, Rouzioux C. 2007. Impact of HIV-1 genetic diversity on plasma HIV-1 RNA quantification: usefulness of the Agence Nationale de Recherches sur le SIDA second-generation long terminal repeat-based real-time reverse transcriptase polymerase chain reaction test. J Acquir Immune Defic Syndr 45:380–388. doi: 10.1097/QAI.0b013e3180640cf5. [DOI] [PubMed] [Google Scholar]
- 26.Avettand-Fenoel V, Chaix ML, Blanche S, Burgard M, Floch C, Toure K, Allemon MC, Warszawski J, Rouzioux C. 2009. LTR real-time PCR for HIV-1 DNA quantitation in blood cells for early diagnosis in infants born to seropositive mothers treated in HAART area (ANRS CO 01). J Med Virol 81:217–223. doi: 10.1002/jmv.21390. [DOI] [PubMed] [Google Scholar]
- 27.Avettand-Fenoel V, Prazuck T, Hocqueloux L, Melard A, Michau C, Kerdraon R, Agoute E, Rouzioux C. 2008. HIV-DNA in rectal cells is well correlated with HIV-DNA in blood in different groups of patients, including long-term non-progressors. AIDS 22:1880–1882. doi: 10.1097/QAD.0b013e32830fbdbc. [DOI] [PubMed] [Google Scholar]
- 28.Dalmau J, Codoner FM, Erkizia I, Pino M, Pou C, Paredes R, Clotet B, Martinez-Picado J, Prado JG. 2012. In-depth characterization of viral isolates from plasma and cells compared with plasma circulating quasispecies in early HIV-1 infection. PLoS One 7:e32714. doi: 10.1371/journal.pone.0032714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prado JG, Prendergast A, Thobakgale C, Molina C, Tudor-Williams G, Ndung'u T, Walker BD, Goulder P. 2010. Replicative capacity of human immunodeficiency virus type 1 transmitted from mother to child is associated with pediatric disease progression rate. J Virol 84:492–502. doi: 10.1128/JVI.01743-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saez-Cirion A, Shin SY, Versmisse P, Barre-Sinoussi F, Pancino G. 2010. Ex vivo T cell-based HIV suppression assay to evaluate HIV-specific CD8+ T-cell responses. Nat Protoc 5:1033–1041. doi: 10.1038/nprot.2010.73. [DOI] [PubMed] [Google Scholar]
- 31.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, Lai J, Blankson JN, Siliciano JD, Siliciano RF. 2013. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laird GM, Bullen CK, Rosenbloom DI, Martin AR, Hill AL, Durand CM, Siliciano JD, Siliciano RF. 2015. Ex vivo analysis identifies effective HIV-1 latency-reversing drug combinations. J Clin Invest 125:1901–1912. doi: 10.1172/JCI80142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darcis G, Kula A, Bouchat S, Fujinaga K, Corazza F, Ait-Ammar A, Delacourt N, Melard A, Kabeya K, Vanhulle C, Van Driessche B, Gatot J-S, Cherrier T, Pianowski LF, Gama L, Schwartz C, Vila J, Burny A, Clumeck N, Moutschen M, De Wit S, Peterlin BM, Rouzioux C, Rohr O, Van Lint C. 2015. an in-depth comparison of latency-reversing agent combinations in various in vitro and ex vivo HIV-1 latency models identified bryostatin-1+JQ1 and ingenol-B+JQ1 to potently reactivate viral gene expression. PLoS Pathog 11:e1005063. doi: 10.1371/journal.ppat.1005063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shuai K, Liu B. 2003. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol 3:900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 35.Williams SA, Chen LF, Kwon H, Fenard D, Bisgrove D, Verdin E, Greene WC. 2004. Prostratin antagonizes HIV latency by activating NF-kappaB. J Biol Chem 279:42008–42017. doi: 10.1074/jbc.M402124200. [DOI] [PubMed] [Google Scholar]
- 36.Contreras X, Barboric M, Lenasi T, Peterlin BM. 2007. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog 3:1459–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E. 2009. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog 5:e1000495. doi: 10.1371/journal.ppat.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bouchat S, Gatot JS, Kabeya K, Cardona C, Colin L, Herbein G, De Wit S, Clumeck N, Lambotte O, Rouzioux C, Rohr O, Van Lint C. 2012. Histone methyltransferase inhibitors induce HIV-1 recovery in resting CD4(+) T cells from HIV-1-infected HAART-treated patients. AIDS 26:1473–1482. doi: 10.1097/QAD.0b013e32835535f5. [DOI] [PubMed] [Google Scholar]
- 39.Archin NM, Espeseth A, Parker D, Cheema M, Hazuda D, Margolis DM. 2009. Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res Hum Retroviruses 25:207–212. doi: 10.1089/aid.2008.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen H, Li C, Huang J, Cung T, Seiss K, Beamon J, Carrington MF, Porter LC, Burke PS, Yang Y, Ryan BJ, Liu R, Weiss RH, Pereyra F, Cress WD, Brass AL, Rosenberg ES, Walker BD, Yu XG, Lichterfeld M. 2011. CD4+ T cells from elite controllers resist HIV-1 infection by selective upregulation of p21. J Clin Invest 121:1549–1560. doi: 10.1172/JCI44539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Connell KA, Brennan TP, Bailey JR, Ray SC, Siliciano RF, Blankson JN. 2010. Control of HIV-1 in elite suppressors despite ongoing replication and evolution in plasma virus. J Virol 84:7018–7028. doi: 10.1128/JVI.00548-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pereyra F, Addo MM, Kaufmann DE, Liu Y, Miura T, Rathod A, Baker B, Trocha A, Rosenberg R, Mackey E, Ueda P, Lu Z, Cohen D, Wrin T, Petropoulos CJ, Rosenberg ES, Walker BD. 2008. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J Infect Dis 197:563–571. doi: 10.1086/526786. [DOI] [PubMed] [Google Scholar]
- 43.Saez-Cirion A, Bacchus C, Hocqueloux L, Avettand-Fenoel V, Girault I, Lecuroux C, Potard V, Versmisse P, Melard A, Prazuck T, Descours B, Guergnon J, Viard JP, Boufassa F, Lambotte O, Goujard C, Meyer L, Costagliola D, Venet A, Pancino G, Autran B, Rouzioux C. 2013. Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLoS Pathog 9:e1003211. doi: 10.1371/journal.ppat.1003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Julg B, Williams KL, Reddy S, Bishop K, Qi Y, Carrington M, Goulder PJ, Ndung'u T, Walker BD. 2010. Enhanced anti-HIV functional activity associated with Gag-specific CD8 T-cell responses. J Virol 84:5540–5549. doi: 10.1128/JVI.02031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crowell TA, Gebo KA, Blankson JN, Korthuis PT, Yehia BR, Rutstein RM, Moore RD, Sharp V, Nijhawan AE, Mathews WC, Hanau LH, Corales RB, Beil R, Somboonwit C, Edelstein H, Allen SL, Berry SA. 2015. Hospitalization rates and reasons among HIV elite controllers and persons with medically controlled HIV infection. J Infect Dis 211:1692–1702. doi: 10.1093/infdis/jiu809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hatano H, Yukl SA, Ferre AL, Graf EH, Somsouk M, Sinclair E, Abdel-Mohsen M, Liegler T, Harvill K, Hoh R, Palmer S, Bacchetti P, Hunt PW, Martin JN, McCune JM, Tracy RP, Busch MP, O'Doherty U, Shacklett BL, Wong JK, Deeks SG. 2013. Prospective antiretroviral treatment of asymptomatic, HIV-1 infected controllers. PLoS Pathog 9:e1003691. doi: 10.1371/journal.ppat.1003691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boufassa F, Saez-Cirion A, Lechenadec J, Zucman D, Avettand-Fenoel V, Venet A, Rouzioux C, Delfraissy JF, Lambotte O, Meyer L. 2011. CD4 dynamics over a 15 year-period among HIV controllers enrolled in the ANRS French observatory. PLoS One 6:e18726. doi: 10.1371/journal.pone.0018726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chun TW, Justement JS, Murray D, Kim CJ, Blazkova J, Hallahan CW, Benko E, Costiniuk CT, Kandel G, Ostrowski M, Kaul R, Moir S, Casazza JP, Koup RA, Kovacs C, Fauci AS. 2013. Effect of antiretroviral therapy on HIV reservoirs in elite controllers. J Infect Dis 208:1443–1447. doi: 10.1093/infdis/jit306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Persaud D, Luzuriaga K. 2014. Absence of HIV-1 after treatment cessation in an infant. N Engl J Med 370:678. doi: 10.1056/NEJMc1315498. [DOI] [PubMed] [Google Scholar]
- 50.Ndhlovu ZM, Stampouloglou E, Cesa K, Mavrothalassitis O, Alvino DM, Li JZ, Wilton S, Karel D, Piechocka-Trocha A, Chen H, Pereyra F, Walker BD. 2015. The breadth of expandable memory CD8+ T cells inversely correlates with residual viral loads in HIV elite controllers. J Virol 89:10735–10747. doi: 10.1128/JVI.01527-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nasi M, Pinti M, Manzini S, Gibellini L, Manzini L, Bisi L, De Biasi S, Del Giovane C, D'Amico R, Borghi V, Mussini C, Cossarizza A. 2010. Predictive value of intracellular HIV-1 DNA levels during CD4-guided treatment interruption in HIV+ patients. AIDS Res Hum Retroviruses 26:553–558. doi: 10.1089/aid.2009.0256. [DOI] [PubMed] [Google Scholar]
- 52.Goujard C, Bonarek M, Meyer L, Bonnet F, Chaix ML, Deveau C, Sinet M, Galimand J, Delfraissy JF, Venet A, Rouzioux C, Morlat P. 2006. CD4 cell count and HIV DNA level are independent predictors of disease progression after primary HIV type 1 infection in untreated patients. Clin Infect Dis 42:709–715. doi: 10.1086/500213. [DOI] [PubMed] [Google Scholar]
- 53.Rouzioux C, Hubert JB, Burgard M, Deveau C, Goujard C, Bary M, Sereni D, Viard JP, Delfraissy JF, Meyer L. 2005. Early levels of HIV-1 DNA in peripheral blood mononuclear cells are predictive of disease progression independently of HIV-1 RNA levels and CD4+ T cell counts. J Infect Dis 192:46–55. doi: 10.1086/430610. [DOI] [PubMed] [Google Scholar]
- 54.Kiselinova M, De Spiegelaere W, Buzon MJ, Malatinkova E, Lichterfeld M, Vandekerckhove L. 2016. Integrated and total HIV-1 DNA predict ex vivo viral outgrowth. PLoS Pathog 12:e1005472. doi: 10.1371/journal.ppat.1005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leng J, Ho HP, Buzon MJ, Pereyra F, Walker BD, Yu XG, Chang EJ, Lichterfeld M. 2014. A cell-intrinsic inhibitor of HIV-1 reverse transcription in CD4(+) T cells from elite controllers. Cell Host Microbe 15:717–728. doi: 10.1016/j.chom.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buckheit RW III, Allen TG, Alme A, Salgado M, O'Connell KA, Huculak S, Falade-Nwulia O, Williams TM, Gallant JE, Siliciano RF, Blankson JN. 2012. Host factors dictate control of viral replication in two HIV-1 controller/chronic progressor transmission pairs. Nat Commun 3:716. doi: 10.1038/ncomms1697. [DOI] [PMC free article] [PubMed] [Google Scholar]