Abstract
Kinase inhibitors are used widely to treat various cancers, but adaptive reprogramming of kinase cascades and activation of feedback loop mechanisms often contribute to therapeutic resistance. Determining comprehensive, accurate maps of kinase circuits may therefore help elucidate mechanisms of response and resistance to kinase inhibitor therapies. In this study, we identified and validated phosphorylatable target sites across human cell and tissue types to generate PhosphoAtlas, a map of 1,733 functionally interconnected proteins comprising the human phospho-reactome. A systematic curation approach was used to distill protein phosphorylation data cross-referenced from 38 public resources. We demonstrated how a catalog of 2,617 stringently verified heptameric peptide regions at the catalytic interface of kinases and substrates could expose mutations that recurrently perturb specific phospho-hubs. In silico mapping of 2,896 nonsynonymous tumor variants identified from thousands of tumor tissues, also revealed that normal and aberrant catalytic interactions co-occur frequently, showing how tumors systematically hijack, as well as spare, particular sub-networks. Overall, our work provides an important new resource for interrogating the human tumor kinome to strategically identify therapeutically actionable kinase networks which drive tumorigenesis.
Keywords: Kinase enzyme, phosphorylation network, non-synonymous tumor mutations, targeted therapy, Cytoscape maps
Introduction
Biological processes are largely mediated by fine-tuned cascades of Protein-Protein Interactions (PPIs). Many PPIs are transient and enzymatic in nature, resulting in highly dynamic and plastic signaling networks capable of propagating information within and between cells. Protein kinases are phosphorylating enzymes that regulate the function of specific protein substrates by interacting with and chemically modifying them (1). Phosphorylation circuits govern most cellular processes essential to tissue homeostasis. Disruption of the activity of kinases or their substrates by mutations or aberrant signaling is associated with cancer and other diseases. As a pivotal cause of disease, disrupted phosphorylation circuits are frequent targets for therapeutic interventions. Kinase inhibitors have proved successful in a number of clinical scenarios, including targeting HER2 in breast cancer, BCR-ABL in chronic myeloid leukemia, and BRAFV600E in melanoma. However, adaptive reprogramming of kinase cascades and feedback loop mechanisms contribute to the development of treatment resistance, the primary obstacle in achieving sustained responses to targeted therapies (2). Therefore, it is imperative to understand not only individual kinase-substrate protein relationships but to acquire in depth knowledge of the inter-connectivity of these networks and the biochemical specificity of phospho-catalytic events in both healthy and diseased states. Such comprehensive kinome resource would greatly facilitate the elucidation of disease mechanisms and help identify the network components responsible for therapeutic responses.
Decades of research and recent powerful advances in biotechnology and systems biology are producing an ever-increasing accumulation of biological information about proteins, phospho-proteins, and proteomic-level regulation of cell processes and diseases (3–10). There are over 500 databases devoted to protein sequence, structure, molecular and chemical interactions, and signaling pathways (11–14). PPI databases such as STRING (15), PINA (16), MINT and IntACT (17), BioGRID (18), HPRD (19), IMEx (13) and numerous others collect and curate, to varying degrees, genetic and protein interaction data from numerous organisms. One limitation of these resources is that they merge known and predicted protein interactions, combine physical with indirect genomic interactions, and include data inferred from evolutionary conservation analysis. Moreover, PPI databases can contain several million interactions, but only about fifty thousand of these are found in human and even fewer relate to phosphorylation events.
Several major efforts integrate publicly available kinase-substrate phosphorylation data. A majority of these resources, such as KSD (20), KinBase (21), KinG (22), and Kinweb (23), provide valuable information about kinases, such as sequence alignment, phylogeny, or regulatory domains. These databases, however, often lack verified information on target residue sites within substrate proteins phosphorylated by kinases. Novel resources have begun to map the relationship between kinases and substrates. For example, PhosphoSitePlus (24), PhosphoPOINT (25), RegPhos 2.0 (26), or Phospho.ELM (27) provide information on phosphorylation events garnered from literature and derived from substrate motif scans and high throughput Mass Spectrometry (MS) approaches. Although such resources are comprehensive, individual results may be compromised by integration systems that rely on highly automated computational pipelines. It often remains difficult for non-expert users to distinguish experimentally validated knowledge from potentially less reliable high-dimensional screens or prediction-based datasets. Finding information about the precise connectivity between a human kinase and its cognate substrates and specific target sites, or the reverse, can be cumbersome. Access to a well-characterized and experimentally validated, albeit smaller data set describing established phosphorylation circuits would enable efficient and reliable exploration of the molecular mechanisms of human diseases, and generate new hypotheses.
Here, we present PhosphoAtlas, a highly curated map of the human phospho-reactome and its mutated, tumor kinome counterpart. We define the phospho-reactome as a complete catalog of verified phosphorylation reactions involving an effector/kinase and a receiver/substrate protein, and their network-level connectivity including details of phosphorylatable residue sites and heptameric target sequences when known. This dataset was developed via a series of computational steps designed to extract protein information from multiple data sources, and identify validated kinase-substrate interactions. We then utilized PhosphoAtlas database to explore the mutational variability of phospho-protein networks in tumors. We identify non-synonymous mutations that most directly and systematically impact the catalytic circuits of human kinase-substrate networks, potentially revealing which somatic mutations in cancer may have functional consequences and thus be therapeutically actionable.
Methods
Computational strategy to build PhosphoAtlas relational database
The Extended Method section describes the computational approach for building a database where human protein kinases are associated with their downstream protein targets and exact HPS’s they phosphorylate.
Identification and analysis of variants in peptide target sequences
PhosphoAtlas’ compendium of HPR’s was analyzed using the Catalogue of Somatic Mutations (COSMIC) database (28). First, to establish the presence of reported variants within HPRs, their genomic locations were calculated using the GENCODE GRCh37/HG19 assembly (29) for all known substrate protein isoforms. Next, the variants within these locations were found by overlapping the variant GRCh37 genomic locations acquired from a complete COSMIC database with the genomic locations of our HPRs. The COSMIC database also provided additional information on type of the mutation, its consequence, and the histology associated with the particular alteration. The tools used for the database build and analysis included Bedtools, R programming language (Vienna, Austria, http://www.R-project.org/), and R packages GenomicFeatures, Biostrings, and RWebLogo.
PhosphoAtlas database access
The PhosphoAtlas dataset is a resource readily available to the public at http://cancer.ucsf.edu/phosphoatlas upon registration. Users may download a ZIP archive that includes CSV and XLS files of the database, and a pre-build Cytoscape CYS session file (Cytoscape version 3). Cytoscape (30) is a very well adapted tool to visualize and explore the networks of kinase-substrate-residue target sites and their tumor variants. Cytoscape provides sophisticated means to search and filter the PhosphoAtlas dataset and access all nodes and edge attributes, and respective relationships for either the entire network or user-selected sub-networks. Once the CSV / XLS / CYS data files are respectively loaded in Excel or Cytoscape, data can be filtered and exported in a variety of additional formats that support further processing in other network analysis and statistics packages. All files can be used to study or query either normal phospho-circuits, or tumor-associated networks.
Results
Strategy to establish a comprehensive map of the validated human phospho-reactome
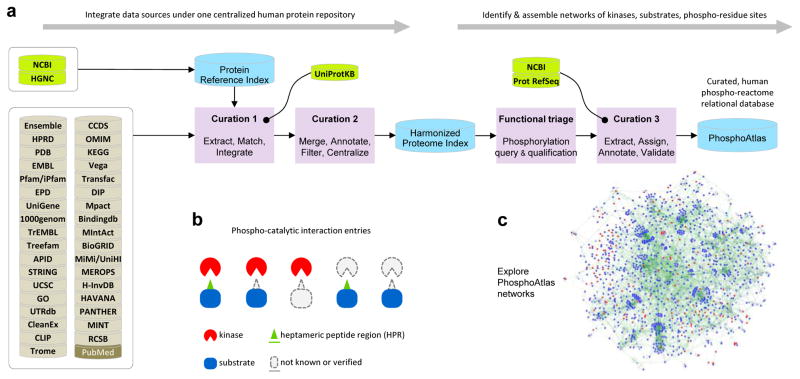
We created a resource for the exploration of human phosphorylation circuits and its aberrant, cancer variations. The flow diagram presented in Figure 1a depicts the key steps of the processing pipeline we employed to acquire, integrate, filter, curate, and analyze pre-existing molecular data from publicly accessible resources (see Extended Methods, and Figure S1). This strategy enabled identifying experimentally verified kinase proteins, protein substrates, phospho-residue sites, and heptameric peptide sequences (HPS).
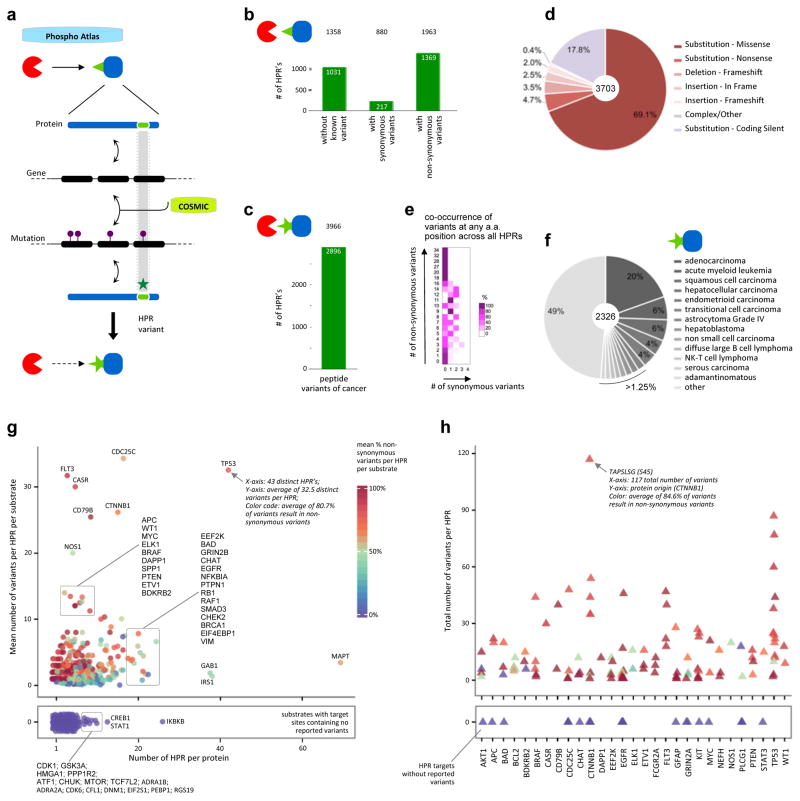
Figure 1. Phospho-reactome curation workflow: creation of a comprehensive and unified relational database that indexes human kinases and their cognate phosphorylation targets.
a. Resources, logical workflow, computational curation processes, and indexation steps are summarized. First, a Harmonized Proteome Index is created as a structured non-redundant repository of all known human proteins through curation steps 1 and 2, enabling the systematic extraction, harmonization and classification of public protein records. HGNC, NCBI and UniProtKB are used as critical databases that serve as blueprints to cross-reference and filter data and annotations extracted across 35 public databases. Second, PhosphoAtlas is build as a relational database of phosphorylation events. Curation ‘functional triage’ and step 3 identify which proteins are kinase enzymes or substrate proteins, how these proteins functionally interact with each other, and whether verifiable phosphorylatable residue sites and surrounding sequences can be defined. This establishes a comprehensive, curated dataset that maps human catalytic phospho-circuits, PhosphoAtlas.
b. Schematic representation of PhosphoAtlas entries. Five groups of complete or partial knowledge of phospho-catalytic interactions are shown. A majority of heptameric peptide regions (HPR’s) is centered on a phospho-residue site and stretch over 3 amino acids up and down, but for phospho-residues located at the N- or C-terminal of a protein, phospho-residues are displaced down or up the heptameric end portion of the protein.
c. PhosphoAtlas networks of kinase–substrate interactions can be explored (visual representation powered by Cytoscape (30)). Searchable CSV and XLS data files, and Cytoscape sessions, are downloadable upon registration at http://cancer.ucsf.edu/phosphoatlas.
Integration of multiple data sources into one harmonized protein repository
First, we generated a ‘Protein Reference Index’ from HGNC and NCBI/Entrez to create a non-redundant inventory of human proteins defined by their standardized symbols, names, RefSeq protein and nucleotide IDs, which established a reliable ‘Primary Identifier’ record for each protein. Next, the Protein Reference Index served as a blueprint to systematically integrate and cross-reference the protein records extracted from other ‘external’ data sources using a curation method analogous to the PPI integration pipeline from (16) (Figure 1a, left). Directly matching external records were imported and structured by their common, unique Primary Identifier. If no matching primary identifier was available in the record, identifiers from external sources were cross-referenced in Curation 1 with the UniProtKB database (31) (Extended Methods). Once an external identifier was successfully cross-referenced, the related record was updated. Unmatched data were excluded. At each step of the assembly of the data, all additional available functional data, annotations and references were compiled as complementary content linked to their Primary Identifier.
To harmonize the heterogeneous content across the various sources of this initial human proteome resource, collected records were curated (Curation 2). Pattern matching computer-implemented methods and sequence alignment algorithms was used to check, remove, or merge any redundant or ambiguous records, and flag discrepancies, as described in detail in Extended Methods. This resulted in a highly curated, non-redundant, comprehensive dataset of known human proteins, referred to as the ‘Harmonized Proteome Index’.
Building a relational database connecting kinases to substrates and phospho-residue sites
Next, we attempted to identify the most complete and rigorously verified phospho-reactome portion of the human proteome. We applied data-mining methods to extract protein records related to phosphorylation events to identify kinase–substrate catalytic interactions, and phosphorylatable amino acids from substrates’ phospho-residue sites. Records from the Harmonized Proteome Index were queried using natural language processing algorithms to identify proteins that function as kinases or phosphorylatable substrates, as described in detail in Extended Methods using a curation method analogous to that in (24) (Figure 1a, Functional triage, and Figure S1, bottom-left section).
Qualified records generated a preliminary repository of functionally interconnected kinase and substrate proteins that was then mined for phosphorylation modification information (Figures 1a and S1, Curation 3). Each qualified substrate with an available phosphorylation (abbreviated as ‘phospho’) -site or -peptide sequence was compared to the latest corresponding curated protein substrate sequence (RefSeq/NCBI) using string- and pattern matching methods, as described in detail in Extended Methods. Once the location of a phosphorylated residue was confirmed, or if the sequence alignment or peptide composition was conclusive, the amino acid sequence surrounding the validated phospho-site was extracted from the protein RefSeq. We collected heptameric peptide sequences around phospho-residue target sites based on previous biochemical observations (32, 33) and our data (Figure S3d). Both residue site location and heptameric peptide were indexed and assigned to their substrate (and kinase(s) when reported) (Figures 1b). Since the repository of phospho-catalytic sites represents the critical cornerstone of all kinase–substrate functional connections, we rigorously ensured its accuracy by excluding all primary records providing candidate targets from data solely based on confidence-based approaches or not cross-referenced or not confirmed using complementary molecular techniques. We further distinguished validated phospho-residue sites with or without an identified upstream kinase, and sub-classified them separately (Figures 1b, two right groups).
The resulting database exclusively contains validated human kinases and substrates and their respective phosphorylation sites. These interactions were integrated into a network of kinase–substrate phospho-catalytic circuits that constitute ‘PhosphoAtlas’ (Figure 1b–c). Flat database files and a Cytoscape session are publicly available via the web portal http://cancer.ucsf.edu/phosphoatlas to facilitate exploration and visualization of molecular networks (see Methods).
Overview of the human phospho-reactome
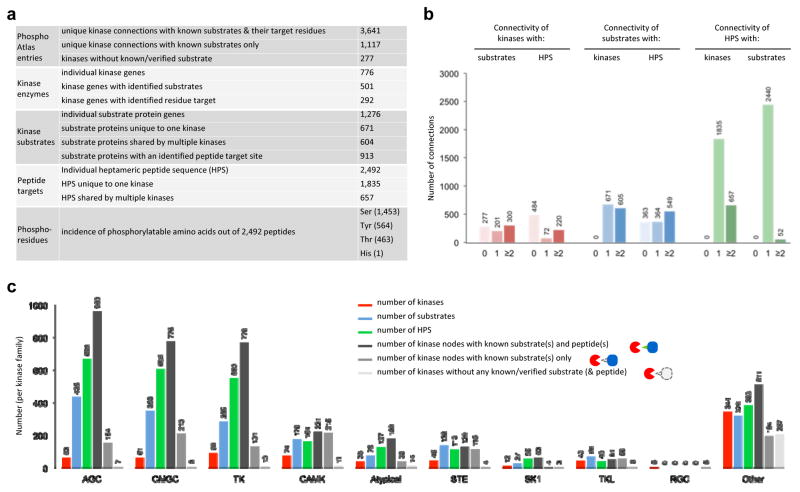
Analysis of PhosphoAtlas suggests that ~11% of all PPIs (16) (Figures S2) represent potentially actionable phospho-catalytic circuits of human cells. A total of 4,758 unique edges connect kinases to a target. PhosphoAtlas catalogues this network as 3,641 kinases with known substrates and residue target sites, and 1,117 with known substrates only. 292 kinases are reported as exclusively or redundantly phosphorylating 1,276 distinct substrate proteins via 2,617 unique heptameric peptide regions (HPR’s) that correspond to 2,492 distinct heptameric peptide sequences (HPS’s), indicating that some HPS’s are ‘shared’ across multiple HPR’s (Figure 2a).
Figure 2. PhosphoAtlas database overview.
a. Table summary of PhosphoAtlas entries.
b. Representation of connections between kinases, substrates and heptameric peptide sequences (HPS’s.). Connecting substrates and HPS’s are depicted for all kinases within PhosphoAtlas depending on the known number of connections for the kinase (left panel), and similarly shown for substrates (center) and HPS’s (right). Data presented for substrates and HPS’s exclude substrates and HPS’s that have yet to be conclusively mapped to kinases.
c. Breakdown of most represented kinase families in PhosphoAtlas by the number of unique kinases per family (red), respective interacting substrates (blue) and their known target peptides (green). Bars in gray shade represent kinase families by status of entries: kinase nodes with known both substrates and peptides, with known substrates only, or without known substrates or target peptides. Kinase enzyme families groups are based on (21).
The connectivity between kinases, substrates and HPS’s (Figure 1b,c) can be categorized based on the number of cognate partners. For example, 300 kinases phosphorylate at least 2 substrates, and 220 kinases phosphorylate at least 2 HPS’s (Figures 2b left panel, and S3). Most highly connected kinases and substrates are shown in Figure S4. Of the 1,276 substrates with identified kinase partners, 53% (n=671) interact with a single unique kinase, while the remaining 47% (n=605) interact with at least two kinases. 913 of these 1,276 substrates contain either one (29%, n=363) or at least two (43%, n=364) unique HPS’s. 39.9% of all substrates in PhosphoAtlas contain only one unique HPS. Of the verified 2,492 HPS’s, 74% (n=1,835) connect with a single unique kinase, and 26% (n=667) are shared by at least two kinases. In contrast, 98% (n=2,440) of the HPS’s are associated with a single substrate, while the remaining 2% are found across multiple substrates. We also found that 58.9% of all kinase-substrate phosphorylation events are achieved via a unique peptide sequence. 24.7% of all kinases in PhosphoAtlas target one unique HPS. 41.8% of all kinases in PhosphoAtlas target one unique substrate. Taken together, these data portray the human phospho-reactome as almost evenly divided between highly specific kinase–substrate pairs and more highly connected nodes. Combinatorial logic of signaling, whereby multiple kinases can phosphorylate a single substrate, is mainly achieved through multiple distinct HSP’s on a single substrate (60.1%), while as few as 26.4% HSP’s are recognized by multiple kinases. Only six kinase-specific HSP’s are found on multiple substrates.
Grouping kinases by gene families (21) shows that 84% of validated substrates and 73% of validated peptides associate with either the AGC (e.g., PKA, PKC; 34.1% of substrates, 26.8% of peptides), CMGC (e.g., MAPK, CDK; 27.4% substrates, 24.3% peptides) or TK (e.g., EGFR, SRC; 22.3% substrates, 22.1% peptides) kinase families (Figures 2c, and S5). The kinase interaction edges for each kinase family can be categorized as fully validated (known kinase, substrate and target site) or partially known (known kinase and substrate only) (Figure 2c, grey bars).
Our curation process also revealed valuable knowledge gaps (Figure 1b, four right groups). The database includes 275 kinase enzymes known to be upstream of 363 substrates, but for which no phospho-residue site was found or definitively established, and 277 identified kinases with no validated substrate. Reciprocally, 632 substrate proteins with 706 confirmed HPS’s have no conclusively established effector kinase, and 74 substrate proteins have no identified or verifiable upstream kinase or phospho-residue sites. These gaps suggest the need to broaden experimental investigation of phosphorylation cascades or circuits.
Target peptides provide insight into the diverse modes of kinase–substrate interactions
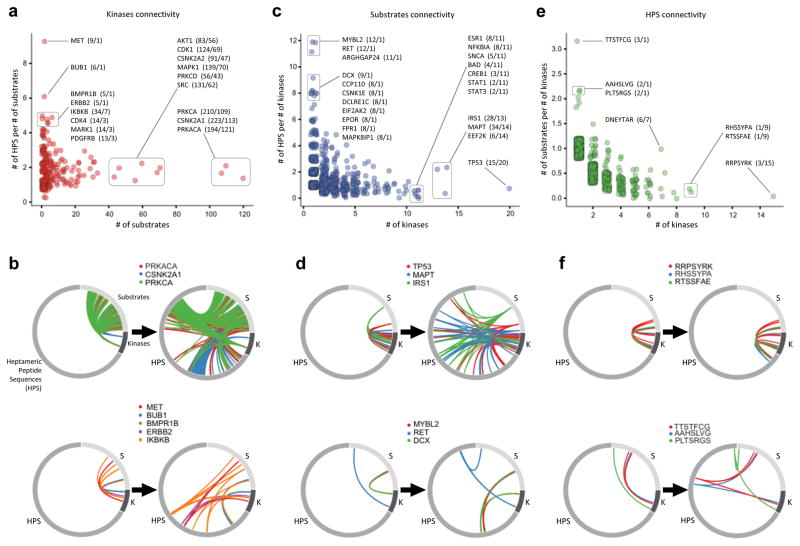
Representing kinase–substrate interactions solely as 1,276 pairs masks the inherent complexity of phospho-catalytic circuits. We further resolved their connectivity using the available 3,641 phospho-residue site-specific distinct interactions, which yielded 4,758 unique kinase–substrate phospho-catalytic connections. Accordingly, Figure 3 shows how incorporating phospho-residue sites increases the resolution of kinase–substrate catalytic interaction networks. Figure 3a,b displays kinases according to the number of identified substrates for each kinase and its number of HPS’s per substrate. There is a continuum from highly substrate-specific kinases with multiple HPS targets, such as MET (9 HPS on 1 substrate) (Figure 3a, top left; 3b, bottom), to broad-spectrum kinases that phosphorylate 10’s to >100 individual substrates at an average of 1–2 HPS targets, such as PRKCA (210 HPS on 109 substrates, averaging 1.9 HSP per substrate) (Figure 3a, right; 3b, top). Most interaction networks are simpler; 91.4% kinases phosphorylate between one and twenty substrates, with a median of 2 unique HPS targets per substrate (mean = 2.804 HPS, st. dev. = 2.813). Figure 3c,d represent a similar spectrum for protein substrates and their connections to kinases and identified HPS targets. At the extreme ends are substrates phosphorylated by multiple kinases at mostly unique phospho-sites on these substrates (e.g. IRS1 (phosphorylated by 13 kinases on 28 HPS, average 2.1 HPS per kinase), MAPT (34 HPS/14 kinases)), and substrates known to be targeted by few kinases, yet phosphorylation occurs at many target sites (e.g. RET (12 HPS/1 kinase). Similarly, when examining modes of action on HPS targets, there emerge sequences that are found in many substrates, but phosphorylated by single kinase, and those that are recognized by multiple kinases, yet are found only in few substrate proteins (Figure 3e,f). Ultimately, this increased resolution facilitates a more detailed understanding of regulatory signaling mechanisms, especially considering that many substrates have more than one phospho-residue site that are specifically and differentially phosphorylated by many kinases.
Figure 3. Biological peptide sequences collected in PhosphoAtlas provide a new depth and unique dimension into kinase–substrate catalytic interactions.
a. Individual kinases (red discs) are plotted based on the number of unique connecting substrates (x-axis) and related HPS target per substrate ratio (y-axis). Some kinases are annotated by name along with number of unique HPS per number of unique substrates in parenthesis.
b. Circos plots visualize the number of kinase-substrate (left) and kinase-HPS-substrate (right) connections for selected kinases.
c. Protein substrates (blue discs) are plotted based on the number of unique connecting kinases and peptide per kinase ratio. Annotated substrates are identified by name followed by the number of known HPS targets within it per number of kinases it is phosphorylated by.
d. Circos plots visualize the number of kinase–substrate (left) and kinase–peptide–substrate (left) connections for selected examples of substrates.
e. HPS’s (green discs) are plotted based on the number of unique interacting kinases (x-axis) and substrate per kinase ratio (y-axis). Every peptide in the plot is an N-term to C-term amino acid sequence annotated in parenthesis with the number of substrates it is found within per number of kinases that it is targeted by.
f. Circos plots visualize the number of kinase–substrate and kinase–peptide–substrate connections for different entries.
Exploring AKT cancer phospho-circuits using PhosphoAtlas
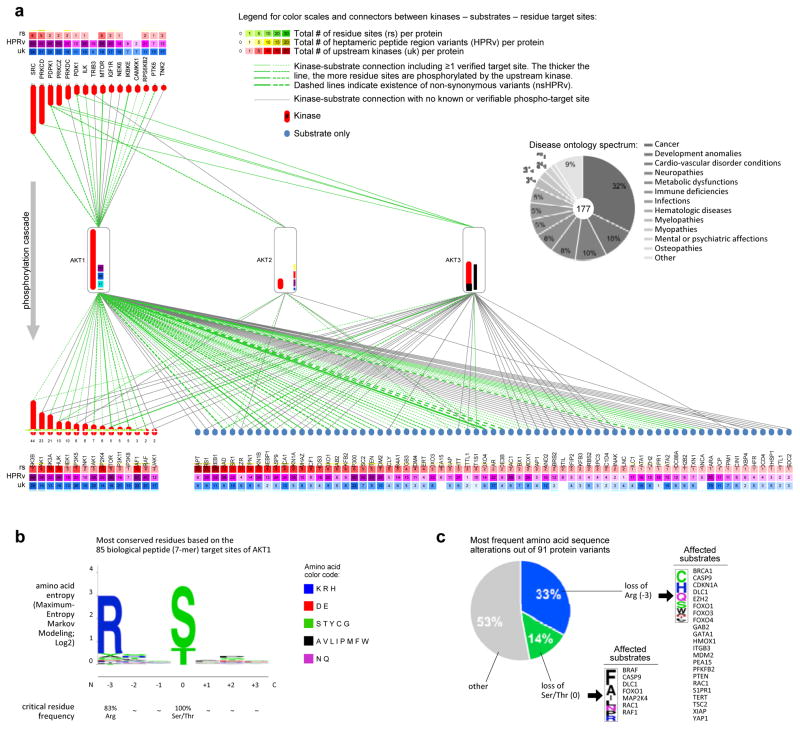
PhosphoAtlas supports the exploration of the human kinome at a global phospho-proteomic level, but also at the level of individual kinase proteins and their sub-networks, interactive partners, enzymatic modalities, and conservation patterns. For example, Figure 4a represents the network of kinases and substrates directly upstream and downstream of the AKT1, AKT2 and AKT3 proteins. This sub-network includes 102 kinase and substrate proteins (red and blue nodes, respectively) that interact via 184 unique kinase–substrate edges (all lines), of which 113 have known phospho-residue site information (green lines) (Figure 4a). Phospho-signaling cascades that funnel through AKTs can further propagate via downstream kinases (Figure 4a, bottom left red) or affect non-kinase substrate proteins (Figure 4a, bottom right blue) that regulate normal and tumor-associated cell processes such as growth, motility, transcription or inflammation. The broad range of disease ontology terms associated with these 102 proteins and mainly associated with cancer, demonstrates the diverse functional impact of AKT-related signals (Figure 4a, pie chart; Figure S6; data mined using Ensembl (Biomart)).
Figure 4. AKT phosphorylation network: a custom query into PhosphoAtlas.
a. Connectivity of AKT family proteins. Proteins that phosphorylate AKT1/2/3, and proteins that are phosphorylated by AKT1/2/3, are respectively listed above and below AKTs. Each connecting line is a directional vector that symbolizes the top-down flow of phosphorylation from a kinase enzyme to a specific substrate. Color and continuity of connectors depict knowledge of phosphorylatable sites within protein targets. All proteins that can function as kinase enzymes are associated with a red bar containing a number that corresponds to the number of different substrate proteins they phosphorylate. The pie chart insert is an example of how the ensemble of AKT-affiliated proteins ramifies into biological processes with human disease implications. Numbers within color-coded boxes include all residue sites, variants, and upstream kinases per protein, beyond the exclusively AKT-related molecular hub.
b. WebLogo alignment of 85 heptameric biological peptide targets of the AKT1 kinase unravels the most conserved residues surrounding the phosphorylation site.
c. Breakdown of amino acid alterations and affected substrate targets resulting from the occurrence of 91 non-synonymous variants found in the genomic locations overlapping with the HPR’s of AKT1.
Considering the number of additional upstream kinases (uk) and distinct residue sites (rs) that exist per protein composing AKT circuits (numbers specified within green and red squares above and below each protein, Figures 4a), reinforces the complexity of AKT circuitry in particular and phospho-circuitry in general, and illustrates the utility of PhosphoAtlas for visualization and exploration.
Differences among the three AKT family members in their number of kinase–substrate edges with or without known phosphorylation sites are apparent (Figures 4a, S7). This in part reflects that, while AKT1 has been thoroughly studied, much less is known about AKT2, AKT3, and their targets. Such differences are repeatedly found across kinase subfamilies (e.g. CDKs or SRCs).
PhosphoAtlas also enables the identification of consensus peptide target sequences that could be used to scan the human proteome to predict new substrate phosphorylatable sites, or design probes for kinase activity assays. For instance, by comparing all HPS’s targets of AKT1, we identified the most conserved amino acid residues within heptameric peptides (Figures 4b, S8), which is supported by other studies. Beyond the required presence of a phosphorylatable Serine (S) or Threonine (T) at the center of all HPS’s, the Arginine (R) at position −3 was highly conserved with a frequency of 83.2%. This result highlights the biochemical importance of Arginine’s presence at this site for AKT1 to phosphorylate its substrates.
Given that much of the human kinome is considered druggable, it is of interest to identify known (pathogenic and/or somatic) variants within target sequences (see below for methodology). Figure 4a shows which substrate target sites of AKT1 contain non-synonymous variations found in tumors. Remarkably, analysis of the cancer-associated variants within AKT1 targets shows that loss of Arginine (−3) accounts for 20.6% of all non-silent mutations reported by COSMIC within these HPS’s (Figures 4c, S8d–e). It is therefore conceivable that this HSP mutation in AKT1 downsteam substrates such as BRCA1, CASP9, PTEN or MDM2, is a causal molecular mechanism resulting in tumorigenic pathway dysregulation in breast, colorectal, brain and other cancers.
Global analysis of oncogenic mutations affecting phospho-target sites reveals the disrupted phospho-circuits of cancer
The collection of phospho-target sites mapped in PhosphoAtlas represents a unique opportunity to identify genetic mutations with functional consequences on kinase circuits. PPI-perturbing mutations are significantly more likely to be deleterious than non-PPI-perturbing mutations (10, 34). To identify cancer mutations that directly impact kinase-substrate catalytic interactions, we designed a reverse-mining process that maps genetic variations onto phosphorylatable substrate regions (Figure 5a; see Methods for details). We analyzed our compendium of HPR’s using the Catalogue of Somatic Mutations In Cancer (COSMIC) database (28), which includes curated information from The Cancer Genome Atlas project (TCGA), the International Cancer Genome Consortium (ICGC), and systematic screens of 1000’s of tumor genomes. We mapped the mutational landscape of cancer phospho-circuits in a tissue- or disease- agnostic fashion.
Figure 5. Impact of non-synonymous variants identified as genetic mutations found in tumors.
a. Reverse mapping approach to reveal the cancer-associated variants embedded within the phospho-reactome. The identification of cancer-associated genetic mutations as reported by COSMIC that exert specific effects on kinase-substrate catalytic interaction sites exposes an additional layer of disturbances caused by mutations found in patients’ tumors.
b. Bar graph depicting the number of heptameric peptide regions (HPR’s) in PhosphoAtlas that contain COSMIC-defined synonymous and non-synonymous variants, or no mutation at all. The number of unique (kinase–HPR–substrate) entries affected by cancer mutations is shown on top.
c. Total number of cancer-associated, non-synonymous heptameric peptide region variants (nsHPRv) derived from COSMIC-mining analysis. The number of potential additional cancer-related predicted PPI entries is indicated on top.
d. Distribution of the protein-coding consequences of genetic mutations on phosphorylatable HPR’s.
e. Distribution of co-occurrence of non-synonymous versus synonymous variants across all amino acid found across all HPRs.
f. Spectrum of malignancies most prone to include mutations affecting phosphorylation circuits.
g. Spectrum of proteins affected by mutations that alter their phosphorylatable target regions. All substrate proteins from PhosphoAtlas are distributed by the mean number of total variants per HPR versus the number of known HPR per given substrate. Each protein is color-coded by the mean percentage of non-synonymous variants per total number of variants in a given substrate (reference color scale 0–100%). The bottom panel contains substrates for which no variants in their HPR’s have been reported.
h. Proteins with the most recurrently mutable peptide sites across all human tumors. The top 50 HPR’s with the highest proportion of non-synonymous variants to total number of variants per HPR were first identified, then the corresponding proteins were selected, and finally all HPR’s from each of these substrates were plotted. Proteins are sorted alphabetically on the x-axis. Each HPR is shown as a triangle and color-coded by the mean percentage of non-synonymous variants out of total reported non-synonymous and synonymous variants per HPR (color scale in (g)).
Protein variants identified through this process were classified based on the consequence of genetic alterations (Figure 5b). Of the original 2,617 distinct substrates’ HPR’s, 39.4% (n=1,031) were not affected by any known mutation, 8.3% (n=217) contained only silent variants (synonymous), and 52.3% (n=1,369) contained at least one non-synonymous variant that altered amino acid composition (‘nsHPRv’). The distinct variants generated a new group of 2,896 mutated sequences (Figure 5c), which perturb 1,963 kinase-substrate interactions (i.e. 46.7% of all kinase circuits) with potential detrimental effects on phospho-catalytic circuits (10).
Among all HPR’s affected by 3,703 synonymous and non-synonymous variations, missense substitutions account for the majority (69.1%) of all variants, followed by 17.8% of coding but silent substitutions (Figure 5d). The differential levels and pattern of co-occurrence of non-synonymous versus synonymous mutations of HPR’s (Figures 5e, S9a–c) imply that phospho-target sites are mutated in a selective fashion in tumors. Only 5% of all HPR variants originate from mutations qualified as germline single nucleotide polymorphisms (Figure S10). Adenocarcinomas are the most prevalent pathology (20%) associated with all variants (Figure 5f), while the highest number (14.4%) of phosphorylation-site-variants originates from tumors of the large intestine (Figure S11).
We next proceeded to identify which substrates are most affected by cancer-associated mutations that alter their phospho-targetability. To do so, each protein was plotted based on its individual ratio of mean number of variants per HPR (Figure 5g, y-axis), against its total number of HPR (x-axis). Each, dot, representing a protein, was subsequently color-coded as the mean percentage of nsHPRv’s per HPR, and averaged per substrate, to indicate whether genomic mutations mainly caused synonymous variants or non-synonymous sequence alterations (non-synonymous high%-red and low%-blue) (Figures 5g, S9d–e). For instance, FLT3 contains few phosphorylatable HPR’s associated with many cancer mutations that mostly result in alternative peptide sequences (high% non-synonymous variants). Conversely, IRS1 display numerous HPR’s that are overall subject to few variants, of which ~50% cause amino acid changes. In this spectrum, TP53 emerges as a substrate with a very high mean ratio of variants per HPR, associated with a large number of known target sites and a high proportion of variants being reported as non-synonymous (80.7%).
This analysis also identified proteins for which the COSMIC database does not report any variant in their phosphorylatable regions (Figure 5g, separated bottom panel; Figure S9d–e). Out of these 214 proteins, IKBKB, CREB1 and STAT1 were especially interesting examples due to their known critical role in regulating phosphorylation cascades involved in cancer development.
When using HPR’s as a way to report which kinases would be most affected by downstream nsHPRv’s in substrates, the spectrum of kinases with most-to-least variable networks emerged (Figure S12). This highlighted kinases that are potentially more frequently affected by non-synonymous mutations in their target sites (e.g. CHUK, GSK3A, or PRKDC), and kinases with genomically stable networks of downstream targets (e.g. MAP3K14, EEF2K).
Next, we investigated the 50 HPR’s with the highest number of reported unique non-synonymous variants. These most diversely altered sequences identified 29 substrate proteins only (Figure 5h). For example, TAPSLSG is a region from CTNNB1 that has the highest number of total variants per HPR, out of which 84.6% are amino acid coding, non-synonymous alterations. Looking at individual proteins, we find that not all target sites show the same mutational susceptibility. Many of the listed proteins are known tumor suppressors or oncogenes, while some are recognized enzymes, including kinases or phosphatases, showing how mutations in their phospho-target sites could globally afflict signaling cascades. For instance, target sites characterized by very high proportion of nsHPRv’s were found in BRAF, EGFR or FLT3, all of which contain activating kinase domain mutations that result in malignancies such as BRAFV600E in melanoma, or EGFRL858R in lung adenocarcinoma.
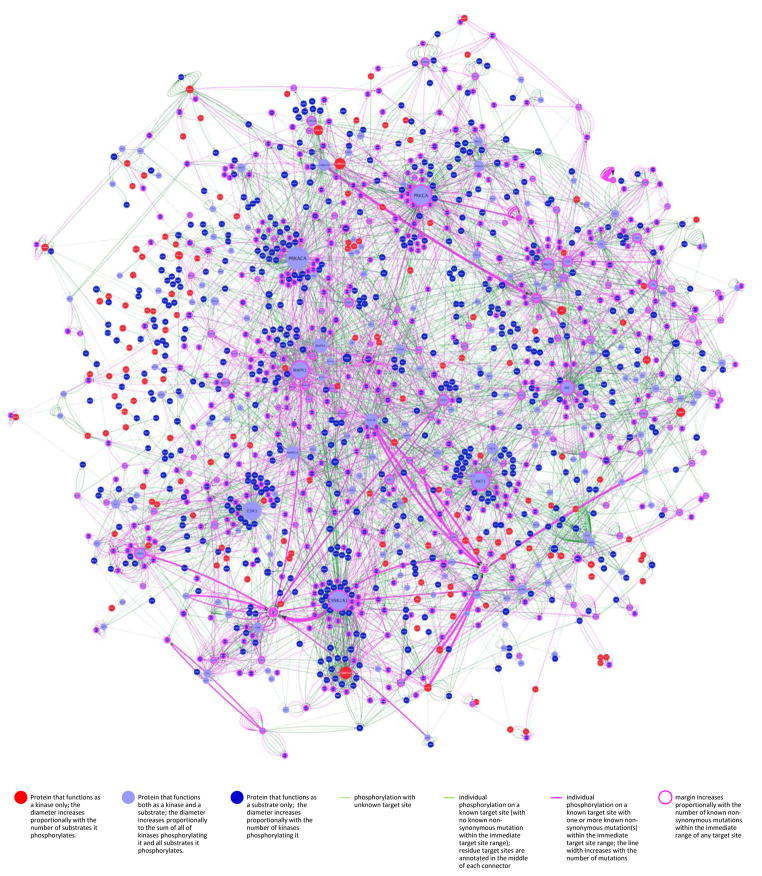
Subsets of distinct mutations may concentrate in pathways implementing cancer hallmark phenotypes (35). As mutations and resulting perturbations in kinase networks are key to cancer biology, we generated a network representation overlaying non-synonymous heptameric peptide region variants (nsHPRv’s), onto the phospho-reactome interaction map using Cytoscape (30) (Figures 6 and S13–S17; purple lines; data file available at http://cancer.ucsf.edu/phosphoatlas upon registration). This network represents all distinct phospho variants in COSMIC associated with tumors of every type. As illustrated, kinase circuits most selectively disrupted (defined by width of purple lines and node margins) across tumor types emerge out of the background of predominantly invariable phospho-circuits (green lines). This mosaic of normal and aberrant catalytic interactions shows how tumors systematically hijack – and spare – particular sub-networks.
Figure 6. The tumor kinome: differential impact of cancer-associated mutations on kinase-substrate phospho-catalytic networks.
Data generated from cross-referencing PhosphoAtlas and COSMIC databases were used to render the differential impact –and potential convergence– of non-synonymous peptide target variants onto substrates of kinases. The complex heterogeneity of mutations across tumor genomes was resolved via the unbiased aggregation of mutations that only and selectively alter substrates’ HPRs. Purple connectors represent nsHPRv’s at the catalytic interface of kinases and substrates that are collapsed into more or less thick edge representing the prevalence of mutations occurring within one heptameric sequence. By overlaying both interconnectedness and mutational impact of tumors on the human phospho-reactome, the most variable nodes visually emerge from the otherwise rarely altered or not mutated network. The absence of any reported mutation for a number of prevalent nodes with high number of interactions is noticeable, such as CDK1, PRKACA, MAPK3, MAPK14, GSK3B, AKT3 or CSNK2A2. The phosphorylation networks were produced using the Cytoscape network analysis platform (30) and is available at http://cancer.ucsf.edu/phosphoatlas.
Discussion
Phosphorylation cascades intimately regulate cell behavior. Availability of a highly curated compendium of phosphorylation data accurately recapitulating the molecular wiring of cells could support elucidation of disease mechanisms and vulnerabilities to therapeutic intervention in cancer. We built PhosphoAtlas, a novel data resource to study human phosphorylation networks, which includes a map of the differential impact of tumor mutations on phospho-target sites and reveals the most variable phospho-signaling hubs of cancer. PhosphoAtlas can support the exploration of a wide range of research questions related to human phospho-circuitry, and is meant to serve as a research hypothesis-generating platform.
To develop PhosphoAtlas, we systematically mined the profusion of available data existing in various formats and stages of verification or completion, found in a large number of state-of-the-art databases. We applied a stringent condensation-like process to filter, select, eliminate, cross-reference, qualify and validate information related to protein phosphorylation in human cells. We did not use prediction algorithms, conditional selection or inferred relationships, but rather relied on mining published data generated and validated by diverse experimental and computational techniques (16–18, 24). Although this approach restricted the number of kinase–substrate interactions represented in PhosphoAtlas, due to the exclusion of unvalidated phospho-catalytic interactions, it is meant to provide researchers with a still-large catalogue of protein kinases, substrates, phosphorylatable sites, and their connectivities from only the most reliable, validated sources. The result of this effort is both a data resource and a coherent map of functionally interconnected proteins comprising the human phospho-reactome (http://cancer.ucsf.edu/phosphoatlas).
The content of PhosphoAtlas can be exploited for at least three broad purposes: integrative in silico analysis and visualization; experimental data analysis; and bioassay development. For all these purposes, a major contribution of PhosphoAtlas is its inclusion of information on the specific phospho-residue sites of substrates, which enables investigation of kinase-substrate interaction networks at a higher level of resolution. As visualized and catalogued using PhosphoAtlas, the human phospho-reactome network appears to be roughly evenly divided between highly specific kinase-substrate pairs (33.2% proteins with single kinase-substrate connection, 19.1% proteins with two kinase-substrate connections), and more highly connected protein nodes (47.7% proteins with three or more kinase-substrate connections).
We identified kinases with preference to target many substrates via unique peptide target sequence versus those that target fewer substrates but can recognize a larger number of peptide target sequences. Such findings suggest, by the sheer number of alternative ways a kinase can phosphorylate a substrate, the essentiality of certain kinase-substrate interactions; in the event of a disruptive mutation in one region, a protein is likely to remain functional due to compensatory rerouting mechanisms. For example, the phosphorylation of GAB1 by MET kinase on 9 different target sites suggests this process may be essential for cell homeostasis. Moreover, kinases with the ability to phosphorylate many substrates on different target peptide sites may be active in multiple networks that regulate functionally similar cellular processes. If such a kinase’s first intended peptide target becomes disabled due to a mutation, a wider range of feedback mechanisms may be expected. Among such proteins, PRKACA can recognize and phosphorylate as many as 194 unique peptide sequences on 121 different substrates with prominent important functions in both the cytoplasm and the nucleus of cells. The diversity of connectedness of peptide targets at the regulatory interface of kinases and substrates emphasizes how the wide spectrum of network topologies of each substrate or kinase relates to their distinctive roles and functional impact on phospho-circuits.
More generally, knowledge of phosphorylation targets’ mutational landscape coupled with a wealth of information on the topology of human kinase networks could advance our understanding of signaling pathways in healthy and diseased states. PhosphoSite Plus (24) contributed to this effort by providing a proteome-wide snapshot of disease-related missense mutations across different post-translational modifications (PTMVar). Investigating the prevalence of these mutations and integrating them into comprehensive signaling networks is still needed to reveal the strategic, therapeutically actionable kinase circuits of tumors. Identifying which cancer mutations functionally impair biological networks remains a challenge, even when using large-scale differential mapping approaches (10, 36–40). Conceptually, the sparse and relatively non-recurrent mutation profiles within and across tumor types may be more comprehensible at the pathway level than at the level of individual genes (35, 41–47). Mutations that directly perturb protein phosphorylation circuits may be especially pathologically relevant to cancer development and eventually guide targeted therapy.
Thus motivated, we used the heptameric peptide library created from our curated kinase-substrate repository as a functional blueprint to filter 100,000’s of candidate somatic mutations found across 1,000’s of different human tumors. We found that over half (54.9%) of the distinct target peptides (2,617) contain at least one non-synonymous variant with altered amino acid composition. We identified which proteins are most affected by non-synonymous variants in phosphorylatable peptide regions (Figure 5g). We depicted the reported variant load for 29 proteins that are most affected by non-synonymous alterations in the target peptide regions (Figure 5h). Cancer phospho-circuits revealed widespread perturbations across the phospho-reactome, but also recurrent aberrations that selectively affect specific phospho-hubs, including both proto-oncogenes and tumor suppressor proteins. The diversity of sub-networks connectivity combined with the differential mutational status of target regions catalogued in PhosphoAtlas is a roadmap to explore the strategic kinase-hot spots of cancer.
Since PhosphoAtlas is meant to be a hypothesis-generating platform, users may –for example– investigate the potential relationship between the impact of oncogenic mutations on kinases and their local signaling topology. For instance, upon examining the number of distinct variants of phosphorylation sites per kinase, users may notice that some of the kinases with a very high endogenous nsHPRv mutational load phosphorylate a relatively low number of substrates, and thus seem to have a low level of signaling pleiotropy (e.g. FLT3, KIT, KDR, PDGFRB, BRAF/RAF1; Figure S15). It is tempting to speculate that such kinases may be especially suitable targets for tumorigenesis, possibly not just because they achieve a particular advantageous hallmark of cancer but also because they minimize “collateral damage” which may otherwise lower the fitness of an emerging cancer cell. This speculation seems consistent with observations of the other end of the spectrum, whereby many kinases with very large number of downstream substrates seem to show relatively few or no non-synonymous variants in their phospho-sites (e.g. PRKCA/G/Z, PRKACA, MAPK1/3/14, CDK1/2/5, GSK3A/B, ATM/ATR, SRC/FYN/LYN; Figure S16). Systematic exploration of such hypothetical relationships could be valuable for understanding tumorigenic mechanisms, and brainstorming new therapeutic strategies. More globally, modeling the cancer kinome by weighing in 2nd degree connectivity of upstream / downstream proteins, and the differential susceptibility of specific tumor tissues to particular nsHPRv mutations, may help identify likely driver mutations and/or functional dependencies of cancer. As such, PhosphoAtlas displays the distinctive cancerous perturbations on normal phospho-catalytic networks with the resolution necessary to begin understanding the differential functional impact and integrative consequences of relevant tumor mutations on signaling networks.
Despite the many tumor genomes reported in COSMIC, we found that for a majority of kinase-substrate catalytic interactions no variants were reported (2,238 versus 1,963; Figure 5b). Furthermore, there are significantly fewer mutational variants per amino acid in HPR’s (mean=0.20) than in the entire protein sequence of their substrate of origin and across all protein substrates catalogued in PhosphoAtlas database and recorded in COSMIC (mean=0.36) (mean-difference [95%CI]= −0.16 variants/amino acid [−0.20–0.11]; t-test p-value = 1.62e–13). Although it is possible that mutations in such phosphorylation sites could be fatal to most cells, these observations inspire the question: are not-mutated target sites not altered because they do not provide any selective advantage to tumors, or because their mutation would be so detrimental that cancer cells could not survive? If the latter, and if the node is not essential to normal cells, these observations could point to potential sources of synthetic lethality that may be exploited to design new, safe combinatorial targeted therapies. More generally, it will be important to investigate whether the spectrum of hyper-mutable-to-never-altered phospho-hubs can predict which oncogenic aberrations are most likely to be actionable in tumors.
The other two broad purposes of PhosphoAtlas, which we do not demonstrate in this study, are to assist proteomic data analysis and to support bioassay development and optimization. Projecting data obtained from RPMA/RPPA assays (3, 8) or MS approaches (5–7, 48–53) onto the curated map of kinase-substrate networks may improve assessment of which phospho-circuits are active in biological samples to identify adaptive reprogramming networks associated with cancer response to therapeutic interventions. Information in PhosphoAtlas, especially that from the phospho-residue site data, may also be leveraged for developing and optimizing epitope mapping for (phospho) antibody development, protein engineering, targeted mutagenic and genetic screens, synthetic phage display/yeast-two-hybrid systems, enzymatic assays to test inhibitor libraries, or utilize HPRs as barcodes of kinase-substrate pairs to infer levels of activity of relevant kinases in MS studies. We are currently in the process of implementing a high throughput kinase activity-mapping platform that utilizes biological peptide sequences as experimental sensors to profile the activity of phosphorylation circuits in biological extracts.
In conclusion, PhosphoAtlas database provides a rich resource for the exploration of the human kinome and its tumor variant, and supports a range of applications from the functional mapping of modular networks for systems biology, to assisting in the development of physical or virtual interfaces for oncology and other diseases.
Supplementary Material
Acknowledgments
We would like to thank computational scientists, engineers and specialists Erin Hayes, Chong Huang, Liyun Luo, Brent Nix, Xuanjun Pan, Neil Park, Alexandra Solomon, Zoe Tribur, Changjun Wang, Kevin Wang, Amanda Wu, Zaidao Yu, Elad Ziv, and support teams of programmers and curators.
Funding.
This work was supported by grants from the Friends for an Earlier Breast Cancer Treatment, UCSF Breast Oncology Program Developmental Research Program, T1 Catalyst Award (UL1 TR000004), NIH UCSF SPORE project 2, National Institute Of General Medical Sciences T32GM067547, the National Science Foundation 1144247, a gift from the “Atwaters”, and the Angela and Shu Kai Chan Endowed chair.
Footnotes
Competing interests.
The authors have declared that no competing interests exist.
Author contributions
ZZ.C and J-P.C designed the computational methods and systems for building the centralized and harmonized protein relational database. ZZ.C supervised the processing of the central PhosphoAtlas database. A.O analyzed the content of the database and created the non-synonymous target variant database. A.O, R.H.N, D.W, C.Y, A.P and J-P.C integrated protein networks and created functional circuit displays. R.H.N and J-P.C integrated PhosphoAtlas with Cytoscape. A.O, D.W, R.H.N, E.P.R.L, L.B-S, L.J.vtV and J-P.C wrote the manuscript.
References
- 1.Hunter T. Signaling--2000 and beyond. Cell. 2000;100(1):113–27. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 2.Bernards R. A missing link in genotype-directed cancer therapy. Cell. 2012;151(3):465–8. doi: 10.1016/j.cell.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 3.Akbani R, et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat Commun. 2014;5:3887. doi: 10.1038/ncomms4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stelzl U, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122(6):957–68. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 5.Linding R, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129(7):1415–26. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rikova K, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131(6):1190–203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 7.Zhang B, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513(7518):382–7. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan Y, et al. Assessing the clinical utility of cancer genomic and proteomic data across tumor types. Nat Biotechnol. 2014;32(7):644–52. doi: 10.1038/nbt.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437(7062):1173–8. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 10.Sahni N, et al. Widespread macromolecular interaction perturbations in human genetic disorders. Cell. 2015;161(3):647–60. doi: 10.1016/j.cell.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niedner RH, et al. Protein kinase resource: an integrated environment for phosphorylation research. Proteins. 2006;63(1):78–86. doi: 10.1002/prot.20825. [DOI] [PubMed] [Google Scholar]
- 12.Croft D, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42(Database issue):D472–7. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orchard S, et al. Protein interaction data curation: the International Molecular Exchange (IMEx) consortium. Nat Methods. 2012;9(4):345–50. doi: 10.1038/nmeth.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Z, et al. GeneSense: a new approach for human gene annotation integrated with protein-protein interaction networks. Sci Rep. 2014;4:4474. doi: 10.1038/srep04474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szklarczyk D, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–52. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu J, et al. Integrated network analysis platform for protein-protein interactions. Nat Methods. 2009;6(1):75–7. doi: 10.1038/nmeth.1282. [DOI] [PubMed] [Google Scholar]
- 17.Licata L, et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40(Database issue):D857–61. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chatr-Aryamontri A, et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43(Database issue):D470–8. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keshava Prasad TS, et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37(Database issue):D767–72. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buzko O, Shokat KM. A kinase sequence database: sequence alignments and family assignment. Bioinformatics. 2002;18(9):1274–5. doi: 10.1093/bioinformatics/18.9.1274. [DOI] [PubMed] [Google Scholar]
- 21.Manning G, et al. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 22.Krupa A, Abhinandan KR, Srinivasan N. KinG: a database of protein kinases in genomes. Nucleic Acids Res. 2004;32(Database issue):D153–5. doi: 10.1093/nar/gkh019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milanesi L, et al. Systematic analysis of human kinase genes: a large number of genes and alternative splicing events result in functional and structural diversity. BMC Bioinformatics. 2005;6(Suppl 4):S20. doi: 10.1186/1471-2105-6-S4-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hornbeck PV, et al. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43(Database issue):D512–20. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang CY, et al. PhosphoPOINT: a comprehensive human kinase interactome and phospho-protein database. Bioinformatics. 2008;24(16):i14–20. doi: 10.1093/bioinformatics/btn297. [DOI] [PubMed] [Google Scholar]
- 26.Huang KY, et al. RegPhos 2.0: an updated resource to explore protein kinase-substrate phosphorylation networks in mammals. Database (Oxford) 2014;2014(0):bau034. doi: 10.1093/database/bau034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dinkel H, et al. Phospho.ELM: a database of phosphorylation sites--update 2011. Nucleic Acids Res. 2011;39(Database issue):D261–7. doi: 10.1093/nar/gkq1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forbes SA, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database issue):D805–11. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22(9):1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huntley RP, et al. The GOA database: gene Ontology annotation updates for 2015. Nucleic Acids Res. 2015;43(Database issue):D1057–63. doi: 10.1093/nar/gku1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brinkworth RI, Breinl RA, Kobe B. Structural basis and prediction of substrate specificity in protein serine/threonine kinases. Proc Natl Acad Sci U S A. 2003;100(1):74–9. doi: 10.1073/pnas.0134224100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar ND, Prakash Nikhil, Mohanty Debasisa. Getting Phosphorylated: Is it Necessary to be Solvent Accessible? Proc Indian Natn Sci Acad. 2015;81(2):493–507. [Google Scholar]
- 34.Miller Martin L, ER, Gauthier Nicholas P, Aksoy Bülent Arman, Korkut Anil, Gao Jianjiong, Ciriello Giovanni, Schultz Nikolaus, Sanderemail Chris. Pan-Cancer Analysis of Mutation Hotspots in Protein Domains. Cell Systems. 2015;1(3):197–209. doi: 10.1016/j.cels.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krogan NJ, et al. The Cancer Cell Map Initiative: Defining the Hallmark Networks of Cancer. Mol Cell. 2015;58(4):690–698. doi: 10.1016/j.molcel.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ciriello G, et al. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45(10):1127–33. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leiserson MDM, et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet. 2015;47(2):106–14. doi: 10.1038/ng.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kandoth C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–9. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Supek F, et al. Synonymous mutations frequently act as driver mutations in human cancers. Cell. 2014;156(6):1324–35. doi: 10.1016/j.cell.2014.01.051. [DOI] [PubMed] [Google Scholar]
- 41.Jerby-Arnon L, et al. Predicting cancer-specific vulnerability via data-driven detection of synthetic lethality. Cell. 2014;158(5):1199–209. doi: 10.1016/j.cell.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 42.Babur Ö, et al. Systematic identification of cancer driving signaling pathways based on mutual exclusivity of genomic alterations. Genome Biol. 2015;16:45. doi: 10.1186/s13059-015-0612-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gatenby RA, Cunningham JJ, Brown JS. Evolutionary triage governs fitness in driver and passenger mutations and suggests targeting never mutations. Nat Commun. 2014;5:5499. doi: 10.1038/ncomms6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoadley KA, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158(4):929–44. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pan Y, et al. Human germline and pan-cancer variomes and their distinct functional profiles. Nucleic Acids Res. 2014;42(18):11570–88. doi: 10.1093/nar/gku772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu J, Li Y, Jiang R. Integrating multiple genomic data to predict disease-causing nonsynonymous single nucleotide variants in exome sequencing studies. PLoS Genet. 2014;10(3):e1004237. doi: 10.1371/journal.pgen.1004237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitra K, et al. Integrative approaches for finding modular structure in biological networks. Nat Rev Genet. 2013;14(10):719–32. doi: 10.1038/nrg3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huttlin EL, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143(7):1174–89. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127(3):635–48. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 50.Wilhelm M, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509(7502):582–7. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 51.Kim MS, et al. A draft map of the human proteome. Nature. 2014;509(7502):575–81. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 53.Huttlin EL, et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell. 2015;162(2):425–40. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.