Abstract
DNA methylation is the most frequent epigenetic alteration. Using methylation-specific polymerase chain reaction (MSP), the methylation status of the adenomatous polyposis coli (APC) and Ras association domain family 1 isoform A (RASSF1A) genes was examined in cell-free circulating DNA from 155 plasma samples obtained from patients with early and advanced colorectal cancer (CRC). APC and RASSF1A hypermethylation was frequently observed in both early and advanced disease, and was significantly associated with a poorer disease outcome. The methylation status of the APC and RASSF1A promoters was investigated in cell-free DNA of patients with CRC. Using MSP, the promoter methylation status of APC and RASSF1A was examined in 155 blood samples obtained from patients with CRC, 88 of whom had operable CRC (oCRC) and 67 had metastatic CRC (mCRC). The frequency of APC methylation in patients with oCRC was 33%. Methylated APC promoter was significantly associated with older age (P=0.012), higher stage (P=0.014) and methylated RASSF1A status (P=0.050). The frequency of APC methylation in patients with mCRC was 53.7%. In these patients, APC methylation was significantly associated with methylated RASSF1A status (P=0.016). The frequency of RASSF1A methylation in patients with oCRC was 25%. Methylated RASSF1A in oCRC was significantly associated with higher stage (P=0.021). The frequency of RASSF1A methylation in mCRC was 44.8%. Methylated RASSF1A in mCRC was associated with moderate differentiation (P=0.012), high levels of carcinoembryonic antigen (P=0.023) and methylated APC status (P=0.016). Patients with an unmethylated APC gene had better survival in both early (81±5 vs. 27±4 months, P<0.001) and advanced disease (37±7 vs. 15±3 months, P<0.001), compared with patients with methylated APC. Patients with an unmethylated RASSF1A gene had better survival in both early (71±6 vs. 46±8 months, P<0.001) and advanced disease (28±4 vs. 16±3 months, P<0.001) than patients with methylated RASSF1A. The observed significant correlations between APC and RASSF1A promoter methylation status and survival may be indicative of a prognostic role for these genes in CRC, which requires additional testing in larger studies.
Keywords: APC, RASSF1A, DNA methylation, cell-free DNA, colon cancer
Introduction
Colorectal cancer (CRC) is a leading cause of morbidity and mortality worldwide, with >600,000 associated mortalities annually (1). Of all CRC cases, 75–80% occur sporadically as the result of complex interactions between susceptibility genes and environmental factors (2). Despite the recent and continuous improvements in diagnosis and treatments, >50% of colon and rectal tumors metastasize to liver, lung and lymph nodes, and the 5-year survival rate remains low (~10%) for patients with metastatic CRC (mCRC) (3). There is, therefore, an urgent requirement for non-invasive, novel molecular biomarkers that could be useful in diagnosis and could also improve prognosis and treatment prediction. The accumulation of in vitro and in vivo evidence suggests that epigenetics exerts a fundamental role in CRC pathogenesis (4). The best known and more frequent epigenetic alteration is DNA methylation, which affects tumor suppressor genes that may be involved in cell cycle control, DNA repair, metabolism of carcinogens, cell-cell interaction, apoptosis and angiogenesis (5). Hypermethylation of CpG islands in the promoter region of tumor suppressor genes leads to their inappropriate silencing, which may trigger cancer initiation and progression (6). The technical advantage of studying DNA methylation is its chemical stability, thus enabling its detection with very high sensitivity (≤1:1,000 molecules) (7). Numerous studies have demonstrated that cancer-specific, methylated DNA may be present in body fluids, suggesting that it could be used as a non-invasive marker (8,9). Blood, plasma or serum constitute the most easy-to-handle samples, and are also a great source of cell-free circulating tumor DNA (10). The mechanism surrounding the origin of tumoral DNA that is released into the circulation is poorly understood, but it is assumed that DNA is released during necrosis and/or apoptosis of tumor cells (11). These circulatory DNA molecules are easily isolated, and may serve for the detection of the methylation status of certain genes (12,13). Several genes with altered levels of methylation have been investigated in respect to their involvement in CRC initiation or progression. A number of those, including human mutL homolog 1, O(6)-methylguanine-DNA methyltransferase (MGMT) and thrombospondin 1, exhibited an increase in methylation during all the stages of the disease, while other genes such as Ras association domain family 1 isoform A (RASSF1A) or tissue inhibitor of metalloproteinase 3 have been reported to be methylated in later disease stage or in metastases (14).
RASSF members belong to a family of putative tumor suppressor RAS effectors, for which epigenetic silencing by promoter methylation has been reported to occur throughout the progression of several types of cancer, including CRC (15,16). Since its identification in 2000, the tumor suppressor RASSF1A gene has been extensively investigated (17). Transcriptional silencing of RASSF1A by inappropriate promoter methylation has been frequently observed in various types of human cancer, including lung, breast, colorectal, gastric and cervical cancer (18). RASSF1A methylation has been reported to range from 12 to 81% in CRC (19,20). In colon primary tumors, Yoon et al (19) detected methylation in the CpG island of the RASSF1A gene in 3 out of 26 (12%) tumor tissues, while none of the available normal tissues were methylated.
Mutations in the human tumor suppressor gene adenomatous polyposis coli (APC) are frequent in both sporadic and familial CRC (21). Wild-type APC protein contributes to the destabilization and degradation of β-catenin, which is a central effector molecule in the Wnt/β-catenin signaling pathway (22). Loss of APC function results in nuclear accumulation of β-catenin, which leads to the transcriptional activation (via the β-catenin/T-cell factor complex) of target genes that may contribute to colorectal tumorigenesis (23,24). The rate of APC promoter methylation in CRC and normal colorectal mucosa has been reported to range from 11 to 62% in different populations, and has been suggested to moderate the Wnt signaling pathway (25–29). Furthermore, hypermethylation of the APC promoter has been demonstrated to be relatively common in other gastrointestinal neoplasms, including those of the stomach, liver, pancreas and oesophagus (30).
In the present study, the methylation status of APC and RASSF1A were investigated in cell-free circulating DNA of patients with operable CRC (oCRC) and mCRC. The aim of the present study was to primarily assess the methylation status of the above genes and to explore their possible prognostic significance in early and advanced disease.
Materials and methods
Study design
The study material consisted of 155 blood samples obtained from patients with CRC between March 2010 and May 2014. Patients were suffering from either early operable (88/155, 56.8%) or metastatic disease (67/155, 43.2%). The clinicopathological data for all patients are presented in Table I. All patients had a performance status (PS) of 0–1 [World Health Organization scale (31)] and provided informed consent. Additionally, 20 blood samples obtained from healthy individuals [friends and non-blood related family members of patients treated at the Department of Medical Oncology of the University Hospital of Alexandroupolis (Alexandroupolis, Greece)] were used as a control group. The majority of controls were men, all age-matched with the patient population, who had no received medical care at the time of sample collection. All patients included in the study had signed an informed consent form along with an agreement to use biological material for experimental purposes.
Table I.
Patients' characteristics in early (n=88) and advanced (n=67) disease.
| Patients' characteristics | All patients, no. (%) | Patients with oCRC, no. (%) | Patients with mCRC, no. (%) |
|---|---|---|---|
| No. of patients | 155 | 88 | 67 |
| Gender | |||
| Males | 89 (57.4) | 51 (58.0) | 38 (56.7) |
| Females | 66 (42.6) | 37 (42.0) | 29 (43.3) |
| Age, years | |||
| ≤70 | 83 (53.5) | 41 (46.6) | 42 (62.7) |
| >70 | 72 (46.5) | 47 (53.4) | 25 (37.3) |
| Dukes' stage | |||
| A+B | 58 (37.4) | 58 (65.9) | – |
| C | 30 (19.4) | 30 (34.1) | – |
| D | 67 (43.2) | – | 67 (100.0) |
| Differentiation | |||
| Well | 48 (31.0) | 30 (34.1) | 18 (26.9) |
| Moderate | 79 (51.0) | 43 (48.9) | 36 (53.7) |
| Poor | 28 (18.0) | 15 (17.0) | 13 (19.4) |
| Presence of metastases | |||
| No | 88 (56.8) | – | – |
| Yes | 67 (43.2) | – | – |
| Tumor location, colon side | |||
| Right | 74 (47.7) | 43 (48.9) | 31 (46.3) |
| Left | 81 (52.3) | 45 (51.1) | 36 (53.7) |
| CEA levels, ng/ml (n=138) | |||
| ≤10 | 57 (41.3) | 40 (48.8) | 17 (30.4) |
| >10 | 81 (58.7) | 42 (51.2) | 39 (69.6) |
| CA 19.9 levels, U/ml (n=137) | |||
| <37 (low) | 95 (69.3) | 67 (81.7) | 28 (50.9) |
| >37 (high) | 42 (30.7) | 15 (18.3) | 27 (49.1) |
CRC, colorectal cancer; oCRC, operable colorectal cancer; mCRC, metastatic colorectal cancer; CEA, carcinoembryonic antigen; CA 19-9, carbohydrate antigen 19-9.
Sample collection and isolation of cell-free DNA
Whole blood was extracted from patients pre-operatively. Blood was collected in serum clot activator tubes. Serum was obtained immediately through centrifugation at 3,000 × g for 10 min and stored at −80°C until DNA extraction. Cell-free DNA from serum samples was isolated using the High Pure Viral Nucleic Acid kit (Roche Diagnostics GmbH, Mannheim, Germany). A total of 300 µl serum were mixed with 300 µl working solution and 60 µl proteinase K (18 mg/ml), and incubated for 10 min at 72°C; DNA isolation was then processed as described in the manufacturer's protocol. DNA concentration was determined with an ND-100 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc., Wilmington, DE, USA).
Carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA 19-9) measurements
CEA and CA 19-9 levels were measured using the Elecsys® CEA kit (Roche Diagnostics GmbH). The cut-offs used were 10 ng/ml for CEA and 37 U/ml for CA 19-9.
Sodium bisulfite conversion
Sodium bisulfite conversion of ≤200 ng cell-free DNA was performed using the EZ DNA Methylation-Gold™ kit (Zymo Research Corporation, Irvine, CA, USA), according to the manufacturer's protocol. The converted DNA was stored at −80°C until used.
Methylation-specific polymerase chain reaction (MSP)
The methylation status of APC and RASSF1A in cell-free circulating serum DNA samples was detected by MSP using specific primer pairs for both the methylated and unmethylated promoter sequences. Each MSP reaction was performed in a total volume of 25 µl. Sodium bisulfite-converted DNA (1 µl) was added into a 24-µl reaction mixture that contained 0.1 µl Taq DNA polymerase (5 U/µl; GoTaq® Hot Start Polymerase; Promega Corporation, Madison, WI, USA), 5 µl 10X buffer, 2.0 µl MgCl2 (50 mmol/l), 0.5 µl deoxynucleotides triphosphate (10 mmol/l; Fermentas; Thermo Fisher Scientific, Inc., Pittsburgh, PA, USA) and 1 µl each of the corresponding forward and reverse primers (10 µmol/l); lastly, distilled H2O was added to a final volume of 25 µl. Sodium bisulfite-treated DNA was amplified in two separate MSP reactions, one with a set of primers specific for methylated DNA, and one for unmethylated promoter sequences. The primer pairs used in this study are as follows: RASSF1A unmethylated, forward GGTTGTATTTGGTTGGAGTG and reverse CTACAAACCTTTACACACAACA; RASSF1A methylated, forward GTTGGTATTCGTTGGGCGC and reverse GCACCACGTATACGTAACG; APC unmethylated, forward GTGTTTTATTGTGGAGTGTGGGTT and reverse CCAATCAACAAACTCCCAACAA; APC methylated, forward TATTGCGGAGTGCGGGTC and reverse TCGACGAACTCCCGACGA. Human placental genomic DNA (gDNA; Sigma-Aldrich, St. Louis, MO, USA) methylated in vitro with M.SssI methylase (New England BioLabs, Inc., Ipswich, MA, USA) was used, following sodium bisulfite conversion, as a fully methylated (100%) MSP positive control. The same unmethylated placental gDNA was used, following sodium bisulfite conversion, as a negative MSP control. The thermocycling conditions used were as follows: i) APC, 1 cycle at 95°C for 5 min, followed by 39 cycles of 95°C for 45 sec, 63°C for 60 sec and 72°C for 60 sec, with a final extension cycle of 72°C for 10 min; and ii) RASSF1A, 1 cycle at 95°C for 5 min, followed by 39 cycles of 95°C for 30 sec, 58°C for 45 sec and 72°C for 45 sec, with a final extension cycle of 72°C for 5 min. MSP products for methylated and unmethylated promoters were fractionated on 2% agarose gels containing 40 mM Tris-acetate/1.0 mM ethylenediaminetetraacetic acid (pH 8.0) and visualized by ethidium bromide staining.
Statistical analysis
Statistical analysis of the data was performed using SPSS version 19.0 (IBM SPSS, Armonk, NY, USA). The methylation status of APC and RASSF1A and all other qualitative variables were expressed as frequencies and percentages. The χ2 test was used to evaluate any potential association of APC and RASSF1A status with patients' demographic and clinicopathological characteristics. Odds ratios (ORs) and their 95% confidence intervals (CIs) were estimated as a measure of the association between APC and RASSF1A status and patients' characteristics. Survival rates were calculated with the Kaplan-Meier method, and the statistical difference between the survival curves was determined with the log-rank test. Multivariate Cox proportional hazards regression (HR) analysis, using a backward selection approach, were performed to explore the independent effect of APC and RASSF1A status on overall survival. Patients' gender, age, clinical stage, tumor differentiation, lymph node status, CEA and CA 19.9 levels were also included in the multivariate model as potential confounders. All tests were two tailed, and P<0.05 was considered to indicate a statistical significant difference.
Results
Characteristics of the study population
The study population consisted of 155 patients with CRC with a median age of 70 years (range, 44–76 years; mean age ± standard deviation, 68.35±9.20 years), 57.4% of whom were males. The patient's clinicopathological characteristics are indicated in Table I. In total, 48 tumors (31.0%) were well differentiated, 79 (51.0%) were moderately differentiated and 28 (18.1%) were poorly differentiated carcinomas. The Dukes' system (32) was used for the classification of patients into different stages. Almost half of the cases (67 patients, 43.2%) were at stage D, 30 (19.3%) cases were at stage C and 58 (37.4%) were at stage A or B. APC and RASSF1A promoters were observed to be methylated in 65 (41.9%) and 52 (33.5%) of the 155 colon cancer samples examined, but in none of the normal control samples (both P<0.001). The association of the patients' demographic and clinicopathological features with APC and RASSF1A methylation status is presented in Tables II and III.
Table II.
Association of APC and RASSF1A methylation status with demographic and clinicopathological characteristics of patients with operable disease.
| Patients' characteristics | APC methylation | P-value | RASSF1A methylation | P-value |
|---|---|---|---|---|
| Gender | 0.711 | 0.170 | ||
| Males | 16 (31.4) | 10 (19.6) | ||
| Females | 13 (35.1) | 12 (32.4) | ||
| Age, years | 0.012 | 0.267 | ||
| ≤70 | 8 (19.5) | 8 (19.5) | ||
| >70 | 21 (44.7) | 14 (29.8) | ||
| Dukes' stage | 0.014 | 0.021 | ||
| A+B | 14 (24.1) | 8 (14.3) | ||
| C | 15 (50.0) | 14 (34.1) | ||
| Differentiation | 0.403 | 0.670 | ||
| Well | 8 (26.7) | 9 (30.0) | ||
| Moderate | 14 (32.6) | 9 (20.9) | ||
| Poor | 7 (46.7) | 4 (26.7) | ||
| Tumor location, colon side | 0.407 | 0.538 | ||
| Right | 16 (37.2) | 12 (27.9) | ||
| Left | 13 (28.9) | 10 (22.2) | ||
| CEA levels, ng/ml (n=82) | 0.088 | 0.697 | ||
| ≤5 | 10 (25.0) | 9 (22.5) | ||
| >5 | 18 (42.9) | 11 (26.2) | ||
| CA 19.9 levels, U/ml (n=82) | 0.083 | 0.372 | ||
| <37 (low) | 20 (29.9) | 15 (22.4) | ||
| >37 (high) | 8 (53.3) | 5 (33.3) | ||
| APC status | – | 0.050 | ||
| Unmethylated | – | 11 (18.6) | ||
| Methylated | – | 11 (37.9) | ||
| RASSF1A status | 0.050 | – | ||
| Unmethylated | 18 (27.3) | – | ||
| Methylated | 11 (50.0) | – |
APC, adenomatous polyposis coli; RASSF1A, Ras association domain family 1 isoform A; CEA, carcinoembryonic antigen; CA 19-9, carbohydrate antigen 19-9.
Table III.
Association of APC and RASSF1A methylation status with demographic and clinicopathological characteristics of patients with metastatic disease.
| Patients' characteristics | APC methylation | P-value | RASSF1A methylation | P-value |
|---|---|---|---|---|
| Gender | 0.076 | 0.994 | ||
| Males | 24 (63.2) | 17 (44.7) | ||
| Females | 12 (41.4) | 13 (44.8) | ||
| Age, years | 0.427 | 0.921 | ||
| ≤70 | 21 (50.0) | 19 (45.2) | ||
| >70 | 15 (60.0) | 11 (44.0) | ||
| Dukes' stage | – | – | ||
| A+B | – | – | ||
| C | – | – | ||
| D | 36 (53.7) | 30 (44.8) | ||
| Differentiation | 0.388 | 0.032 | ||
| Well | 10 (55.6) | 12 (66.7) | ||
| Moderate | 17 (47.2) | 11 (30.6) | ||
| Poor | 9 (69.2) | 7 (53.8) | ||
| Tumor location, colon side | 0.100 | 0.581 | ||
| Right | 20 (64.5) | 15 (48.4) | ||
| Left | 16 (44.4) | 15 (41.7) | ||
| CEA levels, ng/ml (n=56) | 0.159 | 0.023 | ||
| ≤5 | 7 (41.2) | 4 (23.5) | ||
| >5 | 24 (61.5) | 22 (56.4) | ||
| CA 19.9 levels, U/ml (n=55) | 0.883 | 0.898 | ||
| <37 (low) | 15 (53.6) | 13 (46.4) | ||
| >37 (high) | 15 (55.6) | 13 (48.1) | ||
| APC status | – | 0.016 | ||
| Unmethylated | – | 9 (29.0) | ||
| Methylated | – | 21 (58.3) | ||
| RASSF1A status | 0.016 | – | ||
| Unmethylated | 15 (40.5) | – | ||
| Methylated | 21 (70.0) | – |
APC, adenomatous polyposis coli; RASSF1A, Ras association domain family 1 isoform A; CEA, carcinoembryonic antigen; CA 19-9, carbohydrate antigen 19-9.
Correlation between APC methylation status and different tumor parameters
Patients with early oCRC
APC was methylated in 29 of 88 patients with early oCRC (33.0%). χ2 analysis revealed that methylated APC promoter status was associated with age >70 years (OR=3.33, 95% CI=1.27–8.73, P=0.012), higher stage (OR=3.14, 95% CI=1.23–8.00, P=0.014) and methylated RASSF1A status (OR=2.67, 95% CI=1.00–7.22, P=0.050), while a tendency was noticed with high CEA levels (OR=2.25, 95% CI=0.88–5.77, P=0.088) and high CA 19.9 levels (OR=2.69, 95% CI=0.86–8.41, P=0.083) (Table II).
Patients with metastatic disease
APC was methylated in 36 of 67 patients with mCRC (53.7%). Methylated APC promoter status was associated with methylated RASSF1A status (OR=3.42, 95% CI=1.23–9.49, P=0.016) and marginally with male gender (OR=2.43, 95% CI=0.90–6.54, P=0.076). No other significant associations between APC methylation status and other tumor parameters were observed in patients with mCRC (Table III).
Correlation between RASSF1A methylation status and different tumor parameters
Patients with early oCRC
RASSF1A was methylated in 22 of 88 patients with early oCRC (25.0%). Methylated RASSF1A promoter status was significantly associated with higher disease stage (OR=3.11, 95% CI=1.16–8.36, P=0.021). No other significant associations were observed (Table II).
Patients with metastatic disease
RASSF1A was methylated in 30 of 67 patients with mCRC (44.8%). Methylated RASSF1A promoter status was significantly associated with moderate differentiation (OR=3.60, 95% CI=1.31–9.91, P=0.012), high CEA levels (OR=4.21, 95% CI=1.16–15.23, P=0.023) and methylated APC status (OR=3.42, 95% CI=1.23–9.49, P=0.016) (Table III).
APC methylation status and survival in patients with early oCRC
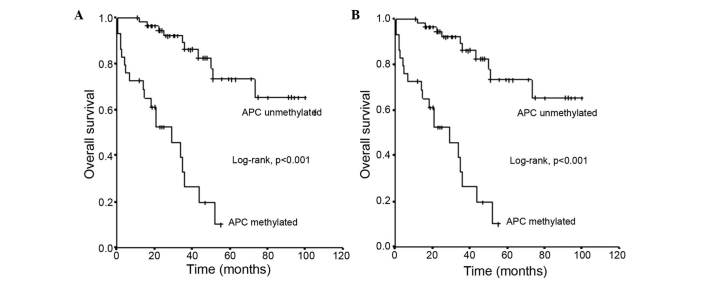
The mean survival time ± standard error (SE) of patients with oCRC and an unmethylated APC promoter status was 81±5 months (95% CI=71–91), which was significantly longer than the mean survival ± SE of 27±4 months (95% CI=19–34) observed for those with a methylated APC promoter status (log-rank test, P<0.001) (Fig. 1A).
Figure 1.
Overall survival of patients with CRC in relation to adenomatous polyposis coli methylation status in (A) early operable CRC and (B) metastatic CRC. CRC, colorectal cancer; APC, adenomatous polyposis coli.
APC methylation status and survival in patients with mCRC
The mean survival time ± SE of patients with mCRC and an unmethylated APC promoter status was 37±7 months (95% CI=23–50), which was substantially longer than the mean survival ± SE of 15±3 months (95% CI=9–20) observed in those with a methylated APC promoter status (log-rank test, P<0.001) (Fig. 1B).
RASSF1A methylation status and survival in patients with early oCRC
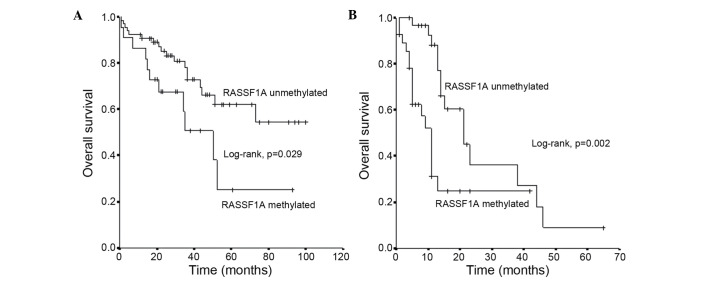
The mean survival time ± SE of patients with oCRC and an unmethylated RASSF1A promoter status was 71±6 months (95% CI=60–81), which was substantially longer than the mean survival ± SE of 46±8 months (95% CI=29–62) observed in those with a methylated RASSF1A promoter status (log-rank test, P<0.001) (Fig. 2A).
Figure 2.
Overall survival of patients with CRC in relation to Ras association domain family 1 isoform A methylation status in (A) early operable CRC and (B) metastatic CRC. CRC, colorectal cancer; RASSF1A, Ras association domain family 1 isoform A.
RASSF1A methylation status and survival in patients with mCRC
The mean survival time ± SE of patients with mCRC and an unmethylated RASSF1A promoter status was 28±4 months (95% CI=19–36) months, which was substantially longer than the mean survival ± SE of 16±3 months (95% CI=9–22) months observed in patients with a methylated RASSF1A promoter status (log-rank test, P<0.001) (Fig. 2B).
Multivariate Cox proportional HR analysis
Upon stratification of the analysis according to the presence or absence of metastases, the negative impact of methylated APC promoter status on patients' survival was more pronounced in patients without metastases [adjusted HR (aHR)=7.88, 95% CI=2.73–22.73, P<0.001] than in patients with metastases (aHR=3.47, 95% CI=1.35–8.92, P=0.017), while the negative impact of methylated RASSF1A promoter status on patients' survival was more pronounced in patients with metastases (aHR=5.76, 95% CI=2.44–14.82, P=0.001) than among patients without metastases (aHR=3.06, 95% CI=1.25–7.50, P=0.038).
Discussion
CRC remains a global burden on world health and economy, despite having a 90% 5-year survival rate when detected and treated early (1,33). Traditional methods cannot sufficiently predict the prognosis of single cancer cases, and clinicians may be not able to accurately decide which patients will be at high risk of recurrence and benefit from chemotherapy. Therefore, it is essential to identify novel biomarkers for improved prognosis, which would aid clinicians to decide which patients should receive adjuvant treatment. The questions of which patients should be treated and why certain patients respond better to chemotherapy than others must be solved, as adjuvant cancer therapy imposes unnecessary toxicity and a huge financial burden on patients. In the present study, the promoter methylation status of the APC and RASSF1A genes in cell-free DNA from patients with CRC was explored, and their incidence and potential correlations with different tumor parameters and survival were examined.
RASSF1A protein is actively involved in microtubule regulation, genomic stability maintenance, cell-cycle regulation, apoptosis modulation, cell motility and invasion control (34–36). A number of studies have identified a high percentage of RASSF1A methylation in CRC samples. Wagner et al (37) observed RASSF1A promoter methylation in 45% (13/29) of the primary CRC and in 80% (4/5) of the CRC cell lines analyzed. Contrarily, in a study with 222 sporadic CRC samples, van Engeland et al (38) detected RASSF1A methylation in 20% (45/222) of the samples, and a mutually exclusive association with the presence of Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations was suggested. Several other studies reported RASSF1A methylation in 17% (8/47), 36% (26/73) and 47% (17/36) of the CRC samples examined (39–41). Notably, the above different studies reporting RASSF1A methylation at different stages, as well as in the corresponding normal mucosa adjacent to the tumor, point to a role of the tumor suppressor gene RASSF1A early in the development of colorectal carcinogenesis. It may mean that methylation possibly occurs in proximal sites of cancer cells due to field effect phenomena; however, the mechanism remains to be determined. The present results are in accordance with the aforementioned reports, as 25% hypermethylated RASSF1A promoter was detected in patients with early oCRC, and 44.8% hypermethylated RASSF1A promoter was detected in patients with mCRC, suggesting that RASSF1A methylation is a frequent event in CRC, although more pronounced at later disease stages. This higher incidence of RASSF1A methylation in metastatic disease may be indicative of a more aggressive tumor behavior, which may explain the association with poorer prognosis observed in the present study. In our previous study, we performed a similar analysis of RASSF1A methylation status in patients with operable gastric cancer (42). In that study, a methylation rate of 68.5% was detected, which underlines the difference in the biological behavior of gastric cancer in comparison with CRC. Which of the multiple roles of RASSF1A is played at different disease stages according to its methylation status is unclear.
Wagner et al (37) investigated whether RASSF1A methylation correlated with tumor-node-metastasis status, but did not detect any significant associations. Similarly, in the study by Van England et al (38), RASSF1A methylation was not associated with gender, Duke's stage or location of the tumor. The only exception was the age at diagnosis, which was slightly higher in RASSF1A methylated CRC cases compared to non-methylated ones.
In a previous study with 36 CRC samples, a significant correlation was observed between RASSF1A methylation and gender, with RASSF1A being more frequently methylated in females (41) In the present study, methylated RASSF1A was associated with higher stages in patients with early oCRC, while in patients with metastatic disease, methylated RASSF1A was significantly associated with moderate differentiations, high CEA levels and APC hypermethylated promoter status. Notably, in other studies, RASSF1A methylation levels were significantly higher in distal CRC, compared with proximal CRC (43,44), as well as in normal mucosae (44). The present data did not support a difference in the profile of methylation between right and left colon. Additionally, a significant difference was observed in the survival of patients with unmethylated RASSF1A promoter status, compared with those with methylated RASSF1A, in patients with and without metastases. Furthermore, the negative impact of methylated RASSF1A promoter status on patients' survival was more pronounced in patients with metastases. Additional studies are required for a better characterization of the subsets of patients with RASSF1A promoter methylation and a better understanding of the role of RASSF1A in CRC development.
Germline mutations in the tumor suppressor APC gene cause familial adenomatous polyposis, and somatic mutations are common in sporadic CRC (45). Hypermethylation of the APC promoter has been reported in early steps of carcinogenesis in several tumors (46). A previous study reported that the frequencies of aberrant promoter methylation were 16% for cadherin 1, 2% for p16, 4% for MGMT and 24% for APC (47). An aberrant methylation of ≥1 of these genes was identified in 45 of 51 (88%) primary tumors (48). In the present cohort, APC was methylated in 33% of patients with early oCRC and in 53.7% of patients with mCRC. In patients with early oCRC, methylated APC promoter status was associated with ages older than 70 years and methylated RASSF1A status, while a tendency was observed with high CEA and CA 19.9 levels. In patients with metastatic disease, APC methylation was associated with methylated RASSF1A status and male gender. Patients with an unmethylated APC promoter status had a substantially longer mean survival than those with a methylated APC promoter status, both in early and metastatic disease. A significant and unexpected finding of the present study was that the negative impact of methylated APC promoter status on patients' survival was more pronounced in patients without metastases than in patients with metastases. The present authors do not have a clear explanation for this observation. It appears that, although APC methylation is most frequently observed at later stages, when present at earlier stages it is indicative of an aggressive tumor phenotype associated with shorter survival. The present findings are in agreement with the proposed roles of APC and RASSF1A as tumor suppressor genes. Their silencing as a result of their methylation is indicative of a more aggressive tumor phenotype with shorter survival. This is also supported by the observed correlations with bad prognostic features, such as higher disease stages and older age for APC, and higher stages, moderate differentiation and high CEA levels for RASSF1A.
In conclusion, the present study demonstrated that serum RASSF1A and APC promoter hypermethylation is a frequent epigenetic event in patients with colon cancer, in both early and metastatic disease, which is indicative of a crucial role for both proteins in colorectal carcinogenesis. In addition, a significant correlation was observed between APC and RASSF1A promoter methylation status and survival. The high percentage of methylation of both proteins in the early and metastatic setting indicates that methylation represents a common event, and when present, it is possibly associated with a more aggressive tumor phenotype. Additional studies in a larger cohort of patients are required to further explore whether these findings could establish the methylation status of APC and RASSF1A as potential biomarkers for early detection and prognosis in CRC.
Glossary
Abbreviations
- MSP
methylation-specific polymerase chain reaction
- OS
overall survival
- PS
performance status
- CRC
colorectal cancer
- oCRC
operable colorectal cancer
- mCRC
metastatic colorectal cancer
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Migliore L, Migheli F, Spisni R, Coppedè F. Genetics, cytogenetics, and epigenetics of colorectal cancer. J Biomed Biotechnol. 2011;2011:792362. doi: 10.1155/2011/792362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segal NH, Saltz LB. Evolving treatment of advanced colon cancer. Annu Rev Med. 2009;60:207–219. doi: 10.1146/annurev.med.60.041807.132435. [DOI] [PubMed] [Google Scholar]
- 4.Bardhan K, Liu K. Epigenetics and colorectal cancer pathogenesis. Cancers (Basel) 2013;5:676–713. doi: 10.3390/cancers5020676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 6.Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2(Suppl 1):S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 7.Nakajima T, Enomoto S, Uschijima T. DNA methylation: A marker for carcinogen exposure and cancer risk. Environ Health Prev Med. 2008;13:8–15. doi: 10.1007/s12199-007-0005-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye T, Chen Y, Fang J. DNA methylation biomarkers in serum for gastric cancer screening. Mini Rev Med Chem. 2010;10:1034–1038. doi: 10.2174/1389557511009011034. [DOI] [PubMed] [Google Scholar]
- 9.Shivapurkar N, Gazdar AF. DNA methylation based biomarkers in non-invasive cancer screening. Curr Mol Med. 2010;10:123–132. doi: 10.2174/156652410790963303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aarthy R, Mani S, Velusami S, Sundarsingh S, Rajkumar T. Role of circulating cell-free DNA in cancers. Mol Diagn Ther. 2015;19:339–350. doi: 10.1007/s40291-015-0167-y. [DOI] [PubMed] [Google Scholar]
- 11.Jahr S, Hentze H, English S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–1665. [PubMed] [Google Scholar]
- 12.Gormally E, Caboux E, Vineis P, Hainaut P. Circulating free DNA in plasma or serum as a biomarker of carcinogenesis: Practical aspects and biological significance. Mutat Res. 2007;635:105–117. doi: 10.1016/j.mrrev.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Jung K, Fleischhacker M, Rabien A. Cell-free DNA in the blood as a solid tumor biomarker-a critical appraisal of the literature. Clin Chim Acta. 2010;411:1611–1624. doi: 10.1016/j.cca.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 14.Bardhan K, Liu K. Epigenetics and colorectal cancer pathogenesis. Cancer (Basel) 2013;5:676–713. doi: 10.3390/cancers5020676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richter AM, Pfeifer GP, Dammann RH. The RASSF proteins in cancer; From epigenetic silencing to functional characterization. Biochim Biophys Acta. 2009;1796:114–128. doi: 10.1016/j.bbcan.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Oliveira C, Velho S, Domingo E, Preto A, Hofstra RM, Hamelin R, Yamamoto H, Seruca R, Schwartz S., Jr Concomitant RASSF1A hypermethylation and KRAS/BRAF mutations occur preferentially in MSI sporadic colorectal cancer. Oncogene. 2005;24:7630–7634. doi: 10.1038/sj.onc.1208906. [DOI] [PubMed] [Google Scholar]
- 17.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 18.Donninger H, Vos MD, Clark GJ. The RASSF1A tumor suppressor. J Cell Sci. 2007;120:3163–3172. doi: 10.1242/jcs.010389. [DOI] [PubMed] [Google Scholar]
- 19.Yoon JH, Dammann R, Pfeifer GP. Hypermethylation of the CpG island of the RASSF1A gene in ovarian and renal cell carcinomas. Int J Cancer. 2001;94:212–217. doi: 10.1002/ijc.1466. [DOI] [PubMed] [Google Scholar]
- 20.Sakamoto N, Terai T, Ajioka Y, Abe S, Kobayasi O, Hirai S, Hino O, Watanabe H, Sato N, Shimoda T, Fujii H. Frequent hypermethylation of RASSF1A in early flat-type colorectal tumors. Oncogene. 2004;23:8900–8907. doi: 10.1038/sj.onc.1207993. [DOI] [PubMed] [Google Scholar]
- 21.Miyoshi Y, Nagase H, Ando H, Horii A, Ichii S, Nakatsuru S, Aoki T, Miki Y, Mori T, Nakamura Y. Somatic mutations of the APC gene in colorectal tumors: Mutation cluster region in the APC gene. Hum Mol Genet. 1992;1:229–233. doi: 10.1093/hmg/1.4.229. [DOI] [PubMed] [Google Scholar]
- 22.Rosin-Arbesfeld R, Townsley F, Bienz M. The APC tumour suppressor has a nuclear export function. Nature. 2000;406:1009–1012. doi: 10.1038/35023016. [DOI] [PubMed] [Google Scholar]
- 23.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 24.Schneikert J, Behrens J. The canonical Wnt signaling pathway and its APC partner in colon cancer development. Gut. 2007;56:417–425. doi: 10.1136/gut.2006.093310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naghibalhossaini F, Zamani M, Mokarram P, Khalili I, Rasti M, Mostafavi-Pour Z. Epigenetic and genetic analysis of WNT signaling pathway in sporadic colorectal cancer patients from Iran. Mol Biol Rep. 2012;39:6171–6178. doi: 10.1007/s11033-011-1434-6. [DOI] [PubMed] [Google Scholar]
- 26.Esteller M, Sparks A, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, Gonzales S, Tarafa G, Sidransky D, Meltzer SJ, et al. Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Res. 2000;60:4366–4371. [PubMed] [Google Scholar]
- 27.Lee S, Hwang KS, Lee HJ, Kim JS, Kang GH. Aberrant CpG island hypermethylation of multiple genes in colorectal neoplasia. Lab Invest. 2004;84:884–893. doi: 10.1038/labinvest.3700108. [DOI] [PubMed] [Google Scholar]
- 28.Lee BB, Lee EJ, Jung EH, Chun HK, Chang DK, Song SY, Park J, Kim DK. Aberrant methylation of APC, MGMT, RASSF2A, and Wif-1 genes in plasma as a biomarker for early detection of colorectal cancer. Clin Cancer Res. 2009;15:6185–6191. doi: 10.1158/1078-0432.CCR-09-0111. [DOI] [PubMed] [Google Scholar]
- 29.Chen SP, Chiu SC, Wu CC, Lin SZ, Kang JC, Chen YL, Lin PC, Pang CY, Harn HJ. The association of methylation in the promoter of APC and MGMT and the prognosis of Taiwanese CRC patients. Genet Test Mol Biomarkers. 2009;13:67–71. doi: 10.1089/gtmb.2008.0045. [DOI] [PubMed] [Google Scholar]
- 30.Clément G, Bosman FT, Fontolliet C, Benhattar J. Monoallelic methylation of the APC promoter is altered in normal gastric mucosa associated with neoplastic lesions. Cancer Res. 2004;64:6867–6873. doi: 10.1158/0008-5472.CAN-03-2503. [DOI] [PubMed] [Google Scholar]
- 31.Oken MM, Creech RH, Tormey DC, Horton J, Davis TE, McFadden ET, Carbone PP. Toxicity and response criteria of the Eastern Cooperative Oncology Group. Am J Clin Oncol. 1982;5:649–655. doi: 10.1097/00000421-198212000-00014. [DOI] [PubMed] [Google Scholar]
- 32.Edge S, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A, editors. AJCC Cancer Staging Manual. 7th. Springer; New York: 2010. [Google Scholar]
- 33.Choong MK, Tsafnat G. Genetic and epigenetic biomarkers of colorectal cancer. Clin Gastroenterol Hepatol. 2012;10:9–15. doi: 10.1016/j.cgh.2011.04.020. [DOI] [PubMed] [Google Scholar]
- 34.Ghazaleh HA, Chow RS, Choo SL, Pham D, Olessen JD, Wang RX, Onyskiw C, Baksh S. 14-3-3 mediated regulation of the tumor suppressor protein, RASSF1A. Apoptosis. 2010;15:117–127. doi: 10.1007/s10495-009-0451-6. [DOI] [PubMed] [Google Scholar]
- 35.Shivakumar L, Minna J, Sakamaki T, Pestell R, White MA. The RASSF1A tumor suppressor blocks cell cycle progression and inhibits cyclin D1 accumulation. Mol Cell Biol. 2002;22:4309–4318. doi: 10.1128/MCB.22.12.4309-4318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dallol A, Agathanggelou A, Fenton SL, Ahmed-Choudhury J, Hesson L, Vos MD, Clark GJ, Downward J, Maher ER, Latif F. RASSF1A interacts with microtubule-associated proteins and modulates microtubule dynamics. Cancer Res. 2004;64:4112–4116. doi: 10.1158/0008-5472.CAN-04-0267. [DOI] [PubMed] [Google Scholar]
- 37.Wagner KJ, Cooper WN, Grundy RG, Caldwell G, Jones C, Wadey RB, Morton D, Schofield PN, Reik W, Latif F, Maher ER. Frequent RASSF1A tumor suppressor gene promoter methylation in Wilms' tumor and colorectal cancer. Oncogene. 2002;21:7277–7282. doi: 10.1038/sj.onc.1205922. [DOI] [PubMed] [Google Scholar]
- 38.van Engeland M, Roemen GM, Brink M, Pachen MM, Weijenberg MP, de Bruïne AP, Arends JW, van den Brandt PA, de Goeij AF, Herman JG. K-ras mutations and RASSF1A promoter methylation in colorectal cancer. Oncogene. 2002;21:3792–3795. doi: 10.1038/sj.onc.1205466. [DOI] [PubMed] [Google Scholar]
- 39.Gonzalo V, Lozano JJ, Muñoz J, Balaguer F, Pellisé M, de Rodríguez Miguel C, Andreu M, Jover R, Llor X, Giráldez MD, et al. Aberrant gene promoter methylation associated with sporadic multiple colorectal cancer. PLoS One. 2010;5:e8777. doi: 10.1371/journal.pone.0008777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miladi-Abdennadher I, Abdelmaksoud-Damak R, Ayadi L, Khabir A, Frikha F, Kallel L, Amouri A, Frikha M, Sellami-Boudawara T, Gargouri A, Mokdad-Gargouri R. Hypermethylation of RARβ2 correlates with high COX-2 expression and poor prognosis in patients with colorectal carcinoma. Tumor Biol. 2010;31:503–511. doi: 10.1007/s13277-010-0063-3. [DOI] [PubMed] [Google Scholar]
- 41.Abouzeid HE, Kassem AM, Wahab Abdel AH, El-mezayen HA, Sharad H, Abdel Rahman S. Promoter hypermethylation of RASSF1A, MGMT, and HIC-1 genes in benign and malignant colorectal tumors. Tumor Biol. 2011;32:845–852. doi: 10.1007/s13277-011-0156-7. [DOI] [PubMed] [Google Scholar]
- 42.Balgkouranidou I, Matthaios D, Karayiannakis A, Bolanaki H, Michailidis P, Xenidis N, Amarantidis K, Chelis L, Trypsianis G, Chatzaki E, et al. Prognostic role of APC and RASSF1A promoter methylation status in cell free circulating DNA of operable gastric cancer patients. Mutat Res. 2015;778:46–51. doi: 10.1016/j.mrfmmm.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 43.Ahlquist T, Bottillo I, Danielsen SA, Meling GI, Rognum TO, Lind GE, Dallapiccola B, Lothe RA. RAS signaling in colorectal carcinomas through alteration of RAS, RAF, NF1, and/or RASSF1A. Neoplasia. 2008;10:680–686. doi: 10.1593/neo.08312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.An B, Kondo Y, Okamoto Y, Shinjo K, Kanemitsu Y, Komori K, Hirai T, Sawaki A, Tajika M, Nakamura T, et al. Characteristic methylation profile in CpG island methylator phenotype-negative distal colorectal cancers. Int J Cancer. 2010;127:2095–2105. doi: 10.1002/ijc.25225. [DOI] [PubMed] [Google Scholar]
- 45.Huang J, Zheng S, Jin SH, Zhang SZ. Somatic mutations of APC gene in carcinomas from hereditary non-polyposis colorectal cancer patients. World J Gastroenterol. 2010;10:834–836. doi: 10.3748/wjg.v10.i6.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim MS, Lee J, Sidransky D. DNA methylation markers in colorectal cancer. Cancer Metastasis Rev. 2010;29:181–206. doi: 10.1007/s10555-010-9207-6. [DOI] [PubMed] [Google Scholar]
- 47.Kamiyama H, Noda H, Takata O, Suzuki K, Kawamura Y, Konishi F. Promoter hypermethylation of tumor-related genes in peritoneal lavage and the prognosis of patients with colorectal cancer. J Surg Oncol. 2009;100:69–74. doi: 10.1002/jso.21291. [DOI] [PubMed] [Google Scholar]
- 48.Kamiyama H, Noda H, Takata O, Suzuki K, Kawamura Y, Konishi F. Promoter hypermethylation of tumor-related genes in peritoneal lavage and the prognosis of patients with colorectal cancer. J Surg Oncol. 2009;100:69–74. doi: 10.1002/jso.21291. [DOI] [PubMed] [Google Scholar]