Abstract
High-throughput sequencing of circulating tumor DNA (ctDNA) promises to facilitate personalized cancer therapy. However, low quantities of cell-free DNA (cfDNA) in the blood and sequencing artifacts currently limit analytical sensitivity. To overcome these limitations, we introduce an approach for integrated digital error suppression (iDES). Our method combines in silico elimination of highly stereotypical background artifacts with a molecular barcoding strategy for the efficient recovery of cfDNA molecules. Individually, these two methods each improve the sensitivity of cancer personalized profiling by deep sequencing (CAPP-Seq) by ~3 fold, and synergize when combined to yield ~15-fold improvements. As a result, iDES-enhanced CAPP-Seq facilitates noninvasive variant detection across hundreds of kilobases. Applied to clinical non-small cell lung cancer (NSCLC) samples, our method enabled biopsy-free profiling of EGFR kinase domain mutations with 92% sensitivity and 96% specificity and detection of ctDNA down to 4 in 105 cfDNA molecules. We anticipate that iDES will aid the noninvasive genotyping and detection of ctDNA in research and clinical settings.
Liquid biopsies have the potential to improve cancer detection, noninvasive tumor genotyping, and disease monitoring1-7. However, in most early- and many advanced-stage solid tumors, ctDNA levels are very low3,8,9, complicating ctDNA detection and analysis. We recently described CAPP-Seq8 as an approach for highly sensitive quantitation of ctDNA in most patients with a given cancer type without the need for patient-specific optimizations. CAPP-Seq combines hybrid affinity capture of hundreds of genomic regions with deep sequencing and a specialized bioinformatics workflow. With a priori knowledge of tumor genotypes from sequencing of tumor biopsies, this method achieved a detection limit of 0.02% in cfDNA, and detected ctDNA in patients with early and advanced stages of the most common human malignancy, non-small cell lung cancer (NSCLC)8.
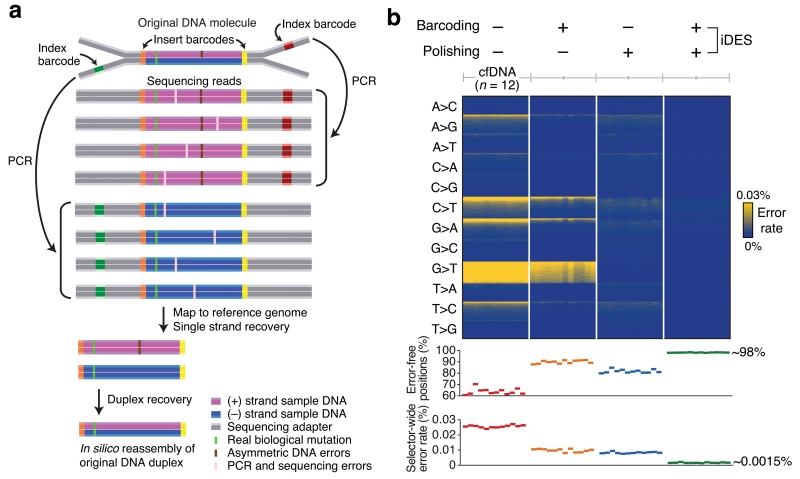
Several factors influence ctDNA detection limits, but recovery of cfDNA molecules and non-biological errors introduced during library preparation and sequencing continue to represent a major obstacle for ultrasensitive ctDNA profiling. To address these challenges and to further improve the performance of CAPP-Seq, we characterized the landscape of background errors observed during deep sequencing of cfDNA. Based on our findings, we developed iDES, an approach that eliminates most artifacts observed in cfDNA sequencing data while maximizing recovery of cfDNA molecules. We found that when incorporated into CAPP-Seq, iDES substantially outperforms other error suppression methods for the analysis of ctDNA from clinically practical blood volumes and allows for efficient non-invasive ctDNA profiling in the absence of available tissue biopsies. By demonstrating biopsy-free tumor genotyping and disease monitoring of NSCLC patients, our results suggest that iDES can significantly improve the sensitivity and specificity of the detection of low frequency alleles in ctDNA.
Results
Barriers to noninvasive genotyping and monitoring of ctDNA
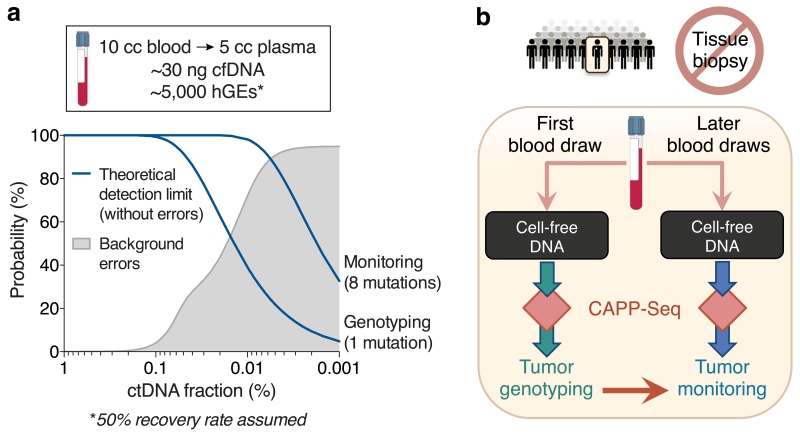
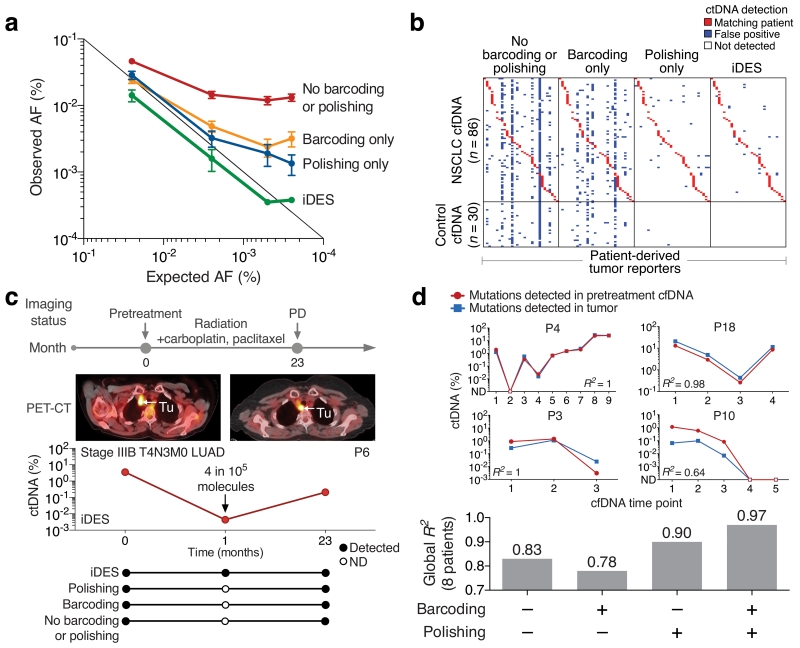
Two important factors that underlie the detection limit of all ctDNA profiling methods are the number of cfDNA molecules that are recovered and the number of mutations in a patient’s tumor that are interrogated. In our original description of CAPP-Seq, we developed strategies to optimize these variables from clinically relevant blood volumes, which are frequently limiting in cancer patients due to poor performance status, anemia, and comorbidities, among other factors. However, the degree to which analytical sensitivity can be further improved, whether through more efficient cfDNA recovery or increased coverage of tumor-specific aberrations, is ultimately limited by technical background (Fig. 1a). In analyzing cfDNA from healthy controls, background errors were increasingly evident below allele fractions (AFs) of ~0.2%. Under 0.02% AF (our previous detection-limit), >50% of sequenced genomic positions had artifacts (Fig. 1a). Therefore, we explored several strategies to combat these errors in order to enhance the ability of CAPP-Seq to perform non-invasive tumor genotyping and monitoring of minimal residual disease (Fig. 1b).
Figure 1. Framework for noninvasive profiling of ctDNA.
(a) Theoretical ctDNA detection limits are shown for single-mutation non-invasive genotyping and multi-mutation monitoring based on a typical cfDNA yield from 10 mL blood (assuming ~50% molecule recovery8; top) (Methods). Background noise of CAPP-Seq is shown to increase as a function of lower ctDNA allele fractions, and was determined using pooled cfDNA sequencing data from 30 healthy adult controls (Methods). (b) Schematic illustrating the potential application of CAPP-Seq to noninvasive (biopsy-free) genotyping and monitoring of ctDNA.
Molecular barcoding
Although a variety of methods for reducing sequencing-related artifacts have been reported10-17, a common approach involves tagging individual DNA molecules with unique identifiers (UIDs, also known as molecular barcodes)10-13,15,17. Such barcodes enable the precise tracking of individual molecules, making it possible to distinguish authentic somatic mutations arising in vivo from artifacts introduced ex vivo. The first descriptions of barcoding strategies reduced errors by tracking single DNA strands10,11, but more recent strategies can track double-stranded ‘duplex’ DNA molecules present in the original sample13,14,17. Although duplex barcoding achieves better error suppression than single-stranded barcoding methods, it is relatively inefficient13 and thus suboptimal for the limited cfDNA quantities obtainable in a clinical setting. We therefore hypothesized that a hybrid strategy leveraging the strengths of both approaches could have advantages for small cfDNA inputs from practical blood collection volumes.
We began by designing DNA sequencing adapters that allow for both single- and double-stranded molecular barcoding (Fig. 2a, Supplementary Fig. 1a). Each strand of the original DNA duplex molecule was tagged using four barcodes: three exogenous barcodes and one endogenous barcode comprising the molecule’s mapped genomic coordinates (Fig. 2a, Supplementary Fig. 1, Methods). For the first exogenous barcode, we replaced part of each sample barcode used for multiplexing with a degenerate 4-base molecular UID, such that each strand of a double-stranded DNA molecule received a separate UID (Supplementary Fig. 1a). This ‘single-stranded’ barcode was sequenced as part of the ‘index read’ and was therefore called the ‘index’ barcode. For the second and third barcodes, we integrated a 2-bp UID adjacent to the ligating side of each adapter. These ‘double-stranded’ UIDs are sequenced as part of the main read of inserted DNA fragments and are therefore called the ‘insert’ barcodes (Fig. 2a). When performing paired-end sequencing, insert barcodes are concatenated in silico into a single 4-base UID for each DNA strand. By matching complementary insert UIDs, this strategy allows for reconstruction of parental double-stranded DNA duplexes (Fig. 2a, Methods). Although considerably longer barcodes have been previously used11-13,17, our 4-bp UIDs yielded sufficient diversity for clinically relevant cfDNA quantities (Supplementary Fig. 2), thereby maximizing sequencing coverage for limited input molecules (Supplementary Note).
Figure 2. Development of integrated digital error suppression (iDES).
(a) Diagram depicting the use of CAPP-Seq barcode adapters to suppress errors. Here, CAPP-Seq adapters are ligated to a double-stranded (duplex) DNA molecule containing a real biological mutation in both strands as well as a non-replicated, asymmetric base change in only one strand (top). The combined application of insert and index barcodes allows for (i) error suppression and (ii) recovery of single stranded (center) and duplex (bottom) DNA molecules (Supplementary Fig. 1a, Methods). (b) Top: Heat map showing position-specific selector-wide error rates parceled into all possible base substitutions (rows) and organized by decreasing mean allele fractions (for each substitution type) across 12 cfDNA samples from healthy controls (columns; Supplementary Table 2). Background patterns are shown for different error suppression methods, including the combined application of barcoding and background polishing. Errors were defined as non-reference alleles excluding germline SNPs. Bottom: Selector-wide error metrics (Methods).
To characterize the performance of our molecular barcoding strategy, we applied a redesigned CAPP-Seq selector (i.e., capture panel) to maximize the number of mutations per NSCLC patient while minimizing panel size and sequencing cost (Methods). The resulting panel was expected to yield at least 8 mutations per tumor in 50% of NSCLC patients (Supplementary Table 1, Methods). We confirmed the expected performance of this panel by applying it to formalin-fixed tumor specimens and peripheral blood leukocyte (PBL) samples from NSCLC patients (Supplementary Tables 1–3).
We next profiled plasma samples from 12 healthy adult subjects with CAPP-Seq, using a median cfDNA input of 32ng. As the number of input molecules (i.e., haploid genomic equivalents, hGEs) is usually limited when analyzing cfDNA samples, we developed a computational pipeline that performs barcode-mediated error suppression in a manner that maximizes molecule retention (details in Methods). With this workflow, using index molecular barcodes alone improved mean error rates by 2.5 fold relative to our original approach8 and improved the fraction of error-free genomic positions by ~50% (Supplementary Fig. 1b). By comparison, insert UIDs outperformed index UIDs by ~1.25 fold, and a strategy combining both UIDs provided the lowest overall error rate (9 ×10−5 errors per base; Supplementary Fig. 1b).
Using 32ng of cell-free DNA, which is commonly obtainable from 1-2 tubes of blood (~10-20 mL; Fig. 1a), we then tested the recovery rates of barcoded cfDNA molecules (Supplementary Fig. 3). Of ~10,000 input hGEs, nearly 60% were present after hybrid capture (Supplementary Fig. 3, Supplementary Note). Notably, this result was independent of overall sequencing depth (Supplementary Fig. 3), which was found to determine final barcode yields in a predictable manner (Supplementary Fig. 4). Because we observed similar post-capture hGE recovery using standard sequencing adapters8, these data indicate that our barcoding strategy reduces errors without decreasing the efficiency of recoverable molecules.
To further explore barcoding performance, we examined nucleotide substitution patterns at each genomic position. We observed low levels of stereotypical background patterns across a subset of genomic locations and nucleotide substitutions prior to barcoding, and these errors were largely resistant to barcode-mediated error suppression (Fig. 2b). Similar patterns of recurrent errors were also present in independently generated cfDNA sequencing data18, suggesting a common artifact (Supplementary Fig. 5a). Thus, although molecular barcoding improves error profiles, it is not equally effective for all nucleotide substitution types encountered in cfDNA, suggesting that additional approaches would be useful to further improve assay sensitivity.
Stereotyped errors in sequencing cell-free DNA
In cfDNA from healthy adults, we observed recurrent background errors across all 12 nucleotide substitution classes (Fig. 2b). However, the majority of errors were due to G>T transversions and, to a lesser extent, C>T or G>A transitions (Fig. 2b). We hypothesized that these errors reflected oxidative damage arising either in vivo or ex vivo, leading to 8-oxoguanine19 and cytosine deamination20. We therefore tested the use of DNA damage repair enzymes to repair cfDNA prior to library preparation. However, such enzymatic treatments yielded no significant benefit in background error rates, suggesting ex vivo oxidative damage arising later in library preparation (Supplementary Fig. 6a). Additionally, when mapped to the reference human genome, G>T changes were much more frequent than reciprocal C>A events (Fig. 2b), and this imbalance was not attributable to strand biases in sequencing (Supplementary Fig. 6b).
These observations suggested that oxidative damage was likely occurring during the hybrid capture step, as CAPP-Seq employs capture baits that exclusively target the plus strand. We therefore examined the effect of varying the time of hybridization from between 0.1 to 3 days and found a progressive increase in the ratio of G>T to C>A errors (Supplementary Fig. 6c). In support of our model, we discovered similar imbalances in sequencing data from two independent studies that also employed hybrid capture for cfDNA7,18, using distinct baits and hybridization conditions, and targeting either the ‘+’ or ‘−’ strand (Supplementary Fig. 6d,e, Supplementary Note). Thus, a large fraction of recurrent background errors in capture-based cfDNA sequencing analyses are likely the result of oxidative damage arising after ligation and during strand-specific target sequence enrichment (Supplementary Fig. 6d).
Since we observed highly stereotypical background errors in cfDNA sequenced using capture-based approaches, we hypothesized that it should be possible to suppress these errors in silico. We therefore devised a computational approach for ’background polishing‘ by modeling position-specific errors in a training cohort of control samples to allow error suppression in independent samples (Supplementary Fig. 7; Methods). By applying a zero-inflated statistical model to capture a broad range of background distributions (Methods), our method addresses each type of recurrent background error in a position-specific and data-driven manner. Importantly, error-prone bases are not indiscriminately eliminated by our approach; rather, candidate errors are only removed if they are indistinguishable from their corresponding null distributions in normal controls.
We next tested the error-suppression capability of background polishing using cfDNA samples from healthy subjects. Notably, background polishing yielded similar global error rates to barcoding alone when applied to cfDNA samples without barcoding (~1.0×10−4 errors per base; Fig. 2b). Moreover, barcoding and background polishing synergized when combined, reducing selector-wide error rates ~15-fold (to 1.5×10−5 errors per base), and increasing error-free positions from ~90% to ~98% (Fig. 2b). The performance of background polishing was confirmed across an extended cohort of 30 healthy controls and 142 NSCLC patient cfDNA samples (Supplementary Fig. 5b). Moreover, differences in performance were marginal when using different training cohorts to build the background database (Supplementary Fig. 7c). We termed this serial application of molecular barcoding and background polishing, “integrated digital error suppression” (iDES).
Integrated digital error suppression
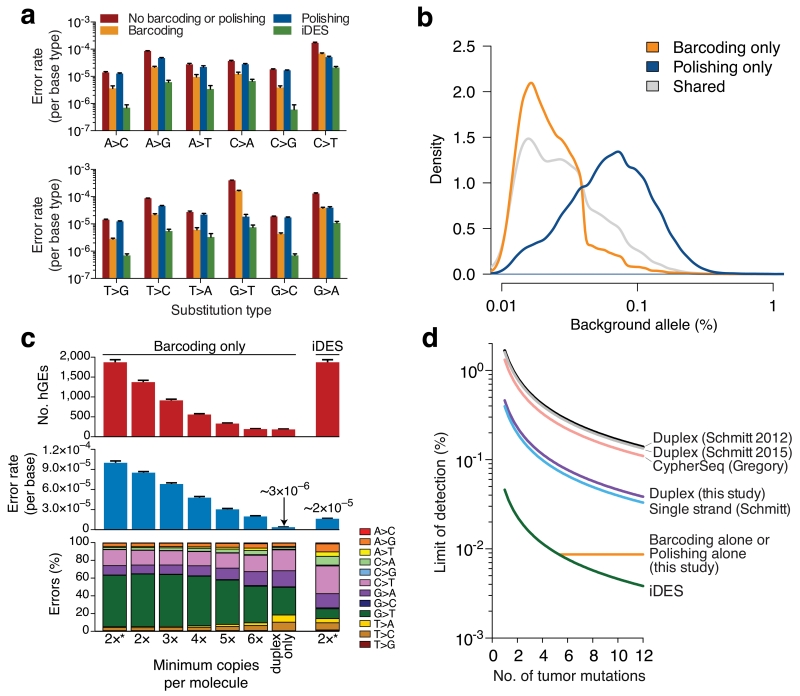
We next investigated the impact of each error suppression strategy on frequencies of each non-reference base (i.e., error) across ~300k genomic positions targeted by our NSCLC panel. Among the methods tested, iDES achieved significantly lower error rates for all 12 nucleotide substitution classes (P < 8.5×10−6, two-sided paired t test comparing iDES with each of the other methods, Fig. 3a). Despite possessing similar global error rates (Fig. 2b), we found barcoding and polishing to work best on different base substitution types, reflecting their distinct modes of action (Fig. 3a). Specifically, polishing preferentially suppressed positions with higher error levels consistent with stereotypical background, whereas barcoding tended to better address positions with lower error levels, characteristic of stochastic background (Fig. 3b). These results highlight the value of a combinatorial approach for error suppression toward maximizing specificity.
Figure 3. Technical performance of iDES.
(a) Impact of alternative error suppression methods on nucleotide substitution classes. Error rates were calculated with respect to each of the four reference bases separately (Methods). (b) Distribution of background alleles uniquely eliminated by barcoding or polishing alone in healthy control cfDNA. (c) Comparison of iDES with various barcoding strategies for selector-wide error profiles and recovered hGEs. The barcoding strategy denoted by ‘2*’ maximizes the retention of sequenced molecules and is the approach used in this work (Methods). Data are presented as means +/− s.e.m. (d) Analytical modeling of detection limits for various error suppression methods12,14,17 as a function of available tumor-derived mutations (90% confidence detection limit; Methods). Sequencing throughput was calibrated to iDES, such that the quantity of reads needed to recover 5,000 hGEs was determined and then used to estimate the number of recovered hGEs for all other methods given their reported efficiencies (Supplementary Fig. 8a). The theoretically maximum detection-limit of a given method, shown as a horizontal line, is bound by the method’s error rate. For additional details, see Supplementary Figure 8. The same 12 normal control samples shown in Fig. 2b were used for the analyses in a–c.
By combining background polishing with a barcoding approach optimized for efficient molecule recovery, we hypothesized that iDES should achieve lower detection limits than other barcoding strategies for clinically-obtainable cfDNA quantities. In support of this hypothesis, we found that barcoding alone required >5 family members (i.e., PCR duplicates) per UID to achieve a comparable error profile to iDES (Fig. 3c). However, as most UIDs were not supported by >5 family members even at high sequencing depths (Fig. 3c, Supplementary Table 2), this resulted in a nearly 10-fold reduction in both initial input molecule recovery and theoretical ctDNA detection-limit (Methods). By contrast, duplex molecules achieved an ultralow error rate of 3×10−6 errors per base in healthy control cfDNA (Fig. 3c). However, because 80–90% of recovered hGEs were single-stranded, nearly all recovered molecules were lost when requiring duplex-supported barcodes (Fig. 3c). These losses were expected based on the recovery rates of single-stranded molecules (Supplementary Fig. 4d, Supplementary Note), but they render duplex sequencing largely inadequate for rare variant detection from limited cfDNA quantities (Fig. 3d, Supplementary Fig. 8). As a result, iDES leverages duplex barcode support when it is observed but primarily relies on efficient recovery of polished single-stranded molecules to simultaneously maximize sensitivity and specificity (Methods). These results demonstrate that for clinically-achievable cfDNA inputs, iDES can achieve lower detection limits than previous approaches (Fig. 3d, Supplementary Fig. 8).
Technical assessment of noninvasive tumor genotyping
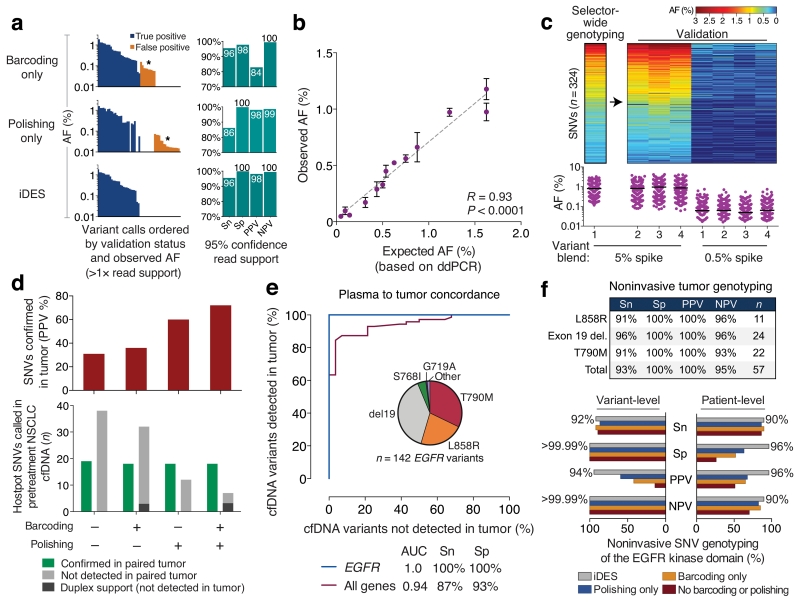
One of the most promising clinical applications of cfDNA analysis is noninvasive tumor genotyping. To evaluate noninvasive genotyping with iDES-enhanced CAPP-Seq, we first assessed its technical performance using a reference blend of cell lines that include known DNA variants within the targeted regions of our panel and that span a broad range of allele frequencies (Supplementary Table 4, Methods). To simulate ctDNA with a clinically-achievable blood volume (~10-20 ml), we created a 32ng mixture containing 5% of this reference blend in cfDNA from a healthy adult, resulting in spiked allele frequencies from 0.05% to 1.6%. We then compared the performance of iDES to barcoding and polishing for genotyping known variants within the reference blend (Supplementary Table 4; Methods). To address specificity, we also assessed 279 somatic alterations (SNVs and indels) that were targeted by our NSCLC selector and that are clinically actionable and/or highly recurrent in NSCLCs, but are not present in the reference blend (Supplementary Table 4, Methods). Consistent with our earlier analysis of cfDNA error profiles of healthy adults (Fig. 3b), barcoding and polishing each suppressed errors in a complementary fashion (Fig. 4a), whereas iDES outperformed both methods, achieving a uniformly high positive predictive value (PPV=98%) and sensitivity (96%) for detecting variants down to 1–3 mutant molecules (Fig. 4a). Notably, ignoring duplex-supported mutations would maintain identical PPV but decrease sensitivity from 96% to 86% (Supplementary Fig. 9a), highlighting the value of including duplex barcodes (Fig. 2a). We observed high consistency and linearity across technical replicates (n=4), and found observed AFs to be highly concordant with their expected fractions based on digital PCR (Fig. 4b).
Figure 4. Noninvasive tumor genotyping with iDES-enhanced CAPP-Seq.
Noninvasive tumor genotyping with iDES-enhanced CAPP-Seq was assessed using technical controls (a–c) and patients with NSCLC (d–f). (a) A DNA reference blend containing known alleles spanning a broad AF range was diluted to 5% in normal cfDNA and analyzed in replicate (n=4) for both known variants (n=29) and 279 negative control variants (Supplementary Table 4, Methods). Left: Differential impact of barcoding, polishing, and iDES on genotyping results for a single representative replicate. Only variant calls with at least 2 supporting reads are shown. Asterisks highlight the complementary background profiles removed by barcoding and polishing. Note that all variant calls are ordered along the x-axis, first by validation status and then by AF. Identical calls are aligned vertically. Right: Performance metrics across all four replicates. Genotyping thresholds were determined as described in Methods. (b) AFs determined by iDES-enhanced CAPP-Seq in the 5% variant blend from panel a (observed) versus their concentrations determined by digital PCR (expected). Only variants in the reference blend with externally validated AFs targeted by our NSCLC selector are shown (n=13; Supplementary Table 4). Data are expressed as means ± s.e.m (n=4 replicates). (c) Heat map (top) and scatter plot (bottom) depicting candidate SNVs identified by noninvasive selector-wide genotyping of the 5% variant blend from panel a (Supplementary Fig. 10, Methods). SNVs were tracked across three additional replicates and a ten-fold lower spike. Horizontal lines depict mean AFs. (d–f) Noninvasive tumor genotyping of NSCLC patients. (d) Bottom: The number of hotspot SNVs noninvasively detected in 24 pretreatment NSCLC cfDNA samples by four methods, including iDES (barcoding + polishing). All queried variants are listed in Supplementary Table 4. Top: Positive predictive value (PPV) of each method (indicated below), based on the number of hotspot SNVs that were later confirmed in matching tumor biopsies. (e) The performance of iDES for noninvasive tumor genotyping of two plasma cohorts was assessed using observed allele fractions with a Receiver Operating Characteristic (ROC) plot. In the first cohort (n=66 plasma samples from patients with matching tumor biopsies), hotspot variants from a predefined list of 292 variants were assessed (Supplementary Table 4). Results are shown for the 46 plasma samples with at least one detectable mutation (‘All genes’, n=24 patients); specificity was assessed using variants that were detected but that could not be verified in the primary tumor. In the second cohort, EGFR hotspot variants were assessed in an extended cohort of 103 plasma samples from 41 EGFR-positive patients with NSCLC (‘EGFR’). Specificity was assessed using 27 EGFR-wildtype subjects (Methods). The pie chart shows the distribution of detected EGFR variants. Only patients with genotyped tumors were analyzed. AUC, area under the curve. (f) Noninvasive genotyping of EGFR mutations in plasma samples from 37 patients with advanced NSCLC and with biopsy-confirmed EGFR mutations. Top: Performance of iDES-enhanced CAPP-Seq for the genotyping of actionable EGFR mutations (n=36 patients; 1 of 37 patients did not have an actionable alteration). All performance metrics were assessed at the variant level. Bottom: Comparison of error-suppression methods for noninvasive tumor genotyping of the entire EGFR kinase domain in all patients with biopsy-confirmed EGFR SNVs (n=29 of 37 patients). Performance metrics were assessed separately at the variant level and patient level (using 27 EGFR-wildtype subjects). Percentages indicate iDES performance only. Further details are provided in Methods. Sn, sensitivity; Sp, specificity; PPV, positive predictive value; NPV, negative predictive value.
We next explored the analytical sensitivity of iDES in a setting less restricted by clinically practical cfDNA inputs, but still within the range of cfDNA obtainable in patients with advanced disease21. Specifically, we prepared a mixture consisting of a 10-fold lower concentration of the same reference blend spiked into 72ng of healthy donor-derived cfDNA, and sequenced this mixture to recover ~12,500 hGEs. When applying the same genotyping approach, 100% of expected variants (0.025% ≤ expected AF ≤ 0.11%) were successfully identified with iDES at a PPV of 100%, representing a substantial improvement over barcoding (56%) and polishing (80%) alone (Supplementary Fig. 9b).
To expand the scope of variant detection beyond a targeted list of candidate mutations, we also tested the performance of iDES for selector-wide genotyping across 300kb of targeted genomic positions. We applied a SNV detection method that leverages hierarchical background modeling and identified 324 SNVs in the 5% reference mixture blend spanning a wide range of AFs (0.12% ≤ AF ≤ 3.3%) (Fig. 4c, Supplementary Fig. 10; Methods). By tracking the same SNVs across four technical replicates and a 10-fold lower dilution of the same blend, we determined which SNVs matched expected AFs and which were likely false positives. This yielded a PPV of 99.4%, highlighting the potential of this approach for noninvasive discovery and characterization of complex mutational profiles in ctDNA (Fig. 4c, Methods). Taken together, these analyses show that iDES can facilitate the simultaneous interrogation of numerous variants without compromising sensitivity or specificity, suggesting it is a robust approach for cfDNA-based noninvasive tumor genotyping.
Biopsy-free tumor genotyping of NSCLC patients
To assess the clinical potential of iDES-enhanced CAPP-Seq for noninvasive plasma genotyping in NSCLC patients, we first evaluated its specificity by querying normal controls for the presence of 292 predefined somatic mutation hotspots (Supplementary Table 4). In all, 94% of these healthy controls (17 of 18) had no detectable mutations, and the mean number of variants per subject was statistically indistinguishable from zero (Supplementary Fig. 11a,b). Only one healthy subject was determined to be positive for a hotspot variant. However, this variant encoded a PIK3CA E542K mutation with a high AF (1.7%) and with independent duplex barcode-supported molecules, suggesting it was a biological variant22 and not a technical artifact. This was consistent with our earlier results showing that most errors in cfDNA sequencing data are introduced ex vivo (Supplementary Fig. 6), and thus, any significantly detectable variants after iDES are likely to reflect true somatic heterogeneity. The specificity attained by either barcoding (17%) or polishing (83%) alone was considerably lower, underscoring the potential clinical utility of iDES (Supplementary Fig. 11a,b).
We next assessed iDES-enhanced CAPP-Seq genotyping using 66 serial plasma samples from patients with NSCLC, all of which had tumor biopsies available (Supplementary Tables 2,3). Applied to pretreatment plasma samples (n=24), iDES achieved a PPV of 72% for detecting hotspot SNVs that could be confirmed in a matched tumor biopsy (Fig. 4d). Notably, this PPV was 20% higher than polishing alone and two-fold better than barcoding alone (Fig. 4d). Moreover, the fraction of mutation-positive pretreatment plasma samples detected after applying iDES was found to increase as a function of disease stage (Supplementary Fig. 11c). When assessing all 66 longitudinal plasma samples for hotspot SNVs and indels, we noninvasively detected tumor-confirmed variants with AFs spanning >2 orders of magnitude (0.1% to 51% for SNVs; 0.08% to 20% for indels). These data yielded an area under the curve (AUC) of 0.94 for the identification of plasma variants that were detectable in matched tumors (Fig. 4e). Notably, in agreement with the high specificity we observed in healthy control samples, the majority of variants observed in plasma but not biopsy samples were either also found in serial blood samples, or in molecules that had duplex support (Supplementary Fig. 11d). This suggests that they were highly enriched for true tumor-derived mutations that were missed in the primary tumor sample due to subclonality, geographic heterogeneity, and/or the acquisition of new mutations between tumor biopsy and plasma sampling23.
Given the importance of EGFR mutations for existing and emerging targeted therapies24-26, we subsequently tested the performance of iDES for noninvasive genotyping of EGFR-mutant NSCLCs (Supplementary Table 2). First, we assessed PPV for candidate hotspot variants within EGFR (Supplementary Table 4). When profiling all available serial plasma samples (n=103) from an EGFR-mutant advanced NSCLC patient cohort (n=41), 142 EGFR variants were detected in 88 plasma samples and 100% of calls were verifiable in tumor biopsies (Fig. 4e, Supplementary Tables 2,4). Moreover, EGFR hotspot variants were never detected in EGFR-wildtype plasma samples, whether from normal controls or from patients with EGFR-wildtype tumors, demonstrating 100% specificity.
Secondary mutations in the EGFR kinase domain, such as T790M, frequently arise in NSCLC patients treated with first generation EGFR TKIs and are drivers of therapeutic resistance27,28. To evaluate the utility of iDES for the noninvasive identification of resistance mutations, we profiled the entire kinase domain of EGFR (885 bp) in 37 patients with advanced EGFR-mutant NSCLC. The detection rates for actionable EGFR variants were high, with an average sensitivity of 94% for activating mutations, and 91% for subclonal T790M resistance mutations emerging in erlotinib-treated patients (Fig. 4f, top). Furthermore, iDES noninvasively identified 92% of all tumor-confirmed EGFR SNVs, doing so with high positive and negative predictive values, regardless of whether data were analyzed at the variant- or patient-level (Fig. 4f, bottom). Although the other error suppression methods were similarly sensitive to iDES, they suffered from considerably higher false positive rates (Fig. 4f, bottom). Collectively, these data demonstrate the promise of iDES-enhanced CAPP-Seq for identifying clinically relevant mutations in cfDNA without prior knowledge of tumor genotypes.
Detection limit of iDES-enhanced CAPP-Seq
By integrating multiple tumor mutations in a single assay, CAPP-Seq can detect ctDNA at lower fractional abundances than methods targeting single mutations in a given patient (Fig. 1a). Therefore, given the superior error rate observed for iDES (Fig. 2b), we sought to experimentally validate its detection-limit for quantitating circulating tumor burden. To establish the detection-limit of iDES-enhanced CAPP-Seq, we designed a “personalized” selector covering 1,502 non-synonymous mutations identified by exome sequencing of a glioblastoma (GBM) tumor. We then created a series of 32ng DNA mixtures in which GBM DNA was spiked into healthy control cfDNA in defined proportions down to 2.5 in 106 molecules. In order to assess reproducibility, library preparation, capture, and sequencing were repeated on the same input. By performing in silico down-sampling to determine the minimum number of mutations needed, we found that iDES-enhanced CAPP-Seq could detect tumor-derived DNA down to 0.0025% (2.5 in 105 molecules) using only 30 tumor mutations and a clinically-achievable ~3,000 hGEs (Fig. 5a, Supplementary Fig. 12a,b, Supplementary Table 2). This detection-limit was consistent with the selector-wide error rate of iDES and was ~10-fold below our original description of CAPP-Seq8.
Figure 5. Ultrasensitive ctDNA detection and monitoring with iDES-enhanced CAPP-Seq.
(a) Analysis of ctDNA detection limits using a hypermutated glioblastoma (GBM) tumor mixed into normal control cfDNA in defined proportions. Here, 30 mutations were randomly selected from a pool of 1,502 total mutations known to be present in the GBM tumor and covered by the sequencing panel. Random sampling of 30 mutations was repeated 50 times and the results are presented as means +/− 95% confidence intervals. For further details, see Supplementary Fig. 12 and Methods. AF, allele fraction. (b) Comparison of error-suppression methods for the detection of ctDNA in pre- and post-treatment plasma from 30 NSCLC patients. Patient-derived somatic variants (columns; n=30 sets) were assessed in every plasma sample (rows; n=116), including 30 normal controls to evaluate specificity. The same samples were analyzed for each method (e.g., iDES) and are identically ordered in the heat map. Red squares denote a genetically matched sample (i.e., patient-derived tumor mutations were significantly detectable in a plasma sample from the same patient). Additional details are provided in Supplementary Fig. 13. (c) Using iDES, but not other methods, ctDNA was detectable prior to clinical progression in a stage IIIB NSCLC patient. (d) Top: Analysis of variants called from tumor biopsies versus variants called directly from pretreatment cfDNA with iDES-enhanced CAPP-Seq. Estimated ctDNA levels were compared by linear regression. Open circles/squares indicate time points without significantly detectable ctDNA. ND, not detected. Time points are shown in chronological order (1, pretreatment; >1, post-treatment). Bottom: Comparison of error suppression methods for the same analysis shown above but across all 8 evaluable patients (Methods). Linear regression was applied globally across all 37 plasma time points profiled for these eight patients.
Notably, when we analyzed the same dilution series considering only duplex-barcoded molecules we were able to accurately detect tumor-derived DNA down to 0.00025% (2.5 in 106 molecules) with high linearity (Supplementary Fig. 12c,d). This detection-limit is 10-fold below iDES and nearly 100-fold below the detection-limit of previous methods5. However, owing to technical limitations in the recovery of double-stranded molecules (Supplementary Fig. 8, Supplementary Note), this was only attainable through use of a personalized selector covering >1,500 mutations. Given 32ng of cfDNA input, duplex sequencing would only be able to outperform iDES when a large number (≥~200) of mutations are available.
Ultrasensitive ctDNA monitoring
Having validated the ctDNA detection-limit of iDES, we next sought to determine its utility for monitoring, which involves quantifying levels of ctDNA across time points. We assembled a cohort of plasma samples from 30 NSCLC patients whose tumor mutations were previously determined (Supplementary Table 3, Methods). We then evaluated the sensitivity and specificity of iDES-enhanced CAPP-Seq for detecting ctDNA using these mutations. When iDES was applied to pretreatment plasma, ctDNA was significantly detectable in 93% of patients, including 3 of 3 stage I tumors (Supplementary Fig. 13). Furthermore, ctDNA was significantly detectable in 73% of pre- and post-treatment plasma samples (n=86), with a specificity of 100% (Fig. 5b, Supplementary Table 5). Polishing alone displayed similar performance to iDES for ctDNA fractions >0.01% (Fig. 5b, Supplementary Fig. 13). However, methods without background polishing exhibited substantially higher false positive rates (Fig. 5b, Supplementary Fig. 13, Supplementary Table 5). Collectively, these data suggest that recurrent background errors are major determinants of specificity in a monitoring context.
Consistent with our GBM spike analysis, iDES also achieved the lowest detection limit among all the methods tested in NSCLC patient samples, enabling quantitation of ctDNA down to 0.004% (4 in 105 cfDNA molecules) in a patient prior to clinical progression (Fig. 5c, Supplementary Table 5, Methods). To our knowledge, this is the lowest amount of ctDNA detected by deep sequencing in any NSCLC patient to date. Thus, by increasing analytical sensitivity without affecting molecule recovery, iDES can improve sensitivity for response monitoring, detection of minimal residual disease, and surveillance.
Direct quantitation of ctDNA without the need for a tumor biopsy would have significant advantages in the clinic. We therefore evaluated noninvasive ctDNA monitoring with iDES by performing selector-wide genotyping directly from plasma, and compared this approach to direct profiling of tumor tissues. We studied all NSCLC patients in our cohort with a known tumor genotype from a tissue biopsy, a matching PBL sample, and at least 3 longitudinal plasma time points for correlation assessments (n=8; Methods). Regardless of whether mutations were identified directly from pretreatment plasma with iDES-enhanced CAPP-Seq or determined from tumor biopsies, ctDNA measurements were highly correlated (Fig. 5d, top; Supplementary Fig. 14). Moreover, mutations detected in plasma were confirmed in a matching primary tumor in 7 of 8 of evaluable patients for ctDNA fractions as low as 0.04%. When comparing the various error-suppression strategies for monitoring ctDNA, iDES continued to outperform the other methods tested (Fig. 5d, bottom). These results demonstrate the substantial advantage of iDES-enhanced CAPP-Seq for noninvasive analysis of disease burden in NSCLC.
Discussion
The analysis of ctDNA is likely to play a major role in personalized cancer therapy by addressing several unmet clinical needs, including (1) noninvasive genotyping of clinically actionable tumor-derived variants to guide therapy selection29,30, (2) the detection of emergent, actionable variants associated with resistance during targeted therapy30-33, (3) the early measurement of therapeutic responses6,34, and (4) the detection of minimal residual disease that is radiographically occult during disease surveillance6,35-37. Each of these applications would benefit from improvements in analytical sensitivity as long as high specificity is maintained.
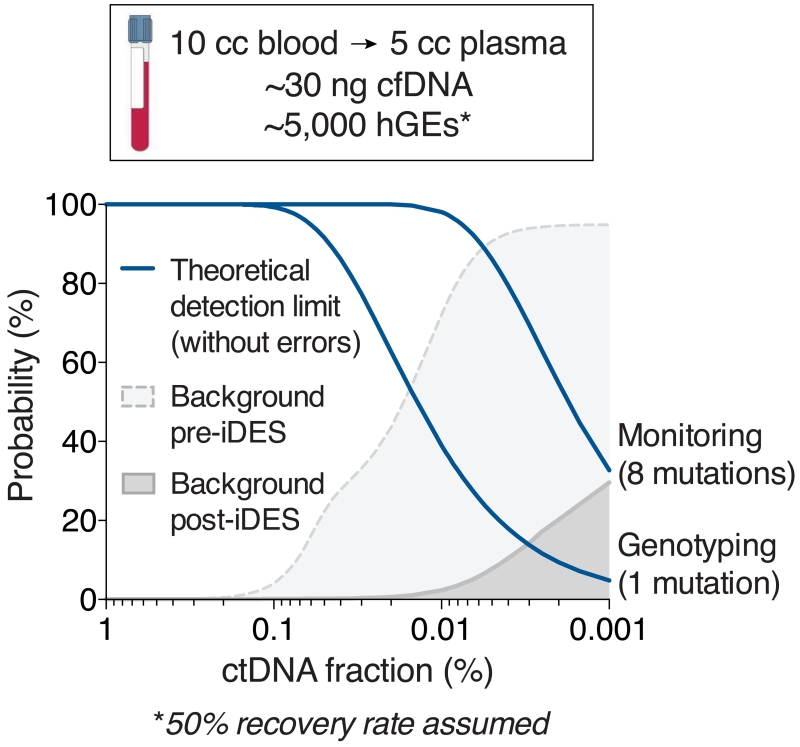
In this study, we present iDES as a sensitive method to improve capture sequencing-based ctDNA detection (Fig. 6). Capture-based enrichment strategies, such as iDES-enhanced CAPP-Seq, have important advantages over alternative methods based on amplicon sequencing11, including increased scalability, flexibility, coverage uniformity, and ability to reliably assess all mutation classes in a single assay38,39. As a result, iDES-enhanced CAPP-Seq achieves similar sensitivity at hotspot alleles as digital PCR or amplicon-based approaches, but can simultaneously interrogate hundreds to thousands of additional genomic positions without affecting sensitivity or specificity.
Figure 6. iDES-enhanced CAPP-Seq.
Same as Fig. 1a, but showing the impact of iDES on the probability of background errors. Post-iDES background data were derived from cfDNA samples pooled from a test cohort of 18 normal donors, none of which were used for learning baseline background distributions. Further details are provided in Methods.
To explore its potential clinical utility, we tested the performance of iDES-enhanced CAPP-Seq for noninvasive genotyping and monitoring of NSCLC. In our analysis of 172 plasma samples from 84 human subjects, we found that iDES facilitates noninvasive detection of multiple somatic variants with improved sensitivity, specificity, and accuracy. In addition to variant-level assessments, we also evaluated biopsy-free genotyping performance at the patient level due to its greater clinical relevance for personalized cancer therapy selection40. Because iDES outperformed other methods in this context (Fig 4f, bottom), it has potential utility for a significant subset of NSCLC patients who cannot be profiled for actionable mutations due to unavailable or inadequate biopsies41-45.
Finally, a CAPP-Seq approach that relies on patient-specific capture panels could allow for greater analytical sensitivity than iDES if >~200 somatic mutations are targeted and only duplex-supported molecules are considered (Supplementary Fig. 12c). However, this approach would require whole genome sequencing of the primary tumor and paired germline in most patients, which is both costly and potentially challenging when using FFPE samples and biopsy specimens with low tumor content. In addition, due to the inherent inefficiency of duplex capture, iDES-enhanced CAPP-Seq will detect a larger fraction of somatic alterations than duplex sequencing in patients with ctDNA levels above the detection limit of iDES. Thus, while personalized CAPP-Seq has potential advantages in certain cases, this approach is currently impractical for broad clinical application.
In summary, molecular barcoding and background polishing synergize to allow for substantially reduced cfDNA error rates without sacrificing hGE recovery (Fig. 6). These qualities improve sensitivity and specificity over other methods for samples with limited DNA content, such as cfDNA from clinically-practical blood collection volumes. Given its advantages, we envision that iDES will prove useful as a general strategy for deep sequencing applications requiring precise digital quantification of low frequency alleles.
Online Methods
Human subjects
All samples in this study including healthy adult subjects and cancer patients were collected with informed consent for research use and were approved by Institutional Review Boards in accordance with the Declaration of Helsinki. All patients were de-identified with samples coded. Demographic and clinical features of the patients profiled are captured in Supplemental Table 2.
Biological specimens
Venous blood was collected in K2EDTA tubes. Tubes were spun at 1800 × g for 10 min and plasma was frozen at −80° C in 1–2 mL aliquots. A portion of the remaining plasma-depleted whole blood was also stored at −80°C for germline DNA isolation. From each phlebotomy specimen, unless otherwise stated, 0.068–20.5 mL (median 2.7 mL) of plasma was profiled to target ~32ng of cfDNA input into CAPP-Seq library preparation.
Archival formalin-fixed, paraffin embedded (FFPE) tumor tissues were available for a subset of patients and used for monitoring analyses and variant calling, as described below.
Sample inventory
Given the large number of patient and technical samples profiled in this work and the diversity of analyses associated with them, a table linking figures to sample identifiers is provided in Supplementary Table 2.
Library preparation and sequencing
DNA isolation, shearing of genomic DNA, preparation of pre-capture sequencing libraries, hybridization-based enrichment, and assessment of library quality and enrichment following hybridization were performed as described previously8, using new adapter designs for molecular barcoding (see Design of sequencing adapters and molecular barcodes below, and Supplementary Fig. 1a). Sequencing was performed using 2×100 or 2×150 paired end reads with an 8-base indexing read on an Illumina MiSeq, NextSeq, or HiSeq 2000, 2500, or 4000. Raw sequencing data have been deposited in the Sequence Read Archive under accession number SRP069806.
Design of sequencing adapters and molecular barcodes
We implemented and evaluated several molecular barcoding strategies to minimize deep-sequencing error rates, accurately enumerate recovered hGEs, and maximize usable sequencing from limited DNA inputs (Figs. 2a, 3c; Supplementary Figs. 1–4, 8; Supplementary Note). For a detailed graphical depiction of all barcoding strategies utilized in this work, see Supplementary Figure 1a.
We began with an “index adapter” design in which a random molecular barcode (called an ‘index’ barcode) was incorporated in the single stranded portion of the adapter immediately adjacent to a sample multiplexing barcode (Fig. 2a, Supplementary Fig. 1a). This strategy liberates the sequencing read to maximize informative sequencing content while allowing for significant error-suppression over non-barcoded data (P = 2.4 × 10−15, paired two-sided t test; Supplementary Fig. 1b). To generate these adapters, we redesigned the standard 8-base sample multiplexing barcode to comprise 4 random bases (‘index barcode’), followed by a defined 4-base sample multiplexing barcode. The random 4-mer allowed for 256 distinct molecular barcodes, which when combined with the distinct start and end positions of sequenced cfDNA fragments, can provide sufficient diversity for clinically-relevant input amounts (Supplementary Note). The 4 remaining bases were used to design 24 sample multiplexing barcodes with pairwise edit distances ≥2. Importantly, index adapters do not alter the efficiency or workflow of library preparation.
Although the use of index adapters results in significant error suppression (Supplementary Fig. 1b), only information from single stranded molecules can be considered since the parental double-stranded ‘duplex’ molecules cannot be reconstituted. Being able to identify which single strands were originally paired in duplexes would allow for additional error suppression, as has been previously demonstrated13. Therefore, we also designed “tandem adapters,” which include two exogenous barcodes: index barcodes for single-strand error suppression along with dedicated barcodes for double-stranded error suppression. The latter were incorporated as 2-base barcodes into the double stranded portion of the adapters13, and were read at the beginning of each main sequencing read (therefore termed ‘insert’ barcodes; Supplementary Fig. 1a). Because insert barcodes were sequenced with the main reads, a dinucleotide insert barcode was obtained from each end of each DNA fragment, yielding a 4 base insert barcode and a maximum diversity of 256 molecules per genomic start/end position (Supplementary Fig. 2, Supplementary Note). We note that in alternate designs, index and/or insert barcodes could be placed in other adapter locations or synthesized with different lengths to accommodate higher/lower molecule diversity.
Previous barcoding strategies for double-stranded (i.e., ‘duplex’) error suppression have incorporated randomers (‘N’s) by synthesizing them as single stranded oligonucleotides, and then enzymatically copying them into double-stranded adapters12,13. In contrast, we chemically synthesized pairs of single-stranded oligonucleotides harboring individual insert barcodes of pre-defined sequence. These pairs were then annealed individually, prior to pooling, to generate a diverse mixture with defined composition and desired diversity. Additional advantages of this approach are described in Supplementary Note. We designed tandem adapters with 12 different sample multiplexing barcodes (with pairwise edit distances ≥3).
We also incorporated a constant 2-bp sequence (GT) at the ligating end of each tandem adapter, immediately adjacent to the insert barcodes. The T was required for ligation, and the G was chosen to maintain the GC “clamp” base pair located at the end of standard Illumina adapters. The GT dinucleotide additionally served as a punctuation mark, allowing for the assessment of proper adapter ligation in sequencing data. For each of the 12 sample multiplexing barcodes, 16 pairs of oligonucleotides were obtained—one for each 2-base insert barcode. Prior to use, tandem adapters were annealed as described in Adapter annealing, below. When using these adapters, we overcame potential issues related to low complexity at the GT constant sequence (read positions 3 and 4) by adding 5-10% PhiX libraries during sequencing (Supplementary Fig. 1c).
We also designed a ‘staggered’ version of insert barcode adapters to increase sequence complexity in the first few base pairs of each main sequencing read and to obviate the need for PhiX. This yielded similar error profiles as the original tandem adapter designs (Supplementary Fig. 1c). Staggered internal barcode sequences were designed with tandem adapters as a starting point—six of the tandem adapters had two bases added immediately distal to the GT dinucleotide at the ligating end of the adapter. For eight of these barcodes, the GT dinucleotide at the end of the adapter was replaced with a CT.
Additional considerations related to barcode performance are provided in Supplementary Note. The adapter sequence used for each sample in this work is provided in Supplementary Table 2.
Adapter annealing
In order to anneal CAPP-Seq barcode adapters, 20 μL of each of two 100 μM adapter oligos were combined in a 50 μL reaction volume with a final concentration of 10 mM Tris/10 mM NaCl pH 7. The adapters were annealed using an Eppendorf VapoProtect Thermocycler (Eppendorf catalog #6321 000.515) using the following protocol: (1) ramp at 0.5° C up to 97.5° C, (2) hold at 97.5° C for 150 seconds, then (3) ramp down at the slowest setting to room temperature. After annealing, the adapters were diluted to 15 μM using 10 mM Tris/10 mM NaCl pH 7.5. For index adapters, the Illumina universal adapter oligonucleotide was ligated with each of 24 index adapter oligonucleotides. For each of the 12 tandem adapters, 16 annealing reactions were performed: one for each dinucleotide barcode at the end of the adapter. These 16 annealing reactions were combined at equal concentrations after annealing, before dilution to 15 μM.
DNA repair enzymes
Related to Supplementary Fig. 6a, we tested enzymatic removal of damaged DNA bases in a representative 32ng cfDNA sample from a healthy adult donor using the following products: (i) uracil DNA-glycosylase (UDG; NEB catalog number M0372S), which leaves an abasic site in place of uracil (a cytosine oxidation product), thereby preventing PCR from continuing through the site of oxidation, eliminating C>T errors due to cytosine oxidation; (ii) 8-oxoguanine DNA glycosylate (FPG; NEB catalog number M0240S), which removes damaged purines and cleaves at the site of the damaged bases, eliminating G>T errors due to guanine oxidation, and (iii) PreCR repair mix (NEB catalog number M0309S), which is designed to remove a variety of damaged bases, including oxidized guanines and cytosines. Before library preparation, the cfDNA sample was directly treated with UDG (1 unit), FPG (8 units), UDG and FPG together, PreCR repair mix (1 μL), or the PreCR repair mix supplemented with 1 mg/mL BSA. Samples were treated for 30 minutes at 37°C, then UDG and FPG were inactivated by heating at 60°C for 10 minutes. Sample clean up was performed using Ampure beads and eluted into 50 μL of water for library preparation.
Reference standards for noninvasive tumor genotyping
For the genotyping analyses shown in Figures 4a–c and Supplementary Figure 9, we used HD500 (Horizon Diagnostics), a DNA diagnostic reference standard consisting of multiple clinically relevant variants spanning a broad range of known AFs (0.94% ≥ AF ≥ 32.5%). To simulate ctDNA, we created 5% and 0.5% dilutions of acoustically shorn HD500 genomic DNA added into cfDNA from a healthy donor. For the analyses in Figures 4a–c, four technical replicates were prepared for each dilution using 32ng of input DNA, and each library was sequenced with 1/4 of a multiplexed HiSeq 2500 lane. For Supplementary Figure 9b, a single dilution of the 0.5% mixture was prepared with 72ng of input DNA and sequenced using ½ of HiSeq 4000 lane. Additional sequencing statistics are provided in Supplementary Table 2. Variants encoding EGFR L858R, KRAS G13D, and BRAF V600E were analyzed by ddPCR (see Droplet digital PCR below) to confirm expected spike concentrations.
HD500 contains 29 variants that are covered by our NSCLC tumor genotyping selector (see NSCLC selector design below) and that are present in a ground truth mutation list provided by Horizon Diagnostics (‘Multiplex Complete Mutation List’). Of these, 13 have AFs that were externally validated by Horizon Diagnostics using ddPCR (Supplementary Table 4). For the analysis of the 5% dilution in Figure 4a, we considered all variants, including those lacking ddPCR-validated AFs. For Figure 4b, we evaluated the concordance between observed and expected AFs within the 5% dilution, and therefore restricted our analysis to the 13 validated alleles. Given the ultra-low range of expected AFs in the 0.5% spike, we restricted this analysis to the 13 alleles with validated AFs. For details related to the calculation of performance metrics, see Performance metrics for noninvasive tumor genotyping, below.
Droplet digital PCR
To verify the expected concentration of a 5% variant blend of HD500 added into control cfDNA (Figure 4b; also see Reference standards for noninvasive tumor genotyping, above), we separately quantified 3 mutant alleles known to be present in the mixture: EGFR, KRAS, and BRAF on a Bio-Rad QX200 ddPCR instrument as previously described46, using reagents, primers and probes obtained from Bio-Rad. We utilized previously validated PrimePCR assays for mutation quantification, where wild-type and mutant assays employed HEX and FAM labels, respectively. For EGFR L858R, wild-type and mutant alleles were profiled within a 73bp amplicon annealed at 56°C, using the corresponding assays (dHsaCP2000021/dHsaCP200002122). For KRAS G13D, wild-type and mutant alleles were profiled within a 57bp amplicon annealed at 53°C, using the corresponding assays (dHsaCP2500598/dHsaCP2500599). For BRAF V600E, wild-type and mutant alleles were profiled within a 91bp amplicon annealed at 56°C using the corresponding assays (dHsaCP2000027/dHsaCP2000028). PCR conditions employed 95°C × 10 min (1 cycle), 40 cycles of 94°C × 30 sec and assay-specific annealing temperature × 1 min, and 4°C hold. A 2°C per second ramping rate was used for all steps.
Hotspot variants
We assembled a list of 292 hotspot and clinically relevant NSCLC variants (SNVs/indels) from external sources including COSMIC47 (release v70; top 1% of COSMIC variants by tumor frequency that intersect our NSCLC tumor genotyping selector), My Cancer Genome (http://www.mycancergenome.org/), and a previously reported SNaPshot panel48 (Supplementary Table 4). These variants were used to genotype cfDNA from NSCLC patients (Fig. 4d,e, Supplementary 11c) and to address specificity in (i) cfDNA from normal controls (Supplementary Fig. 11a,b) and (ii) technical experiments involving the HD500 variant blend (Fig. 4a, Supplementary Fig. 9).
Performance metrics for noninvasive tumor genotyping
Biopsy-free genotyping was evaluated using both technical controls and cfDNA samples obtained from NSCLC patients and normal donors. For analyses related to the former (Fig. 4a, Supplementary Fig. 9), we calculated sensitivity and positive predictive value (PPV) using either all known HD500 alleles (n = 29; Fig. 4a), or the subset of alleles with externally validated AFs (n = 13; Supplementary Fig. 9b) (see Reference standards for noninvasive tumor genotyping, above). We also calculated detection-limit-adjusted sensitivity and PPV for the analysis in Supplementary Figure 9b using the subset of variants predicted to be detectable given the number of recovered hGEs (detailed in Reference standards for noninvasive tumor genotyping, above, and the caption of Supplementary Figure 9b). Specificity and negative predictive value (NVP) for both analyses were determined using a list of candidate mutational hotpots applied to the same mixture samples (see Hotspot variants, above), but only considering variants unique to the hotspot list (n = 279 variants; Supplementary Table 4). We also examined the specificity of HD500 alleles in 18 cfDNA samples from the test cohort of normal controls (as in Supplementary Fig. 11a); iDES produced no false positives.
The complete list of 292 hotspot mutations in Supplementary Table 4 was also used to evaluate biopsy-free clinical genotyping in assessing (i) specificity in 18 normal controls (Supplementary Table 2, Supplementary 11c) and (ii) PPV using 24 NSCLC patients with matching tumor biopsies (Fig. 4e, ‘All genes’).
Noninvasive tumor genotyping of EGFR was tested from two perspectives (Fig. 4e,f). First, using all mutations in EGFR from our list of hotspot mutations (Supplementary Table 4), we evaluated the PPV of biopsy-free variant calls in an extended cohort of 103 cfDNA samples from all 41 available patients with tumor-confirmed EGFR mutations (Fig. 4e, Supplementary Table 2). We evaluated variant-level specificity by profiling pretreatment cfDNA samples from patients with EGFR-wildtype tumors (n = 9) and cfDNA from normal controls (n = 18) (Supplementary Table 2). Second, we evaluated noninvasive genotyping of the entire EGFR kinase domain (exons 18–24 = 885 bp; Fig. 4f). For this analysis, we genotyped all available patient samples with advanced disease (i.e., stage IIIB/IV) and with biopsy-confirmed EGFR variants (n = 37 patients). We initially focused on pretreatment cfDNA samples from the subset of patients with actionable EGFR mutations (L858R, T790M, exon 19 deletions; n = 36 patients). Because some patients harbored more than 1 actionable variant, these data were analyzed at the variant-level, not the patient-level (Fig. 4f, top). To prevent the inflation of specificity and NPV, we considered all observed deletions in exon 19 as a single variant type when enumerating true negatives. We separately performed SNV genotyping of the entire EGFR kinase domain in the subset of advanced stage patients with at least 1 biopsy-confirmed non-synonymous SNV (Fig. 4f, bottom; n = 29 patients). To assess specificity, we analyzed the same cohort of 27 cfDNA samples from EGFR-wildtype subjects. This analysis was evaluated at both the variant level and at the patient level (Fig. 4f, bottom). When assessing the former, all possible base substitutions within the 885bp kinase domain were used to enumerate true negatives (n=2,655 SNVs). Samples used in each analysis are provided in Supplementary Table 2.
Finally, to ensure unbiased specificity assessments in Fig. 4e,f and Supplementary Fig. 11a,b, we only analyzed normal control cfDNA samples that were not used to train the iDES background database (see Background polishing, below, and Supplementary Fig. 6b).
Statistical methods for ctDNA detection
To calculate the probability of detecting ctDNA given the number of hGEs recovered and number of tumor mutations covered, we used the following model. Let n = number of sequenced haploid genome equivalents, d = detection limit (fraction of ctDNA molecules), and k = number of tumor reporters. The probability of observing a single tumor reporter in cfDNA is Poisson with mean λ = n × d, where λ denotes the expected number of mutant allele copies. Therefore, given 1 reporter, the probability x of detecting ≥1 ctDNA molecule is equal to 1 – Poisson(λ), which simplifies to:
| (1) |
Generalizing to k independent tumor reporters, the cumulative distribution function of a geometric distribution can be used to model the probability of observing a success (i.e., detection of ≥1 ctDNA molecule). Thus, the probability p of detecting ≥1 ctDNA molecule given k reporters is 1 – (1 – x)k. Plugging in (1) for x yields:
| (2) |
This equation can be used to solve for any parameter if the other three are specified. For example, when using a single ddPCR assay (k = 1) on a plasma sample from which 2,000 hGEs can be recovered (n = 2,000), the detection limit d is equal to 0.12% at 90% confidence (i.e., d = ln(1 – p)/(–nk)), assuming the assay has a background error level below this detection limit. Finally, the number of tumor reporters needed to observe one reporter in cfDNA is equal to 1/x (mean of a geometric distribution) and the number of expected reporters in plasma is equal to k × x. This model was used to calculate detection-limits in Figures 1a, 3d, and 6, and Supplementary Figures 8 and 9. We also used this model to establish thresholds for read support in noninvasive tumor genotyping (see Noninvasive tumor genotyping of hotspot alleles and selected regions, below).
To evaluate the statistical significance of ctDNA quantitation in Figures 5b–d, Supplementary Figures 13 and 14, and Supplementary Table 5, we used a previously described approach based on Monte Carlo sampling8, with the following modifications. For a given collection of k tumor-associated SNVs and for a given input cfDNA sample, we tested the null hypothesis that μ and s are not jointly above selector-wide background, where μ is defined as the mean SNV fraction and s is defined as the number of SNVs with AF>0. To test the null hypothesis, we randomly sampled k genomic positions and 1 of 3 possible non-reference alleles per position. We then computed μ* and s* for the random collection of k reporters, and repeated this process for 10,000 iterations, recording the number of times t in which μ* ≥ μ and s* ≥ s. An empirical p-value was calculated as t/10,000. Importantly, due to base substitution-specific differences in background patterns, each random sampling of k reporters was performed to preserve the base substitution distribution found in the original reporter pool. Only ctDNA levels with P < 0.05 were considered detectable (Supplementary Table 5).
Error rates and efficiencies of previous barcoding methods
Data from previous methods12,14,17 shown in Supplementary Figure 8a were obtained as follows: For Schmitt and colleagues (2012), error rates were obtained directly from the main text, while efficiency was derived from Supplementary Table 112 using the formula, SSCS or DCS nucleotides / initial nucleotides. For Schmitt and colleagues (2015), error rates were obtained from the main text, while the efficiency data were derived from Supplementary Table 217 using the following relationship: efficiency = duplex nucleotides / (reads * (101-17)), where 101 is the length (in bases) of reads from the HiSeq data described in the paper, and 17 is the length of the barcodes at the beginning of the corresponding reads. For Gregory and colleagues (2015), the error rate and efficiency were obtained directly from Supplementary Figure S214.
NSCLC selector design
Two distinct, but overlapping NSCLC selectors were applied in this work. The first was developed to maximize analytical sensitivity in NSCLC cfDNA samples (‘cfDNA’ selector) and consisted of a 175kb panel that was designed to target a median of at least 8 mutations per NSCLC patient. To improve coverage for non-smokers (whose tumors generally have lower mutation burdens) and to improve clinical applicability, additional clinically relevant regions were included in the cfDNA selector in order to target actionable fusions in ALK, ROS1, and RET using intronic probes, and to ensure coverage of selected regions containing other actionable variants. The final panel spanned 203kb, consisting of regions from 207 exons, 4 introns, and 134 genes. We also designed a larger ~300kb NSCLC ‘tumor genotyping’ selector, which contained the entire cfDNA selector, but also targeted additional regions including complete exon coverage of selected genes and the addition of other regions of clinical or biological interest. The specific selector used for each sample is provided in Supplementary Table 2. Details of the selector source data and coordinate selection algorithm are provided in Supplementary Note (Additional details related to NSCLC selector design).
Biotinylated oligonucleotide capture pools for both selectors were designed using the NimbleDesign portal (Roche NimbleGen) and genome build hg19 (NCBI Build 37.1/GRCh37). Design criteria included Preferred Close Matches of 1 and Maximum Close Matches of 2. Selector coordinates are provided in Supplementary Table 1.
Sequencing data analysis
Mapping and quality control
Mapping and quality control analyses for all sequencing libraries were performed as previously described8 and detailed mapping results are provided in Supplementary Table 2.
Analysis of molecular barcodes
Because cfDNA molecules are often a key limiting resource in the setting of clinically practical blood volumes, we developed a barcode analysis pipeline that performs error suppression while maximizing molecule retention. An important feature of our approach is its treatment of variant alleles found in singleton barcode families (i.e., those without PCR duplicates). Previous techniques typically require 2, 3, or more copies per molecule12-14. This requirement is potentially costly for clinically practical cfDNA quantities, since (1) a large fraction of input hGEs would likely be lost without considerable over-sequencing, and (2) all variants in molecules without PCR duplicates would be eliminated. Therefore, our approach was implemented to perform barcode-mediated error suppression on UID families with >1 copy, but to also allow singleton variants to be rescued if they are found in at least one non-singleton barcode family. Details follow below:
As a first step, we processed all unaligned reads to extract 4-bp index and/or insert barcode sequences. Because insert barcodes flank ligated DNA sequences, they were concatenated to create a composite 4bp UID prior to analysis (Fig. 2a, Supplementary Fig. 1a). To recover duplex sequences using insert barcodes, we used the following criteria, illustrated by way of example: Suppose AT and CG insert barcodes are observed in read 1 and 2, respectively, and their corresponding DNA fragment F1 aligns to the positive strand of the reference genome. If AT and CG barcodes are then respectively observed in read 2 and read 1 from another fragment F2 aligned to the minus strand, and if the two fragments share genomic coordinates, then F1 and F2 likely represent reciprocal strands of a single double-stranded DNA molecule (i.e., duplex). All insert barcodes were analyzed accordingly. Otherwise, both barcode types were treated in an identical fashion, requiring a perfect match between UIDs. Prior to grouping sequences into barcode families, raw reads were mapped to the reference genome and filtered for proper pairs. All single base substitutions were then subjected to Phred quality filtering using a threshold Q of 30. After base quality filtering, each barcode family with identical start/end coordinates and with ≥2 members was analyzed separately to identify and eliminate sequence errors as follows:
-
1)
For every genomic position i in a given barcode family, enumerate the number of barcode family members (i.e., reads) vi supporting a given non-reference allele, considering only non-reference alleles that passed base quality filtering. If there is >1 distinct non-reference allele with Q≥30 at position i, set vi to the frequency of the most abundant non-reference allele, or in the event of a tie, arbitrarily select one of the variants.
-
2)
For each position harboring a candidate variant from step 1 (i.e., vi > 0), adjust the number of barcode family members ni by subtracting the number of non-reference alleles qi that failed the Phred quality filter. Therefore, set ni* = ni – qi.
-
3)
Eliminate non-reference alleles from positions where vi < (f × ni*), where f = 1 (by default).
-
4)
Consolidate all barcode family members into a single consensus sequence, and only retain candidate variants that passed step 3 with ≥2 copies. To eliminate positions with an error-suppressed allele from further analysis, set their Phred quality scores to 0.
As a final error suppression step, all non-reference alleles in singleton barcode families (i.e., families with no PCR duplicates) were excluded from further analysis unless observed in at least one independent DNA molecule with ≥2 family members supporting that variant. We applied this de-duplication strategy to all cfDNA samples in this work (“2×*” in Fig. 3c) since it facilities maximal hGE recovery while still benefiting from barcode-mediated error suppression. Conversely, owing to a broader distribution of DNA fragment lengths, singleton variants in tumor and PBL samples were not suppressed; however, barcode de-duplication was still used to remedy discordant alleles in families with an identical UID and matching start/end coordinates.
Background polishing
To explicitly model position-specific background distributions in cfDNA, we employed the following novel approach, which alternates between two statistical models depending on available information content. Using a training cohort of pre-duplication removal data from healthy normal control cfDNA samples with high background (n = 12, Supplementary Fig. 6b, Supplementary Table 2), we iterated through every possible SNV in the NSCLC tumor genotyping selector (Supplementary Table 1). After excluding germline SNPs, for each of these candidate SNVs we populated a position- and base substitution-specific one-dimensional vector v with all AFs observed in the set of normal cfDNA controls. To mitigate the impact of outliers on overfitting, we removed the maximum AF from v. If the total number of non-zero AFs in v was <5, we used a Gaussian distribution to model the entire vector, and calculated the mean μ and standard deviation σ using all remaining AFs. Otherwise, we fit a Weibull distribution to the set of non-zero AFs in v using fitdist from the fitdistrplus package in R, and the resulting shape and scale parameters were saved to disk. Since v is often zero-inflated, we also saved the fraction of non-zero AFs in v in order to incorporate the frequency of zero-valued observations into the final model. We selected the Weibull distribution owing to its robust performance in fitting position-specific non-zero background errors (Supplementary Fig. 7a). Moreover, we observed high concordance between recurrent errors in pre- and post-duplication removal data (Supplementary Fig. 7b), suggesting that stereotypical background is not reliably suppressed by molecular barcoding. We therefore used pre-duplication removal data to model baseline distributions, yielding a background database Φ.
To eliminate (i.e., “polish”) stereotypical errors in alignment data from an independent cfDNA sample s, we assessed the fractional abundance f of each candidate background allele in s using its corresponding background model in Φ. If the model was Gaussian, we evaluated f with a one-sided z-test, yielding a p-value. Otherwise, shape and scale parameters from the Weibull distribution were used to calculate the cumulative probability p* that a given AF generated by the model was below f (using the pweibull function in R). To account for zero-inflated training data, we then adjusted p* using the fraction δ of non-zero AFs from the training set. Specifically, we used the following formula, p-value = 1 – ((1 – δ) + (δ × p*)), which is analogous in structure to the two-component zero-inflated Poisson model49. Whether calculated by the z-test or zero-inflated Weibull distribution, p-values were then adjusted for multiple hypotheses using a stringent Bonferroni correction (where n = all base substitutions in the background database). Among candidate background alleles occurring in at least 2 normal controls and in at least 20% of normal controls in the training cohort, we eliminated a given candidate if and only if (i) it was statistically indistinguishable from background (adjusted P ≥ 0.05), (ii) it was not present with duplex support, and (iii) f was less than 5% AF or the number of supporting molecules was ≤10.
We tested the reproducibility of our approach using two cohorts of genetically distinct healthy adult controls processed independently to assess batch effects (Supplementary Table 2). Each control cohort was used to build a different background database, which was subsequently applied to polish all NSCLC cfDNA samples analyzed in this work. While the original training cohort had the highest background (Supplementary Fig. 6b), and thus yielded the greatest reduction in selector-wide error rates, differences were marginal overall (Supplementary Fig. 7c). This result is consistent with the highly stereotypical nature of most background patterns observed across cfDNA samples (Supplementary Fig. 6), and was also not dependent on the selector synthesis batch (cfDNA versus tumor genotyping NSCLC selectors; Supplementary Table 2, Supplementary Fig. 6b).
Of note, we used the same approach described above to build a background database for cfDNA from 12 normal controls profiled by a personalized GBM selector (see ctDNA detection limits for iDES, duplex sequencing, and other methods, below).
Calculation of selector-wide error profiles
For Figures 1a and 6, the probability of background errors was calculated as the fraction of all selector positions with at least 1 read supporting a non-reference allele. For all selector-wide assessments of error rate, we restricted the scope of analysis to targeted positions with ≥200× and ≥1000× total reads for data with and without barcode de-duplication, respectively. Non-reference bases with >5% AF were excluded. For positions and alleles that met the above criteria, selector-wide error rate was defined as the number of all non-reference bases divided by all sequenced bases.
Noninvasive tumor genotyping of hotspot alleles and selected regions
Given the ultralow error rate of iDES and the ability to enumerate individual molecules with CAPP-Seq barcode adapters, we used the following cfDNA genotyping approach to detect both hotspots variants (Supplementary Table 4) and variants within a defined genomic region (Fig. 4f). The following order of precedence was used for calling SNVs: duplex support (≥1×), strand support (≥2×), no strand support (≥3×). For SNVs without duplex support, the minimum required AF f was determined based on the following formula, f = ln(1 – p)/–n, where p = probability/confidence of detection and n = total depth at a given genomic position (see Statistical methods for ctDNA detection, above). We set p equal to 0.95 for all analyses involving the interrogation of hotspot variants from Supplementary Table 4. For EGFR kinase domain profiling in Figure 4f, we set p equal to 0.95 for hotspot SNVs present in Supplementary Table 4 in order to emphasize sensitivity for mutations with known significance; for all other SNVs, p was set to 0.99 to emphasize specificity. Only non-synonymous SNVs were further analyzed. Small insertions and deletions were also called if present with ≥1× read support. In all cases, noninvasive genotyping was performed directly on cfDNA samples without prior knowledge of tumor biopsies. For analyses in Figure 4d,e, variants were excluded from further analysis if present in paired PBLs with >0.5% AF; however, regardless of whether paired PBLs were considered, genotyping results were nearly identical (data not shown).
The aforementioned approach was applied to all barcode de-duplicated data, with or without background polishing (Fig. 4a,d,f, Supplementary Fig. 11a,b). To extend this approach to SNV genotyping in pre-duplication removal data, we multiplied the minimum number of reads by the mean UID family size for each sample (Supplementary Table 2). This strategy would not be applicable to non-barcoded data without a reasonably accurate estimate of the cfDNA duplication rate, but was applied here to compare genotyping performance among error suppression methods in a uniform manner (i.e., +/− barcoding and +/− polishing).
Selector-wide genotyping
Deep-sequencing error rates are heterogeneous, differing in magnitude across genomic intervals and between base substitution classes. Moreover, sequencing depths typically vary within and across samples. Collectively, these issues complicate the selection of thresholds for variant calling, leading to suboptimal tradeoffs between sensitivity and specificity. To improve the detection rate of low frequency alleles, we developed a general genotyping approach that adaptively considers local and global variation in background error rates, enabling automatic determination of position-specific variant calling thresholds in each sample. In this work, we explored the utility of this approach for genotyping a 5% variant blend (Fig. 4c) and for noninvasive monitoring of NSCLC patients (Fig. 5d, Supplementary Fig. 14). Methodological details are provided in Supplementary Note (Additional details related to selector-wide genotyping).
ctDNA monitoring analysis
To assess the sensitivity and specificity of iDES in a monitoring context, somatic variants identified in 30 NSCLC patients were queried in 116 cfDNA samples (Supplementary Table 5). All patients had a pretreatment plasma sample and fell into one of three groups: Group 1 (n = 19) had evaluable tumor biopsies with paired PBLs; group 2 (n = 5) had tumor samples without paired PBLs. We also analyzed a third group (n = 6) that did not have tumor biopsies, but had at least one post-treatment plasma sample and a paired PBL sample. Variant lists for each patient were enumerated using adaptive SNV genotyping (Supplementary Fig. 10) applied to either tumors with paired germline (group 1), tumors without paired germline (group 2), or pretreatment plasma samples with paired germline (group 3). To filter calls in the second group, we retained only non-synonymous SNVs found in the Catalog Of Somatic Mutations In Cancer database (COSMIC47, v70), omitting those found in at least 1% of the population in the Exome Aggregation Consortium database (ExAC, release 0.2; http://exac.broadinstitute.org). Positions with a germline SNP in any patient or control were removed from all variant lists. Germline SNPs and tumor-associated indels were called as previously described8. Since group 3 lacked paired PBLs, indels were restricted to COSMIC hotspots, and common indels were excluded using the same filter applied for SNVs (Methods). Tumor variants were determined using thetumor sample closest in time to the pretreatment plasma sample (within 1 year of the plasma sample). For two patients, P28 and P29, we profiled two tumor biopsies, each obtained within 3 months of corresponding pretreatment plasma samples. In those cases, reporters from both tumor samples were pooled into one variant list. Finally, as a quality control check for group 3 variants, which were called from pretreatment cfDNA, we examined individual calls across all sampled time points. Dynamic changes in corresponding allele fractions were observed following therapy (Supplementary Table 5). This observation, coupled with the absence/depletion of group 2 variants from paired PBLs, suggested that group 3 variants were not artifacts resulting from clonal hematopoiesis.
To assess the statistical significance of ctDNA detection, we used our previously described ctDNA detection index8 coupled with a Monte Carlo approach with modifications (Statistical methods for ctDNA detection, above). In addition to cfDNA samples from NSCLC patients, we analyzed cfDNA from 30 healthy controls to evaluate specificity (Supplementary Table 2). To avoid miscalls, variants in common between two patients were not considered. Among pretreatment time points, genetically matched samples were treated as cancer positive, whereas all other samples, including controls, were treated as negatives. To include post-treatment time points, we considered genetically matched samples as positive for the purposes of assessing detection rate. The P value threshold that maximized both sensitivity (detection rate) and specificity was determined for pretreatment (and pre/post-treatment) datasets. Results for optimized p-value thresholds and for P<0.05 are provided in Supplementary Table 5.
Noninvasive ctDNA monitoring
For the analysis of noninvasive ctDNA monitoring (Fig. 5b, Supplementary Fig. 14), alternative error suppression approaches were each applied to 37 plasma samples from 8 patients, all of whom had a matching tumor biopsy and PBL sample and at least 3 longitudinal plasma time points available for correlation assessments. For each error suppression method, adaptive SNV genotyping (Supplementary Fig. 10) was applied to call SNVs directly from pretreatment plasma. The mean SNV fraction for each time point was then calculated using all mutations called directly from plasma and compared to the mean SNV fraction derived from mutations called in the matching primary tumor. This was done separately for plasma samples processed by each error suppression method. Of note, to call SNVs in plasma samples processed by polishing alone or without barcoding or polishing, we performed de-duplication by position (start/end coordinates) prior to analysis.
ctDNA detection limits for iDES, duplex sequencing, and other methods
We designed a spike experiment spanning two orders of magnitude to evaluate the detection-limit of each error-suppression method described in this work. Based on observed error rates (Fig. 3c), we tested the detection-limit of iDES down to 2.5 in 105 cfDNA molecules and duplex sequencing down to 2.5 in 106 cfDNA molecules. We also included a spike fraction of 2.5 in 104 molecules at the upper end to reproduce the detection-limit of our original description of CAPP-Seq8. A key consideration for this analysis was to determine the minimum number of mutations required for each method to reach its detection-limit given a practical cfDNA input. For each concentration of interest, we estimated both the number of independent mutations and the number of recovered hGEs that would be needed to detect tumor DNA above expected background levels (using modeling described in Statistical methods for ctDNA detection above).
Based on our modeling, and given considerations of cost and molecule recovery, we used a glioblastoma (GBM) tumor that was found to harbor ~1,500 coding mutations by whole exome sequencing (Supplementary Table 2). We designed a 157kb “personalized” selector to cover these mutations, employing the same biotinylated oligonucleotide synthesis process as described for NSCLC selector design, above. We then applied this selector to perform deep re-sequencing of the GBM tumor (1/2 of a MiSeq lane), and confirmed 97% of expected SNVs, 99.7% of which had duplex support (n = 1,502).
Next, we designed a dilution series using our analytical model to ensure sufficient GBM mutation counts at the lowest dilution. The following concentrations, mass inputs, and lane shares were split into two technical replicates prior to library preparation, and were independently processed and sequenced: 2.5 in 104 molecules using 32ng in 1/8 of a lane, 2.5 in 105 molecules using 32ng in 1/8 of a lane, 5.0 in 106 molecules using 32ng in 1/4 of a lane, and 2.5 in 106 molecules using 2 × 32ng, each in 1/4 of a lane. To evaluate iDES and polishing alone, cfDNA samples from 12 normal controls were also captured and sequenced using the GBM panel. A background database was created (as described in Background polishing, above) and used to polish the GBM dilution series. Fractional levels of tumor-derived DNA were calculated by averaging AFs over the entire set of interrogated tumor mutations. Results are shown in Figure 5a and Supplementary Figure 12.
Supplementary Material
Acknowledgments
This work was supported by grants from the Department of Defense (A.M.N., M.D., A.A.A.), the National Cancer Institute (A.M.N., 1K99CA187192-01A1; M.D., A.A.A., R01CA188298), the US National Institutes of Health Director’s New Innovator Award Program (M.D., 1-DP2-CA186569), a US Public Health Service/National Institutes of Health U01 CA194389 (A.A.A.), the Ludwig Institute for Cancer Research (M.D., A.A.A.), a Stanford Cancer Institute-Developmental Cancer Research Award (M.D., A.A.A.), the CRK Faculty Scholar Fund (M.D.), V-Foundation (A.A.A.), Damon Runyon Cancer Research Foundation (A.A.A.) and a grant from both the Siebel Stem Cell Institute and the Thomas and Stacey Siebel Foundation (A.M.N.).
Footnotes
Author Contributions
A.M.N., A.F.L., D.M. Klass, M.D., and A.A.A. developed the concept, designed the experiments, and analyzed the data. A.M.N., A.F.L., M.D., and A.A.A. wrote the manuscript. A.F.L. and D.M. Klass performed the molecular biology experiments with assistance from D.M. Kurtz, J.J.C., F.S., S.V.B., and L.Z. Bioinformatics analyses were performed by A.M.N. with assistance from A.F.L., H.S. and C.L.L. Patient specimens were provided by C.S., J.N.C., R.B.W., G.W.S, J.B.S., B.W.L. Jr., J.W.N., H.A.W., and M.D. All authors commented on the manuscript at all stages.
Competing Financial Interests
A.M.N., D.M.Klass, S.V.B., M.D., and A.A.A. are co-inventors on patent applications related to CAPP-Seq. A.M.N., M.D., and A.A.A. are consultants for, and A.F.L. and D.M.Klass are employed by, Roche Molecular Systems.
Code availability. Source code is available for non-profit research use at http://cappseq.stanford.edu/ides.
References
- 1.Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem. 2015;61:112–123. doi: 10.1373/clinchem.2014.222679. [DOI] [PubMed] [Google Scholar]
- 2.Diehl F, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bettegowda C, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra224. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bratman SV, Newman AM, Alizadeh AA, Diehn M. Potential clinical utility of ultrasensitive circulating tumor DNA detection with CAPP-Seq. Expert Rev Mol Diagn. 2015;15:715–719. doi: 10.1586/14737159.2015.1019476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diaz LA, Bardelli A. Liquid Biopsies: Genotyping Circulating Tumor DNA. Journal of Clinical Oncology. 2014 doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurtz DM, et al. Noninvasive monitoring of diffuse large B-cell lymphoma by immunoglobulin high-throughput sequencing. Blood. 2015;125:3679–3687. doi: 10.1182/blood-2015-03-635169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butler TM, et al. Exome Sequencing of Cell-Free DNA from Metastatic Cancer Patients Identifies Clinically Actionable Mutations Distinct from Primary Disease. PLoS One. 2015;10:e0136407. doi: 10.1371/journal.pone.0136407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newman AM, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taniguchi K, et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res. 2011;17:7808–7815. doi: 10.1158/1078-0432.CCR-11-1712. [DOI] [PubMed] [Google Scholar]
- 10.Jabara CB, Jones CD, Roach J, Anderson JA, Swanstrom R. Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proc Natl Acad Sci U S A. 2011;108:20166–20171. doi: 10.1073/pnas.1110064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmitt MW, et al. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A. 2012;109:14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kennedy SR, et al. Detecting ultralow-frequency mutations by Duplex Sequencing. Nat Protoc. 2014;9:2586–2606. doi: 10.1038/nprot.2014.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregory MT, et al. Targeted single molecule mutation detection with massively parallel sequencing. Nucleic Acids Research. 2015 doi: 10.1093/nar/gkv915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kukita Y, et al. High-fidelity target sequencing of individual molecules identified using barcode sequences: de novo detection and absolute quantitation of mutations in plasma cell-free DNA from cancer patients. DNA Research. 2015 doi: 10.1093/dnares/dsv010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lou DI, et al. High-throughput DNA sequencing errors are reduced by orders of magnitude using circle sequencing. Proc Natl Acad Sci U S A. 2013;110:19872–19877. doi: 10.1073/pnas.1319590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmitt MW, et al. Sequencing small genomic targets with high efficiency and extreme accuracy. Nat Methods. 2015;12:423–425. doi: 10.1038/nmeth.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Mattos-Arruda L, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015;6:8839. doi: 10.1038/ncomms9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costello M, et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Research. 2013;41:e67. doi: 10.1093/nar/gks1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen G, Mosier S, Gocke CD, Lin MT, Eshleman JR. Cytosine deamination is a major cause of baseline noise in next-generation sequencing. Mol Diagn Ther. 2014;18:587–593. doi: 10.1007/s40291-014-0115-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–650. [PubMed] [Google Scholar]
- 22.Hafner C, et al. Oncogenic PIK3CA mutations occur in epidermal nevi and seborrheic keratoses with a characteristic mutation pattern. Proc Natl Acad Sci U S A. 2007;104:13450–13454. doi: 10.1073/pnas.0705218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higgins MJ, et al. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin Cancer Res. 2012;18:3462–3469. doi: 10.1158/1078-0432.CCR-11-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sequist LV, et al. Rociletinib in EGFR-mutated non-small-cell lung cancer. N Engl J Med. 2015;372:1700–1709. doi: 10.1056/NEJMoa1413654. [DOI] [PubMed] [Google Scholar]
- 25.Oxnard GR, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res. 2014;20:1698–1705. doi: 10.1158/1078-0432.CCR-13-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pao W, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pao W, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sequist LV, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Douillard JY, et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol. 2014;9:1345–1353. doi: 10.1097/JTO.0000000000000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mok T, et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin Cancer Res. 2015;21:3196–3203. doi: 10.1158/1078-0432.CCR-14-2594. [DOI] [PubMed] [Google Scholar]
- 31.Misale S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murtaza M, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 33.Thress KS, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21:560–562. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchetti A, et al. Early Prediction of Response to Tyrosine Kinase Inhibitors by Quantification of EGFR Mutations in Plasma of NSCLC Patients. J Thorac Oncol. 2015;10:1437–1443. doi: 10.1097/JTO.0000000000000643. [DOI] [PubMed] [Google Scholar]
- 35.Dawson SJ, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Murillas I, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7:302ra133. doi: 10.1126/scitranslmed.aab0021. [DOI] [PubMed] [Google Scholar]
- 37.Roschewski M, et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: a correlative biomarker study. Lancet Oncol. 2015;16:541–549. doi: 10.1016/S1470-2045(15)70106-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samorodnitsky E, et al. Evaluation of Hybridization Capture Versus Amplicon-Based Methods for Whole-Exome Sequencing. Hum Mutat. 2015;36:903–914. doi: 10.1002/humu.22825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drilon A, et al. Broad, Hybrid Capture-Based Next-Generation Sequencing Identifies Actionable Genomic Alterations in Lung Adenocarcinomas Otherwise Negative for Such Alterations by Other Genomic Testing Approaches. Clin Cancer Res. 2015;21:3631–3639. doi: 10.1158/1078-0432.CCR-14-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rehm HL, et al. ACMG clinical laboratory standards for next-generation sequencing. Genet Med. 2013;15:733–747. doi: 10.1038/gim.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ellis PM, Verma S, Sehdev S, Younus J, Leighl NB. Challenges to implementation of an epidermal growth factor receptor testing strategy for non-small-cell lung cancer in a publicly funded health care system. J Thorac Oncol. 2013;8:1136–1141. doi: 10.1097/JTO.0b013e31829f6a43. [DOI] [PubMed] [Google Scholar]
- 42.Leighl NB, et al. Molecular testing for selection of patients with lung cancer for epidermal growth factor receptor and anaplastic lymphoma kinase tyrosine kinase inhibitors: American Society of Clinical Oncology endorsement of the College of American Pathologists/International Association for the study of lung cancer/association for molecular pathology guideline. J Clin Oncol. 2014;32:3673–3679. doi: 10.1200/JCO.2014.57.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lim C, et al. Biomarker testing and time to treatment decision in patients with advanced nonsmall-cell lung cancer. Ann Oncol. 2015;26:1415–1421. doi: 10.1093/annonc/mdv208. [DOI] [PubMed] [Google Scholar]
- 44.Shiau CJ, et al. Sample features associated with success rates in population-based EGFR mutation testing. J Thorac Oncol. 2014;9:947–956. doi: 10.1097/JTO.0000000000000196. [DOI] [PubMed] [Google Scholar]
- 45.Yatabe Y, et al. EGFR mutation testing practices within the Asia Pacific region: results of a multicenter diagnostic survey. J Thorac Oncol. 2015;10:438–445. doi: 10.1097/JTO.0000000000000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hindson BJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forbes SA, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Su Z, et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. J Mol Diagn. 2011;13:74–84. doi: 10.1016/j.jmoldx.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lambert D. Zero-Inflated Poisson Regression, with an Application to Defects in Manufacturing. Technometrics. 1992;34:1–14. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.