Abstract
The aim of the present study was to investigate the status of speckle-type POZ (pox virus and zinc finger protein) protein (SPOP) gene located on chromosome 17q21 in ovarian cancer (OC). The present study evaluated a tissue microarray, which contained 90 samples of ovarian cancer and 10 samples of normal ovarian tissue, using fluorescence in situ hybridization (FISH). FISH is a method where a SPOP-specific DNA red fluorescence probe was used for the experimental group and a centromere-specific DNA green fluorescence probe for chromosome 17 was used for the control group. The present study demonstrated that a deletion of the SPOP gene was observed in 52.27% (46/88) of the ovarian cancer tissues, but was not identified in normal ovarian tissues. Simultaneously, monosomy 17 was frequently identified in the ovarian cancer tissues, but not in the normal ovarian tissues. Furthermore, the present data revealed that the ovarian cancer histological subtype and grade were significantly associated with a deletion of the SPOP gene, which was assessed by the appearance of monosomy 17 in the ovarian cancer samples; the deletion of the SPOP gene was observed in a large proportion of serous epithelial ovarian cancer (41/61; 67.21%), particularly in grade 3 (31/37; 83.78%). In conclusion, deletion of the SPOP gene on chromosome 17 in ovarian cancer samples, which results from monosomy 17, indicates that the SPOP gene may serve as a tumor suppressor gene in ovarian cancer.
Keywords: ovarian cancer, speckle-type pox virus and zinc finger protein, tissue microarray, fluorescence in situ hybridization
Introduction
Ovarian cancer is one of three most common gynecological malignant tumors, along with cervical cancer and endometrial carcinoma, with the highest morbidity and mortality among these types of tumors. In 2008, it was estimated that 225,500 women were diagnosed with ovarian cancer and 140,200 women succumbed to this disease worldwide; however, these figures vary geographically (1). The fourth most common type of cancer among gynecological malignant tumors worldwide is epithelial ovarian cancer (EOC) (2). Due to the unfavorable anatomical location of ovarian cancer, ~70% of ovarian cancer patients are diagnosed at advanced stages, and the 5-year survival rate of these patients is 20–30% (2). At present, surgery and chemotherapy are the major treatment modalities of ovarian cancer, but the effectiveness of treatment is poor. The prognosis of patients with ovarian cancer is poor, due to late diagnosis, since there are non-specific symptoms and a lack of effective screening methods (2). Furthermore, the etiology and early events in the progression of ovarian cancer are poorly understood, and the genetic mechanism of ovarian cancer development is extremely complex. Deletion, mutation and/or altered expression of oncogenes and tumor suppressor genes are important in ovarian cancer development (3). Speckle-type POZ (pox virus and zinc finger protein) protein (SPOP) gene deletion or loss of heterozygosity (LOH) in its loci has been reported in 57.8% of breast cancer patients (4). In addition, in 24 types of cancer, including ovarian cancer, copy number analysis of SPOP amplification, LOH, deletion and mutation revealed that LOH frequently appears in the SPOP locus (4).
SPOP is an E3 ubiquitin ligase adaptor protein, which promotes substrate ubiquitination, leading to proteolysis of the substrate via the 26S proteasome. In addition, the SPOP gene is located on chromosome 17q21.33, which has frequent deletions and mutations (5). Recent studies on prostate and endometrial cancer demonstrated that the SPOP gene was frequently mutated and its locus was observed to undergo LOH, which indicated that SPOP may act as a tumor suppressor gene (5–8). In breast cancer, the SPOP gene frequently undergoes copy number loss, which also indicates that SPOP functions as a tumor suppressor gene (4). Similarly in ovarian cancer, the SPOP gene has been revealed to frequently undergo copy number loss (4–6). However, the role of SPOP gene in ovarian cancer has not been confirmed.
A number of studies have revealed that SPOP is frequently altered in human cancer. However, there are few studies that focus on the genetic aberrations of the SPOP gene in ovarian cancer. The aim of the present study was to investigate the role of the SPOP gene in ovarian tumorigenesis, and to evaluate numerical aberrations of chromosome 17 and SPOP gene alterations in an ovarian cancer tissue microarray (TMA) with fluorescence in situ hybridization (FISH).
Materials and methods
TMA
A TMA of ovarian cancer was purchased from Xi'an Ailina Biotechnology Co., Ltd. (catalog no. BC11115a; Xi'an, China), which consisted of 100 cases and 100 spots, including 5 cases of clear cell carcinoma, 63 cases of serous EOC, 10 cases of mucous EOC, 2 cases of endometrial carcinoma, 10 cases of metastatic adenocarcinoma and 10 cases of normal ovarian tissue. Evaluation of the TMA by the present study was approved by Xi'an Ailina Biotechnology Ethics Committee (The Second Affiliated Hospital of Chongqing Medical University, Chongqing, China).
FISH
FISH technique was used on the TMA, with a SPOP-specific DNA fluorescence probe used as the experimental group and a centromere-specific DNA fluorescence probe for chromosome 17 [chromosome enumeration probe 17 (CEP17)] for the control group. Fluorescence in situ hybridization Detection kit (catalog no., G 100984R-8; included SPOP probe, CEP17 probe, SSC buffer, RNase A, pepsin and hybridization buffer) was purchased from Agilent Technologies, Inc. (Cedar Creek, TX, USA). The SPOP probe, labeled red (rhodamine), covers a 190 Kb region of 17q21.33, which includes the whole SPOP genome. The CEP17 control probe was labeled green [fluorescein isothiocyanate (FITC)]. The FISH technique was performed according to the manufacturer's protocol. Briefly, the TMA samples were dewaxed, and underwent proteolysis for 10 min. The probes were then added, followed by protein denaturation for 10 min at 85°C and hybridization for 15–17 h at 37°C. The hybridization was observed using a fluorescence microscope (BX51; Olympus Corporation, Tokyo, Japan) equipped with selective filters for the fluorochromes used.
Fluorescence signals were scored using a well-established criteria (9). The interpretation of the signals followed the same interpretation criteria as previous studies using human epidermal growth factor (HER-2) gene (3) and p16 gene (10). A normal cell has two copies of chromosome 17 (shown by two green spots) and two copies of SPOP gene (shown by two red spots). For the SPOP FISH test the FISH ratio was calculated as follows: FISH ratio (SPOP/CEP17) = number of red rhodamine fluorescent SPOP signals / number of green FITC fluorescent CEP17 signals. FISH was reported using the following ratios: Deletion of SPOP gene, SPOP/CEP17 <0.7 or SPOP signals/nucleus <1.5; normal SPOP gene, 0.7≤SPOP/CEP17≤2.0; amplification of SPOP gene, SPOP/CEP17 >2.0; monosomy, >10% cells with CEP17 signals/nucleus <1.5; polysomy, >10% cells with CEP17 signals/nucleus >2.5; disome, CEP17 signals/nucleus = 1.5–2.5. The results were reviewed by two pathologists (The Center of Pathological Diagnosis, Chongqing Medical University, Chongqing, China).
Statistical analysis
SPSS version 19.0 software for Windows (IBM SPSS, Armonk, NY, USA) was used. Bivariate analysis was performed using χ2 test or likelihood-ratio test, and Fisher's exact probability method when required. P<0.05 was considered to indicate a statistically significant difference.
Results
Status of SPOP gene and CEP17
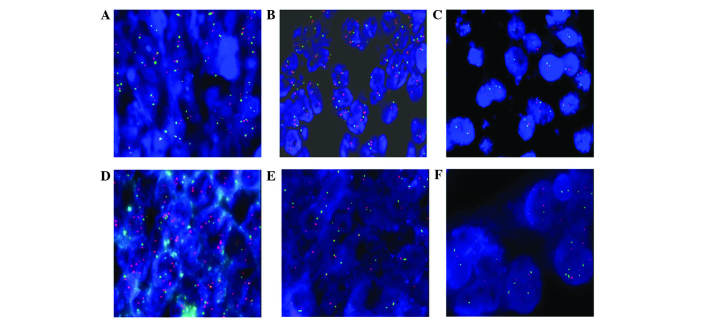
In total, 98 out of 100 cases from the TMA were evaluated; 2 cases of serous EOC could not be analyzed, due to unclear fluorescence with FISH. No alterations in the SPOP gene or CEP17 were observed in normal ovarian tissue using FISH (Fig. 1A). In ovarian cancer, 37 cases (37/88; 42.05%) were classified as disome and the SPOP genes were normal (Fig. 1B). Comparatively, the SPOP genes in 51 cases (51/88; 57.95%) were abnormal as follows: deletion, 1 case (Fig. 1C); amplification, 1 case (Fig. 1D); monosomy, 45 cases (Fig. 1E); and polysomy, 4 cases (Fig. 1F).
Figure 1.
SPOP gene alterations and numerical abnormalities of chromosome 17 in ovarian tissue. (A) In normal ovarian tissue, signals/nucleus of CEP17 all showed two green spots, and signals of SPOP gene showed two red spots. (B) In certain ovarian cancer tissues, signals/nucleus of CEP17 showed two green spots, and signals of SPOP gene showed two red spots. (C) Deletion of the SPOP gene had a SPOP/CEP17 signal ratio of <0.7. (D) Amplification of the SPOP gene had a SPOP/CEP17 signal ratio of >2.0. (E) Monosomy of CEP17 had a CEP17 signals/nucleus ration of <1.5 in >10% cells. (F) Polysomy of CEP17 had a CEP17 signals/nucleus ration of >2.5 in >10% cells. SPOP, speckle-type POZ (pox virus and zinc finger protein) protein; CEP17, chromosome enumeration probe 17.
Differences in SPOP genes between histological types and grades of ovarian cancer
The status of SPOP genes in ovarian cancer are shown in Table I. The status of the SPOP gene in ovarian cancer and normal ovarian tissues differed significantly (P<0.01; Table II). In addition for ovarian cancer, the status of the SPOP gene in serous EOC and clear cell carcinoma differed significantly (P<0.01), while the status of the SPOP gene of serous EOC and other histological subtypes did not differ significantly (P>0.05; Tables III and IV). The present data revealed that the SPOP gene differed significantly between grade 3 and grade 1 serous EOC (P<0.01). Similarly, the status of the SPOP gene differed significantly between grade 3 and grade 2 serous EOC (P<0.01). However, the status of the SPOP gene of grade 1 and grade 2 did not differ significantly in serous EOC (P=0.104) (Tables V and VI).
Table I.
SPOP/CEP17 in ovarian cancer and normal ovarian tissues.
| Ratio (SPOP/CEP17) | Status of SPOP gene | Status of CEP17 | Ovarian cancer tissue, n | Normal ovarian tissue, n |
|---|---|---|---|---|
| Total | 88 | 10 | ||
| Ratio >2 | Amplification | – | 1 | 0 |
| 0.7≤ratio≤2; CEP17<1.5 | – | Monosomy | 45 | 0 |
| 0.7≤ratio≤2; 1.5≤CEP17≤2.5 | – | Disome | 37 | 10 |
| 0.7≤ratio≤2; CEP17 >2.5 | – | Polysomy | 4 | 0 |
| Ratio <0.7 | Deletion | – | 1 | 0 |
SPOP/CEP17, fluorescence in situ hybridization ratio of speckle-type POZ (pox virus and zinc finger protein) protein to chromosome enumeration probe 17.
Table II.
SPOP gene status between ovarian cancer and normal ovarian tissues.
| SPOP gene status | |||
|---|---|---|---|
| Tissue type | Total, n | Abnormal, n | Normal, n |
| Ovarian cancera | 88 | 51 | 37 |
| Normal ovarian | 10 | 0 | 10 |
P<0.01 vs. normal ovarian tissues. SPOP, speckle-type POZ (pox virus and zinc finger protein) protein.
Table III.
SPOP gene status between different types of EOC.
| SPOP gene status | |||
|---|---|---|---|
| Histological type | Total, n | Abnormal, n | Normal, n |
| Total | 88 | 51 | 37 |
| Serous EOC | 41 | 20 | 61 |
| Mucinous EOC | 4 | 6 | 10 |
| Clear cell carcinoma | 0 | 5 | 5 |
| Endometrioid carcinoma | 0 | 2 | 2 |
| Metastatic adenocarcinoma | 6 | 4 | 10 |
SPOP, speckle-type POZ (pox virus and zinc finger protein) protein; EOC, epithelial ovarian cancer.
Table IV.
Bivarate analysis of speckle-type POZ (pox virus and zinc finger protein) gene status between different types of EOC.
| Histological type | χ2 | P-value |
|---|---|---|
| Serous EOC vs. mucinous EOC | 2.635 | 0.105a |
| Serous EOC vs. clear cell | 0.006b | |
| Serous EOC vs. endometrioid | 0.118b | |
| Serous EOC vs. metastatic | 0.195 | 0.658a |
Likelihood-ratio test
Fisher exact probability method. EOC, epithelial ovarian carcinoma.
Table V.
SPOP gene status between pathological grades of serous epithelial ovarian cancer.
| SPOP gene status | |||
|---|---|---|---|
| Grade | Total | Abnormal, n | Normal, n |
| Total | 61 | 41 | 20 |
| 1 | 2 | 8 | 10 |
| 2 | 8 | 6 | 14 |
| 3 | 31 | 6 | 37 |
SPOP, speckle-type POZ (pox virus and zinc finger protein) protein.
Table VI.
Bivarate analysis of speckle-type POZ (pox virus and zinc finger protein) protein gene status between pathological grades of serous epithelial ovarian cancer.
Fisher exact probability method
χ2 test
likelihood-ratio test.
Discussion
The present study demonstrated that there is a significant difference in the status of SPOP genes between ovarian cancer and normal ovarian tissues using FISH assay on a TMA. The present results reveal that in ovarian cancer, SPOP gene copies undergo deletion and amplification, and chromosome 17 undergoes monosomy, disome and polysomy. In addition, out of 88 cases of ovarian cancer, there were 45 cases of monosomy 17 (45/88; 51.14%); monosomy 17 results in the deletion of SPOP gene. By contrast, the present study observed that the SPOP gene and chromosome 17 were unaltered in normal ovarian tissues (Tables I–III).
The data gathered in the present study indicated that the deletion of the SPOP gene, in association with monosomy 17, may contribute to ovarian carcinogenesis. In addition, the present results demonstrate that there is a high proportion (45/88; 51.14%) of monosomy 17 in ovarian cancer (Table I), and indicates that chromosome 17 has undergone loss of a certain number of tumor suppressor genes. Furthermore, there may be a positive association between histological subtypes and tumor grade and the deletion of the SPOP gene in ovarian cancer (Table IV and VI, respectively). The deletion of the SPOP gene appeared in a large proportion of serous EOC tissues (41/61; 67.21%), particularly in grade 3 EOC (31/37; 83.78%) (Tables III–VI).
Previously, several studies concerning the genetic alterations in ovarian carcinogenesis have identified specific molecular markers that may provide a novel method of diagnosing ovarian cancer; therefore, these markers may potentially increase the prognosis of ovarian cancer patients and provide novel therapeutic strategies (11–14). The present study demonstrated that the SPOP gene loses its function when it is inactive, since the inactivation of the SPOP gene is primarily caused by deletion, LOH or mutation of the gene (4,6). However, investigation concerning the SPOP protein indicated that SPOP protein function may also alter when it translocates between the cell nucleus and the cytoplasm in clear cell renal cell carcinoma (ccRCC), which indicates that the SPOP protein serves as a regulatory hub to promote ccRCC tumorigenesis (15).
In addition to the inactivation of the SPOP gene, certain studies have identified that inactivation of certain genes located in chromosomal region 17q21, including breast cancer 1 and putative oncoprotein nm23, may contribute to ovarian carcinogenesis (16). Additionally, the present study data may indicate that a number of oncogenes that are associated with polysomy 17, including HER-2 and growth factor receptor bound protein 7 gene, may contribute to ovarian carcinogenesis (4). Therefore, the present study hypothesizes that alterations in chromosome 17 may be important in ovarian carcinogenesis.
SPOP is an adaptor for E3 ligase Cullin3, a nuclear protein, which promotes substrate protein ubiquitination and degradation by the 26S proteasome (4,6). Sequence analysis indicates that the SPOP protein contains a nuclear localization signal (NLS), which locates the SPOP protein to the cell nucleus leading to a promotion of cell apoptosis (15). The function of the SPOP protein is altered when it translocates from the nucleus to the cytoplasm, and in this situation it is referred to as SPOP-cyto, since it lacks the NLS (15).
Recent studies concerning the structures of SPOP-substrate complexes have revealed that SPOP-substrates require SPOP-binding consensus motifs φ-π-S-S/T-S/T (φ, nonpolar; π, polar) as a prerequisite for binding to the SPOP protein (17). Certain protein substrates, including death-domain-associated-protein (Daxx), are involved in transcription, cell-cycle regulation and apoptosis, and may bind to the MATH domain of the SPOP protein, leading to ubiquitination and degradation in the proteasome of the substrate. Therefore, the SPOP protein serves as a regulator of cellular function (6). At present, known SPOP substrates include macro H2A, Puckered, Daxx, glioma-associated-oncogene and phosphatase and tensin homolog (17).
Several studies concerning SPOP gene function indicate that the mutation or LOH of the SPOP gene is critical in prostate and breast cancer (4,5). In addition, the SPOP protein is key in ubiquitin-mediated degradation of steroid receptor coactivator-3 by 26S proteasome in breast cancer (4). These results suggest that SPOP may act as a tumor suppressor in breast and prostate cancers. The SPOP protein is primarily located in the cell nucleus, but SPOP-cyto and hypoxia drive the SPOP protein to become accumulated in the cytoplasm in kidney cancer, where the function of the SPOP protein may alter from proapoptotic to antiapoptotic and induce proliferation (15). This indicates that the SPOP protein may act as a tumor promoter in ccRCC.
The contradiction of tumor-promoting and tumor-suppressing roles of SPOP may be partially attributed to the following factors: Difference in substrate function due to different subcellular localization of the SPOP protein, and a difference in the expression of SPOP substrates owing to diverse cell and cancer varieties. In breast cancer, the SPOP protein binds to substrates that are tumor-promoters and therefore plays a tumor-suppressing role (4). In prostate cancer, the mutated SPOP gene does not bind to substrates that are tumor-suppressors, and so SPOP may play a tumor-suppressing role (5,6). In conclusion, the role of SPOP is associated with the status of the SPOP protein and the function of substrates (6).
TMA is a cost- and time-efficient method for high-throughput analysis. The TMA used in the present study contained 90 spots of ovarian cancer and 10 spots of normal ovarian tissue. Combined with the FISH technique, TMA limits the damage to specimens and creates consistent experimental conditions, thus ensuring the stability of experimental results.
In conclusion, the FISH technique is an experimental method that uses a SPOP DNA probe (red) and a CEP17 probe (green). It is a valuable method that concurrently detects numerical abnormalities in chromosome 17 and numerical alterations in SPOP gene copies. The results of the present study indicate that the deletion of the SPOP gene was observed in a large proportion of serous EOC cases, particularly in grade 3 serous EOC. The use of molecular diagnosis technologies allows for a high-throughput analysis of genes, therefore providing novel insights into gene expression profiles in ovarian cancer. Consequently, it may promote the development of novel biomarkers or aid in the identification of a novel therapy for human cancer. The present study has revealed that investigating the molecular mechanisms of the deletion of the SPOP gene in ovarian cancer is an important direction for future study.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (Beijing, China; grant no. 81100443), Chongqing Yuzhong District Science and Technology Plan projects (Chongqing, China; grant no. 20110303) and Chongqing Municipal Health Bureau Scientific Research Project (Chongqing, China; grant no. 20121039).
References
- 1.Lowe KA, Chia VM, Taylor A, O'Malley C, Kelsh M, Mohamed M, Mowat FS, Goff B. An international assessment of ovarian cancer incidence and mortality. Gynecol Oncol. 2013;130:107–114. doi: 10.1016/j.ygyno.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 2.Mayr D, Kanitz V, Amann G, Engel J, Burges A, Löhrs U, Diebold J. HER-2/neu amplification in ovarian tumours: A comprehensive immunohistochemical and FISH analysis on tissue microarrays. Histopathology. 2006;48:149–156. doi: 10.1111/j.1365-2559.2005.02306.x. [DOI] [PubMed] [Google Scholar]
- 3.Chao WR, Lee MY, Lin WL, Chen CK, Lin JC, Koo CL, Sheu GT, Han CP. HER2 amplification and overexpression are significantly correlated in mucinous epithelial ovarian cancer. Hum Pathol. 2014;45:810–816. doi: 10.1016/j.humpath.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 4.Li C, Ao J, Fu J, Lee DF, Xu J, Lonard D, O'Malley BW. Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene. 2011;30:4350–4364. doi: 10.1038/onc.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geng C, He B, Xu L, Barbieri CE, Eedunuri VK, Chew SA, Zimmermann M, Bond R, Shou J, Li C, et al. Prostate cancer-associated mutations in speckle-type POZ protein (SPOP) regulate steroid receptor coactivator 3 protein turnover. Proc Natl Acad Sci USA. 2013;110:6997–7002. doi: 10.1073/pnas.1304502110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mani RS. The emerging role of speckle-type POZ protein (SPOP) in cancer development. Drug Discovery Today. 2014;19:1498–1502. doi: 10.1016/j.drudis.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim MS, Je EM, Oh JE, Yoo NJ, Lee SH. Mutational and expressional analyses of SPOP, a candidate tumor suppressor gene, in prostate, gastric and colorectal cancers. APMIS. 2013;121:626–633. doi: 10.1111/apm.12030. [DOI] [PubMed] [Google Scholar]
- 8.Theurillat JP, Udeshi ND, Errington WJ, Svinkina T, Baca SC, Pop M, Wild PJ, Blattner M, Groner AC, Rubin MA, et al. Ubiquitylome analysis identifiesdysregulation of effector substrates inSPOP-mutant prostate cancer. Science. 2014;346:85–89. doi: 10.1126/science.1250255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hopman AH, Ramaekers FC, Raap AK, Beck JL, Devilee P, van der Ploeg M, Vooijs GP. In situ hybridization as a tool to study numerical chromosome aberrations in solid bladder tumors. Histochemistry. 1988;89:307–316. doi: 10.1007/BF00500631. [DOI] [PubMed] [Google Scholar]
- 10.Aravidis C, Panani AD, Kosmaidou Z, Thomakos N, Rodolakis A, Antsaklis A. Detection of numerical abnormalities of chromosome 9 and p16/CDKN2A gene alterations in ovarian cancer with fish analysis. Anticancer Res. 2012;32:5309–5313. [PubMed] [Google Scholar]
- 11.Puig S, Ruiz A, Lázaro C, Castel T, Lynch M, Palou J, Vilalta A, Weissenbach J, Mascaro JM, Estivill X. Chromosome 9p deletions in cutaneous malignant melanoma tumors: The minimal deleted region involves markers outside the p16 (CDKN2) gene. Am J Hum Genet. 1995;57:395–402. [PMC free article] [PubMed] [Google Scholar]
- 12.Caldas C, Hahn SA, da Costa LT, Redston MS, Schutte M, Seymour AB, Weinstein CL, Hruban RH, Yeo CJ, Kern SE. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet. 1994;8:27–32. doi: 10.1038/ng0994-27. [DOI] [PubMed] [Google Scholar]
- 13.Williamson MP, Elder PA, Shaw ME, Devlin J, Knowles MA. p16 (CDKN2) is a major deletion target at 9p21 in bladder cancer. Hum Mol Genet. 1995;4:1569–1577. doi: 10.1093/hmg/4.9.1569. [DOI] [PubMed] [Google Scholar]
- 14.Panani AD, Maliaga K, Babanaraki A, Bellenis I. Numerical abnormalities of chromosome 9 and p16CDKN2A gene deletion detected by FISH in non-small cell lung cancer. Anticancer Res. 2009;29:4483–4487. [PubMed] [Google Scholar]
- 15.Li G, Ci W, Karmakar S, Chen K, Dhar R, Fan Z, Guo Z, Zhang J, Ke Y, Wang L, et al. SPOP promotes tumorigenesis by acting as a key regulatory hub in kidney cancer. Cancer Cell. 2014;25:455–468. doi: 10.1016/j.ccr.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato H, Arakawa A, Suzumori K, Kataoka N, Young SR. FISH analysis of BRCA1 copy number in paraffin-embedded ovarian cancer tissue samples. Exp Mol Pathol. 2004;76:138–142. doi: 10.1016/j.yexmp.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 17.Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, Miller DJ, Walden H, Duda DM, Seyedin SN, et al. Structures of SPOP-substrate complexes: Insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell. 2009;36:39–50. doi: 10.1016/j.molcel.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]