Abstract
An improved pre-clinical cardiac chemical exchange saturation transfer (CEST) pulse sequence (cardioCEST) was used to selectively visualize paramagnetic CEST (paraCEST)-labeled cells following intramyocardial implantation. In addition, cardioCEST was used to examine the effect of diet-induced obesity upon myocardial creatine CEST contrast. CEST pulse sequences were designed from standard turbo-spin-echo and gradient-echo sequences, and a cardiorespiratory-gated steady-state cine gradient-echo sequence. In vitro validation studies performed in phantoms composed of 20mM Eu-HPDO3A, 20mM Yb-HPDO3A, or saline demonstrated similar CEST contrast by spin-echo and gradient-echo pulse sequences. Skeletal myoblast cells (C2C12) were labeled with either Eu-HPDO3A or saline using a hypotonic swelling procedure and implanted into the myocardium of C57B6/J mice. Inductively coupled plasma mass spectrometry confirmed cellular levels of Eu of 2.1 × 10−3 ng/cell in Eu-HPDO3A-labeled cells and 2.3 × 10−5 ng/cell in saline-labeled cells. In vivo cardioCEST imaging of labeled cells at ±15ppm was performed 24 h after implantation and revealed significantly elevated asymmetric magnetization transfer ratio values in regions of Eu-HPDO3A-labeled cells when compared with surrounding myocardium or saline-labeled cells. We further utilized the cardioCEST pulse sequence to examine changes in myocardial creatine in response to diet-induced obesity by acquiring pairs of cardioCEST images at ±1.8 ppm. While ventricular geometry and function were unchanged between mice fed either a high-fat diet or a corresponding control low-fat diet for 14 weeks, myocardial creatine CEST contrast was significantly reduced in mice fed the high-fat diet. The selective visualization of paraCEST-labeled cells using cardioCEST imaging can enable investigation of cell fate processes in cardioregenerative medicine, or multiplex imaging of cell survival with imaging of cardiac structure and function and additional imaging of myocardial creatine.
Keywords: MRI, chemical exchange saturation transfer, cell tracking, metabolic imaging, obesity
INTRODUCTION
Fluorescence microscopy has developed into a ubiquitous tool for multi-color reporter gene and cell tracking imaging in ex vivo tissue sections; however, light scattering, limited depth of penetration, and the need to register molecular information to anatomical images limit in vivo cardiac application. Chemical exchange saturation transfer (CEST) MRI has emerged over the last decade as a novel method for in vivo molecular imaging based upon the exchange of saturated protons with surrounding mobile water protons (1–3). The frequency-specific saturation of endogenous (e.g. fibrotic substrate, glucose, creatine (4–8)) or exogenous CEST targets (e.g. paramagnetic CEST (paraCEST) contrast agents (9–14) or MRI reporter genes (15–18)) and subsequent exchange enables the selective activation and visualization of contrast from multiple CEST targets without disruption of underlying image integrity. CEST-MRI is non-invasive, does not require ionizing radiation, is not limited by light scattering or depth of penetration, and enables the acquisition of molecular images that are automatically registered to anatomical detail. CEST-MRI has been performed almost exclusively in stationary organs and tissues with two dominant themes focused on (i) imaging of endogenous targets such as amine proton transfer or creatine (6,10,19–21) or (ii) imaging and tracking of populations of CEST active cells in pre-clinical models (9,11,16,17,22). The application of CEST-MRI to cardiac research has great potential for tracking cell fate decisions in cell therapy, or for non-invasive tissue and metabolic characterization. However, conventional CEST imaging pulse sequences overwhelmingly utilize spin-echo image acquisitions, which are rendered inapplicable in the rapidly beating small animal heart.
In a prior study, we developed a free-breathing retrospectively cardiorespiratory-gated CEST pulse sequence (cardioCEST) and described its application to imaging of endogenous fibrotic substrate and the myocardial redistribution kinetics of the exogenous paraCEST contrast agent Eu-HPDO3A (7). In the current study, we refine the pulse sequence design for improved cardioCEST imaging and demonstrate its utility for the prevailing themes in CEST-MRI: endogenous creatine imaging and CEST cell tracking. We validate our new sequence against the spin-echo standard in paraCEST phantoms and demonstrate similar CEST contrast using the two methods. We subsequently utilize cardioCEST to image Eu-HPDO3A-labeled cells following cardiac transplantation in mice. Finally, we perform cardiac creatine CEST imaging to image the impact of diet-induced obesity on myocardial creatine in a model of preserved systolic function.
METHODS
CardioCEST pulse sequence design
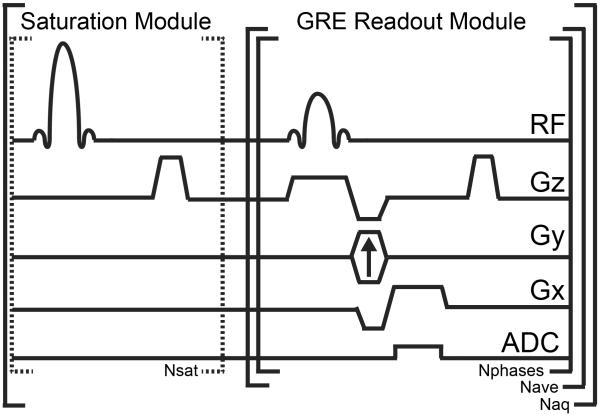
A pulse sequence diagram for cardioCEST is shown in Fig. 1. CEST encoding used a train of frequency selective and spatially non-selective Gaussian saturation pulses (bandwidth = 200 Hz, duration = 8.8 ms, number of pulses = 196, time between pulses = 2.03 ms; for additional information regarding optimization of saturation module see Supplementary Methods). Immediately after the conclusion of saturation, RF excitation pulses at a constant repetition time are used to encode the change in initial longitudinal magnetization due to saturation transfer into the steady state longitudinal magnetization. Combined respiratory and electrocardiogram gating is employed to trigger the acquisition of cine gradient echo images, with dummy pulses (same flip angle and repetition time as excitation pulses) continuing at the end of the cardiac interval in order to maintain steady state conditions between triggers and during periods of respiratory motion (see Supplementary Methods for more detail). The number of images (Nphases) acquired is set dynamically based on the cardiac interval at the time of imaging, with subsequent RF excitation performed at a constant repetition time without ADC activation until the subsequent ECG trigger (i.e. dummy pulses). In order to ensure consistent CEST weighting across all lines in k-space, n averages of one phase-encode step are acquired for each cardiac phase after each saturation period (Nave). All imaging was performed on a 7 T horizontal dedicated small animal scanner (ClinScan, Bruker BioSpin MRI, Ettlingen, Germany) running the Siemens VB15 front end (Siemens Medical Solutions, Erlangen, Germany) and using a cylindrical volume coil for excitation and a four-channel phased array surface coil for signal detection.
Figure 1.
Pulse sequence diagram for cardioCEST. CEST contrast is generated via a train (Nsat) of spatially non-selective saturation pulses tuned to a specific offset frequency. The gradient echo readout module is used to acquire cine images with N cardiac phases (Nphases) following each electrocardiogram trigger. After N phases, the readout module is repeated at a constant repetition time but data is not acquired until the subsequent electrocardiogram trigger (dummy pulses). Following execution of the saturation module, the GRE readout module is executed Nave times for one line of k-space and the results subsequently averaged prior to image reconstruction. The saturation and readout modules are repeated Naq times to acquire all phase-encode steps. Both the offset frequency and the power of the saturation module can be adjusted to image specific molecular targets.
Phantom validation experiments
Eu-HPDO3A and Yb-HPDO3A complexes (provided as white crystalline solids by Professor Silvio Aime, Molecular Imaging Center, University of Torino, Italy) were diluted in distilled H2O to a concentration of 20 mM, and 50 μL aliquots were collected in 5mm borosilicate glass tubes. The tubes were suspended in 3% low-gelling-temperature agar (Sigma Aldrich, St. Louis, MO, USA) and positioned horizontally for imaging. Z spectra were obtained using both a modified turbo-spin-echo pulse sequence (TR/TE = 3 s/12 ms, echo train length following each saturation period = 41 using a centric-out encoding scheme) and a modified gradient-echo FLASH sequence (TR/TE = 10 ms/2.2 ms, flip angle = 15°, number of readouts/saturation = 50) with identical CEST preparation (flip angle = 1080°, B1average=20 μT, bandwidth = 200 Hz, pulse duration = 8.8 ms, number of pulses=196, inter-pulse duration=2.03 ms, total duration=2123 ms). The total acquisition time was 139 s per image for the modified spin-echo sequence, and 18 s per image for the modified gradient-echo sequence. For all acquired images across both sequences, imaging parameters were maintained including FOV=2.50×2.50 cm, matrix=192×192, slice thickness=1mm, and number of averages=3 (Nphases=1, Nave=3, Naq=192). Images were obtained with saturation offsets of 0, ±[1, 2, 3, 5, 9, 12, 15, 17 – 20 (increment 0.2ppm), 21, 23, 25, 30, 40, 50, 60, 64, 65 – 67 (increment 0.2ppm), 68, 69, 70, 75, 85, 90, 94 – 96 (increment 0.2 ppm), 97, 99, 100, 110, 120], and were normalized to an image acquired with identical imaging parameters except a saturation flip angle of 1° and offset of −300 ppm.
Cell culture
Immortalized mouse skeletal myoblasts (C2C12) were grown at 37 °C and 5% CO2 in 10 mL Dulbecco's modified Eagle's medium (DMEM) (Life Technologies, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals, Lawrenceville, GA, USA) and 1% penicillin–streptomycin (10 000 U/mL) (Life Technologies). Cultures were grown to 80–90% confluence in 100mm tissue culture dishes to avoid differentiation of C2C12 myoblasts into myotubes.
Cell labeling
C2C12 cells were labeled with either Eu-HPDO3A or saline using a hypotonic swelling technique as described by Di Gregorio et al. (23). Briefly, cells were washed twice with sterilized (1×) phosphate-buffered saline (PBS), harvested using 1.5mL trypsin–EDTA solution (Life Technologies), and isolated following centrifugation (1000 rpm for 5min). Afterwards, cells were counted, divided as 3 × 106 cells per 1mL (Falcon tube Corning, NY, USA), and again centrifuged with subsequent removal of supernatant. Afterwards, 500 μL (160mOsm) of a hypotonic solution (100 μL of Ln-HPDO3A stock (500mM), 107 μL 1× PBS, and 293 μL distilled H2O) was added to each Falcon tube. Cells were incubated in a dry bath at 37 °C for 30min with agitation every 10min, after which 500 μL of hypertonic/restoring solution (400mOsm/L 1× PBS) was added and incubated at 37 °C for 30 min with agitation every 10min. Unincorporated contrast agent was removed by washing the cells three times in 1× PBS, after which cells were re-suspended in 50 μL 1× PBS prior to further use.
Inductively coupled plasma mass spectrometry analysis
Freeze-dried cells were digested in concentrated trace-metal-free HNO3 using a MARS Xpress microwave digestion system (CEM, Matthews, NC, USA) according to U.S. Environmental Protection Agency (USEPA) (1996). Total Eu concentrations in the digestates were determined by inductively coupled plasma mass spectrometry (ICP-MS) following USEPA (1998) using an Agilent 7500cx (Agilent Technologies, Santa Clara, CA, USA). Digestion sets included cells labeled with Eu-HPDO3A, cells exposed to hypotonic swelling with saline, and control cells. Duplicate digestions and reagent blanks were also included with each digestion set. Analytical runs included duplicate dilutions, spike recovery samples, and inter-calibration–cross-calibration verification samples using traceable standards from an independent lot. The ICP-MS was calibrated using a certified reference standard containing Eu from Ultra Scientific (catalogue no IMS-101, Kingstown, RI, USA). The calibration curve was validated by comparison to an independent lot of the same standard. The internal standard (Tb, catalogue no MSTB-10PPM-125ML) was a certified reference standard obtained from Inorganic Ventures (Christiansburg, VA, USA).
In vivo cardioCEST cell tracking
Eight male C57BL6/J mice (8 weeks of age) were purchased from the Jackson Laboratory (Bar Harbor, ME, USA). Experiments were carried out according to the NIH Guidelines on the Use of Laboratory Animals, and all procedures were approved by the University of Kentucky's Institutional Animal Care and Use Committee. Cardiac implantation of the labeled cells was carried out using the “pop-out” technique as described by Gao and Koch (24). Briefly, mice were anesthetized and maintained with 2% isoflurane in O2 and a small skin incision was made over the left chest. After dissection of the pectoral muscles to expose the fourth intercostal space, a small hole was generated with a mosquito clamp to open the pleural membrane. The heart was gently pressed towards the surface while maintaining pressure on the thorax. Cells were injected into the anterior left ventricular mid-wall using a 27 gauge needle and an injection volume of 10 μL. Following cell injection, the heart was immediately placed back into the intra-thoracic space, followed by evacuation of the air and closure of the skin incision to prevent pneumothorax. The mice were removed from anesthesia and allowed to recover. Using this technique, about 1 × 106 cells labeled with either Eu-HPDO3A (n = 4) or saline (n = 4, control) were implanted into the anterior wall of the left ventricle.
In vivo cardioCEST imaging was performed 24 h after cell implantation. Anesthesia was maintained using 1.5% isoflurane in oxygen and core body temperature was maintained using circulating constant-temperature water. ECG and respiratory gating used a Small Animal Instruments (Stony Brook, NY, USA) system. Once core temperature was stabilized at 37 °C, pairs of cardioCEST images were acquired with saturation offsets of ±15 ppm in two slices: one at the site of injection and one as an internal control 2mm apical to the injection site as identified on long axis images based on disruption of the pericardium. Specific imaging parameters included FOV = 2.56 × 2.56 cm, matrix=256×128 (Naq=128), slice thickness=1mm, TR/TE=6.92ms/3.42 ms, number of averages (Nave) = 4, and flip angle = 15°. The average value for Nphases was 15 ± 3 across all acquisitions. Saturation parameters were identical to those of in vitro studies. The total acquisition time was 2.5–3 min per cine image, or 5–6 min per image pair.
Myocardial creatine CEST
Ten C57BL6/J male mice randomly assigned at 8 weeks of age to either a high-fat diet (HFD, n = 5) consisting of 60% kilocalories from fat (Research Diets no D12492, New Brunswick, NJ, USA) or a low-fat diet (LFD, n = 5) consisting of 10% kilocalories from fat (Research Diets no D12450B) were examined using cardioCEST imaging. Imaging of myocardial creatine CEST contrast was performed via acquisition of pairs of cardioCEST images with saturation offsets of ±1.8 ppm in one mid-ventricular slice. All imaging parameters were identical to those used for cell tracking; however, based on preliminary optimization results (see supplemental methods), and based on the findings of Haris et al. (4), the saturation flip angle was reduced to 270° (B1average = 4.9 μT) to minimize indirect saturation of water. The average value of Nphases was 18 ± 2. Separately, a reference image was acquired without CEST preparation, from which septal wall thickness (h), the ratio of h to inner ventricular radius (h/r), and fractional shortening measured as the percent change in inner diameter between diastole and peak systole were calculated.
Analysis of CEST images
All image processing was performed in MATLAB (MathWorks, Natick, MA, USA) using custom-written software. CEST contrast was quantified as the asymmetric magnetization transfer ratio (MTRasym): MTRasym = (Sconjugate−SCESTresonance)/Sconjugate × 100(%), where S represents the signal intensity of a given pixel when saturation is applied at either the resonant frequency of the CEST target (CESTresonance) or the conjugate frequency (conjugate). In order to account for differences in heart position, MTRasym maps were calculated from matching end-diastolic phases. For creatine-CEST imaging, histograms of myocardial MTRasym values were calculated in MATLAB encompassing all voxel MTRasym values within the myocardium.
Statistical analysis
Statistical analysis was performed in JMP 11.1.1 (SAS Institute, Cary, NC, USA). MTRasym values in phantom studies were compared using a two-way analysis of variance (ANOVA). Differences in MTRasym, fractional shortening, wall thickness, h/r ratio, body mass, heart rate, respiratory rate, and body temperature between HFD and LFD mouse cohorts were assessed using a one-way ANOVA. For analysis of cell tracking data, the mice were treated as experimental units, and percent voxel measurements from slices within the same mouse and group were averaged. Although there were repeated observations within a single mouse between the slices containing Eu-labeled cells and the adjacent control slice, a repeated measures ANOVA could not be performed due to small sample sizes. Instead, assuming normality of the data, two-sample t-tests with unequal group variances were used to compare the control–Eu-labeled and control–Eu-control groups at each of the three potential threshold values of MTRasym for the detection of Eu-HPDO3A-labeled cells. To compare observations containing Eu-labeled cells with adjacent control slices in the same mice, we performed a paired t-test at each percent voxel cutoff using the three observations. These analyses were performed in SAS (SAS Institute, Cary, NC, USA).
RESULTS
Phantom validation experiments
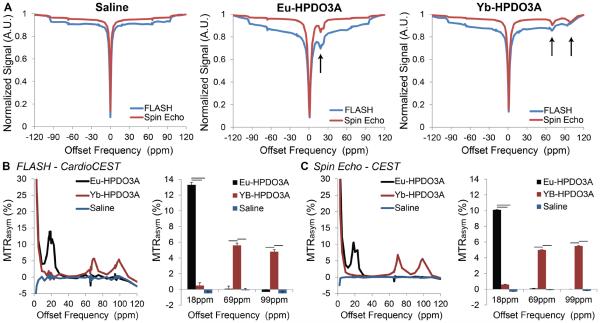
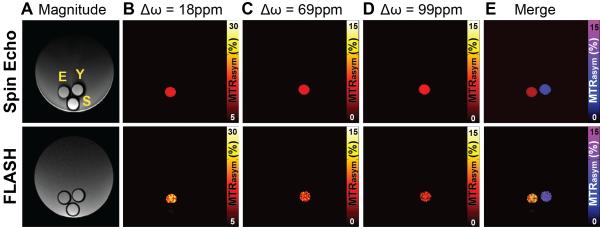
Z spectra acquired using FLASH–cardioCEST and spin-echo CEST sequences in paraCEST and saline phantoms are shown in Fig. 2. Spectra from both sequences are uniform for saline and demonstrated asymmetric saturation transfer for Eu-HPDO3A between 15 and 20 ppm, as well as dual asymmetric saturation transfer for Yb-HPDO3A in the range of 70–100 ppm (Fig. 2). Spectra of MTRasym demonstrate similar identification of CEST signatures for Eu-HPDO3A and Yb-HPDO3A using both sequences (Fig. 2). A significantly elevated MTRasym, when CEST preparation was performed at ±18 ppm, was observed only in Eu-HPDO3A phantoms in both sequences (Fig. 2). Similarly, significantly elevated MTRasym was observed only in Yb-HPDO3A phantoms when CEST preparation was performed at ±69 ppm and ±99 ppm (Fig. 2). Representative MTRasym maps generated using both sequences (Fig. 3) further demonstrate the ability to selectively visualize each paraCEST agent at its labile proton's resonant frequency (Fig. 3). The signal to noise ratio for Eu-HPDO3A, Yb-HPDO3A, and saline phantoms was significantly lower for cardioCEST (74.2, 78.5, 98.4, respectively) compared with spin-echo CEST (665.3, 701.1, 949.3, respectively) measurements. By assigning arbitrary colors to each offset, similar multi-color merged MTRasym maps were constructed using both pulse sequences (Fig. 3).
Figure 2.
Phantom validation experiments. (A) Z spectra acquired using conventional spin-echo CEST imaging (red) and FLASH–cardioCEST imaging (blue) in phantoms containing either saline (left), 20 mM Eu-HPDO3A (middle), or 20 mM Yb-HPDO3A (right). Saturation module parameters and the duration of readout following saturation were identical for both methods. Asymmetry in Z spectra reflecting CEST contrast in phantoms containing Eu-HPDO3A or Yb-HPDO3A is highlighted by black arrows. Slight deviations near the resonant frequencies of Eu/Yb-HPDO3A in non-corresponding phantoms likely result from increased sampling density about those frequencies. (B, C) MTRasym data from spectra acquired using FLASH–cardioCEST (B) and spin-echo CEST (C) methods revealed similar unique peaks for Eu-HPDO3A and Yb-HPDO3A, with no CEST contrast in saline phantoms. Data in bar graphs is represented as mean + standard deviation; black bars indicate p<0.05.
Figure 3.
Spin-echo and FLASH–cardioCEST imaging of multiple paraCEST agents. (A) Magnitude reconstructed images acquired using spin-echo and FLASH sequences reveal the location of phantoms containing Eu-HPDO3A (E), Yb-HPDO3A (Y), or saline (S). The larger external phantom is composed of 3% agarose. (B) MTRasym maps generated from saturation offsets (Δω) of 18 ppm selectively reveal the phantom containing Eu-HPDO3A for both acquisition methods. (C, D) Similarly, MTRasym maps generated at offsets of 69 ppm (C) and 99 ppm (D) selectively reveal the phantom containing Yb-HPDO3A. (E) The merger of MTRasym maps at 18 ppm (red–yellow color bar, same as B) and 69 ppm (blue–purple color bar) demonstrate similar multi-color imaging capabilities using conventional spin-echo CEST and FLASH–cardioCEST.
CardioCEST cell tracking
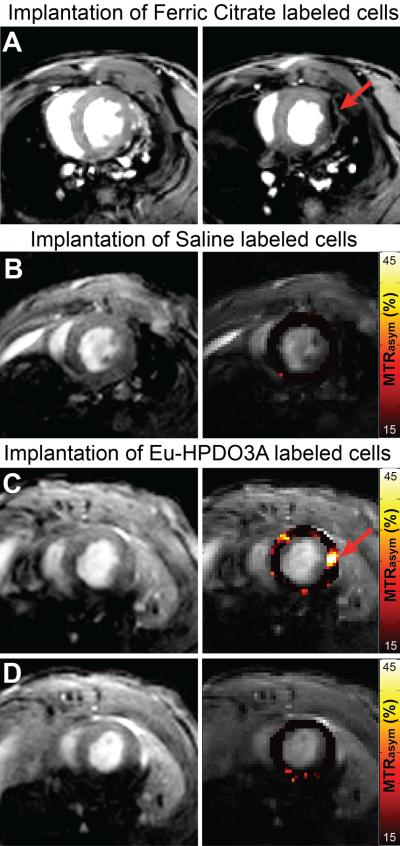
We examined the suitability of cardioCEST imaging to visualize paraCEST-labeled cells following myocardial implantation. Representative MTRasym maps generated following CEST preparation at ±15 ppm are shown in Fig. 4 for mice receiving saline-labeled cells and Eu-HPDO3A-labeled cells. Significantly heightened MTRasym values compared with surrounding myocardium (red arrow), including adjacent slices without injected cells (Fig. 4), were generated by cells labeled with Eu-HPDO3A. The location of elevated MTRasym generated by Eu-HPDO3A-labeled cells was confirmed using ex vivo histological staining in all samples (Fig. 5). In juxtaposition, the significant CEST contrast in cardioCEST-generated MTRasym maps was not generated by saline-labeled cells (Fig. 4). The fraction of voxels demonstrating elevated MTRasym values was significantly higher in slices containing Eu-HPDO3A-labeled cells compared with saline-labeled cells and adjacent control slices across a number of MTRasym threshold values (Table 1). ICP-MS revealed cellular levels of Eu of 2.1 × 10−3 ng/cell in Eu-HPDO3A-labeled cells and 2.3 × 10−5 ng/cell in saline-labeled cells, indicating a background error of 1% in labeled cells.
Figure 4.
In vivo cardioCEST tracking of Eu-HPDO3A-labeled cells. (A) End-diastolic images acquired following the intramyocardial implantation of ferric-citrate-labeled (see supplemental data) C2C12 cells are shown for reference purposes. In comparison with a mid-ventricular slice without labeled cells (left), the adjacent slice containing labeled cells (right) demonstrates severe signal loss (red arrow) at the location of labeled cells along the lateral wall of the left ventricle. (B) Magnitude (left) and superimposed left ventricular MTRasym maps (right) generated using cardioCEST reveal uniformly low MTRasym patterns in a mouse receiving saline-labeled cells. (C) In a mouse receiving cells labeled with Eu-HPDO3A, the location of transplanted cells (red arrow) demonstrates significantly elevated MTRasym values. The location of labeled cells was confirmed using ex vivo histological staining (see Supplementary Figure 1). (D) Similar mapping of MTRasym in an adjacent slice 2 mm towards the apex reveals the absence of Eu-HPDO3A-labeled cells. All MTRasym maps are displayed using a threshold of MTRasym >15%.
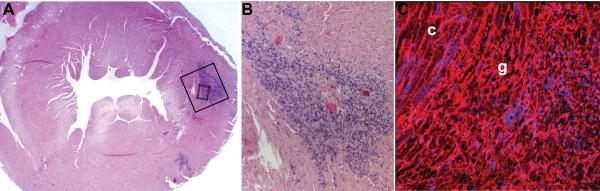
Figure 5.
Histological confirmation of transplanted cell location. (A) Hematoxylin and eosin staining of a mid-ventricular tissue section (corresponding to the MTRasym map shown in Figure 4C) reveals the presence of implanted C2C12 cells in the lateral wall of the left ventricle. (B) Higher magnification of the area in the solid black box in A reveals a dense area of implanted cells (dark blue) between areas of preserved myocardium (pink). (C) Wheat germ agglutinin staining with nuclear counter-staining of the area in the small dashed box in A reveals the presence of disorganized engrafted cells (g) adjacent to areas of preserved cardiomyocyte structure (c).
Table 1.
Distribution of MTRasym values in cardioCEST cell tracking study. The fraction of voxels with MTRasym above each threshold value was significantly higher in slices containing Eu-HPDO3A-labeled cells than in corresponding control slices 2mm away within the same heart at a threshold value of MTRasym>30%, or in mid-ventricular slices in hearts with transplanted saline-labeled cells across all thresholds. The use of a high saturation flip angle likely contributed to the fraction of voxels with MTRasym>5% in all hearts. The significant elevation in slices containing Eu-HPDO3A-labeled cells is likely caused by the release of Eu-HPDO3A from apoptotic cells and the subsequent retention in the interstitial space. All values are presented as mean ± SD
| Fraction of voxels with MTRasym>5% | Fraction of voxels with MTRasym>15% | Fraction of voxels with MTRasym>30% | |
|---|---|---|---|
| Slice containing saline-labeled cells | 19.1 ± 10.6 (%) | 8.1 ± 6.3 (%) | 0.7 ± 1.4 (%) |
| Slice adjacent to Eu-HPDO3A-labeled cells | 25.0 ± 8.1 (%) | 8.1 ± 2.7 (%) | 0.4 ± 0.5 (%) |
| Slice containing Eu-HPDO3A-labeled cells | 61.3 ± 6.1 (%)* | 34.2 ± 4.4 (%)* | 8.9 ± 2.3 (%)*† |
P<0.05 versus saline-labeled cells,
P<0.05 versus adjacent slice
Myocardial creatine CEST imaging
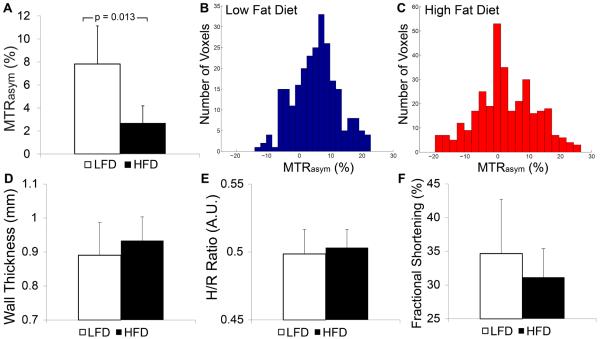
Mice fed an HFD had significantly elevated body mass at 14 weeks after initiation of the diet in comparison with mice fed a control LFD (Table 2). Representative MTRasym maps acquired using cardioCEST with preparation at ±1.8 ppm shown in Fig. 6 reveal broadly reduced creatine MTRasym in the heart of a mouse on an HFD in comparison with the heart of a mouse on an LFD control. Imaging of myocardial creatine using cardioCEST revealed significantly reduced creatine MTRasym in the hearts of mice fed an HFD for 14 weeks compared with those fed a corresponding control LFD (Fig. 7). Representative histograms of MTRasym distribution in the hearts of mice fed an LFD and an HFD are shown in Fig. 7. In mice on an HFD, the distribution of MTRasym values was consistently greater, with a lower median value, and a larger percentage of negative values compared with these values for mice on an LFD (Fig. 7). Finally, despite differences in creatine MTRasym, septal wall thickness, the h/r ratio, and fractional shortening were all similar between mice fed an HFDand an LFD (Fig. 7).
Table 2.
Mouse characteristics and imaging hemodynamics during creatine CEST imaging. Mice fed an HFD demonstrated significantly elevated body mass and respiratory rate but reduced heart rate compared with LFD mice
| LFD (n=5) | HFD (n=5) | ||
|---|---|---|---|
| Body mass (g) | 31 ± 2 | 45 ± 2 | P<0.05 |
| Heart rate (beats/min) | 459 ± 40 | 379 ± 12 | P<0.05 |
| Respiratory rate (breaths/min) | 86 ± 11 | 108 ± 16 | P<0.05 |
| Temperature (°C) | 36.3 ± 0.9 | 36.7 ± 0.4 | P=NS |
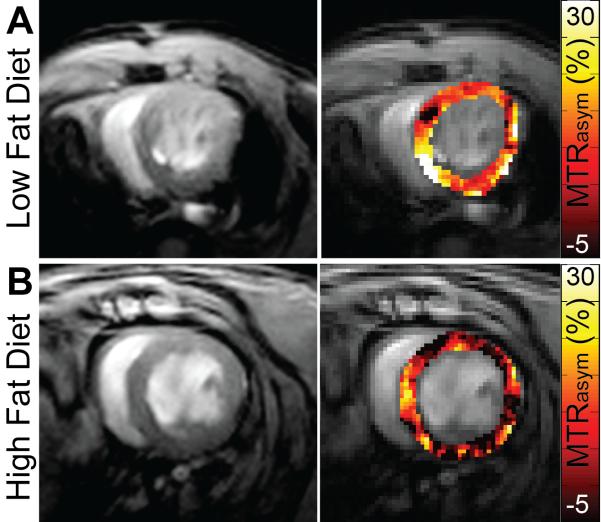
Figure 6.
CardioCEST imaging of creatine in the setting of diet-induced obesity. Representative magnitude reconstructed images (left) and MTRasym maps (right) acquired using cardioCEST with saturation offsets of ±1.8 ppm reflect CEST contrast generated by creatine. (A) Mice fed a control LFD demonstrate robust creatine CEST contrast throughout the left ventricle. (B) In comparison, mice fed an HFD for 14 weeks demonstrate similar ventricular structure but attenuated creatine CEST contrast as observed through reduced MTRasym at 1.8 ppm.
Figure 7.
Metabolic changes in response to diet-induced obesity occur in the absence of structural or functional changes. (A) Average myocardial creatine MTRasym values (1.8 ppm) were significantly reduced in mice fed an HFD for 14 weeks compared with LFD control mice. (B, C) Representative histograms of voxel MTRasym values from an LFD control mouse (B) and an HFD mouse (C) reveal a greater distribution of MTRasym values in mice fed an HFD. (D–F) Changes in myocardial creatine CEST were not accompanied by changes in important parameters of ventricular structure including septal wall thickness (D, p = NS) and the ratio of septal wall thickness to the inner left ventricular radius (E, h/r, p = NS), or ventricular function as assessed by fractional shortening (F, p = 0.4). All data is represented as mean + standard deviation.
DISCUSSION
In this study, an improved cardioCEST pulse sequence design for CEST imaging in the rapidly beating mouse heart was designed and validated with in vitro detection of Eu-HPDO3A and Yb-HPDO3A compared with a conventional spin-echo CEST approach. The major findings of this study are that (i) cardioCEST enabled the selective visualization of cells labeled with paraCEST agents following myocardial implantation in a mouse model of cardiac cell therapy, and (ii) cardioCEST imaging of creatine CEST contrast identified reduced myocardial creatine in response to diet-induced obesity, and prior to the onset of morphological and functional changes in the left ventricle. These findings respond to the dominant themes in the rapidly growing CEST-MRI field of imaging of endogenous markers such as creatine, and the use of CEST-MRI for cell tracking by demonstrating application in pre-clinical cardiac models.
In a previous cardioCEST study, we used a free-breathing and free-beating acquisition scheme with retrospective filtering of k-space data based on reconstructed respiratory and cardiac waveforms (7). Although CEST weighting of the initial longitudinal magnetization was encoded into the steady-state longitudinal magnetization, the effective saturation time varied between lines of k-space and led to potential image blurring. In addition, retrospective reconstruction resulted in slight differences in the number of averages between pairs of cardioCEST images. In the current study, several key modifications to the pulse sequence design of cardioCEST were implemented in order to eliminate such problems. First, although dummy pulses were used immediately after the conclusion of saturation and repeated at constant repetition times in order to maintain steady-state encoding of CEST weighting, the acquisition of image data was triggered prospectively based on respiratory and ECG waveforms. Retrospective triggering was obviated using this approach, which ensured a consistent number of averages for each line of k-space across all images. Second, the same phase-encode position was acquired Nave times over consecutive heart beats. This feature was used to ensure a consistent mean saturation time for each line of k-space across all cardiac phases and between CEST image pairs. More consistent CEST encoding between image pairs and a decrease in the potential confounding influence of cardiac and respiratory rate instability between acquisitions were afforded by these two modifications.
We compared our improved cardioCEST sequence with widely used spin-echo CEST methods in phantoms containing the paraCEST contrast agents Eu-HPDO3A and Yb-HPDO3A. Z spectra were obtained from Eu-HPDO3A, Yb-HPDO3A, and saline phantoms using spin-echo CEST. Similar spectral behaviors of these Z spectra were demonstrated relative to those generated in a prior study by Ferrauto et al. (11). Differences in normalized signal intensity as a function of frequency and the degree of saturation transfer in Eu/Yb-HPDO3A phantoms between our study and that of Ferrauto et al. (11) was caused by differences in saturation power between studies. However, spectra of MTRasym generated from spin-echo CEST data were shown to agree closely in both studies. Both Z spectra and MTRasym spectra obtained using FLASH–cardioCEST revealed similar characteristics for Eu-HPDO3A and Yb-HPDO3A when compared with spin-echo CEST imaging (Fig. 2). In addition, similar MTRasym maps for both phantoms at specific offset frequencies (Fig. 3) were generated by the two techniques, and enabled multi-color imaging. Finally, the mean MTRasym measured by FLASH–cardioCEST in a phantom containing Eu-HPDO3A was shown to be moderately higher than that measured using spin-echo CEST in the range of 15–20 ppm. This difference is likely attributable to preservation of the initial CEST weighting in the steady-state longitudinal magnetization in FLASH–cardioCEST, compared with a loss of CEST weighting with T2 relaxation in spin-echo CEST (for additional explanation see Supplementary Methods). However, it is important to note that the use of a gradient-echo readout resulted in a larger standard deviation in MTRasym measurements compared with a spin-echo readout (Fig. 3).
MRI tracking of cells transplanted into the murine heart in models of cell therapy has been performed in a number of prior studies, primarily using cells labeled with a variety of iron oxide nanoparticles (25–28). Although signal voids generated by such cells were easily visualized on T2/T2*-weighted images, the negative contrast was difficult to distinguish from susceptibility artifact and obstructed the possibility of acquiring additional imaging information. In contrast, labeling of cells with certain paraCEST agents was not found to disrupt the underlying image contrast (Fig. 4). In addition, the entrapment of iron oxide nanoparticles within tissues following cell death has been previously demonstrated to lead to prolonged false-positive signals of cell survival (29). In contrast, chelated lanthanide metals wash out over time, similarly to chelated gadolinium salts (20). The selective visualization of paraCEST-labeled cells can potentially enable imaging of multiple cell populations, and allow for additional MRI characterization of tissue surrounding implanted cells (11). In this study, cardioCEST imaging was used to visualize Eu-HPDO3A-labeled cells following implantation into the murine heart (Fig. 4). Regions of myocardium containing Eu-HPDO3A-labeled cells demonstrated significantly elevated MTRasym values compared with adjacent slices and compared with myocardium containing saline-labeled cells (Fig. 4). We observed a significant number of voxels with MTRasym above 5% but below 15% in all hearts (Table 1). This likely results from the use of a high saturation flip angle (1080°), which caused higher indirect saturation of myocardial tissue than was observed in our prior study. Subsequently, small differences in signal intensity between images manifested as a higher proportion of voxels with slightly elevated MTRasym values. In addition, the use of a high flip angle induced noise in the ECG waveform, which may have led to slight inaccuracies in ECG triggering. In particular, differences between pairs of images in areas of early motion such as the right ventricular insertion points would cause aberrantly high MTRasym values due to the comparison of myocardium to right ventricular blood, as is observed in Fig. 4C. Interestingly, we observed a higher fraction of voxels with MTRasym>5% in slices containing Eu-HPDO3A-labeled cells (Table 1), possibly caused by the leakage of Eu-HPDO3A from apoptotic cells. We have previously demonstrated that Eu-HPDO3A accumulates in the interstitial space of disrupted tissue and leads to elevated MTRasym values (7). In future studies, we will employ a lower saturation flip angle to reduce non-specific MTRasym elevation and to reduce noise in the ECG waveform. In addition, it will be important to examine in future studies whether particular MTRasym threshold values can be used to discriminate between labeled cells and non-specific MTRasym elevation.
The creatine kinase reaction governs the conversion of phosphocreatine (PCr) to creatine, playing a key role in maintaining myocardial energetics and contractile function. Reductions in metabolites associated with the creatine kinase reaction are characteristic of the failing heart (30), and are typically detected at late stages of heart failure using spectroscopic techniques (31,32). In a recent study by Haris et al. (4), cardiac CEST imaging of creatine contrast (crEST) identified reductions in myocardial creatine in infarcted swine and sheep myocardium when compared with non-infarcted and healthy myocardium. Haris et al. (7) separately correlated creatine contrast by crEST to measurements of myocardial creatine concentration, and confirmed that saturation transfer from the amine proton of creatine, and not other metabolites, is the primary contributor to MTRasym at ±1.8 ppm (4). Mouse models of diet-induced obesity are characterized by prolonged periods of preserved cardiac contractile function and are considered to resemble clinical heart failure with preserved ejection fraction (33,34). We utilized cardioCEST imaging of myocardial creatine to examine whether altered creatine contrast could be used as an early in vivo biomarker of heart failure. We observed significantly reduced creatine CEST contrast in mice fed an HFD for 14 weeks in the absence of overt structural or functional remodeling (Figs. 5 and 6). Our results mirror the clinical findings of Rider et al. (31), who used 31P spectroscopic imaging to reveal significantly reduced resting PCr to adenosine triphosphate ratio (PCr/ATP) in obese adults with preserved systolic function when compared with healthy controls. Interestingly, in a prior study by Abdurrachim et al. (33), 31P spectroscopy did not reveal any difference in PCr/ATP between HFD and LFD mice at 20 weeks after diet initiation. However, the use of a single spectroscopic voxel covering the majority of the left ventricle and blood pool in this study may have reduced the sensitivity to decreased metabolite concentrations. We were not able to perform cardiac 31P spectroscopy at our institution to examine the association between 31P measurements and creatine CEST contrast. In addition, prior studies of cardiac mechanics in similar mice have identified changes in myocardial strains (35) or diastolic strain rates (33) at only 20 weeks after diet initiation. These findings suggest that further studies using cardiac CEST imaging of myocardial creatine are warranted to examine potential metabolic changes in heart failure associated with obesity.
A limitation to the current design of the cardioCEST pulse sequence is that, during the brief periods of rapid inhalation and exhalation, the imaging slice moves far out of the excitation plane. While k-space data is not acquired during this period, which is significantly shorter than the T1-relaxation times of tissue, the effective steady-state encoding of the initial CEST weighting is reduced and can be influenced by changes in respiratory rates between image pairs. Future efforts to improve cardioCEST sequence design will include a real-time feedback mechanism that switches the dummy pulses from slice selective to non-selective during respiratory motion in order to maintain steady-state conditions in the imaging slice. An additional limitation to the current study is that we did not employ post-mortem staining for apoptosis in tissue sections from mice undergoing cell implantation. However, we chose to implant C2C12 cells derived from C3H mice into C57B6 mice, which, when combined with attrition due to the implantation procedure, would likely contribute to apoptosis in a modest fraction of injected cells. Further studies comparing ICP-MS of excised cell implants with MTRasym values will help to elucidate a potential association between labeled cell density and CEST contrast. Separately, care must be taken when interpreting the results of cardioCEST creatine imaging. As demonstrated in Fig. 6C, MTRasym values in the hearts of obese mice demonstrate wider distributions and higher fractions of voxels with negative MTRasym values. The appearance of negative values must be interpreted within the context of the nuclear Overhauser effect (NOE), which is present at −1.8 ppm, as described by Cai et al. (8).
Specifically, negative MTRasym values occur when creatine CEST at +1.8 ppm is less than the NOE at −1.8 ppm, and reflects a lower limit to the sensitivity of creatine CEST imaging, not the absence of myocardial creatine. In this context, the significant reductions in mean MTRasym values in obese mice should not be interpreted as representing a fourfold reduction in myocardial creatine. While this limitation could be overcome using the Z-spectral fitting approach suggested by Cai et al. (8), such an approach would require scan times of the order of 1–1.5 h, which are not practical for pre-clinical cardiac imaging. Finally, we did not perform water saturation shift referencing (WASSR) correction (36) in our calculations of MTRasym. Preliminary WASSR scans during sequence optimization (see Supplementary Methods) confirmed previous findings that the spread of resonant frequencies across the mouse heart is significantly lower than the bandwidth of the saturation pulse used to generate CEST contrast. Importantly, care was taken to position the mouse heart precisely at the magnet isocenter in order to limit the effects of field inhomogeneity (see Supplementary Methods), and higher-order respiratory-gated shimming protocols were used.
CONCLUSIONS
An improved pulse sequence design for pre-clinical cardiac CEST imaging was developed and applied to the tracking of paraCEST-labeled cells and for the imaging of endogenous metabolic markers such as myocardial creatine. In future studies, the selective visualization of paraCEST-labeled cells could enable multi-color imaging of distinctly labeled cell populations, and could be further multiplexed with additional important measurements of myocardial function, perfusion, and tissue characterization. In addition, application of myocardial creatine CEST imaging can be used to better understand the complex interplay between metabolic and contractile changes in obesity-associated heart disease.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Silvio Aime for providing paraCEST agents. This work was supported by NIH CTSA UL1TR000117 (AAL and MV), and by funding from the Slomo and Cindy Silvian Foundation to MV. This work was also supported by P20 GM103527 to BF, MV, and AAL and 1S10RR029541-01. AAL is also supported by R56 HL124266.
Abbreviations used
- CEST
chemical exchange saturation transfer
- paraCEST
paramagnetic CEST
- MTRasym
asymmetric magnetization transfer ratio
- ppm
parts per million
- TR/TE
repetition time/echo time
- PBS
phosphate-buffered saline
- ICP-MS
inductively coupled plasma mass spectrometry
- HFD
high-fat diet
- LFD
low-fat diet
- ANOVA
analysis of variance
- PCr
phosphocreatine
- crEST
CEST imaging of creatine contrast
- ATP
adenosine triphosphate
- NOE
nuclear Overhauser effect
- WASSR
water saturation shift referencing
Footnotes
SUPPORTING INFORMATION Additional supporting information may be found in the online version of this article at the publisher's web site.
REFERENCES
- 1.Ward KM, Aletras AH, Balaban RS. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST) J. Magn. Reson. 2000;143(1):79–87. doi: 10.1006/jmre.1999.1956. [DOI] [PubMed] [Google Scholar]
- 2.Liu G, Song X, Chan KWY, McMahon MT. Nuts and bolts of chemical exchange saturation transfer MRI. NMR Biomed. 2013;26(7):810–828. doi: 10.1002/nbm.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bar-Shir A, Bulte JWM, Gilad AA. Molecular engineering of nonmetallic biosensors for CEST MRI. ACS Chem. Biol. 2015;10(5):1160–1170. doi: 10.1021/cb500923v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haris M, Singh A, Cai K, Kogan F, McGarvey J, DeBrosse C, Zsido GA, Witschey WRT, Koomalsingh K, Pilla JJ, Chirinos JA, Ferrari VA, Gorman JH, Hariharan H, Gorman RC, Reddy R. A technique for in vivo mapping of myocardial creatine kinase metabolism. Nat. Med. 2014;20:209–214. doi: 10.1038/nm.3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haris M, Singh A, Cai K, Nath K, Crescenzi R, Kogan F, Hariharan H, Reddy R. MICEST: a potential tool for non-invasive detection of molecular changes in Alzheimer's disease. J. Neurosci. Methods. 2013;212(1):87–93. doi: 10.1016/j.jneumeth.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu G, Qin Q, Chan KWY, Li Y, Bulte JWM, McMahon MT, van Zijl PCM, Gilad AA. Non-invasive temperature mapping using temperature-responsive water saturation shift referencing (T-WASSR) MRI. NMR Biomed. 2014;27(3):320–331. doi: 10.1002/nbm.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vandsburger M, Vandoorne K, Oren R, Leftin A, Mpofu S, Castelli DD, Aime S, Neeman M. Cardiac-CEST MRI reveals molecular signatures of endogenous fibrosis and exogenous contrast media. Circulation Cardiovasc. Imaging. 2014;8(1):e002180. doi: 10.1161/CIRCIMAGING.114.002180. [DOI] [PubMed] [Google Scholar]
- 8.Cai K, Singh A, Poptani H, Li W, Yang S, Lu Y, Hariharan H, Zhou XJ, Reddy R. CEST signal at 2 ppm (CEST@2 ppm) from Z-spectral fitting correlates with creatine distribution in brain tumor. NMR Biomed. 2015;28(1):1–8. doi: 10.1002/nbm.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aime S, Carrera C, Delli Castelli D, Geninatti Crich S, Terreno E. Tunable imaging of cells labeled with MRI-PARACEST agents. Angew. Chem. Int. Ed. 2005;44(12):1813–1815. doi: 10.1002/anie.200462566. [DOI] [PubMed] [Google Scholar]
- 10.Delli Castelli D, Ferrauto G, Cutrin JC, Terreno E, Aime S. In vivo maps of extracellular pH in murine melanoma by CEST–MRI. Magn. Reson. Med. 2014;71(1):326–332. doi: 10.1002/mrm.24664. [DOI] [PubMed] [Google Scholar]
- 11.Ferrauto G, Castelli DD, Terreno E, Aime S. In vivo MRI visualization of different cell populations labeled with PARACEST agents. Magn. Reson. Med. 2013;69(6):1703–1711. doi: 10.1002/mrm.24411. [DOI] [PubMed] [Google Scholar]
- 12.Terreno E, Delli Castelli D, Cabella C, Dastrù W, Sanino A, Stancanello J, Tei L, Aime S. Paramagnetic liposomes as innovative contrast agents for magnetic resonance (MR) molecular imaging applications. Chem. Biodivers. 2008;5(10):1901–1912. doi: 10.1002/cbdv.200890178. [DOI] [PubMed] [Google Scholar]
- 13.Hancu I, Dixon WT, Woods M, Vinogradov E, Sherry AD, Lenkinski RE. CEST and PARACEST MR contrast agents. Acta Radiol. 2010;51(8):910–923. doi: 10.3109/02841851.2010.502126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Viswanathan S, Kovacs Z, Green KN, Ratnakar SJ, Sherry AD. Alternatives to gadolinium-based metal chelates for magnetic resonance imaging. Chem. Rev. 2010;110(5):2960–3018. doi: 10.1021/cr900284a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bar-Shir A, Liu G, Chan KWY, Oskolkov N, Song X, Yadav NN, Walczak P, McMahon MT, van Zijl PCM, Bulte JWM, Gilad AA. Human protamine-1 as an MRI reporter gene based on chemical exchange. ACS Chem. Biol. 2013;9(1):134–138. doi: 10.1021/cb400617q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bar-Shir A, Liu G, Greenberg MM, Bulte JWM, Gilad AA. Synthesis of a probe for monitoring HSV1-tk reporter gene expression using chemical exchange saturation transfer MRI. Nat. Protoc. 2013;8(12):2380–2391. doi: 10.1038/nprot.2013.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bar-Shir A, Liu G, Liang Y, Yadav NN, McMahon MT, Walczak P, Nimmagadda S, Pomper MG, Tallman KA, Greenberg MM, van Zijl PCM, Bulte JWM, Gilad AA. Transforming thymidine into a magnetic resonance imaging probe for monitoring gene expression. J. Am. Chem. Soc. 2013;135(4):1617–1624. doi: 10.1021/ja312353e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilad AA, McMahon MT, Walczak P, Winnard PT, Raman V, van Laarhoven HWM, Skoglund CM, Bulte JWM, van Zijl PCM. Artificial reporter gene providing MRI contrast based on proton exchange. Nat. Biotech. 2007;25(2):217–219. doi: 10.1038/nbt1277. [DOI] [PubMed] [Google Scholar]
- 19.Singh A, Haris M, Cai K, Kassey VB, Kogan F, Reddy D, Hariharan H, Reddy R. Chemical exchange saturation transfer magnetic resonance imaging of human knee cartilage at 3 T and 7 T. Magn. Reson. Med. 2012;68(2):588–594. doi: 10.1002/mrm.23250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crich SG, Biancone L, Cantaluppi V, Duò D, Esposito G, Russo S, Camussi G, Aime S. Improved route for the visualization of stem cells labeled with a Gd-/Eu-chelate as dual (MRI and fluorescence) agent. Magn. Reson. Med. 2004;51(5):938–944. doi: 10.1002/mrm.20072. [DOI] [PubMed] [Google Scholar]
- 21.Chen LQ, Howison CM, Jeffery JJ, Robey IF, Kuo PH, Pagel MD. Evaluations of extracellular pH within in vivo tumors using acidoCEST MRI. Magn. Reson. Med. 2014;72(5):1408–1417. doi: 10.1002/mrm.25053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilad AA, Ziv K, McMahon MT, van Zijl PCM, Neeman M, Bulte JWM. MRI reporter genes. J. Nucl. Med. 2008;49(12):1905–1908. doi: 10.2967/jnumed.108.053520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Gregorio E, Ferrauto G, Gianolio E, Aime S. Gd loading by hypotonic swelling: an efficient and safe route for cellular labeling. Contrast Media Mol. Imaging. 2013;8(6):475–486. doi: 10.1002/cmmi.1574. [DOI] [PubMed] [Google Scholar]
- 24.Gao E, Koch W. A novel and efficient model of coronary artery ligation in the mouse. In: Gourdie RG, Myers TA, editors. Wound Regeneration and Repair. Humana; New York, NY: 2013. pp. 299–311. [DOI] [PubMed] [Google Scholar]
- 25.Chen I, Greve J, Gheysens O, Willmann J, Rodriguez-Porcel M, Chu P, Sheikh A, Faranesh A, Paulmurugan R, Yang P, Wu J, Gambhir S. Comparison of optical bioluminescence reporter gene and superparamagnetic iron oxide MR contrast agent as cell markers for noninvasive imaging of cardiac cell transplantation. Mol. Imaging Biol. 2009;11(3):178–187. doi: 10.1007/s11307-008-0182-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank JA, Miller BR, Arbab AS, Zywicke HA, Jordan EK, Lewis BK, Bryant LH, Bulte JWM. Clinically applicable labeling of mammalian and stem cells by combining superparamagnetic iron oxides and transfection agents. Radiology. 2003;228(2):480–487. doi: 10.1148/radiol.2281020638. [DOI] [PubMed] [Google Scholar]
- 27.Kraitchman D, Bulte JM. Imaging of stem cells using MRI. Basic Res. Cardiol. 2008;103(2):105–113. doi: 10.1007/s00395-008-0704-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraitchman D, Kedziorek D, Bulte JM. MR Imaging of transplanted stem cells in myocardial infarction. In: Shah K, editor. Molecular Imaging. Humana; 2011. pp. 141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winter EM, Hogers B, van der Graaf LM, Gittenberger-de Groot AC, Poelmann RE, van der Weerd L. Cell tracking using iron oxide fails to distinguish dead from living transplanted cells in the infarcted heart. Magn. Reson. Med. 2010;63(3):817–821. doi: 10.1002/mrm.22094. [DOI] [PubMed] [Google Scholar]
- 30.Cavalera M, Wang J, Frangogiannis NG. Obesity, metabolic dysfunction, and cardiac fibrosis: pathophysiological pathways, molecular mechanisms, and therapeutic opportunities. Transl. Res. 2014;164(4):323–335. doi: 10.1016/j.trsl.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rider OJ, Francis JM, Ali MK, Holloway C, Pegg T, Robson MD, Tyler D, Byrne J, Clarke K, Neubauer S. Effects of catecholamine stress on diastolic function and myocardial energetics in obesity. Circulation. 2012;125(12):1511–1519. doi: 10.1161/CIRCULATIONAHA.111.069518. [DOI] [PubMed] [Google Scholar]
- 32.Holloway CJ, Cochlin LE, Emmanuel Y, Murray A, Codreanu I, Edwards LM, Szmigielski C, Tyler DJ, Knight NS, Saxby BK, Lambert B, Thompson C, Neubauer S, Clarke K. A high-fat diet impairs cardiac high-energy phosphate metabolism and cognitive function in healthy human subjects. Am. J. Clin. Nutr. 2011;93(4):748–755. doi: 10.3945/ajcn.110.002758. [DOI] [PubMed] [Google Scholar]
- 33.Abdurrachim D, Ciapaite J, Wessels B, Nabben M, Luiken JJFP, Nicolay K, Prompers JJ. Cardiac diastolic dysfunction in high-fat diet fed mice is associated with lipotoxicity without impairment of cardiac energetics in vivo. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2014;1841(10):1525–1537. doi: 10.1016/j.bbalip.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 34.Pascual M, Pascual DA, Soria F, Vicente T, Hernández AM, Tébar FJ, Valdés M. Effects of isolated obesity on systolic and diastolic left ventricular function. Heart. 2003;89(10):1152–1156. doi: 10.1136/heart.89.10.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kramer SP, Powell DK, Haggerty CM, Binkley C, Mattingly A, Cassis L, Epstein F, Fornwalt BK. Obesity reduces left ventricular strains, torsion, and synchrony in mouse models: a cine displacement encoding with stimulated echoes (DENSE) cardiovascular magnetic resonance study. J. Cardiovasc. Magn. Reson. 2013;15(109):1. doi: 10.1186/1532-429X-15-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim M, Gillen J, Landman BA, Zhou J, van Zijl PCM. Water saturation shift referencing (WASSR) for chemical exchange saturation transfer (CEST) experiments. Magn. Reson. Med. 2009;61(6):1441–1450. doi: 10.1002/mrm.21873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.