Abstract
As a member of the p53 gene family, the p73 gene can affect an individual's susceptibility to cancer through a p53-like manner. DNA sequence variation in the p73 gene has been reported to be associated with cancer risk. The present study aimed to identify whether the p73 gene G4C14-to-A4T14 single nucleotide polymorphism (SNP) is associated with risk of cervical cancer in a Chinese population. The p73 G4C14-to-A4T14 polymorphism was genotyped in 175 cervical cancer and 189 healthy control peripheral blood DNA samples using high resolution melting, polymerase chain reaction with confronting two-pair primers and direct DNA sequencing. The results demonstrated that carriers of the AT/AT genotype were associated with a significantly increased risk of cervical cancer (P=0.042; χ2=4.122; odds ratio = 2.241; 95% confidence interval = 1.013–4.956) compared with the GC/GC genotype carriers. In addition, there was a significant association between p73 genotypes and tumor size in patients with cervical cancer (P=0.014; χ2=8.607). However, no association was identified between p73 genotypes and tumor stage, histological type or lymph node metastasis in patients with cervical cancer. These results suggest that the p73 G4C14-to-A4T14 SNP may function as a marker of genetic susceptibility to cervical cancer in the Chinese population.
Keywords: p73, polymorphism, susceptibility, high resolution melting, polymerase chain reaction with confronting two-pair primer, cervical cancer
Introduction
Cervical cancer is the third most prevalent malignancy in women worldwide, and is a predominant cause of cancer-associated mortality in women in developing countries; with an estimated 527,600 novel cases and 265,700 mortalities worldwide in 2012 (1–5). In addition, cervical cancer accounts for 45% of the total number of cancer incidence in the south of China (6). Epidemiological studies have identified major risk factors of the disease, and include human papilloma virus (HPV), sexual behavior, diet, smoking and genetic factors (7–10). In the majority of cases, cervical cancer results from genital infection with the human carcinogen, HPV, with ~90% of HPV infections clearing within a few months to a few years with no complications (11). Only a small proportion of HPV infections advance to cancer, thus indicating that genetic susceptibility may be important in cervical carcinogenesis (12). Identification of a genetic variant associated with cervical cancer may aid the elucidation of fundamental mechanisms underlying its formation and development, and also identify possible therapeutic targets.
p73 is located on chromosome 1p36.33, and encodes a member of the p53 transcription factor family involved in development and cellular responses to stress (13–16). p73 activates various p53-responsive gene promoters, including that of p21, which participates in DNA repair, cell-cycle arrest and apoptosis, and inhibits cell growth in a p53-like manner by inducing G1 cell-cycle arrest or apoptosis (17). Although p73 gene mutations occur in <2% of all cancer cases, loss of heterozygosity at the p73 locus has been reported at varying frequencies for different tumors (18–20). For example, the loss of heterozygosity, allele silencing and the decreased expression of the p73 gene can lead to changes in the prevalence of alterations in breast cancer (21).
At present, >19 single nucleotide polymorphisms (SNPs) have been identified in the p73 gene, either in exons or introns (22). However, only two SNPs located in exons have been determined to cause amino acid changes; these SNPs are naturally-linked at positions 4 (G to A) and 14 (C to T) in the 5′-untranslated region of exon 2 of the p73 gene, and are known as G4C14-to-A4T14 (rs2273953, rs1801173) (23). The G4C14-to-A4T14 SNP forms a stem-loop structure, which may affect gene expression by altering the efficiency of translation initiation; thus, this polymorphism may lead to functional consequences (24). The p73 gene G4C14-to-A4T14 SNP has been reported to correlate with various forms of cancer (25), including breast, lung and epithelial ovarian cancer (26–29). However, genetic studies have not yet attempted to identify the role of the p73 G4C14-to-A4T14 polymorphism, which may be associated with cervical cancer in the Chinese population.
The present study aimed to investigate whether this SNP was associated with risk of cervical cancer in a Chinese population using high-resolution melting (HRM) and polymerase chain reaction with confronting two-pair primers (PCR-CTPP), in addition to direct DNA sequencing.
Materials and methods
Study subjects and samples
Peripheral blood samples were obtained between January 2013 and October 2014 from 175 newly diagnosed patients with cervical cancer and 189 healthy controls at Xi'an No. 4 Hospital (Xi'an, China). Once collected, the samples were stored at −80°C. Final diagnoses were confirmed by routine histopathological examination. The selection criteria for the healthy controls included no individual history of cancer, and the frequency of cervical cancer cases was matched on age. All subjects were unrelated, ethnic Han Chinese. At recruitment, written informed consent was obtained from all participants, and each individual was interviewed to obtain information regarding demographic characteristics. The research protocol was approved by the Institutional Review Board of Xi'an No. 4 Hospital.
DNA extraction
The primary reagents used for DNA extraction included potassium iodide, 0.9% NaCl, chloroform/isoamyl alcohol (dilution, 24:1), isopropanol and 70% ethanol, which were all obtained from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Genomic DNA was extracted from ethylenediaminetetraacetic acid anticoagulated peripheral blood samples by phenol-chloroform extraction (30). Extracted DNA was stored at −80°C until use.
Genotyping of polymorphisms by HRM
Firstly, samples were genotyped by HRM (31). The sequences of the p73 primers used were as follows: Forward, 5′-CAGGAGGACAGAGCACGAG-3′ and reverse, 5′-CGAAGGTGGCTGAGGCTAG-3′. Quantitative PCR was performed with a 25 µl reaction mixture containing 12.5 µl SGExcel FastSYBR Mixture (Sangon Biotech, Shanghai, China), 10.5 µl sterilized water, 0.5 µl each primer and 1 µl DNA template. The PCR conditions consisted of an initial denaturation step of 94°C for 5 min, followed by 30 cycles of 94°C for 30 sec, 65°C for 30 sec and 72°C for 30 sec, and a final extension step of 72°C for 10 min. Melting curves were obtained following a denaturation period for 60 sec at 95°C and a holding period of 60 sec at 40°C; the start temperature was 75°C and the final temperature was 85°C, with a temperature gradient of 0.05°C/sec. PCR and melting procedures were analyzed using a LightCycler® Nano Instrument (Roche Diagnostics, Shanghai, China), and the melting curve (Fig. 1) and accuracy of genotyping data was validated by direct sequencing (Fig. 2).
Figure 1.
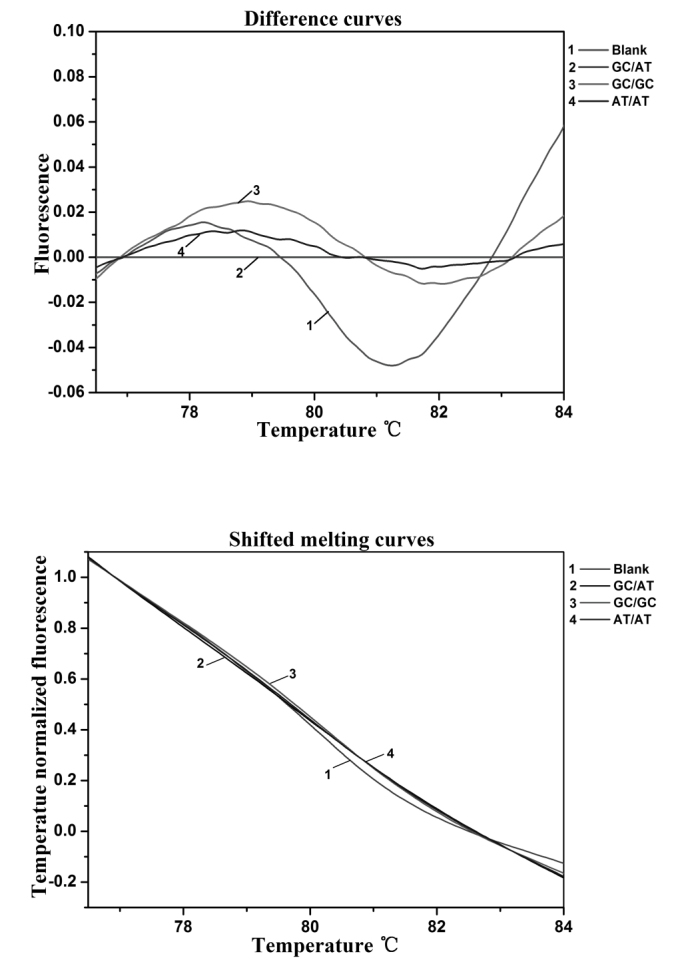
Melting curve of p73 G4C14-to-A4T14 polymorphism. Curve 1, blank; curve 2, GC/AT genotype; curve 3, GC/GC genotype; and curve 4, AT/AT genotype.
Figure 2.
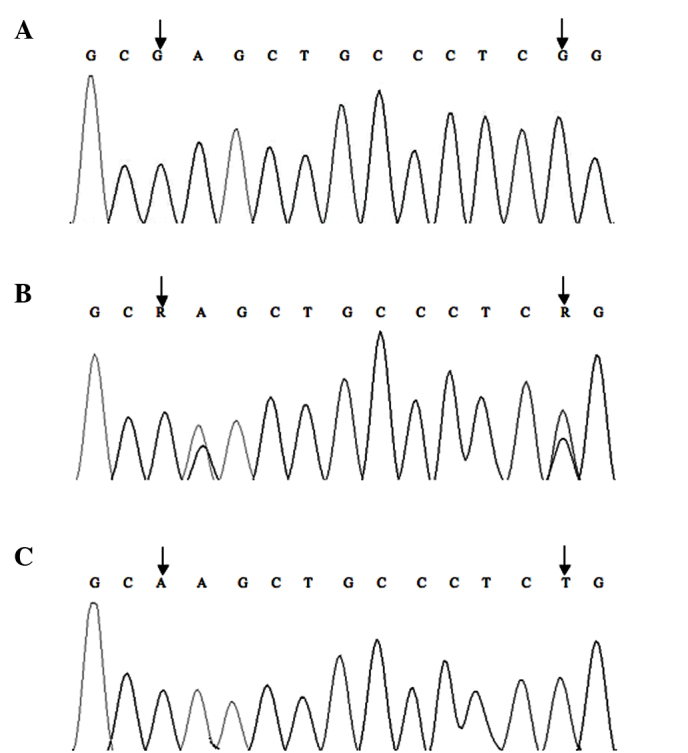
Direct-sequencing analysis for genotypes of p73 G4C14-to-A4T14 polymorphism. (A) GC/GC genotype; (B) GC/AT genotype; and (C) AT/AT genotype.
Genotyping of polymorphism by PCR-CTPP
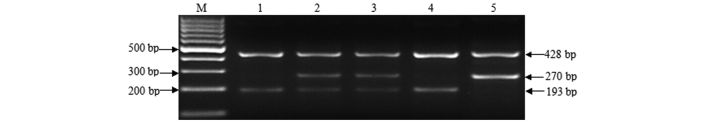
Secondly, the samples were analyzed for the p73 G4C14-to-A4T14 SNP genotype by PCR-CTPP. The PCR assay was performed using the GeneAmp™ PCR system 9700 (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The two-pair primers were purchased from Sangon Biotech Co., Ltd. (Shanghai, China) and the sequences were as follows: Forward 1, 5′-CCACGGATGGGTCTGATCC-3′; and reverse 1, 5′-GGCCTCCAAGGGCAGCTT–3′; forward 2, 5′-CCTTCCTTCCTGCAGAGCG-3′; and reverse 2, 5′-TTAGCCCAGCGAAGGTGG-3′. The melting temperature values of the forward 1, forward 2, reverse 1 and reverse 2 primers were 61.88°C, 61.88°C, 61.86°C and 59.54°C, respectively. PCR was performed in a 25 µl reaction mixture containing 12.5 µl DreamTaq Green PCR master mixture (Sangon Biotech), 7.5 µl double-distilled water, 1 µl each primer and 1 µl DNA template. PCR conditions consisted of an initial denaturation step of 95°C for 5 min, followed by 35 cycles of 95°C for 40 sec, 60°C for 40 sec and 72°C for 40 sec, and a final extension step of 72°C for 10 min. An aliquot (9 µl) of PCR product was visualized on 2% agarose gel. GC/GC yields 2 bands of 428 and 193 bp, AT/AT yields 2 bands of 428 and 270 bp, and GC/AT heterozygosity yields 3 bands of 428, 270 and 193 bp. Genotyping by PCR-CTPP (Fig. 3) was confirmed by DNA sequencing. The results of PCR-CTPP genotyping and sequencing analysis were consistent.
Figure 3.
Polymerase chain reaction with confronting two-pair primer analyses of p73 G4C14-to-A4T14 polymorphism. M, 100 bp marker; lane 1 and 4, GC/GC genotype (428 and 193 bp); lane 2 and 3, GC/AT genotype (428, 270 and 193 bp); lane 5, AT/AT genotype (428 and 270 bp).
Statistical analysis
Comparisons of clinical variables between patients with cancer and healthy controls and the distribution differences of genotypes and alleles were analyzed by the χ2 test. The association between p73 gene polymorphism and genetic susceptibility to cervical cancer was examined by multiple logistic regression analyses, and the P-values, odds ratio (OR) and 95% confidence intervals (CI) were calculated. χ2 tests were used to assess the association between p73 genotypes and clinicopathological characteristics of patients with cervical cancer. All statistical tests were two-sided, and P<0.05 was considered to indicate a statistically significant difference. SPSS software version 13.0 (SPSS, Inc., Chicago, IL, USA) was used to perform all statistical analyses.
Results
Characteristics of study subjects
The frequency distribution of selected characteristics of patients with cancer and healthy controls are presented in Table I. There was no significant difference in the frequency distributions of age (P=0.212), alcohol consumption (P=0.625) or family history (P=0.216) in the patients with cancer and the healthy controls. However, the frequency of smoking was higher in the patients with cancer (17.7%) compared with the controls (6.3%) (P=0.001; χ2=11.266; OR = 3.175, 95% CI = 1.574–6.405), which indicated that smoking is a significant risk factor for cervical cancer.
Table I.
Demographic characteristics of cervical cancer cases and controls.
| Characteristics | Patients with cancer, n (%) | Controls, n (%) | P-value | χ2a |
|---|---|---|---|---|
| Total | 175 | 189 | ||
| Age, years | 0.212 | 1.555 | ||
| ≤45 | 83 (47.4) | 102 (54.0) | ||
| >45 | 92 (52.6) | 87 (46.0) | ||
| Cigarette smoking | 0.001 | 11.266 | ||
| No | 144 (82.3) | 177 (93.7) | ||
| Yes | 31 (17.7) | 12 (6.3) | ||
| Alcohol consumption | 0.625 | 0.239 | ||
| No | 140 (80.0) | 155 (82.0) | ||
| Yes | 35 (20.0) | 34 (18.0) | ||
| Family history | 0.216 | 1.529 | ||
| No | 160 (91.4) | 179 (94.7) | ||
| Yes | 15 (8.6) | 10 (5.3) | ||
| Histology | ||||
| Adenocarcinoma | 158 (90.3) | |||
| Squamous cell | 14 (8.0) | |||
| Other | 3 (1.7) |
Two-sided test.
Distribution of polymorphisms
As is shown in Figs. 1 and 3, the allele and genotype frequencies of the p73 G4C14-to-A4T14 were obtained by HRM and PCR-CTPP. The results of these two methods, which are proved consistently by the DNA direct sequencing company (Sangon Biotech), are presented in Fig. 2. Furthermore, the HRM requires a short test time, but PCR-CTPP is low-cost. Allele and genotype frequencies of the p73 G4C14-to-A4T14 SNP are presented in Table II. The observed genotype frequencies of the p73 gene G4C14-to-A4T14 SNP in healthy controls did not significantly deviate from those predicted by the Hardy-Weinberg equilibrium. The frequencies of the p73 GC allele and AT allele were 74.3 and 25.7% among patients with cancer and 76.2 and 23.8% among controls, respectively. There was no significant association between carriers of the AT allele and risk of cervical cancer compared with GC allele carriers (P=0.552). The frequencies of the p73 GC/GC, GC/AT and AT/AT genotypes were 61.1, 26.3 and 12.6% in patients with cancer, and 57.7, 37.0 and 5.3% in controls, respectively. A significant association was observed between the AT/AT genotype and an increased risk of cervical cancer (P=0.042; χ2=4.122; OR = 2.241; 95% CI = 1.013–4.956) when the p73 GC/GC genotype was applied as a reference group.
Table II.
Genotype and allele frequencies of p73 among patients with cancer and controls and their association with the risk of cervical cancer.
| Genotype | Patients with cancera, n (%) | Controlsb, n (%) | P-value | χ2 | ORd (95% CI) |
|---|---|---|---|---|---|
| GC/GC | 107 (61.1) | 109 (57.7) | – | – | 1 (reference) |
| GC/AT | 46 (26.3) | 70 (37.0) | 0.085 | 2.966 | 0.669 (0.424–1.058) |
| AT/AT | 22 (12.6) | 10 (5.3) | 0.042c | 4.122 | 2.241 (1.013–4.956) |
| GC allele | 260 (74.3) | 288 (76.2) | – | – | 1 (reference) |
| AT allele | 90 (25.7) | 90 (23.8) | 0.552 | 0.354 | 1.108 (0.791–1.551) |
n=175
n=189
P<0.05
ORs were adjusted for age, cigarette smoking, alcohol consumption and family history. OR, odds ratio; CI, confidence interval.
Association of p73 gene G4C14-to-A4T14 SNP with clinicopathological characteristics of patients with cervical cancer
Associations between p73 G4C14-to-A4T14 genotypes and clinicopathological characteristics of patients with cervical cancer are presented in Table III. No correlation was identified between p73 G4C14-to-A4T14 genotype and tumor histological type, tumor grade or lymph node metastasis (P>0.05). However, a significant association was observed between p73 genotypes and tumor size in patients with cervical cancer (P=0.014; χ2=8.607).
Table III.
Association between p73 gene polymorphisms and clinicopathological parameters of patients with cervical cancer.
| Characteristics | n | GC/GC, n (%) | GC/AT, n (%) | AT/AT, n (%) | P-value |
|---|---|---|---|---|---|
| Histological type | 172 | ||||
| Squamous cell | 97 (61.4) | 41 (25.9) | 20 (12.7) | – | |
| Adenocarcinoma | 8 (57.1) | 5 (35.7) | 1 (7.2) | 0.665 | |
| Tumor stage | 175 | ||||
| I | 40 (59.7) | 20 (29.9) | 7 (10.4) | – | |
| II | 54 (62.1) | 20 (23.0) | 13 (14.9) | 0.519 | |
| III | 13 (61.9) | 6 (28.6) | 2 (9.5) | 0.983 | |
| Lymph node metastasis | 175 | ||||
| Yes | 35 (51.5) | 20 (29.4) | 13 (19.1) | – | |
| No | 72 (67.3) | 26 (24.3) | 9 (8.4) | 0.052 | |
| Tumor size, cm | 175 | ||||
| ≤2 | 57 (73.1) | 15 (19.2) | 6 (7.7) | – | |
| >2 | 50 (51.5) | 31 (32.0) | 16 (16.5) | 0.014 |
Discussion
In the present case-control study, the effect of the p73 gene G4C14-to-A4T14 SNP on the risk of cervical cancer was investigated in a Chinese population. Genotype distribution was significantly different between 175 patients with cervical cancer and 189 healthy controls, and indicated that the p73 G4C14-to-A4T14 AT/AT genotype was associated with an increased risk of cervical cancer.
The p73 gene, located in chromosome 1p36.33, is a novel member of the p53 family. The gene is frequently deleted in tumors, and is considered to possess multiple tumor suppressor properties (23). The gene has two naturally-linked SNPs in exon 2, consisting of a double-nucleotide substitution of G4C14 to A4T14 that form a stem-loop structure, which subsequently affects gene expression (31). Recently, several studies have demonstrated that the p73 G4C14-to-A4T14 SNP was significantly associated with different tumors and susceptibility of cancer (32–34). However, the number of these studies is limited. It has been demonstrated that patients with cervical cancer carrying the p73 AT allele in a southwestern European population have a two-fold increased susceptibility of developing high-grade squamous intraepithelial lesions (35). By contrast, an unconditional logistic model performed risk estimation for each p73 genotype and identified a possible association between the p73 A4T14 variant and risk of cervical cancer in a Japanese population (P=0.053; OR = 1.57; 95% CI = 0.99–2.48) (36). Furthermore, a previous study observed that the AT variant was associated with significantly decreased risk of cervical cancer compared with p73 GC homozygotes (37). The results of the present study are consistent with Craveiro et al (35) and demonstrated that individuals carrying the p73 G4C14-to-A4T14 AT/AT genotype are associated with a higher risk of cervical cancer than individuals with the GC/GC genotype in a Chinese population. In addition, a significant association was observed between p73 G4C14-to-A4T14 SNP genotypes and tumor size in patients with cervical cancer (P=0.014; χ2=8.607). No association was identified between the p73 G4C14-to-A4T14 SNP and tumor histological type, tumor grade or lymph node metastasis in patients with cervical cancer. According to these results, there are conflicting consequences of the p73 gene G4C14-to-A4T14 SNP on the risk of cervical cancer. Therefore, further studies with an increased number of samples from different ethnic populations are required to identify the mechanisms underlying this varying risk of cancer.
There are a number of available SNP detection methods, which include PCR-CTPP, PCR-restriction fragment length polymorphism, HRM, PCR-single strand conformational polymorphism and direct DNA sequencing. HRM analysis is a relatively new method of post-PCR examination that allows direct characterization of PCR amplicons in a closed system (38). Probe-free HRM quantitative PCR is performed in a closed-tube system, has no manual post-PCR processing, does not require the multiplex method and has a low reaction cost relative to other methods for rapid screening and detection of closely-related species in a laboratory. Therefore, the present study investigated the p73 G4C14-to-A4T14 SNP genotypes according to the difference in melting curves established by HRM.
In conclusion, the current study detected the genotypes of 175 patients with cervical cancer and 189 healthy controls in a Chinese population using HRM and PCR-CTPP, and a significant association was observed between the p73 G4C14-to-A4T14 SNP and risk of cervical cancer. These results suggest that the p73 G4C14-to-A4T14 SNP may function as a marker of genetic susceptibility to cervical cancer in the Chinese population. If confirmed by additional studies, the underlying mechanisms of genetic variations in the p73 gene G4C14-to-A4T14 SNP as presented here may be valuable for the treatment and diagnosis of this disease.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (Beijing, China; grant nos. 61371042, 61171061 and 31401456), the Technology Planning Project of Hunan Province (Changsha, China; grant no. 2014SK2019), the Natural Science Foundation of Hunan Province of China (Changsha, China; grant nos. 14JJ2149, 12JJ4032 and 12JJ4082).
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Akinyemiju TF. Socio-economic and health access determinants of breast and cervical cancer screening in low-income countries: Analysis of the world health survey. PLoS One. 2012;7:e48834. doi: 10.1371/journal.pone.0048834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ginsberg GM, Lauer JA, Zelle S, Baeten S, Baltussen R. Cost effectiveness of strategies to combat breast, cervical, and colorectal cancer in sub-Saharan Africa and South East Asia: Mathematical modelling study. BMJ. 2012;344:e614. doi: 10.1136/bmj.e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shuai HL, Luo X, Yan RL, Li J, Chen DL. XRCC1 polymorphisms are associated with cervical cancer risk and response to chemotherapy: A systematic review and meta-analysis. Asian Pac J Cancer Prev. 2012;13:6423–6427. doi: 10.7314/APJCP.2012.13.12.6423. [DOI] [PubMed] [Google Scholar]
- 5.Gocze K, Gombos K, Juhasz K, Kovacs K, Kajtar B, Benczik M, Gocze P, Patczai B, Arany I, Ember I. Unique microRNA expression profiles in cervical cancer. Anticancer Res. 2013;33:2561–2567. [PubMed] [Google Scholar]
- 6.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 7.Moore EE, Wark JD, Hopper JL, Erbas B, Garland SM. CeCaGeEn Study Group: The roles of genetic and environmental factors on risk of cervical cancer: A review of classical twin studies. Twin Res Hum Genet. 2012;15:79–86. doi: 10.1375/twin.15.1.79. [DOI] [PubMed] [Google Scholar]
- 8.Atalah E, Urteaga C, Rebolledo A, Villegas RA, Medina E, Csendes A. Diet, smoking and reproductive history as risk factor for cervical cancer. Rev Med Chile. 2001;129:597–603. (In Spanish) [PubMed] [Google Scholar]
- 9.Bayo S, Bosch FX, De SS, Munoz N, Combita AL, Coursaget P, Diaz M, Dolo A, van den Brule AJ, Meijer CJ. Risk factors of invasive cervical cancer in Mali. Int J Epidemiol. 2015;31:202–209. doi: 10.1093/ije/31.1.202. [DOI] [PubMed] [Google Scholar]
- 10.Vaccarella S, Lortet-Tieulent J, Plummer M, Franceschi S, Bray F. Worldwide trends in cervical cancer incidence: Impact of screening against changes in disease risk factors. Eur J Cancer. 2013;49:3262–3273. doi: 10.1016/j.ejca.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 11.Lippman A, Melnychuk R, Shimmin C, Boscoe M. Human papillomavirus, vaccines and women's health: Questions and cautions. CMAJ. 2007;177:484–487. doi: 10.1503/cmaj.070944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turati F, Negri E, Vecchia CL. Family history and the risk of cancer: Genetic factors influencing multiple cancer sites. Expert Rev Anticancer Ther. 2014;14:1–4. doi: 10.1586/14737140.2014.863713. [DOI] [PubMed] [Google Scholar]
- 13.Asher G, Tsvetkov P, Kahana C, Shaul Y. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. 2005;19:316–321. doi: 10.1101/gad.319905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramadan S, Terrinoni A, Catani MV, Sayan AE, Knight RA, Mueller M, Krammer PH, Melino G, Candi E. p73 induces apoptosis by different mechanisms. Biochem Biophys Res Commun. 2005;331:713–717. doi: 10.1016/j.bbrc.2005.03.156. [DOI] [PubMed] [Google Scholar]
- 15.Murray-Zmijewski F, Lane DP, Bourdon JC. p53/p63/p73 isoforms: An orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006;13:962–972. doi: 10.1038/sj.cdd.4401914. [DOI] [PubMed] [Google Scholar]
- 16.Zawacka-Pankau J, Kostecka A, Sznarkowska A, Hedström E, Kawiak A. p73 tumor suppressor protein: A close relative of p53 not only in structure but also in anti-cancer approach? Cell Cycle. 2010;9:720–728. doi: 10.4161/cc.9.4.10668. [DOI] [PubMed] [Google Scholar]
- 17.Massagué J. G1 cell-cycle control and cancer. Nature. 2004;432:298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- 18.Moll UM, Slade N. p63 and p73: Roles in development and tumor formation. Mol Cancer Res. 2004;2:371–386. [PubMed] [Google Scholar]
- 19.Tomasini R, Mak TW, Melino G. The impact of p53 and p73 on aneuploidy and cancer. Trends Cell Biol. 2008;18:244–252. doi: 10.1016/j.tcb.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Rufini A, Agostini M, Grespi F, Tomasini R, Sayan BS, Niklison-Chirou MV, Conforti F, Velletri T, Mastino A, Mak TW, et al. p73 in Cancer. Genes Cancer. 2011;2:491–502. doi: 10.1177/1947601911408890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahomadegbe JC, Tourpin S, Kaghad M, Zelek L, Vayssade M, Mathieu MC, Rochard F, Spielmann M, Tursz T, Caput D, et al. Loss of heterozygosity, allele silencing and decreased expression of p73 gene in breast cancers: Prevalence of alterations in inflammatory breast cancers. Oncogene. 2000;19:5413–5418. doi: 10.1038/sj.onc.1203914. [DOI] [PubMed] [Google Scholar]
- 22.Schabath MB, Wu X, Wei Q, Li G, Gu J, Spitz MR. Combined effects of the p53 and p73 polymorphisms on lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2006;15:158–161. doi: 10.1158/1055-9965.EPI-05-0622. [DOI] [PubMed] [Google Scholar]
- 23.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, et al. Monoallelically expressed gene related to p53 at lp36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/S0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Dong W, Mou Q, Leng Y, Zhang L, Duan L. Impact of p73 gene polymorphism on cancer susceptibility: A meta analysis. Int J Clin Exp Pathol. 2014;7:6820–6825. [PMC free article] [PubMed] [Google Scholar]
- 25.Deng B, Liu F, Wei Y, Luo L, Chen X, Yan L, Li B. Association of a p73 exon 2 G4C14-to-A4T14 polymorphism with risk of hepatocellular carcinoma in a Chinese population. Tumour Biol. 2013;34:293–299. doi: 10.1007/s13277-012-0550-9. [DOI] [PubMed] [Google Scholar]
- 26.Yuan P, Miao XP, Zhang XM, Wang ZH, Tan W, Zhang XR, Sun Y, Xu BH, Lin DX. Association of the responsiveness of advanced non-small cell lung cancer to platinum-based chemotherapy with p53 and p73 polymorphisms. Zhonghua Zhong Liu Za Zhi. 2006;28:107–110. (In Chinese) [PubMed] [Google Scholar]
- 27.Kang S, Wang DJ, Li WS, Wang N, Zhou RM, Sun DL, Duan YN, Li SZ, Li XF, Li Y. Association of p73 and MDM2 polymorphisms with the risk of epithelial ovarian cancer in Chinese women. Int J Gynecol Cancer. 2009;19:572–577. doi: 10.1111/IGC.0b013e3181a130ab. [DOI] [PubMed] [Google Scholar]
- 28.Zhou X, Wu C. Association of p73 G4C14-to-A4T14 polymorphisms with genetic susceptibilities to breast cancer: A case-control study. Med Oncol. 2012;29:3216–3221. doi: 10.1007/s12032-012-0240-x. [DOI] [PubMed] [Google Scholar]
- 29.Wang SS, Guo HY, Dong LL, Zhu XQ, Ma L, Li W, Tang JX. Association between a p73 gene polymorphism and genetic susceptibility to non-small cell lung cancer in the south of China. Asian Pac J Cancer Prev. 2014;15:10387–10391. doi: 10.7314/APJCP.2014.15.23.10387. [DOI] [PubMed] [Google Scholar]
- 30.Zhang HQ, Lu SJ, Tang MX, Chen LQ, Liu SH, Guo CF, Wang XY, Chen J, Xie L. Association of estrogen receptor beta gene polymorphisms with susceptibility to adolescent idiopathic scoliosis. Spine. 2009;34:760–764. doi: 10.1097/BRS.0b013e31818ad5ac. [DOI] [PubMed] [Google Scholar]
- 31.De Feo E, Simone B, Kamgaing RS, Gallì P, Hamajima N, Hu Z, Li G, Li Y, Matsuo K, Park JY, et al. p73 G4C14-to-A4T14 gene polymorphism and interaction with p53 exon 4 Arg72Pro on cancer susceptibility: A meta-analysis of the literature. Mutagenesis. 2012;27:267–273. doi: 10.1093/mutage/ger065. [DOI] [PubMed] [Google Scholar]
- 32.Xia S, Fang L, Zhao Z, Xie F, Li H. Genetic association between p73 G4C14-A4T14 polymorphism and risk of squamous cell carcinoma. Clin Exp Med. 2016;16:49–55. doi: 10.1007/s10238-014-0331-4. [DOI] [PubMed] [Google Scholar]
- 33.Zhou X. p73 G4C14-to-A4T14 polymorphisms are positively correlated with triple-negative breast cancer in southwestern China. Med Oncol. 2013;30:515. doi: 10.1007/s12032-013-0515-x. [DOI] [PubMed] [Google Scholar]
- 34.Wang DJ, Yan LI, Zhou RM, Wang N, Duan Y, Sun D, Li S, Li X, Kang S. Association of p73 and MDM2 polymorphisms with the risk of epithelial ovarian cancer. Tumor. 2009;29:892–897. doi: 10.1111/IGC.0b013e3181a130ab. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 35.Craveiro R, Bravo I, Catarino R, Teixeira AL, Sousa H, Pereira D, Pereira H, Medeiros R. The role of p73 G4C14-to-A4T14 polymorphism in the susceptibility to cervical cancer. DNA Cell Biol. 2012;31:224–229. doi: 10.1089/dna.2011.1294. [DOI] [PubMed] [Google Scholar]
- 36.Niwa Y, Hamajima N, Atsuta Y, Yamamoto K, Tamakoshi A, Saito T, Hirose K, Nakanishi T, Nawa A, Kuzuya K, Tajima K. Genetic polymorphisms of p73 G4C14-to-A4T14 at exon 2 and p53 Arg72Pro and the risk of cervical cancer in Japanese. Cancer Lett. 2004;205:55–60. doi: 10.1016/j.canlet.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 37.Liu S, Chan K, Leung R, Tsang P, Wei N, Ngan H. Genetic polymorphisms of p73 is associated with the risk of cervical cancer. Cancer Res. 2007;67:3434. [Google Scholar]
- 38.Vossen RH, Aten E, Roos A, den Dunnen JT. High-resolution melting analysis (HRMA): More than just sequence variant screening. Hum Mutat. 2009;30:860–866. doi: 10.1002/humu.21019. [DOI] [PubMed] [Google Scholar]