Each year, more than 400,000 cases (>1,000 per day) of sudden cardiac death (SCD) are reported in the United States.1 The majority of these SCD cases and more than 10% of all natural deaths are due to cardiac arrhythmias.2 One condition that affects approximately 1 in 2,000 individuals and is known to cause these potentially lethal arrhythmias is long QT syndrome (LQTS).3
LQTS, a disorder of ventricular myocardial repolarization, is characterized by the prolongation of the heart rate-corrected QT interval (QTc) on an ECG, which can precipitate torsades de pointes (a subtype of polymorphic ventricular tachycardia) and as a result syncope, seizures, or SCD, in the setting of a structurally normal heart.2 LQTS is an inherited disorder, and approximately 80% of LQTS has been elucidated genetically. Accordingly, its genetic basis remains elusive in as many as 20% of patients.4
Here, we describe a case of severe LQTS in a young subject exhibiting profound QT prolongation (QTc > 650 ms) in which commercial genetic testing was negative. Recently, whole exome sequencing (WES) analysis has been used to successfully pinpoint the novel genetic cause of a variety of diseases when the genetic cause is still unknown.4, 5 Due to a negative family history, seemingly healthy parents, and normal parental ECGs, WES was performed on the trio (index case and his parents), and the results were filtered to include either novel sporadic or rare recessive variants. A sporadic p.D130G mutation in CALM3 was identified as the most likely cause of the patient’s LQTS.
CASE REPORT
The proband was the first child born to a Caucasian 34-year-old female and a Hispanic 35-year-old male. The mother had Factor V Leiden deficiency which was treated with aspirin throughout pregnancy. She has had no known miscarriages or spontaneous abortions and has never had syncope. There is no family history of sudden death. His father is adopted and there is little known about this family history; however he has never had syncope.
A fetal ultrasound at 27 weeks was reported to be normal with no mention of fetal bradycardia. He was born at 38.2 weeks gestational age via spontaneous vaginal delivery with reported decelerations and meconium stained fluid at the time of delivery. Apgar scores were 7 at one minute and 8 at five minutes. He was noted to have heart rates into the 60s and had hypotonia during the first eight hours of life. At ~8 hours of age, he was noticed to be slightly dusky, and the preductal saturations were 96–97 and the postductal saturations were 70–80. Chest x-ray demonstrated slight cardiomegaly.
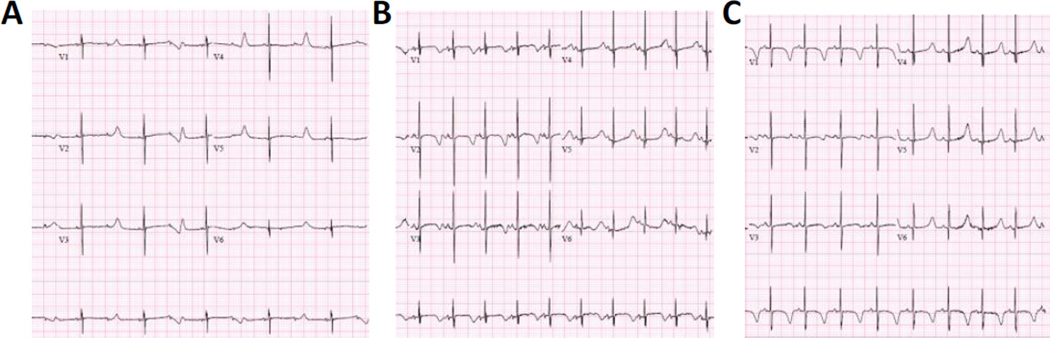
At this point, the proband was airlifted to a Children’s Hospital where his first ECG demonstrated bradycardia with sinus rhythm, functional 2:1 second-degree AV block, T-wave alternans, and profound QT prolongation, with a corrected QT interval of 690 ms (Figure 1A). An echocardiogram demonstrated a moderate sized perimembranous ventricular septal defect (VSD), a moderate patent ductus arteriosus (PDA), right ventricular dilatation with normal function, and normal left ventricular size and systolic function.
Figure 1. Electrocardiograms of the Proband.
(A) ECG taken at 1 day of age showing extreme QT prolongation (QTc = 690 ms), 2:1 atrioventricular block, bradycardia (59 bpm), and T wave alternans. (B) ECG at 5 weeks of age showing QT prolongation (QTc = 586 ms) at a normal heart rate (111 bpm) and resolution of the 2:1 AV block. (C) ECG at 6 months of age showing marked QT prolongation (QTc = 556 ms).
He continued to have ECGs with evidence of functional second-degree AV block due to extremely prolonged QT intervals. He was monitored at the hospital for about one month, during which time propranolol treatment was initiated and a single chamber pacemaker was placed. He had no arrhythmias, however he was treated for pulmonary hypertension. An ECG at five weeks revealed that the QTc had stabilized at 579 ms, with a normal heart rate and the 2:1 AV block had resolved (Figure 1B).
At seven weeks of age his weight was 5 kg (14th percentile) and height was 54.0 cm (1st percentile). At his most recent follow-up at six months, his marked QT prolongation persisted (Figure 1C); however no arrhythmias have been detected by ECG or device interrogation. In addition, the pulmonary hypertension resolved, his PDA has closed, and his VSD is small.
METHODS and RESULTS
When commercially available LQTS genetic testing (AKAP9, ANK2, CACNA1C, CAV3, KCNE1, KCNE2, KCNH2, KCNJ2, KCNQ1, SCN4B, SCN5A, and SNTA) was unable to identify any variants to determine the cause of the patient’s LQTS, this family was directed to the Mayo Clinic Windland Smith Rice Sudden Death Genomics Laboratory for further research-based genetic testing. After written consent for this Institutional Review Board-approved study, WES was initiated in attempt to determine the root cause of the proband’s LQTS.
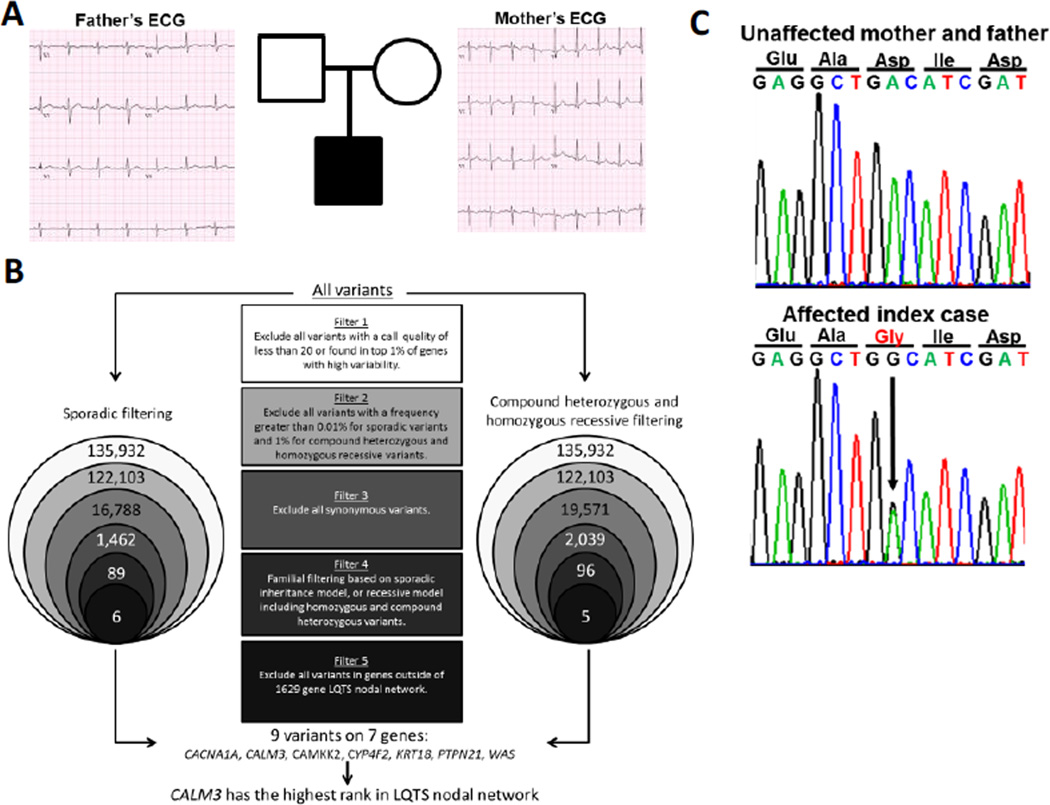
Due to the normal ECG status of each parent (Figure 2A), dominant sporadic or recessive inheritance patterns were suspected. Therefore, a case-parent trio based WES approach was utilized in an attempt to determine the patient’s underlying LQTS phenotype, and two distinct filtering strategies were employed based on those two hypothesized inheritance patterns (Figure 2B). Ingenuity Variant Analysis (Qiagen, Valencia, CA) was utilized to complete the following filtering strategy. In total, 135,932 single nucleotide variants and insertions/deletions (In/Dels) were called within the trio. In both strategies, the variants were filtered to remove poor quality variants by eliminating variants with a call quantity of less than 20 and variants found in the top 1% of genes with high variability. Second, variants were filtered for rarity using the NHLBI GO Exome Sequencing Project (http://evs.gs.washington.edu/EVS/) and 1,000 Genomes Project (http://browser.1000genomes.org/index.html) databases. Third, all synonymous variants (encoding for the same amino acid residue) were excluded to focus on non-synonymous (protein altering) variants or In/Dels which commonly cause LQTS.6
Figure 2. Familial Electrocardiograms, WES Filtering Strategy, and Mutation Confirmation.
(A) Pedigree of family utilized for WES, including ECGs for the mother and father of the proband, showing normal corrected QT intervals. (B) Filtering strategy utilized for WES, separated by inheritance pattern, to include filtering for sporadic inheritance (left) and recessive inheritance (right). Some variants were identified in both sequencing strategies; please see Table 1 for more information regarding the identified variants. (C) DNA Sanger sequencing confirmation of the p.D130G mutation in the proband (lower) and the p.D130G mutation’s absence in the parents (upper), supporting a sporadic inheritance pattern.
Next, familial filtering strategies utilizing the case-parent trio were employed. The first familial sequencing strategy was dominant sporadic, in which only rare variants unique to the child were kept as viable candidates. The second familial sequencing strategy was recessive, focusing on either homozygous variants or compound heterozygous variants. Homozygous variants occur when the same mutant allele is inherited from each parent, and compound heterozygous variants exist when two distinct variants are inherited from each parent, or when a sporadic variant is acquired in combination with a distinct inherited variant in the same gene. Dominant sporadic variants had to have a frequency of less than 0.01%, to identify only those variants that were extremely rare. Compound heterozygous and homozygous recessive variants were required to have a frequency of less than 1%. Compound heterozygous and homozygous recessive variants had a higher background frequency because these variants are likely not deleterious by themselves and therefore may be present at a higher frequency in the general population.
The final filtering step utilized the 1629 gene LQTS nodal network, which includes all genes which are significantly related to 13 known LQTS-susceptibility genes in terms of protein-protein interactions and signaling pathways, and are ranked based on association.7 Focusing specifically on genes within this network, we identified nine variants in seven genes (CACNA1A, CALM3, CAMKK2, CYP4F2, KRT18, PTPN21, and WAS). Overall, CALM3 had the highest ranking in the 1629 gene LQTS nodal network of the seven genes identified through our filtering process (Table 1). Within CALM3, we identified a variant, c.389A>G, which is predicted to encode for an aspartic acid to glycine change at protein position 130 (p.D130G). DNA Sanger sequencing was used to confirm that the mutation was present in the proband and absent in both of the unaffected parents, matching the sporadic inheritance pattern (Figure 2C).
Table 1.
Variants Identified through WES after Filtration and Ranked Using the LQTS Nodal Network.
| Gene | Variant | Rank in LQTS Nodal Network |
Inheritance Pattern |
|---|---|---|---|
| CALM3 | p.D130G | 88 | Sporadic |
| CACNA1A | p.H2219del | 150 | Sporadic |
| WAS | p.H180N | 349 | X-linked recessive |
| KRT18 | p.I67M | 383 | Sporadic |
| CAMKK2 | p.G539fs*5 | 822 | Sporadic |
| PTPN21 | p.Y569H | 1367 | Sporadic/compound heterozygous |
| PTPN21 | p.A725T | 1367 | Compound heterozygous |
| CYP4F2 | p.T472A | 1507 | Compound heterozygous |
| CYP4F2 | p.A483G | 1507 | Sporadic/compound heterozygous |
DISCUSSION
WES can be a very useful tool to uncover the genetic cause of a diagnosed disease. In this report, we described a severe case of LQTS in which clinical genetic testing was negative, and familial WES elucidated nine variants in seven potential LQTS-associated genes within our patient. Three independent algorithms ranked the CALM3 missense mutation as the most likely causative mutation. Although an in-frame deletion was identified in CACNA1A, the encoded pore-forming P/Q calcium channel subunit, this channel has negligible expression in the heart in contrast to the cardiac specific CACNA1C-encoded CaV1.2 L-type calcium channel. Thus, based on the previously published literature and its overall importance in the heart,7–8, 12–14 the sporadic p.D130G variant in CALM3-encoded calmodulin (CaM) is the most likely root cause for the patient’s phenotype.
Overall, there are three calmodulin genes, CALM1 (chr14q31), CALM2 (chr2p21), and CALM3 (chr19q13), that all encode for an identical calmodulin protein (CaM), which have been demonstrated to have differential expression levels in the human heart.8 CaM is a Ca2+ sensing, signal-transducing protein that modulates several ion channels. CaM is most well-known for its direct interaction with the Cav1.2 L-type calcium channel; deactivating Cav1.2 when CaM binds to free Ca2+.13 Mutations in two of the three calmodulin genes (CALM1 and CALM2) have been discovered recently in arrhythmia syndromes. The first discovery, in 2012, identified a CaM mutation in a large pedigree of catecholaminergic polymorphic ventricular tachycardia (CPVT).9 Subsequent cohort analysis identified an additional individual with sporadic CaM mutation. Since then, mutations have also been identified in cases of LQTS,8, 10 LQTS/CPVT overlap phenotypes,10 as well as a pedigree of idiopathic ventricular fibrillation (IVF).11 However, to date, no CALM3 mutations have been identified in any sudden death predisposing disorder.
This severe case of LQTS presented with several distinctive identifiers: presentation at birth, an extremely prolonged QTc of 690 ms, fetal bradycardia, T-wave alternans, and a 2:1 AV block. Much like the case described here, this precise missense mutation, p.D130G, was described originally in the CALM1-encoded CaM protein, and was identified in a 6-month-old female who had suffered from cardiac arrest, and was subsequently found to have a QTc of 630 ms, frequent episodes of T-wave alternans, and intermittent 2:1 AV block. Other LQTS cases with identified CaM mutations generally harbor a similar set of characteristics including extremely prolonged QTc intervals (often > 550 ms) and young age at presentation (typically < 10 years of age).6, 11
Many functional assays have been completed on a subset of the identified CaM mutations, including calcium binding assays, calcium transient assays, as well as interactions with ion channels such as Cav1.2 and RyR2.12–14 Because p.D130G has been identified previously in CALM1, functional assays using both mouse fetal ventricular myocytes and adult guinea pig cardiomyocyte models have been used to characterize this mutation. D130G-CaM reduces CaM's Ca2+ binding affinity, causes a calcium cycling disturbance in the form of increased calcium transients, results in electrical alternans, and prolongs the action potential.8, 12, 13 Because CALM1 and CALM3 encode a completely identical CaM, we confidently conclude that the pathophysiology of D130G-CALM3 equals that of D130G-CALM1.
One of the most interesting paradoxes surrounding the concept of CaM-mediated arrhythmogenesis centers around the peculiar finding that there exist 3 distinct CALM genes on 3 different chromosomes that nevertheless encode an identical calmodulin protein, and that disruption of only one of six possible CALM alleles could have such substantial phenotypic implications in human disease. Novel studies have begun to elucidate the important intricate balance of mutated and wildtype CaM proteins, and how 1/6th mutant product is sufficient to disrupt the L-type calcium channel’s deactivation.12 However, future research will be necessary to continue to understand the role of CaM, their differential expression in vivo, and how these mutated proteins contribute to sudden death predisposing diseases.
CONCLUSION
Overall, the identification of the genetic substrate responsible for sudden death predisposing disorders can help provide proper genotype specific therapies for affected patients. In addition, these genotypes can prevent sudden cardiac death by both identifying at risk individuals who could benefit from prophylactic treatment strategies and help generate a molecular substrate for novel therapies. Based on the findings of this report, in addition to screening both CALM1 and CALM2 for genetic mutations in cases of LQTS, CPVT and IVF, CALM3 should also be genetically interrogated for LQTS and possibly other sudden death related disorders.
Acknowledgments
Funding Sources:
This work was supported by the Mayo Clinic Windland Smith Rice Comprehensive Sudden Cardiac Death Program, the Sheikh Zayed Saif Mohammed Al Nahyan Fund in Pediatric Cardiology Research, the Dr. Scholl Fund, and the Hannah M. Wernke Memorial Fund. This project was also supported in part by funding from Mayo Clinic’s Center for Individualized Medicine (CIM). In addition, N.J. Boczek was supported by a CTSA Grant (UL1 TR000135) from the National Center for Advancing Translational Science (NCATS), a component of the NIH. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest:
Michael J. Ackerman is a consultant for Boston Scientific, Gilead Sciences, Medtronic, and St. Jude Medical and receives sales based royalties from Transgenomic for their FAMILION-LQTS and FAMILION-CPVT genetic tests. The other authors have no conflicts of interest relevant to this article to disclose.
REFERENCES
- 1.Go AS, Mozaffarian D, Roger VL, et al. Executive summary: heart disease and stroke statistics-2014 update: a report from the American Heart Association. Circulation. 2014 Jan 21;129:399–410. doi: 10.1161/01.cir.0000442015.53336.12. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz PJ, Ackerman MJ. The long QT syndrome: a transatlantic clinical approach to diagnosis and therapy. Eur Heart J. 2013 doi: 10.1093/eurheartj/eht089. [DOI] [PubMed] [Google Scholar]
- 3.Schwartz PJ, Stramba-Badiale M, Crotti L, et al. Prevalence of the congenital long-QT syndrome. Circulation. 2009;120:1761–1767. doi: 10.1161/CIRCULATIONAHA.109.863209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boczek NJ, Best JM, Tester DJ, Giudicessi JR, Middha S, Evans JM, Kamp TJ, Ackerman MJ. Exome sequencing and systems biology converge to identify novel mutations in the L-type calcium channel, CACNA1C, linked to autosomal dominant long QT syndrome. Circ Cardiovasc Genet. 2013;6:279–289. doi: 10.1161/CIRCGENETICS.113.000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szperl AM, Ricano-Ponce I, Li JK, et al. Exome sequencing in a family segregating for celiac disease. Clin Genet. 2011;80:138–147. doi: 10.1111/j.1399-0004.2011.01714.x. [DOI] [PubMed] [Google Scholar]
- 6.Kapplinger JD, Tester DJ, Salisbury BA, Carr JL, Harris-Kerr C, Pollevick GD, Wilde AA, Ackerman MJ. Spectrum and prevalence of mutations from the first 2,500 consecutive unrelated patients referred for the FAMILION long QT syndrome genetic test. Heart Rhythm. 2009;6:1297–1303. doi: 10.1016/j.hrthm.2009.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger SI, Ma'ayan A, Iyengar R. Systems pharmacology of arrhythmias. Sci Signal. 2010;3 doi: 10.1126/scisignal.2000723. ra30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crotti L, Johnson CN, Graf E, et al. Calmodulin mutations associated with recurrent cardiac arrest in infants. Circulation. 2013;127:1009–1017. doi: 10.1161/CIRCULATIONAHA.112.001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nyegaard M, Overgaard Michael T, Sondergaard Mads T, et al. Mutations in Calmodulin Cause Ventricular Tachycardia and Sudden Cardiac Death. Am J Hum Genet. 2012;91:703–712. doi: 10.1016/j.ajhg.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makita N, Yagihara N, Crotti L, et al. Novel calmodulin mutations associated with congenital arrhythmia susceptibility. Circ Cardiovasc Genet. 2014;7:466–474. doi: 10.1161/CIRCGENETICS.113.000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marsman RF, Barc J, Beekman L, Alders M, Dooijes D, van den Wijngaard A, Ratbi I, Sefiani A, Bhuiyan ZA, Wilde AA, Bezzina CR. A mutation in CALM1 encoding calmodulin in familial idiopathic ventricular fibrillation in childhood and adolescence. J Am Coll Cardiol. 2014;63:259–266. doi: 10.1016/j.jacc.2013.07.091. [DOI] [PubMed] [Google Scholar]
- 12.Limpitikul WB, Dick IE, Joshi-Mukherjee R, Overgaard MT, George AL, Jr, Yue DT. Calmodulin mutations associated with long QT syndrome prevent inactivation of cardiac L-type Ca(2+) currents and promote proarrhythmic behavior in ventricular myocytes. J Mol Cell Cardiol. 2014;74:115–124. doi: 10.1016/j.yjmcc.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin G, Hassan F, Haroun AR, Murphy LL, Crotti L, Schwartz PJ, George AL, Satin J. Arrhythmogenic calmodulin mutations disrupt intracellular cardiomyocyte Ca2+ regulation by distinct mechanisms. J Am Heart Assoc. 2014;3:e000996. doi: 10.1161/JAHA.114.000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hwang HS, Nitu FR, Yang Y, et al. Divergent regulation of ryanodine receptor 2 calcium release channels by arrhythmogenic human calmodulin missense mutants. Circ Res. 2014;114:1114–1124. doi: 10.1161/CIRCRESAHA.114.303391. [DOI] [PMC free article] [PubMed] [Google Scholar]