Abstract
Summary: PathwaysWeb is a resource-based, well-documented web system that provides publicly available information on genes, biological pathways, Gene Ontology (GO) terms, gene–gene interaction networks (importantly, with the directionality of interactions) and links to key-related PubMed documents. The PathwaysWeb API simplifies the construction of applications that need to retrieve and interrelate information across multiple, pathway-related data types from a variety of original data sources. PathwaysBrowser is a companion website that enables users to explore the same integrated pathway data. The PathwaysWeb system facilitates reproducible analyses by providing access to all versions of the integrated datasets. Although its GO subsystem includes data for mouse, PathwaysWeb currently focuses on human data. However, pathways for mouse and many other species can be inferred with a high success rate from human pathways.
Availability and implementation: PathwaysWeb can be accessed via the Internet at http://bioinformatics.mdanderson.org/main/PathwaysWeb:Overview.
Contact: jmmelott@mdanderson.org
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Although the inclusion of pathway and gene interaction information into novel bioinformatics analysis methods is widely expected to result in more powerful and sensitive bioinformatic analyses, we found it surprisingly difficult to assemble a comprehensive database of pathway information in a form suitable for our requirements. Specific challenges we faced included:
Finding a single comprehensive source of pathway-related data that maintains consistent approved identifiers for genes and biological pathways across multiple data sources.
Finding data sources that provide coordinated snapshots of data from multiple sources at a given time to enable replication of results.
Finding data licensed in a way that would allow us to make our tools freely available, not just to academic institutions or requiring an additional license agreement for use.
Although many databases and application programming interfaces (APIs) exist for retrieving pathway-related data for use in biological research, they provide data that are incomplete, use different data identifiers or include databases that are not in sync with one another. Those incompatibilities make the data difficult to merge. Furthermore, the APIs and data formats change often and without warning. Many systems have no API and require either the download of an entire database or parsing of web pages to access the data using an automated system. Some have severe limitations as to how much information can be retrieved using a single search.
Because of the issues listed above, we have developed the PathwaysWeb system. To harmonize the data from various sources, we ensure that gene identifiers are based on gene symbols approved by the Human Gene Nomenclature Committee (HGNC) where possible. The system presents the combined harmonized data via an integrated web API. Each set of data loaded from the various sources is made available as an archived version that provides a snapshot of data from all of the sources at one point in time. That pattern of organization assists in reproducibility of analyses. The system is divided into two parts: (i) PathwaysWeb, which provides data in XML or JSON format for use by automated software systems, and (ii) PathwaysBrowser which provides human friendly access via HTML pages viewable via standard web browsers. The code base and database are not currently available for download online but arrangements could be made available to provide this data. A new version of the data will be added every 6 months.
2 Methods and implementation
PathwaysWeb system provides two interfaces for the user:
A web service implemented in Java that provides data in Extensible Markup Language (XML) and JavaScript Object Notation (JSON) for use by automated systems.
A web application that transforms data from the web service to a user-friendly HTML format.
3 Source datasets
The PathwaysWeb system collects data from the following open-license publicly available sources (with references to original publications on the sources):
Pathways: Reactome Pathways and Genes (Joshi-Tope et al., 2005); NCI-Nature Pathways and Genes and sub pathways (Schaefer et al., 2009).
Genes: HUGO Gene Nomenclature Committee (Gray et al., 2015).
Interactions: NCBI GeneRIFS; Pathway Commons Extended SIF (All) (Cerami et al., 2011); Predictive Networks Interactions (Haibe-Kains et al., 2012).
Other: Custom List of Interaction Types; NCBI Gene to PubMed (Maglott et al., 2007); The Gene Ontology (GO) (Ashburner et al., 2000).
4 Data processing pipeline
Processing of data from the above sources is extensive. A Java application downloads and harmonizes the data and adds them to an Oracle relational database of 26 tables (see Supplementary Material). A detailed diagram of the process is provided in the Supplementary Material.
For data sources generated manually or no longer updated (e.g. NCI Nature Pathways, Predictive Networks) some data are downloaded and processed once rather than with each data version.
New versions of data will be collected from all sources approximately every 6 months. The steps below are performed by an automated process that does not require direct manual curation of the data.
Steps repeated for each data load include the following:
Download latest files from sources and decompress if needed.
Extract subset of tables from databases as individual text files.
Filter text files by removing incomplete or invalid data, unneeded fields and information for non-homo sapiens species. (‘house mouse’ is also retained for GO data).
Fill Entrez IDs or HGNC gene symbol in records if missing.
Update gene symbols to latest official HGNC version.
Merge interactions records from various sources and assign custom interaction types.
Assign consistent IDs to pathways across versions.
Process data from The GO by dropping obsolete and duplicated terms, calculating all paths from root terms to leaf terms and extending mapping of genes-to-terms to include all parent terms.
5 Request and response formats
The Pathways system provides APIs for use in retrieving data through standard HTML-based GET requests. For cases in which gene (or other) lists in the request might result in a URL longer than permitted in GET requests, HTTP POST requests are also supported. The PathwaysWeb API, by default, provides data in an XML format but can also return data in JSON format. PathwaysBrowser has the same request methods as the PathwaysWeb API but returns the data in a human-usable HTML format that includes menus, a search page and other HTML objects to assist in navigation among related pages.
6 API resources
The PathwaysWeb API is REST-like and resource-based. The resources include information on pathways (and sources), interactions (custom types, directions and related PubMed documentation links), data versions, genes (name and symbol synonyms and associated PubMed links) and various data associated with The Gene Ontology. See Figure 1 for relationships between these resources.
Fig. 1.
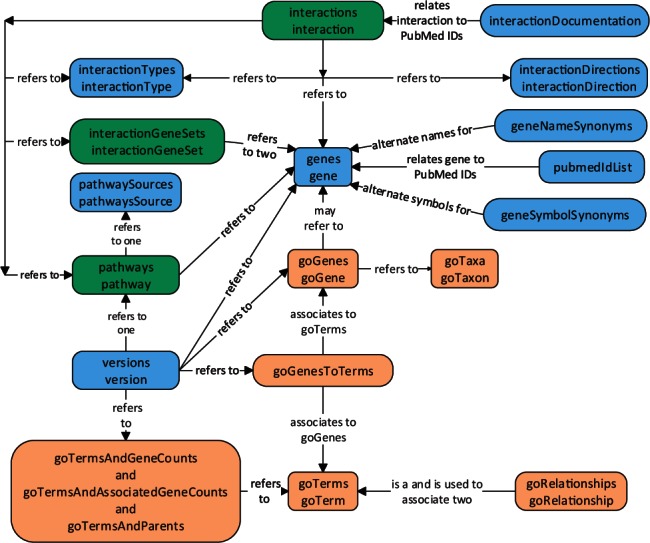
PathwaysWeb resource relationship diagram
The latest documentation can be found via links at: bioinformatics.mdanderson.org/main/PathwaysWeb:Overview.
Supplementary Material
Acknowledgement
The authors thank David Richards for proofreading the manuscript.
Funding
U.S. National Cancer Institute (NCI; MD Anderson TCGA Genome Data Analysis Center) grant numbers CA143883 and CA083639, the Cancer Prevention Research Institute of Texas (CPRIT) grant number RP130397, the Mary K. Chapman Foundation, the Michael & Susan Dell Foundation (honoring Lorraine Dell) and MD Anderson Cancer Center Support Grant P30 CA016672 (the Bioinformatics Shared Resource).
Conflict of Interest: none declared.
References
- Ashburner M., et al. (2000) Gene ontology: tool for the unification of biology. Nat. Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E., et al. (2011) Pathway commons, a web resource for biological pathway data. Nucleic Acids Res., 39, D686–D690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray K., et al. (2015) genenames.org: the HGNC resources in 2015. Nucleic Acids Res., 43, D1079–D1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haibe-Kains B., et al. (2012) Predictive networks: a flexible, open source, web application for integration and analysis of human gene networks. Nucleic Acids Res., 40, D866–D875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi-Tope G., et al. (2005) Reactome: a knowledgebase of biological pathways. Nucleic Acids Res., 33, D428–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglott D., et al. (2007) Entrez gene: gene-centered information at NCBI. Nucleic Acids Res., 35, D26–D31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer C., et al. (2009) PID: the pathway interaction database. Nucleic Acids Res., 37, D674–D679. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


