Abstract
Background. Chronic inflammation and composition of the colon microbiota have been associated with colorectal cancer in humans. The human commensal enterotoxigenic Bacteroides fragilis (ETBF) is linked to both inflammatory bowel disease and colorectal cancer and, in our murine model, causes interleukin 17A (IL-17A)–dependent colon tumors. In these studies, we hypothesized that persistent colonization by ETBF is required for tumorigenesis.
Methods. We established a method for clearing ETBF in mice, using the antibiotic cefoxitin. Multiple intestinal neoplasia mice were colonized with ETBF for the experiment duration or were cleared of infection after 5 or 14 days. Gross tumors and/or microadenomas were then evaluated. In parallel, IL-17A expression was evaluated in wild-type littermates.
Results. Cefoxitin treatment resulted in complete and durable clearance of ETBF colonization. We observed a stepwise increase in median colon tumor numbers as the duration of ETBF colonization increased before cefoxitin treatment. ETBF eradication also significantly decreased mucosal IL-17A expression.
Conclusions. The timing of ETBF clearance profoundly influences colon adenoma formation, defining a period during which the colon is susceptible to IL-17A–dependent tumorigenesis in this murine model. This model system can be used to study the microbiota-dependent and molecular mechanisms contributing to IL-17A–dependent colon tumor initiation.
Keywords: enterotoxigenic Bacteroides fragilis, colitis, colon cancer, microbiota
The risk of colorectal cancer (CRC) increases with the duration of colitis, the anatomic extent of disease, and the degree of inflammation. Patients with ulcerative colitis and Crohn's disease have an estimated 2–3-fold higher incidence of CRC than the general population [1]. The microbiota, or the diverse population of microbes (primarily bacteria) in the gut, plays a crucial role in intestinal health, and, alternatively, dysbiosis, or the disruption of the microbiota, is suspected to contribute to the development of CRC [2, 3].
A key player in dysbiosis and the development of CRC may be the bacterial symbiont enterotoxigenic Bacteroides fragilis (ETBF). ETBF is a molecular subset of Bacteroides fragilis distinguished by the secretion of a sole pathogenic factor, the B. fragilis toxin (BFT) [4, 5]. Increased detection of ETBF has been reported in patients with inflammatory bowel disease and those with CRC [6–8].
Murine ETBF infection causes brief, acute colitis characterized by rapid and robust colonic activation of the signal transducer and activator of transcription 3 protein (Stat3), which drives a T-helper type 17 (Th17) immune response and evolves into chronic colitis [9, 10]. Further, ETBF colonization of multiple intestinal neoplasia (Min) mice induces numerous distal colon adenomas. Min mice are heterozygous for the wild-type gene encoding adenomatous polyposis coli (Apc), a tumor suppressor gene mutated in the majority of human CRC cases; however ApcMin-driven tumors in these mice develop predominantly in the small bowel, with very few tumors in the colon unless the mice are colonized with ETBF [9, 11]. Notably, ETBF colonization of germ-free mice results in rapid death (within 1 day) due to overwhelming colitis, suggesting that, in this model, the microbiota temper ETBF virulence to enable persistent asymptomatic colonization of the mouse colon [12].
Here we determined whether persistent colon colonization by ETBF is essential for tumorigenesis in the MinApcΔ716 mouse. B. fragilis is known to favor mucosal colonization, and tools to selectively remove any B. fragilis strain from the complex mucosal or luminal microbiota are not yet available. Thus, we used ETBF infection time courses to assess whether the acute colonic response to ETBF is sufficient to drive tumorigenesis after ETBF (and microbiota) clearance with the antibiotic cefoxitin. Further, we assessed whether the cefoxitin-induced clearance of ETBF would result in loss of the interleukin 17A (IL-17A) immune response or whether the elevated IL-17A response would persist and continue to drive disease. Patients with strong, chronic Th17 type (ie, IL-17A–driven) intestinal inflammation with CRC have a worse prognosis [13, 14]. Therefore, this model could aid in developing therapies for these high-risk patients.
MATERIALS AND METHODS
Bacteriology
ETBF strain 86-5443-2-2 (a clindamycin-resistant isolate from piglet; kindly provided by Lyle Myers, Montana State University) was grown anaerobically overnight at 37°C in brain heart infusion (BHI) medium (BD Diagnostic Systems) supplemented with 5 g/L yeast extract (Difco), 0.1 mg/L vitamin K (Sigma-Aldrich), 0.5 mg/L hemin (Sigma-Aldrich), 50 mg/L L-cysteine, and 6 g/L clindamycin (Sigma-Aldrich).
Antibiotic Minimum Inhibitory Concentrations (MICs) for ETBF
ETBF organisms in suspension were added to 2 mL of BHI such that the bacteria were at a concentration of 5 × 105 colony-forming units (CFU)/mL. Antibiotics (metronidazole, doxycycline, or cefoxitin) were added to final concentrations of 0.5, 1, 2, 4, 8, 16, and 32 µg/mL. Metronidazole, doxycycline, or cefoxitin were present during growth for 48 hours at 37°C in an anaerobic chamber (AS-580 Anaerobe Chamber, Anaerobe Systems). The MIC end point was defined as the lowest concentration without visible growth.
Animals
MinApcΔ716 (kindly provided by Dr David Huso [Johns Hopkins]) and C57BL/6J (Jackson Laboratories) Helicobacter species–free and specific-pathogen-free mice, kept in conventional housing, were genotyped and inoculated with bacteria as described previously [11, 12] Mice were inoculated with approximately 108 CFU ETBF in phosphate-buffered saline (PBS; approximately 0.1 mL) by oral gavage. As described in “Results” section, ETBF-colonized mice were given antibiotics in the drinking water (their sole water source) as follows: cefoxitin (Apotex), 0.5 mg/mL in water; doxycycline (Hospira), 1 mg/mL in water; and metronidazole (Hospira), 2 mg/mL in 2% grape Kool-Aid (to mask the bitter taste; Dr T. Stappenbeck [Washington University, St. Louis, MO], personal communication); for metronidazole experiments, control mice were also given the flavored water. Antibiotics were given for 14 days unless otherwise stated. A group of ETBF-infected mice were also treated by gavage with metronidazole, 22.5 mg/kg once per day for 7 days. To limit fecal matter as a source of reinfection, cages were changed when starting antibiotic-containing water, 2 days after starting antibiotic-containing water, and then at least weekly.
To prepare tissue specimens for colon polyp analysis, mice were euthanized by CO2 asphyxiation, and colons and ceca were removed, fixed, and stained as described previously [9]. Briefly, to count visible tumors, colons were opened longitudinally and laid flat for fixation with 10% buffered formalin. Colons were then stained with methylene blue for tumor visualization and counted under a Leica ES2 dissecting scope. For microadenoma counts, colons were flushed, swiss-rolled into cryomolds (Tissue Tek), and submerged in 10% buffered formalin for 48 hours prior to histopathologic processing. Microadenoma counts were averaged from two 5-µm sections obtained at 75-µm intervals and stained with hematoxylin and eosin.
All mouse protocols were approved by the Johns Hopkins University Animal Care and Use Committee in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care International.
Fecal Analysis of Colonic ETBF Colonization
ETBF Colony-Forming Units (CFU) per Gram of Stool
Colonization was quantified by plating serial dilutions of stool specimens suspended in PBS on BHI-clindamycin agar plates and incubating them for 48 hours in anaerobic conditions at 37°C. Colonization was generally within the range of 108–1010 CFU/g stool at 3–5 days after inoculation. ETBF colonies are phenotypically recognizable as opaque and domed, compared with other opportunistic colonies; however, putative ETBF colonies were also spot checked for the presence of bft2, using standard polymerase chain reaction (PCR) analysis (forward primer, 5′-TCC CTC TTT GGC GTC GCC-3′; and reverse primer, 5′-CGC TCG GGC AAC TAT-3′) as described previously [15].
Stool DNA Isolation
DNA was isolated from fecal pellets, using the Qiagen QIAamp DNA Stool Mini Kit according to the manufacturer's instructions.
Peptone Yeast Glucose With Bile (PYGB) Liquid Broth Amplification
To ensure clearance of ETBF, stool bacteria were enriched using Bacteroides species–selective PYGB liquid broth (Anaerobe Systems) and incubated anaerobically at 37°C for 48 hours followed by PCR analysis for the bft gene (forward, 5′-GAA CCT AAA ACG GTA TAT GT-3′; and reverse, 5′-GTT GTA GAC ATC CCA CTG GC-3′).
Mucosal Colonization
To evaluate ETBF mucosal colonization, the most distal 2 cm of the colon was washed twice in 1 mL of 0.016% DTT in saline to remove loosely adherent bacteria. Tissue pieces were weighed, homogenized, and serially diluted in PBS. Serial dilutions were plated on BHI-clindamycin agar plates to detect ETBF colonies, and the number of CFU per gram of tissue was calculated.
TaqMan qPCR Analysis
Distal colon tissues for RNA isolation were snap frozen in RNAlater (Ambion). RNA was extracted using the Qiagen Qiacube according to the manufacturer's instructions. TaqMan primers (Life Technologies) were used according to the manufacturer's instructions. Fold-changes from values for sham controls were calculated; threshold cycle values were normalized to those for 18S.
Statistical Analysis
To compare measurements across experimental conditions, we used the Mann–Whitney U test or Kruskal–Wallis test as indicated for continuous variables and the Pearson χ2 test for the categorical variable of tumor size distribution, using Prism 6, version 6.04.
RESULTS
Antibiotic Susceptibility in Culture Does Not Translate to Fecal or Mucosal Clearance in Mice Infected With ETBF
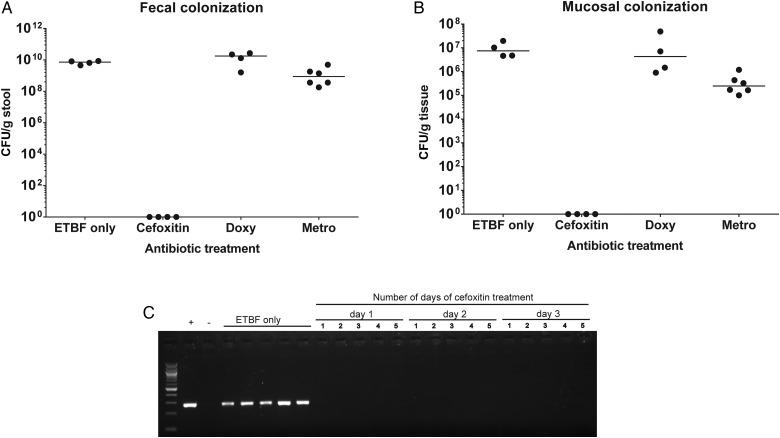
To assess the susceptibility of ETBF to antibiotics commonly used to treat infections with Bacteroides species, we first established the ETBF-specific MICs of cefoxitin, doxycycline, and metronidazole to be 8 µg/mL, 8 µg/mL, and 2 µg/mL, respectively, with concentrations of >16 µg/mL (for cefoxitin and doxycycline) and >2 µg/mL (for metronidazole) indicative of resistance to these antibiotics. Although ETBF was susceptible to all 3 antibiotics in vitro, only peroral cefoxitin consistently cleared ETBF when measured by fecal or mucosal colonization (Figure 1A and 1B). In contrast, fecal and mucosal colonization confirmed persistent ETBF colonization in doxycycline- and metronidazole-treated mice (Figure 1A and 1B). Treatment of ETBF infection with daily metronidazole for 1 week by gavage also did not clear ETBF colonization (data not shown).
Figure 1.
Peroral cefoxitin clears enterotoxigenic Bacteroides fragilis (ETBF) colonization. Wild-type C57Bl/6 mice were colonized with ETBF (for 2 weeks) and then treated for 14 days with cefoxitin (0.5 mg/mL), metronidazole (metro; 2 mg/mL), and doxycycline (doxy; 1 mg/mL) via drinking water. A and B, ETBF colonization was assessed by stool plating (A) and mucosal adherence (B). C, Cefoxitin clearance of fecal ETBF, tested using polymerase chain reaction for detection of bft after enrichment with fecal Bacteroides-selective peptone yeast glucose with bile liquid broth. Stool specimens from ETBF alone mice and mice from the first, second, and third days of cefoxitin treatment are shown. Each lane represents an individual mouse. −, negative control (no template); +, positive control (ETBF DNA). Abbreviation: CFU, colony-forming units.
To ensure cefoxitin-induced ETBF clearance was complete and durable, we amplified the Bacteroides species present in the stool by using PYGB broth and performed PCR to detect the presence of bft. PYGB broth amplification is 2–4 orders of magnitude more sensitive for detecting ETBF colonization than PCR analysis to detect bft in DNA isolated directly from stool (Supplementary Figure 1). Unexpectedly, we found that ETBF was undetectable after the first day of cefoxitin treatment (Figure 1C). Consistent with ETBF clearance by cefoxitin, cecal contraction, an indication of inflammation, was reversed by 2 days after cefoxitin treatment of ETBF-colonized mice (Supplementary Figure 2). Additional experiments also revealed that cefoxitin-treated mice quickly (within 72 hours) reassemble a microbiota (C. Craig and C, Sears, unpublished observations). Further, ETBF was not detectible at the latest time point assayed, 37 days after cefoxitin removal. Thus, we used oral cefoxitin treatment to test the hypothesis that ETBF clearance would diminish ETBF-induced tumorigenesis.
Timing of ETBF Clearance Profoundly Influences Macroscopic Colon Tumor Numbers
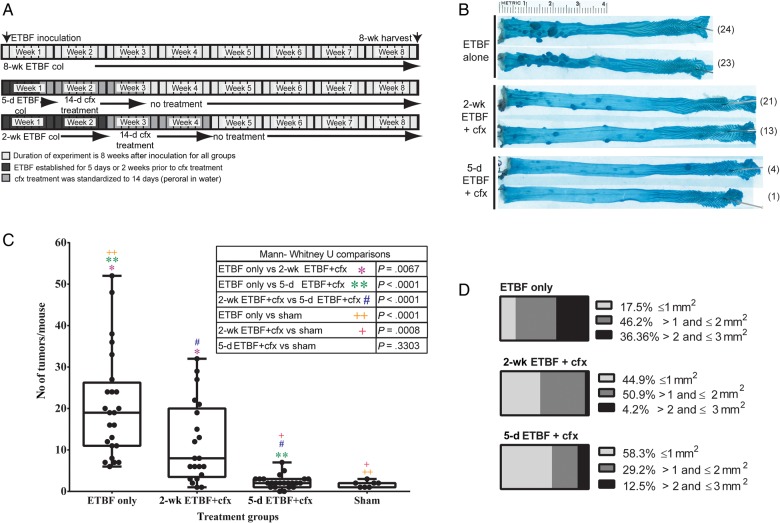
We next examined the impact of ETBF clearance by cefoxitin on colon tumorigenesis in MinApcΔ716 mice (Figure 2A). Briefly, ETBF colonization for either 5 or 14 days was followed by cefoxitin treatment for 14 days (hereafter, the 5-day ETBF + cfx group and the 2-week ETBF + cfx group, respectively), and the 5-day ETBF + cfx and 2-week ETBF + cfx groups were compared to parallel MinApcΔ716 mouse groups colonized with ETBF and not given cefoxitin (3 independent experiments were performed, with 17–23 mice/group). Cefoxitin treatment decreased the number of colon tumors significantly (Figure 2B and 2C). However, the timing of cefoxitin commencement significantly influenced ETBF-driven colon tumor numbers, with a stepwise increase in the median number of colon tumors that paralleled the number of days of ETBF colonization (median number of colon tumors, 2 in the 5-day ETBF + cfx group, 8 in the 2-week ETBF + cfx group, and 19 in the ETBF alone group). The distribution of tumor sizes was also affected, implying that tumors that do form after cefoxitin treatment may grow or progress more slowly (Figure 2D).
Figure 2.
Reduction in adenoma formation in cefoxitin (cfx)–treated mice is dependent on duration of enterotoxigenic Bacteroides fragilis (ETBF) colonization. A, Experimental time line. B, Representative photographs of colons from mice colonized with ETBF for 8 weeks without treatment (ETBF alone), mice colonized with ETBF for 2 weeks and then treated with cfx for 2 weeks (2-wk ETBF + cfx), and mice colonized with ETBF for 5 days and then treated with cfx for 2 weeks (5-d ETBF + cfx). Total number of tumors for the colon shown are in parentheses next to image. C, Total number of macroscopic colonic tumors for each mouse. The number of tumors in the ETBF alone group was significantly higher than in the 2-wk ETBF + cfx group (*P = .0067) and the 5-d ETBF + cfx group (#P < .0001). The difference in tumor numbers between the 2-wk ETBF + cfx group and the 5-d ETBF + cfx group was also significant (**P < .0001). D, Colon adenomas grouped according to size: ≤1 mm2, >1 to ≤2 mm2, and ≥2 mm2. The χ2 statistic is 54.58, and the P value is <.00001. For size assessment, the number of mice per group was as follows: ETBF alone group, 7; 2-wk ET + cfx group, 14; and 5-d ETBF + cfx group, 12. Abbreviation: col, colonization.
Cefoxitin Treatment Reduces the Number of Microadenomas in ETBF-Colonized MinApcΔ716 Mice
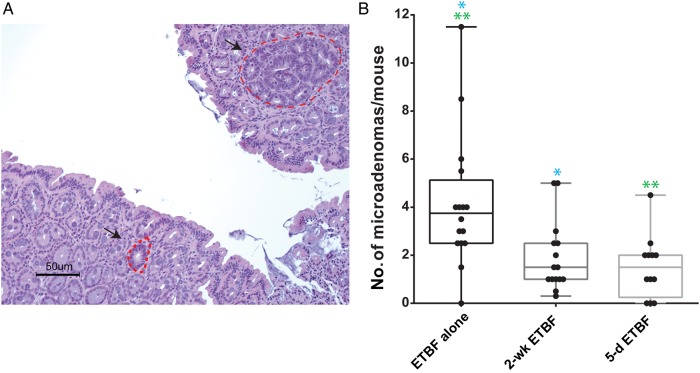
Microadenomas are adenomas that are not grossly visible, are not polyploid, and are characterized by dysplasia with pronounced nuclear atypia and a loss of nuclear polarity (Figure 3A) [16]. Microadenoma formation precedes the formation of macroadenomas. Because cefoxitin treatment reduced the number of macroadenomas formed, we sought evidence that cefoxitin treatment changes the initiation of microadenomas or their progression to macroadenomas by counting the number of microadenomas present, using histopathologic analysis, at multiple time points after ETBF inoculation (Figure 3).
Figure 3.
A, Representative images of microadenomas after enterotoxigenic Bacteroides fragilis (ETBF) colonization. B, Microadenoma formation in the colons of MinApcΔ716+/− mice in groups colonized with ETBF without treatment (ETBF alone) or with cefoxitin treatment; average microadenomas are counted for 2 hematoxylin-eosin–stained sections per mouse. Analysis of the 3 groups was performed by the Kruskal–Wallis test (P = .0017). Pair-wise comparisons were performed by the Mann–Whitney U test for comparison of mice in the ETBF alone group to mice colonized with ETBF for 2 weeks and then treated with cfx for 2 weeks (2-wk ETBF + cfx; *P = .0038); comparison of mice in the ETBF alone group to mice colonized with ETBF for 5 days and then treated with cfx for 2 weeks (5-d ETBF + cfx; **P = .0009); and comparison of mice in the 5-d ETBF + cfx group to mice in the 2-wk ETBF + cfx group (P = .4857).
Data for microadenoma counts at day 19 and 28 after inoculation (pooled) are representative of the pattern that significantly fewer microadenomas were detected in the cefoxitin groups, compared with the ETBF alone group (Figure 3B). No microadenomas were observed in sham MinApcΔ716 mice harvested at the same ages as the ETBF-infected group, supporting the critical contribution of ETBF colonization to microadenoma initiation.
IL-17A Cytokine Expression in the Distal Colon Decreases Significantly Only in Mice Treated With Cefoxitin After 5 Days of ETBF Colonization
Colonic tumor formation in ETBF-colonized MinApcΔ716 mice is dependent on a Th17 immune response [9]. Thus, we examined the impact of ETBF clearance by cefoxitin on IL-17A expression in the most distal 2 cm of the colon at time points from 5 to 28 days after ETBF colonization for all 3 treatment groups (Table 1). Here, we used C57Bl/6 wild-type mice (background mouse of MinApcΔ716) to avoid confounding IL-17A expression by the high levels of IL-17A detected in ETBF-induced colon microadenomas or tumors (Housseau, Wu, DeStefano Shields, Sears, unpublished data). In C57Bl/6 mice, IL-17A expression is elevated by day 5 of ETBF colonization, reaching a peak at approximately 10 days after which colonic IL-17A expression declines but remains significantly elevated, compared with that in controls up to 1 year later [10]. In contrast, compared with wild-type ETBF-colonized mice, IL-17A expression was significantly decreased in the 5-day ETBF + cfx group 7, 10, and 14 days after colonization. Mice in the 2-week ETBF + cfx group also had decreased IL-17A expression upon ETBF eradication but did not differ significantly from time-matched ETBF only controls.
Table 1.
Interleukin 17A Expression in the Distal Colon of Wild-Type C57Bl/6 Mice
| Variable | Harvest Day |
||||||
|---|---|---|---|---|---|---|---|
| 5 | 7 | 10 | 14 | 16 | 19 | 28 | |
| ETBF alone | |||||||
| Condition | 5 d of ETBF col | 7 d of ETBF col | 10 d of ETBF col | 14 d of ETBF col | 16 d of ETBF col | 19 d of ETBF col | 28 d of ETBF col |
| IL-17A level | 76.4 (37.3–230.2) | 106.3 (52.7–295.9) | 369.3 (211.1–560.3) | 234.9 (57.4–369.4) | 124.1 (83.6–178.2) | 42.8 (20.0–97.4) | 74.0 (21.1–151.0) |
| Mice, no. | 6 | 6 | 4 | 6 | 5 | 5 | 4 |
| 5-d ETBF+cfx | |||||||
| Condition | 5 d of ETBF col | 5 d of ETBF col plus 2 d of cfx | 5 d of ETBF col plus 5 d of cfx | 5 d of ETBF col plus 9 d of cfx | 5 d of ETBF col plus 11 d of cfx | 5 d of ETBF col plus 14 d of cfx | 5 d of ETBF col, 14 d of cfx, plus 8 d after cfx end |
| IL-17A level | ND | 25.9 (8.6–108.8) | 25.6 (10.2–33.7) | 7.3 (5.4–35.3) | ND | ND | 43.0 (14.8–98.6) |
| Mice, no. | … | 6 | 5 | 5 | … | … | 4 |
| Pa | .026 | .0159 | .0043 | .4857 | |||
| 2-wk ETBF+cfx | |||||||
| Condition | 5 d of ETBF col | 7 d of ETBF col | 10 d of ETBF col | 14 d of ETBF col | 2 wk of ETBF col plus 2 d of cfx | 2 wk of ETBF col plus 5 d of cfx | 2 wk of ETBF col plus 14 d of cfx |
| IL-17A level | ND | ND | ND | ND | 28.1 (17.5–133.1) | 36.4 (33.5–58.7) | 11.6 (4.0, 19.2) |
| Mice, no. | … | … | … | … | 4 | 5 | 2 |
| Pa | .111 | .9004 | .1333 | ||||
RNA isolated from the most distal centimeter of the colon was analyzed for IL-17A expression by TaqMan quantitative polymerase chain reaction at the times indicated. IL-17A expression was normalized to that of 18S, and changes are expressed as the fold-change from median sham control values (range).
Abbreviations: col, colonization; ETBF, enterotoxigenic Bacteroides fragilis; ETBF alone, mice colonized with ETBF without treatment; ND, not done; 2-wk ETBF+cfx, mice colonized with ETBF for 2 weeks and then treated with cefoxitin for 2 weeks; 5-d ETBF+cfx, mice colonized with ETBF for 5 days and then treated with cefoxitin for 2 weeks.
a By the Mann–Whitney U test, for comparison with the ETBF alone group at the specified harvest time.
DISCUSSION
Previously we demonstrated that ETBF colonization induces inflammation and IL-17A–dependent colon adenomas in MinApcΔ716 mice [9]. Early acute inflammation may also play an important role in disease progression, given that epigenetic silencing complexes have been shown to be recruited to chromatin within 48 hours of ETBF inoculation [17]. However, whether these early events are sufficient for tumor formation and whether the continued presence of ETBF is essential for colon tumor initiation and/or progression are unknown.
Our key finding is that median adenoma formation decreased by almost 10-fold when ETBF was cleared within 5 days of colonization but only approximately 2-fold when cleared after 14 days of colonization by using this cefoxitin model. These results suggests that key disease drivers occur within a small interval in this model and that focusing on this interval may yield important mechanisms for procarcinogenic inflammatory, epigenetic, and genetic studies. Our results do not specifically discern whether the colon tumorigenesis observed results from ETBF colonization alone or requires interaction with other microbiota species [18]. However, we note that ETBF is rapidly lethal in germ-free mice [12], suggesting that, at least in our model, the murine microbiota dampens ETBF pathogenicity. Additionally, Min mice colonized with normal, or conventional, gut flora form very few if any tumors in the colon; thus, ETBF acts as a driver for tumorigenesis. Treatment with the broad-spectrum antibiotic cefoxitin to eradicate ETBF also changes the gut flora of the experimental mice. However, given the very low tumor burden of this genetic background, it is experimentally challenging to discern the contribution of the microbiota to baseline tumorigenesis in this Min mouse model. Additional data indicate that an in-frame bft deletion mitigates ETBF pathogenicity (F. Housseau and S. Wu et al, unpublished data), confirming the importance of BFT to the colitic and tumorigenic potential of ETBF. However, tools to induce time-dependent BFT neutralization or bft excision would be helpful to examine with precision the selective contribution of BFT (vs other microbiota members or molecules) to the development of colitis and colon tumors.
Of note, although ETBF eradication by cefoxitin therapy was accompanied by an abrupt reduction in colonic IL-17A levels, expression did not decrease to the levels of sham controls. We hypothesize that, in this IL-17A–dependent model, clearance of ETBF blunts the IL-17A response. However, colon adenoma formation in the 2-week ETBF + cfx group suggests that, in many mice, tumorigenesis had progressed and/or a procarcinogenic mucosal immune environment was established such that ETBF clearance was not sufficient to prevent either tumor initiation or its progression. Namely, the majority of the 2-week ETBF + cfx mice exhibited tumor numbers exceeding those of sham controls. Notably, we used wild-type mice for our IL-17A studies, to avoid confounding influences by the IL-17A–rich tumor microenvironment of the Min mouse, with or without ETBF colonization [19]. Further studies are warranted to define the immune, molecular, and/or environmental signals initiated by ETBF colonization that are required for altering IL-17A homeostasis in the colon, as well as the accelerated colon adenoma initiation in the ETBF model.
Treatment of or immunization against infectious agents (eg, Helicobacter pylori, hepatitis B and C viruses, and human papillomavirus) can have a dramatic effect on cancer development. Courses of antibiotics to clear asymptomatic colonization by enteric bacteria are currently not advisable, particularly due to the risks of development of antibiotic resistance, as well as disruption of the normal microbiota that can precipitate short-term complications of antibiotic therapy, as occurs with Clostridium difficile colitis [20]. In addition, there is rising concern that microbiota disruptions result in long-term, disease-inducing microbiota dysbiosis [21]. Further, bacteria proposed as initiators/promoters of CRC (eg, ETBF, Fusobacterium nucleatum, and Escherichia coli [pks]) are not routinely sought as pathogens in clinical medicine, and it is unclear whether detection and clearance of chronic colonizing, putatively oncogenic, bacteria will modify long-term risk for CRC. Our ETBF colon tumorigenesis model is markedly accelerated (approximately 100-fold) as compared to our understanding of the protracted time course for the development of human colon neoplasias. Nonetheless, our results suggest that early interventions in infection-related procarcinogenic colitis associated with induction of mucosal IL-17A could blunt colon polyp/adenoma formation, a known precursor lesion of CRC.
Our work presented here is novel because we have established an early, brief interval (5–14 days after ETBF colonization) in which ETBF colonization induces the majority of the IL-17A–dependent colon tumorigenesis burden in mice. ETBF-induced inflammation is sufficient to produce microadenomas in 5 days, yet very early clearance of ETBF allows for the regression of disease. Thus, this model provides an opportunity to define the molecular sequence of events crucial to IL-17A–dependent colon adenoma initiation and progression, with the potential to define testable biomarkers for early detection of carcinogenic potential in the colon.
Supplementary Data
Supplementary materials are available at http://jid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Acknowledgments. We are deeply grateful for all of the contributions that D. L. H. made to our work over many years and celebrate his remarkable collegial spirit.
Financial support. This work was supported by the National Institute of Environmental Health Sciences (NIEHS) (grant ES07141 to C. E. D. S.), the NIEHS (grant RO1ES023183 to H. M. O. H.), and the National Institutes of Health (National Cancer Institute) (grant R01CA 151325 to C. L. S.).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Bernstein CN, Blanchard JF, Kliewer E, Wajda A. Cancer risk in patients with inflammatory bowel disease: a population-based study. Cancer 2001; 91:854–62. [DOI] [PubMed] [Google Scholar]
- 2.Fite A, Macfarlane S, Furrie E et al. Longitudinal analyses of gut mucosal microbiotas in ulcerative colitis in relation to patient age and disease severity and duration. J Clin Microbiol 2013; 51:849–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe 2014; 15:317–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu S, Lim KC, Huang J, Saidi RF, Sears CL. Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proc Natl Acad Sci U S A 1998; 95:14979–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sears CL, Geis AL, Housseau F. Bacteroides fragilis subverts mucosal biology: from symbiont to colon carcinogenesis. J Clin Invest 2014; 124:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basset C, Holton J, Bazeos A, Vaira D, Bloom S. Are Helicobacter species and enterotoxigenic Bacteroides fragilis involved in inflammatory bowel disease? Dig Dis Sci 2004; 49:1425–32. [DOI] [PubMed] [Google Scholar]
- 7.Toprak NU, Yagci A, Gulluoglu BM et al. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect 2006; 12:782–6. [DOI] [PubMed] [Google Scholar]
- 8.Boleij A, Hechenbleikner EM, Goodwin AC et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis 2015; 60:208–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu S, Rhee KJ, Albesiano E et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med 2009; 15:1016–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wick EC, Rabizadeh S, Albesiano E et al. Stat3 activation in murine colitis induced by enterotoxigenic Bacteroides fragilis. Inflamm Bowel Dis 2014; 20:821–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oshima M, Oshima H, Kitagawa K, Kobayashi M, Itakura C, Taketo M. Loss of Apc heterozygosity and abnormal tissue building in nascent intestinal polyps in mice carrying a truncated Apc gene. Proc Natl Acad Sci U S A 1995; 92:4482–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhee KJ, Wu S, Wu X et al. Induction of persistent colitis by a human commensal, enterotoxigenic Bacteroides fragilis, in wild-type C57BL/6 mice. Infect Immun 2009; 77:1708–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tosolini M, Kirilovsky A, Mlecnik B et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res 2011; 71:1263–71. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Duan Y, Cheng X et al. IL-17 is associated with poor prognosis and promotes angiogenesis via stimulating VEGF production of cancer cells in colorectal carcinoma. Biochem Biophys Res Commun 2011; 407:348–54. [DOI] [PubMed] [Google Scholar]
- 15.Rabizadeh S, Rhee KJ, Wu S et al. Enterotoxigenic bacteroides fragilis: a potential instigator of colitis. Inflamm Bowel Dis 2007; 13:1475–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boivin GP, Washington K, Yang K et al. Pathology of mouse models of intestinal cancer: consensus report and recommendations. Gastroenterology 2003; 124:762–77. [DOI] [PubMed] [Google Scholar]
- 17.O'Hagan HM, Wang W, Sen S et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell 2011; 20:606–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sears CL, Pardoll DM. Perspective: alpha-bugs, their microbial partners, and the link to colon cancer. J Infect Dis 2011; 203:306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chae WJ, Gibson TF, Zelterman D, Hao L, Henegariu O, Bothwell AL. Ablation of IL-17A abrogates progression of spontaneous intestinal tumorigenesis. Proc Natl Acad Sci U S A 2010; 107:5540–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lessa FC, Mu Y, Bamberg WM et al. Burden of Clostridium difficile infection in the United States. N Engl J Med 2015; 372:825–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ridaura VK, Faith JJ, Rey FE et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013; 341:1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.