Abstract
Purpose: The rcellminer R package provides a wide range of functionality to help R users access and explore molecular profiling and drug response data for the NCI-60. The package enables flexible programmatic access to CellMiner’s unparalleled breadth of NCI-60 data, including gene and protein expression, copy number, whole exome mutations, as well as activity data for ∼21K compounds, with information on their structure, mechanism of action and repeat screens. Functions are available to easily visualize compound structures, activity patterns and molecular feature profiles. Additionally, embedded R Shiny applications allow interactive data exploration.
Availability and implementation: rcellminer is compatible with R 3.2 and above on Windows, Mac OS X and Linux. The package, documentation, tutorials and Shiny-based applications are available through Bioconductor (http://www.bioconductor.org/packages/rcellminer); ongoing updates will occur according to the Bioconductor release schedule with new CellMiner data. The package is free and open-source (LGPL 3).
Contact: lunaa@cbio.mskcc.org or vinodh.rajapakse@nih.gov
1 Introduction
The NCI-60 cancer cell line panel has been used over the course of several decades as an anti-cancer drug screen. This panel was developed as part of the Developmental Therapeutics Program (DTP, http://dtp.nci.nih.gov/) of the U.S. National Cancer Institute (NCI). Thousands of compounds have been tested on the NCI-60, which have been further characterized by numerous platforms for gene and protein expression, DNA copy number, whole exome mutation and others (Reinhold et al., 2012). Furthermore, most NCI-60 cell lines are included in the larger panels of the Broad Cancer Cell Line Encyclopedia (CCLE) and the Sanger Cancer Genome Project (CGP) (Reinhold et al., 2014).
CellMiner (http://discover.nci.nih.gov/cellminer) integrates NCI-60 compound activity and molecular profiling data derived from multiple platforms. With its additional data relative to the NCI-DTP site, stringent quality control and powerful online tools, CellMiner is unmatched for exploration of NCI-60 data by biologists. Still, there is currently no convenient programmatic interface to its wealth of data for computational biologists seeking to develop specialized analyses. Related projects, such as The Cancer Genome Atlas (TCGA), provide such functionality through the Firehose and R packages including RTCGAToolbox and MSKCC Cancer Genomics Data Server (CGDSR) (Gao et al., 2013; Samur, 2014), and several packages enable use of structural information (Cao et al., 2008; Guha, 2007; Wang et al., 2013). Here we present an R package providing convenient access to CellMiner data, with tools to facilitate deeper exploration and use by researchers.
2 Implementation
This work exists as two packages: (i) rcellminer, a software R package that provides functionality and (ii) rcellminerData, a data package, which is a dependency of rcellminer. Within the rcellminer package, numerous accessor functions are provided together with Shiny-based user interfaces.
2.1 Available rcellminer data
The available data include mRNA expression, whole exome mutation and DNA copy number. The data are retrieved directly from the CellMiner project. We refer readers to Reinhold et al. and package documentation for information on the processing of the data; all R scripts used for CellMiner data pre-processing are provided within the package (Abaan et al., 2013; Reinhold et al., 2012, 2014).
2.2 Data representation
Data in rcellminerData are organized within two S4 class objects: molData and drugData. These are instances of general-purpose classes that are appropriate for R-based storage of pharmacogenomic datasets involving multiple assay types. These two classes build on existing base Bioconductor classes to give developers both flexibility and access to existing functionality. molData contains results for molecular assays (e.g. genomics, proteomics, etc.) that have been performed on the NCI-60 and drugData contains results for drug response assays. molData is an instance of the MolData S4 class that is composed of two slots: eSetList and sampleData; eSetList is a list of eSet objects that can be of different dimensions; this is conceptually similar to a single Bioconductor eSet object, but differs in that there is no requirement for data matrices to have equal dimensions. The second slot, sampleData, returns a data.frame containing information for each sample. Drug activity (response) data is provided in the rcellminerData package using the DrugData S4 class, which is composed of three slots: act, repeatAct and sampleData. Both act (summary response data across multiple repeats) and repeatAct (individual repeat data) are eSet objects. Additionally, rcellminerData provides a large amount of information on drugs, including structure information, clinical testing status, etc.
2.3 Accessing CellMiner data
In addition to providing access to the data within CellMiner, rcellminer provides basic descriptive statistics and plotting functionality for both the molecular and drug response data. rcellminer also makes available the structures of compounds screened against the NCI-60 and allows users to create and compare structural fingerprints between compounds. Tutorials in the form of a vignette with several use cases related to recent CellMiner publications are provided. Many of these tasks can also be performed using embedded interactive tools.
2.4 Interactive exploration of CellMiner data
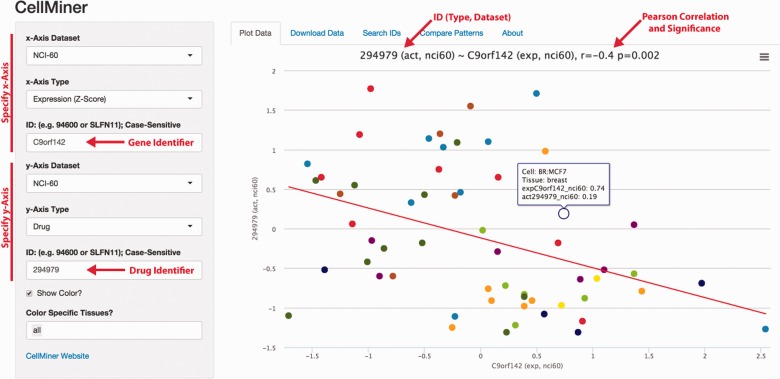
While the main purpose of rcellminer is to provide a programmatic interface to CellMiner data, interactive interfaces allow users to quickly explore available data and develop insight for deeper analyses. To support this aim, rcellminer contains several interactive tools that can be run by users locally. The ‘2D-Plot’ application, shown in Figure 1, allows users to quickly assess relationships between any two features in the available data. Additionally, this application allows any of the available features to be used as a pattern for retrieving correlated features. The ‘Structure Comparison’ application allows users to identify structurally similar compounds either by using an NSC identifier or by providing a simplified molecular-input line-entry system (SMILES) formatted chemical structure. The ‘Compound Information’ application allows visualization of compound structures and drug response profiles, together with available metadata and information about repeat screens.
Fig. 1.
rcellminer simplifies the exploration of CellMiner data in R. Scatterplot of PAXX (C9orf142) gene expression and Bleomycin (NSC294979) activity within the NCI60. PAXX interacts with Ku to mediate DNA double-strand break repair, and bleomycin acts by inducing DNA double-strand breaks. High PAXX expression is associated with bleomycin resistance. In the application, point colors represent tissues of origin for the cell lines. Drug activity (act), and gene expression (exp) are indicated as standardized (z-score) values, as described by (Reinhold et al., 2012)
3 Conclusion
The rcellminer R package is designed to allow R users to explore molecular profiling and drug response data on the NCI-60. rcellminer complements the online CellMiner tools with ones that can be run locally to enhance access to the CellMiner data. We anticipate that these tools will allow researchers to build powerful analyses of the NCI-60 data. We showcase some possibilities with the embedded R Shiny applications allowing quick exploration of the available data. We plan to extend this R package to include additional compound data made available by DTP, and other molecular profiling data for the NCI-60 from assays conducted by the Developmental Therapeutics Branch of the NCI and other groups. With its extensive data and flexible underlying functionality, rcellminer will make the NCI-60 accessible to a larger segment of the community, enabling development of novel analyses. It additionally provides useful guidance for organizing data from other cell line pharmacogenomic databases as R packages.
Acknowledgements
We thank Margot Sunshine who developed CellMiner, Marc Carlson and the Bioconductor community, and the NCI-DTP team for the drug data and support for data acquisition. (Dr. Jerry Collins and Dr. James H. Doroshow).
Funding
This work was supported by the NIH grants: MSKCC Genome Data Analysis Center grant (U24 CA143840), the Ruth L. Kirschstein National Research Service Award (F32 CA192901) and by the Center for Cancer Research, Intramural Program of the National Cancer Institute (Z01 BC006150).
Conflict of Interest: none declared.
References
- Abaan O.D. et al. (2013) The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology. Cancer Res., 73, 4372–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y. et al. (2008) ChemmineR: a compound mining framework for R. Bioinformatics, 24, 1733–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J. et al. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal., 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha R. (2007) Chemical informatics functionality in R. J. Stat. Softw., 18, 1–6. [Google Scholar]
- Reinhold W.C. et al. (2012) CellMiner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res., 72, 3499–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold W.C. et al. (2014) Using drug response data to identify molecular effectors, and molecular “omic” data to identify candidate drugs in cancer. Hum. Genet, 3, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samur M.K. (2014) RTCGAToolbox: a new tool for exporting TCGA Firehose data. PloS One, 9, e106397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. (2013) fmcsR: mismatch tolerant maximum common substructure searching in R. Bioinformatics, 29, 2792–2794. [DOI] [PubMed] [Google Scholar]