Abstract
Motivation: Recent advances in single molecule real-time (SMRT) and nanopore sequencing technologies have enabled high-quality assemblies from long and inaccurate reads. However, these approaches require high coverage by long reads and remain expensive. On the other hand, the inexpensive short reads technologies produce accurate but fragmented assemblies. Thus, a hybrid approach that assembles long reads (with low coverage) and short reads has a potential to generate high-quality assemblies at reduced cost.
Results: We describe hybridSPAdes algorithm for assembling short and long reads and benchmark it on a variety of bacterial assembly projects. Our results demonstrate that hybridSPAdes generates accurate assemblies (even in projects with relatively low coverage by long reads) thus reducing the overall cost of genome sequencing. We further present the first complete assembly of a genome from single cells using SMRT reads.
Availability and implementation: hybridSPAdes is implemented in C++ as a part of SPAdes genome assembler and is publicly available at http://bioinf.spbau.ru/en/spades
Contact: d.antipov@spbu.ru
Supplementary information: supplementary data are available at Bioinformatics online.
1 Introduction
While de novo sequencing from long and inaccurate SMRT reads results in accurate assemblies (Berlin et al., 2015; Chin et al., 2013), these projects require high coverage of a genome by reads and thus remain expensive. Moreover, for Oxford Nanopore technology (with even higher error rates than in SMRT reads), accurate de novo assemblies remain challenging even in high coverage sequencing projects (Goodwin et al., 2015; Loman et al., 2015). For example, the highest reported accuracy of assemblies from Oxford Nanopore reads (99.5%) is significantly below the acceptable standards for finished genomes.
On the other hand, recently developed hybrid approaches for assembling long (and inaccurate) and short (and accurate) reads proved to be useful in generating high-quality assemblies at a relatively low cost (Deshpande et al., 2013; Koren et al., 2012; Ribeiro et al., 2012). In some complex applications (e.g. metagenomics and single cell genomics), the hybrid approaches may represent an attractive alternative to de novo assembly for long reads.
We describe hybridSPAdes—a hybrid assembly approach that benefits from synergy between accurate short and error-prone long reads. hybridSPAdes uses the same algorithms for both Pacific Biosciences reads (about % error rate) and Oxford Nanopore reads (an even higher error rate), e.g. recently, hybridSPAdes was applied to studies of bacterial plankton using assembly of Illumina and Pacific Biosciences reads (Labonté et al., 2015) and for analyzing antibiotics resistance using assembly of Illumina and Oxford Nanopore reads (Ashton et al., 2015).
We benchmark hybridSPAdes against other hybrid assembly tools (Deshpande et al., 2013; Koren et al., 2012) and demonstrate that it enables accurate assemblies even in the case when the number of long reads is relatively small. We further show that hybridSPAdes works well even in the difficult case of single cell genome assembly resulting in the complete circular chromosome assembly of the elusive Candidate Phylum TM6 (McLean et al., 2013) that remains uncultivated.
2 Approach
While the de Bruijn graph approach currently dominates the short reads sequencing projects, its applications to assembling long reads faces various challenges. Indeed, high error rate in long reads makes it difficult to construct the de Bruijn graph from long reads for any reasonable choice of the k-mer size. As a result, the existing de novo long read assemblers use the overlap-layout-consensus approach instead of the de Bruijn graph approach (Berlin et al., 2015; Chin et al., 2013).
Thus, one has to choose between the de Bruijn graph and the overlap-layout-consensus approaches while assembling short and long reads. SPAdes constructs the de Bruijn graph from short reads and transforms it into an assembly graph (Bankevich et al., 2012; Nurk et al., 2013). The assembly graph is defined as the condensed and simplified de Bruijn graph (Pevzner et al., 2001) of k-mers in reads after removal of bulges, tips and chimeric edges. After SPAdes constructs the assembly graph, hybridSPAdes uses long reads for gap closure and repeat resolution in this graph.
exSPAnder (Prjibelski et al., 2014; Vasilinetc et al., 2015) is a module of SPAdes that utilizes various sources of data (e.g. multiple paired-end or mate-pair libraries) for resolving repeats and closing gaps in assembly. exSPAnder is a modular and easily extendable algorithm based on the path extension framework (Boisvert et al., 2010; Bresler et al., 2012; Zhu et al., 2014). Given a path in the assembly graph, exSPAnder iteratively attempts to grow it by choosing one of its extension edges (all the edges that start at the terminal vertex of this path). The choice of the extension edge is controlled by the exSPAnder decision rule (Prjibelski et al., 2014) that evaluates how well this extension edge is supported by data (e.g. paired reads). Thus, in order to incorporate the repeat resolution by long reads in the exSPAnder framework, one has to represent each long read as a read-path, the path in the assembly graph that spells out the error-free version of the long read. hybridSPAdes uses a new decision rule in exSPAnder that is based on the analysis of these read-paths.
In addition to resolving repeats in the assembly graph constructed from short reads, long reads can also contribute to closing the coverage gaps in this graph. In the case when a coverage gap is spanned by multiple long reads, one can fill in the gap by constructing the consensus of long reads within the gap’s span (Chin et al., 2013).
Overall, hybridSPAdes includes the following steps:
Constructing the assembly graph from short reads using SPAdes;
Mapping long reads to the assembly graph and generating read-paths;
Closing gaps in the assembly graph using the consensus of long reads that span the gaps;
Resolving repeats in the assembly graph by incorporating long read-paths into the decision rule of exSPAnder.
3 Methods
3.1 Mapping long reads to the assembly graph
Given a set of short reads ShortReads, is the de Bruijn graph constructed on all k-mers from this set. SPAdes uses various graph simplification procedures (Bankevich et al., 2012; Nurk et al., 2013) to transform the de Bruijn graph into the assembly graph . In this section we describe an algorithm for analyzing how each long read Read traverses the graph resulting in a read-path Path(Read).
Similarly to BLASR tool for SMRT reads alignment (Chaisson and Tesler, 2012), hybridSPAdes selects a seed length t (the default value t = 13) and maps t-mers in long reads to edges in the assembly graphs that contain these t-mers. This information is used to find out how each long read traverses . To answer this question, we first find out how a long read traverses edges in the assembly graph.
3.1.1 Mapping t-mers from reads to the assembly graph
hybridSPAdes transforms each long read into a set of t-mers and finds positions of these t-mers on the edges of the assembly graph. Note that t-mers starting at the first positions or ending at the last positions of an edge map to a vertex in the assembly graph. Thus, since such t-mers may be assigned to multiple edges incident to these vertices, we exclude them from further consideration.
Given a t-mer shared by a read and an edge in the assembly graph, we define its t-mer mapping as a triple (e, i, j) where e is an edge in the assembly graph where the t-mer is mapped, and i and j are the positions of the t-mer on this edge and in the read, respectively.
Since there are many spurious t-mer mappings, the fact that a t-mer in a read Read maps to an edge in the assembly graph does not necessarily mean that the read-path Path(Read) traverses this edge. However, our analysis revealed that for nearly all reads, if more than MinSeedNumber t-mers in a read map to an edge in the assembly graph then the read-path traverses this edge (the default value MinSeedNumber = 8). We therefore say that an edge in the assembly graph is supported by a read if at least MinSeedNumber t-mers in this read map to this edge.
3.1.2 Mapping long reads to edges of the assembly graph
Consider mappings and of two t-mers from a given read. Define and dgraph as the distances between these t-mers in the read and in the assembly graph, respectively. dgraph is defined as follows: if the mappings share the same edge and i1 < i2, the distance is , otherwise it is the length of the shortest path in the assembly graph from the position i1 on edge e1 to position i2 on edge e2.
A mapping is a predecessor of mapping if
;
, i.e. the distance along the read is not too small as compared to the distance in the assembly graph;
if these t-mers map to the same edge in the assembly graph, i.e. the distance along the read is not too large as compared to the distance along a single edge in the assembly graph. This condition is not enforced if two t-mers map to different edges in the assembly graph since the read-path between these edges is not necessarily the shortest path in the assembly graph;
both dread and dgraph are smaller than c3.
The default values of parameters c1, c2 and c3 are 0.7, 1.3 and 500, respectively.
We further construct a directed graph using the set of all t-mer mappings from the read Read as the vertex-set. We connect vertices (t-mer mappings) in this graph by a directed edge if the first one is a predecessor of the second. Since the resulting graph is acyclic (every edge connects a mapping with a smaller read coordinate to a vertex with a larger read coordinate), we can find a longest path in this graph using a fast dynamic programming algorithm.
Next, we determine how the found path through t-mer mappings in the graph traverses long edges of the assembly graph. Since there are many spurious t-mer mappings, we limit attention to the sequence of edges EdgeSequence(Read) in this path that are supported by the given read Read. Note that EdgeSequence(Read) may have some missing edges as compared to the correct read-path Path(Read). Our goal now is to reconstruct these missing edges that often aggregate into complex subgraphs in the assembly graph.
3.1.3 Mapping long reads to complex subgraphs of the assembly graph
Consider two consecutive edges in the sequence EdgeSequence(Read) that are not consecutive in the assembly graph. Our goal is to figure out how the correct read-path Path(Read) traverses the assembly graph between these edges.
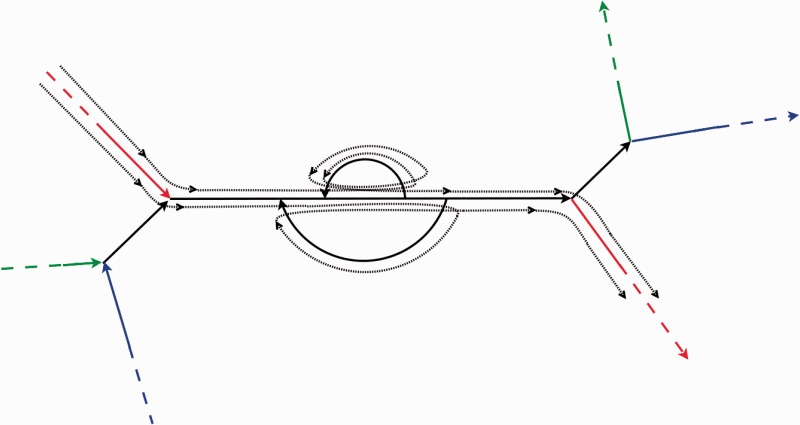
Figure 1 depicts two consecutive edges from EdgeSequence(Read) (shown in red) that are separated by a complex subgraph in the assembly graph. We need to determine which of the alternative paths between these edges in the assembly graph in Figure 1 are traversed by Path(Read).
Fig. 1.
Three pairs of long edges in the assembly graph (corresponding to unique regions in the genome and shown as colored edges) separated by short edges that represent repeats in the genome (shown in black). The genome path traverses edges of the same color in the consecutive fashion. Two dotted paths represent two different options for a long read (with fixed length and alignment to long edges) to traverse this repetitive region. The goal is to figure which of these dotted paths is correct
Given a path Path in a directed graph with edges labeled by characters from a given alphabet, we define String(Path) as the concatenation of labels of the edges from Path. We define the edit distance between strings String1 and String2 as the minimum total cost of substitutions and indels needed to transform one string into another (we assume that every substitution has cost μ and every indel has cost σ). Our goal is to solve the following problem:
Graph Alignment Problem: Find a path between two given vertices of the labeled directed graph that spells out the string with minimal edit distance to the given string (among all possible paths between these vertices).
Input: A string String and a labeled directed graph Graph with vertices source and sink.
Output: A path Path in Graph minimizing over all possible paths in Graph from source to sink.
A brute-force solution of this problem (in the context of hybrid assembly) is to enumerate all possible paths between two long edges (within a certain range of lengths) and to find a path with the minimum edit distance to the long read. While this approach works for bacterial genomes and is used in the current hybridSPAdes implementation, the number of paths may be exponential in the number of vertices of the assembly graph. Below we describe a polynomial algorithm for solving the Graph Alignment Problem.
Given a labeled directed graph Graph and a string String, we define a graph Graph(String) with the vertex-set corresponding to the pairs where v is a vertex in Graph and . In order to define the edge-set of Graph(String), we specify the incoming edges to the vertex as follows:
edge of length σ for each edge (w, v) in Graph;
edge of length σ for each vertex v in Graph;
edge for each edge (v, w) in Graph. The length of this edge is defined as zero if the label of the edge (v, w) in Graph is equal to the ith symbol of String, and μ otherwise.
It is easy to see that each series of edit operations with total cost score between String and a string spelled by a path from source to sink in Graph corresponds to a path of length score between and in Graph(String).
Therefore, in order to solve the Graph Alignment Problem, we need to find a shortest path between and in Graph(String). Since this graph may have directed cycles, we use the Dijkstra algorithm (Cormen et al., 2001) with the worst case running time , where and are the vertex-set and edge-sets of the graph Graph(String), respectively.
In the case of the hybrid assembly, since there are at most 4 outgoing edges for each vertex in the assembly graph, there are at most outgoing edges for each vertex in the graph Graph(String). Thus, since the total number of edges in Graph(String) is , the running time of the algorithm is , where V is the vertex-set of the assembly graph.
In the context of a typical assembly graph, is much larger than . Also, for the majority of reads, there exists a path of length approximately between source and sink in the assembly graph. Therefore, we can ignore all the vertices of that are farther than from both source and sink while searching for a path with the minimum edit distance.
3.2 Closing coverage gaps in the assembly graph
Although the coverage by short reads is rather uniform in most assembly projects, there are sometimes significant drops in k-mer coverage (Bankevich et al., 2012) and even regions where the k-mer coverage drops to zero. However, these drops in the k-mer coverage rarely affect repetitive edges in the assembly graph since it is unlikely that they occur in all copies of a repeat. Below we focus on gaps in the k-mer coverage that occur within a unique (non-repetitive) region of a genome corresponding a single edge in the assembly graph.
A coverage gap breaks this edge into two edges that we refer to as a sink edge (ending in a vertex without outgoing edges) and a source edge (starting in a vertex without incoming edges). If a long read maps to both a sink and a source edge, then this read can potentially close a gap in the assembly graph. However, a single error-prone long read spanning the gap does not allow one to accurately close a gap, i.e. to reconstruct the nucleotide sequence of the gap. We thus collect the set of all long reads spanning the same pair of sink and source edges (forming the set of reads SpanningReads) and close the coverage gap using the consensus sequence of all these reads.
For each read from SpanningReads, we align this read against the sink edge (ending at position p of the read) and the source edge (starting at position q of the read). The segment of the read from position p + 1 to q – 1 represents an error-prone sequence of the gap. hybridSPAdes fills in the gap by solving the Multiple String Consensus Problem (Sim and Park, 2003) for all such segments derived from SpanningReads. To solve this problem, we apply the Partial Order Graph approach (Lee et al., 2002) and use its ConsensusCore library implementation from Pacific Biosciences that proved to work well for SMRT reads (Chin et al., 2013).
3.3 Repeat resolution in the assembly graph
The read-against-graph alignment algorithm described above allows one to map each long read to a read-path in the assembly graph. During the repeat resolution stage of hybridSPAdes, we limit attention to paths traversing at least two long edges in the assembly graph. Our goal is to transform this set of paths into contigs that represent the genome assembly. Below we explain how to achieve this goal using the exSPAnder repeat resolution framework (Prjibelski et al., 2014; Vasilinetc et al., 2015). exSPAnder iteratively constructs a set of paths Paths that represent contiguous segments of the genome. In the beginning, Paths is formed by paths consisting of single long edges in the assembly graph. exSPAnder attempts to iteratively extend each path in Paths using its decision rule (see Section 2). If multiple extension edges pass the decision rule for a given path (which usually implies that this path ends in a difficult-to-resolve repeat), exSPAnder stops the extension process for this path.
Given a path P and its extension edge e, exSPAnder defines the scoring function and bases its decision rule on analyzing all values for all extension edges. Below we describe how hybridSPAdes defines .
Read-paths and overlap if a suffix of (i.e. the path formed by the last i edges of ) coincides with a prefix of (i.e. the path formed by the first i edges of ). We define as the longest suffix of that coincides with a prefix of .
A read-path is called trivial if it consists of a single edge and non-trivial otherwise. Since trivial read-paths do not contribute to the repeat resolution, we exclude them from further consideration. Note that there are typically multiple reads with the same read-path, at least in projects with high coverage by SMRT reads. We define multiplicity of a read-path as the number of long reads resulting in this read-path and classify a read-path as reliable if its multiplicity exceeds 1 (SMRT datasets have many chimeric reads that typically have multiplicity 1).
Let ReadPaths be the set of all non-trivial reliable read-paths and ReadPaths(e) be its subset formed by all read-paths containing an edge e. An edge e in is called non-repetitive if
all pairs of read-paths in ReadPaths(e) overlap and their overlap contains e.
edge e appears at most once in each read-path from ReadPaths;
For the datasets with relatively even coverage by Illumina reads (e.g. reads generated from cultured cells but not single cells amplified with MDA) we impose an additional condition—an edge is called non-repetitive if it is sufficiently long (exceeds 500 bp in the default setting) and its coverage does not significantly exceeds the median coverage of the entire dataset (does not exceed the median coverage by more than 20% in the default setting).
A read-path ReadPath follows a path P in the assembly graph if there exists a path such that its prefix coincides with P, its suffix coincides with ReadPath, and at least one of edges from is non-repetitive.
Given a path P and a set of read-paths ReadPaths, we define ReadPathsP as the set of all read-paths from ReadPaths that follow P. Given an extension edge e of a path P, we define as the total multiplicity of read-paths in the set , where is the path P extended by the edge e.
If a path P has an extension edge e whose score dominates scores of all other extension edges (i.e. exceeds them by a factor of at least c), hybridSPAdes extends P by e (the default value c = 2). Otherwise, the extension procedure stops. If the highest scoring extension edge does not dominate the scores of all other extension edges, exSPAnder applies the standard extension rules based on read-pairs (Prjibelski et al., 2014; Vasilinetc et al., 2015).
4 Results
4.1 Datasets
We analyzed datasets combining short and long reads from E.coli str. K12 (datasets ECOLI100, ECOLI200 and ECOLI-NANO), M.ruber (dataset MRUBER), Streptomyces sp. PAMC26508 (dataset STREPTO) and candidate division TM6 bacterium TM6SC1 (dataset TM6). The reads in the latter dataset were generated from single cells amplified with the Multiple Displacement Amplification (MDA) technology (Lasken, 2007). Prior to this study, the genome of TM6SC1 was only partially assembled (see McLean et al., 2013 for details).
ECOLI200 dataset contains SMRT reads with 200 × coverage and P6/C4 enzyme/chemistry (average read length 5280 bp). ECOLI100 dataset contains SMRT reads with 100 × coverage and older P4/C2 enzyme/chemistry (average read length 10 598 bp). ECOLI-NANO dataset contains Oxford Nanopore reads (average read length 6060 bp). All three E.coli str. K12 datasets contain Illumina reads of length 100 bp, mean insert length 215 bp and coverage 230 × obtained with Illumina Genome Analyzer IIx.
Mapping of Illumina reads to E.coli str. K12 genome revealed that the strain used for generating these datasets differs from the reference sequence of E.coli str. K12 (three insertions of mobile elements about 1 kbp in length). These differences result in six breakpoints that are reported as six assembly errors by the assembly evaluation tool QUAST (Gurevich et al., 2013). We thus ignored these six (pseudo) errors while benchmarking various assemblers.
MRUBER dataset contains SMRT reads with 120 × coverage (average read length 2430 bp). Illumina reads for this dataset were generated using Illumina Nextera Mate Pair technology (there were no paired-end reads in this dataset) with read length 150 bp, mean insert length 3500 bp and low 20 × coverage.
STREPTO dataset contains SMRT reads with 25 × coverage (average read length 1410 bp). Illumina reads were generated with Illumina HiSeq 2000 with read length 150 bp, mean insert length 280 bp and coverage ×. Streptomyces sp. PAMC26508 genome has high (71%) GC content.
TM6 dataset contains both SMRT reads (45 × coverage) and Illumina reads (265 × coverage) generated from MDA-amplified single cells. The Illumina reads were generated with Genome Analyzer IIx (read length 100 bp, mean insert length 270 bp). Note that MDA-based single cell approaches result in highly uneven genome coverage by reads (Bankevich et al., 2012).
The links to all the datasets and reference genomes are available in supplementary materials.
4.2 Software tools
We benchmarked hybridSPAdes (as a part of SPAdes 3.6 release), Cerulean (Deshpande et al., 2013) and PBcR (Koren et al., 2012) (version wgs-8.3rc1). In the hybrid mode (that we refer to as hybridPBcR), PBcR uses short Illumina reads to error-correct the long (SMRT or Nanopore) reads. In the self-correction mode (that we refer to as selfPBcR) PBcR only uses long reads for assembly. We used the QUAST assembly evaluation tool (Gurevich et al., 2013) for benchmarking. While QUAST reports many assembly metrics, the benchmarking tables below are limited to NG50, NG75, LG50, the length of the longest contig, and the number of misassemblies (MA), where NG50 is the length for which the collection of all contigs of that length or longer covers at least half of the reference genome, NG75 is defined similarly to NG50 with 75% of reference genome instead of 50%. LG50 is the number of contigs longer or equal than NG50.
Although AllPaths-LG (Ribeiro et al., 2012) has a hybrid mode for assembling short and long reads, we did not have an opportunity to benchmark it since none of the datasets described above satisfy the strict constraints on the insert sizes imposed by AllPaths-LG.
The field of hybrid assembly has been rapidly developing in the last year when the Oxford Nanopore assembly pipeline Nanocorrect (Loman et al., 2015), hybrid Nanopore & Illumina assembly pipeline NanoCorr (Goodwin et al., 2015) and hybrid scaffolder LINKS (Warren et al., 2015) were added to the arsenal of tools for assembling Oxford Nanopore reads. However, Nanocorrect and NanoCorr focused on Oxford Nanopore reads rather than Pacific Biosciences reads. We and others (Ashton et al., 2015; Liao et al., 2015; Utturkar et al., 2014) demonstrated that hybridSPAdes works well for hybrid assembly with both Pacific Biosciences and Oxford Nanopore reads.
4.3 Benchmarking
hybridSPAdes and selfPBcR assembled both ECOLI100 and ECOLI200 datasets in a single contig (Table 1). As expected, both hybridSPAdes and selfPBcR resulted in six (pseudo) assembly errors caused by the known differences between the analyzed and the reference strains (three insertions of mobile elements). selfPBcR produced two additional (real) misassemblies and hybridSPAdes produced one. Cerulean and hybridPBcR generated more fragmented assembly and, in case of Cerulean, more misassemblies for ECOLI100 dataset. For ECOLI200 dataset, both Cerulean and hybridPBcR generated inferior assemblies.
Table 1.
Benchmarking of hybridSPAdes, PBcR and Cerulean on E.coli datasets
| LG50 | NG50 | NG75 | longest | MA | |
|---|---|---|---|---|---|
| ECOLI200 | |||||
| hybridSPAdes | |||||
| (Illumina + SMRT) | 1 | 4652737 | 4652737 | 4652737 | 7 |
| hybridPBcR | – | – | – | 34501 | 1 |
| selfPBcR | 1 | 4680888 | 4680888 | 4680888 | 8 |
| Cerulean | 16 | 108914 | 61790 | 225438 | 87 |
| ECOLI100 | |||||
| hybridSPAdes | |||||
| (Illumina + SMRT) | 1 | 4652737 | 4652737 | 4652737 | 7 |
| hybridPBcR | 14 | 109938 | 38778 | 311375 | 4 |
| selfPBcR | 1 | 4661789 | 4661789 | 4661789 | 8 |
| Cerulean | 2 | 1238378 | 1215680 | 1258795 | 10 |
| ECOLI-NANO | |||||
| hybridSPAdes | |||||
| (Illumina + Nanopore) | 1 | 4477336 | 4477336 | 4477336 | 7 |
For each parameter we boldfaced the best results. Longest contig, NG50 and NG75 were compared with reference (4639675 bp). We did not run various genome polishing tools like Quiver (Chin et al., 2013) since our benchmarking focused on assembly errors rather than basecalling errors.
In addition to hybrid assembly of Illumina and SMRT reads, hybridSPAdes also assembled ECOLI-NANO dataset into a single contig. All other tested assemblers failed to assemble this dataset.
We have also investigated how the performance of hybridSPAdes and PBcR deteriorates when the coverage by long reads is reduced. To perform this analysis, we retained a fixed fraction of randomly chosen SMRT reads resulting in coverage varying from 200× to 6.25×. As Table 2 illustrates, even with low 12.5 × coverage by SMRT reads, hybridSPAdes generates a high-quality assembly (better than PBcR with 50 × coverage). The quality of PBcR assemblies deteriorates when the coverage falls below 50×.
Table 2.
Benchmarking of hybridSPAdes and selfPBcR on down-sampled ECOLI200 datasets with reduced coverage by long reads
|
ECOLI200 |
|||||
|---|---|---|---|---|---|
| LG50 | NG50 | NG75 | longest | MA | |
| hybridSPAdes 200× | 1 | 4652737 | 4652737 | 4652737 | 7 |
| selfPBcR 200× | 1 | 4680888 | 4680888 | 4680888 | 8 |
| hybridSPAdes 100× | 1 | 4652556 | 4652556 | 4652556 | 7 |
| selfPBcR 100× | 1 | 4677843 | 4677843 | 4677843 | 8 |
| hybridSPAdes 50× | 1 | 4652375 | 4652375 | 4652375 | 7 |
| selfPBcR 50× | 3 | 758477 | 494886 | 876582 | 9 |
| hybridSPAdes 25× | 1 | 2643623 | 1817256 | 2643623 | 7 |
| selfPBcR 25× | – | – | – | 82951 | 6 |
| hybridSPAdes 12.5× | |||||
| (Illumina + PacBio) | 1 | 3398297 | 746845 | 3398297 | 9 |
| selfPBcR 12.5× | – | – | – | 15884 | 0 |
| hybridSPAdes 6.25× | 5 | 356505 | 210465 | 692018 | 9 |
| selfPBcR 6.25× | – | – | – | – | – |
The coverage was downsampled from 200× to 6.25×. NG50, LG50 and NG75 are not defined for PBcR assembly with coverage 25× and lower because the total assembly length is less than half of the genome length. For coverage 6.25×, PBcR failed to generate an assembly.
selfPBcR assembled MRUBER dataset into a single contig with a single misassembly, while hybridSPAdes assembled this dataset into three error-free contigs with zero misassemblies (Table 3). hybridSPAdes failed to assemble this dataset into a single contig because long reads in this dataset do not span over a long 7 Kbp repeat. hybridPBCR produced an assembly with quite similar stats. Cerulean produced lower quality assembly, hybridSPAdes generated a high-quality assembly of STREPTO dataset with Kbp (2 misassemblies), while Cerulean generated an assembly with Kbp and 10 misassemblies (Table 4). hybridPBcR failed on this dataset while selfPBcR produced a low-quality assembly due to the low coverage by SMRT reads.
Table 3.
Benchmarking of hybridSPAdes, PBcR and Cerulean on M.ruber dataset
|
MRUBER |
|||||
|---|---|---|---|---|---|
| LG50 | NG50 | NG75 | longest | MA | |
| hybridSPAdes | 1 | 1709645 | 1387667 | 1709645 | 0 |
| Cerulean | 3 | 305771 | 262734 | 1117272 | 4 |
| hybridPBcR | 1 | 1753481 | 766814 | 1753481 | 0 |
| selfPBcR | 1 | 3100304 | 3100304 | 3100304 | 1 |
Table 4.
Benchmarking of hybridSPAdes, PBcR and Cerulean on Streptomyces dataset
|
STREPTO |
|||||
|---|---|---|---|---|---|
| LG50 | NG50 | NG75 | longest | MA | |
| hybridSPAdes | 4 | 903095 | 679085 | 1366650 | 1 |
| Cerulean | 4 | 645600 | 388134 | 1240002 | 10 |
| hybridPBcR | 12 | 225991 | 124032 | 452318 | 1 |
In contrast to the previous assemblies of SMRT reads in single cell genomics (Labonté et al., 2015; Swan et al., 2014) that came short of closing the assemblies, application of hybridSPAdes to TM6 dataset resulted in a single circular contig of length 1089 Kbp (which contains all previously sequenced seven long contigs with total length 1075 Kbp (McLean et al., 2013)). To the best of our knowledge, it is the first assembly of SMRT reads in single cell genomics that resulted in a complete genome.
Since prior to this study, TM6 genome was incomplete, we used the genome assembled by hybridSPAdes to evaluate performance of other assemblers on this dataset.
Cerulean generated an assembly with the largest contig of length 774 Kbp and 1 misassembly (Table 5). hybridPBCR failed on this dataset while selfPBcR produced a low-quality assembly.
Table 5.
Benchmarking of hybridSPAdes, PBcR and Cerulean on TM6 dataset
|
TM6 |
|||||
|---|---|---|---|---|---|
| LG50 | NG50 | NG75 | longest | MA | |
| hybridSPAdes | 1 | 1088795 | 1088795 | 1088795 | – |
| Cerulean | 1 | 773677 | 221583 | 773677 | 1 |
| selfPBcR | 7 | 41009 | 18600 | 146018 | 26 |
The initial TM6 assembly by SPAdes had total size exceeding 4 Mb that greatly exceeds the genome length reported in McLean, 2013 (McLean et al., 2013). This is caused by contaminants since TM6 dataset represents a mini-metagenome. See (McLean et al., 2013) for details.
Our benchmarking demonstrated that hybridSPAdes improves on the state-of-the-art hybrid assemblers on all datasets we have analyzed (on two of these datasets with a high SMRT read coverage, selfPBcR showed similar results).
5 Conclusions
Early tools for hybrid assembly combined Illumina and Sanger reads or Illumina and 454 reads (Boisvert et al., 2010; Chevreux et al., 1999; Zimin et al., 2013). However, hybrid assembly of Illumina and SMRT reads presents new algorithmic challenges since SMRT reads have higher error rates than Sanger reads or 454 reads.
Our benchmarking demonstrated that hybridSPAdes assembles short accurate and long error-prone reads into long and accurate contigs. The resulting low-cost high-quality assemblies are important for accurate genome annotations and comparative genomics. Moreover, hybridSPAdes opens a possibility to complete genomes assembled from single cells. Although 1000 s of bacterial genomes have been assembled from single cells in the last 3 years using specialized single cell assemblers SPAdes (Bankevich et al., 2012) and IDBA-UD (Peng et al., 2012), finishing genomes amplified from single cells is often viewed as an impossible task (Lasken and McLean, 2014). Moreover, sequencing single cell genomes from SMRT reads is likely to be excessively expensive due to highly non-uniform coverage characteristic of the MDA-amplified datasets. Hybrid assembly of short and long reads, on the other hand, turns complete genome assembly from single cells into reality.
While the detailed analysis of the relative market costs and trade-offs of various sequencing technologies remained beyond the scope of this article, we anticipate that many future sequencing projects will use hybrid assembly of reads generated by various technologies.
Supplementary Material
Acknowledgements
We thank Roger Lasken, Mark Novotny and Cheryl Heiner for their expertise that helped to generate the SMRT reads for the MDA amplified TM6 genome. We are grateful to Alla Lapidus, Kira Vyatkina and SPAdes developing team for many thoughtful discussions, suggestions and comments that improved the article. We are also grateful to the anonymous referees whose comments have benefited the article greatly.
Funding
This work was supported by the Russian Science Foundation [grant 14-50-00069].
Conflict of Interest: none declared.
References
- Ashton P.M., et al. (2015) MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat. Biotechnol., 33, 296–300. [DOI] [PubMed] [Google Scholar]
- Bankevich A., et al. (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol., 19, 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin K., et al. (2015) Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol., 33, 623–630. [DOI] [PubMed] [Google Scholar]
- Boisvert S., et al. (2010) Ray: Simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J. Comput. Biol., 17, 1519–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresler M., et al. (2012) Telescoper: de novo assembly of highly repetitive regions. Bioinformatics, 28, 311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson M.J., Tesler G. (2012) Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics, 13, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevreux B., et al. (1999) Genome sequence assembly using trace signals and additional sequence information. In: German Conference on Bioinformatics, pp. 45–56. [Google Scholar]
- Chin C.-S., et al. (2013) Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods, 10, 563–569. [DOI] [PubMed] [Google Scholar]
- Cormen T.H., et al. (2001) Introduction to Algorithms. MIT Press, Cambridge, MA. [Google Scholar]
- Deshpande V., et al. (2013) Cerulean: a hybrid assembly using high throughput short and long reads. In: Algorithms in Bioinformatics. LNBI 8126 Springer, Berlin-Heidelberg, pp. 349–363. [Google Scholar]
- Goodwin S., et al. (2015) Oxford Nanopore sequencing and de novo assembly of a eukaryotic genome. BioRxiv, pp. 013490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A., et al. (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics, 29, 1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S., et al. (2012) Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat. Biotechnol., 30, 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonté J.M., et al. (2015) Single-cell genomics-based analysis of virus–host interactions in marine surface bacterioplankton. ISME J., 9, 2386–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasken R.S. (2007) Single-cell genomic sequencing using multiple displacement amplification. Curr. Opin. Microbiol., 10, 510–516. [DOI] [PubMed] [Google Scholar]
- Lasken R.S., McLean J.S. (2014) Recent advances in genomic DNA sequencing of microbial species from single cells. Nat. Rev. Genet., 15, 577–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., et al. (2002) Multiple sequence alignment using partial order graphs. Bioinformatics, 18, 452–464. [DOI] [PubMed] [Google Scholar]
- Liao Y.-C., et al. (2015) Completing bacterial genome assemblies: strategy and performance comparisons. Sci. Rep., 5, 8747 doi: 10.1038/srep08747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loman N.J., et al. (2015) A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods, 12, 733–735. [DOI] [PubMed] [Google Scholar]
- McLean J.S., et al. (2013) Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum. Proc. Natl Acad. Sci., 110, E2390–E2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S., et al. (2013) Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol., 20, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., et al. (2012) IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics, 28, 1420–1428. [DOI] [PubMed] [Google Scholar]
- Pevzner P., et al. (2001) An Eulerian path approach to DNA fragment assembly. Proc. Natl Acad. Sci. USA, 98, 9748–9753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prjibelski A.D., et al. (2014) ExSPAnder: a universal repeat resolver for DNA fragment assembly. Bioinformatics, 30, i293–i301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro F.J., et al. (2012) Finished bacterial genomes from shotgun sequence data. Genome Res., 22, 2270–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim J.S., Park K. (2003) The consensus string problem for a metric is NP-complete. J. Discret. Algorithms, 1, 111–117. [Google Scholar]
- Swan B.K., et al. (2014) Genomic and metabolic diversity of marine group i thaumarchaeota in the mesopelagic of two subtropical gyres. PLoS One, 9, e95380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utturkar S.M., et al. (2014) Evaluation and validation of de novo and hybrid assembly techniques to derive high-quality genome sequences. Bioinformatics, 30, 2709–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilinetc I., et al. (2015) Assembling short reads from jumping libraries with large insert sizes. Bioinformatics, btv337. [DOI] [PubMed] [Google Scholar]
- Warren R.L., et al. (2015) LINKS: Scalable, alignment-free scaffolding of draft genomes with long reads. GigaScience, 4, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., et al. (2014) PERGA: a paired-end read guided de novo assembler for extending contigs using SVM and look ahead approach. PLoS ONE, 9, e114253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin A.V., et al. (2013) The MaSuRCA genome assembler. Bioinformatics, 29, 2669–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.